Abstract
CRIPT, the cysteine-rich PDZ-binding protein, binds to the third PDZ domain of PSD-95 (postsynaptic density protein 95) family proteins and directly binds microtubules, linking PSD-95 family proteins to the neuronal cytoskeleton. Here, we show that overexpression of full length CRIPT leads to a modest decrease and knockdown of CRIPT leads to an increase in dendritic branching in cultured rat hippocampal neurons. Overexpression of truncated CRIPT lacking the PDZ-domain binding motif, which does not bind to PSD-95, significantly decreases dendritic arborization. Conversely, overexpression of full length CRIPT significantly increases the number of immature and mature dendritic spines, and this effect is not observed when CRIPTΔPDZ is overexpressed. Competitive inhibition of CRIPT binding to the third PDZ domain of PSD-95 with PDZ3-binding peptides resulted in differential effects on dendritic arborization based on origin of respective peptide sequence. These results highlight multifunctional roles of CRIPT during development and underscore the significance of the interaction between CRIPT and the third PDZ domain of PSD-95.
Keywords: CRIPT, Neuron, Dendrite, Dendritic Spine, Morphology, Arborization
Introduction
The highly branched morphology of the neuronal dendritic arbor allows for the induction of synaptic plasticity and enables regulation of information input, integration, and ultimately, output, to the rest of the neural circuit [1–3]. Proper regulation of these processes is crucial for neuronal function and synaptic transmission, and abnormal development of the dendritic arbor contributes to a host of neurodevelopmental and neuropsychiatric disorders [4–7]. Additionally, alterations in spine density and morphology and abnormal synaptic plasticity contribute to aberrant synaptic and neural signaling as the distribution pattern of synapses and properties of each synapse determine the input and output parameters of an individual neuron [8,9]. Thus, understanding the molecular mechanisms that regulate dendritic arborization (the number and placement of dendrites) and spine formation (formation of dendritic protrusions where excitatory synaptic input occurs) during development is essential for future advances in treatments for developmental disorders.
Microtubules serve as the structural building blocks for dendrites [10–12]. They rapidly polymerize and depolymerize, and tight regulation of rapid spatial and temporal reorganization of the cellular cytoskeleton is essential for rapid remodeling of dendrites and spines to promote dendritic and synaptic plasticity [13–15]. Microtubule dynamics are regulated by microtubule assembly, promoting, stabilizing, and destabilizing factors, microtubule severing proteins, and kinesin and dynein family microtubule motor proteins [13]. Many of these factors have been implicated in regulating dendritic outgrowth and branching and spinogenesis [16]. Postsynaptic density 95 protein/synapse-associated protein 90 (PSD-95/SAP90) is a member of the membrane-associated guanylate kinase (MAGUK) superfamily of PDZ-domain containing scaffold proteins, including PSD-93/chapsyn-110, SAP-97, and SAP-102. [17] PSD-95 is involved in the synaptic targeting of receptors and signaling proteins and in regulation of dendritic spine morphology [18,19]. We recently reported that PSD-95 negatively regulates dendritic arborization by altering microtubule dynamics [20,21]. PSD-95 binds to the +TIP microtubule end-binding protein 3 (EB3) and +TIP binding protein adenomatous polyposis coli [22] to disrupt microtubule assembly [23,21] and cause microtubule bundling [24,20]. Furthermore, overexpression of PSD-95 increases spine maturation [18], supporting a role for PSD-95 during multiple phases of neuronal development.
To understand how PSD-95 acts to shape the dendritic arbor and spines during development via microtubule-dependent mechanisms, we performed experiments to determine whether the interaction of PSD-95 with the cysteine-rich PDZ-binding protein (CRIPT) regulates PSD-95 action during neuronal development. CRIPT, a 12 kDa protein that binds to the third PDZ domain (PDZ3) of PSD-95 and family members, directly binds microtubules, linking the microtubule cytoskeleton and PSD-95 family scaffolding proteins [25,26,17]. Currently, the precise functional role of CRIPT has not been completely elucidated. Transgenic mice homozygous for the CRIPTtm1.1(KOMP)Vlcg null allele die in the period between fertilization and weaning, supporting the significance of CRIPT during development [27]. To date, three patients with defects in the CRIPT gene have been reported. Two unrelated patients with primordial dwarfism (PD) were found to have different truncating mutations in CRIPT [28]. This study did not focus on brain alterations; however, the authors proposed a proliferation deficit. A recent study also reported on a third patient with biallelic mutations in the CRIPT gene, presenting growth retardation, microcephaly, and developmental delay [29]. This patient was heterozygous for c.8G>A (p.C3Y) variant in exon 1 and an exon 1 deletion. Based on our data, a lack of CRIPT could result in aberrant dendritic arborization and spines, and hence, neuronal connectivity, leading to functional abnormalities, such as language delay and complex partial seizures observed in the patient. Further examination of CRIPT function during development is necessary to definitively determine whether mutant CRIPT plays a role in aberrant dendritic arborization and dendritic spine formation in these clinical cases.
It was recently reported that CRIPT is necessary, but not sufficient, for promoting dendritic arborization in mammalian spinal cord neurons, and nematodes that lack CRIPT show a decreased dendritic branching phenotype [30]. Here, in contrast to spinal cord neurons and Caenorhabditis elegans neurons, we demonstrate the novel finding that CRIPT overexpression in mammalian hippocampal neurons modestly decreases proximal dendrite branching, while overexpression of CRIPT lacking the PDZ-domain binding motif substantially decreases dendritic arborization. Additionally, we show that CRIPT overexpression increases the number of dendritic spines, and we demonstrate that these effects are dependent on the PDZ-domain binding motif of CRIPT. Furthermore, we show that competitive inhibition of CRIPT binding to PSD-95 with linear peptide ligands based on the carboxyl terminal of CRIPT, but not on a general PDZ3 binding consensus sequence, decreases dendritic branching in hippocampal neurons. Taken together, our data support a regulatory role of the interaction between PSD-95 and CRIPT in dendritic arborization and spine number.
Results
Overexpression of CRIPT alters dendrite arborization in a PDZ domain-binding dependent manner
Previous studies by our group show that PSD-95 negatively regulates dendritic arborization by altering microtubule dynamics via an association between PSD-95 and the +TIP EB3 [20,21]. Furthermore, immunostaining studies show that CRIPT, PSD-95 and MAP2 co-localize in immature neurons (Supplementary Fig. 1). As CRIPT directly interacts with the PDZ3 domain of PSD-95 and directly binds to microtubules, we asked if CRIPT also plays a functional role in the regulation of dendritic arborization [24,25]. To test this, we overexpressed CRIPT in primary rat hippocampal neurons on day in vitro (DIV) 7 and assessed dendritic arborization on DIV 12 using Sholl analysis [31–33]. We quantified CRIPT expression by immunostaining transfected neurons with antibody against CRIPT and confirmed that neurons transfected with CRIPT cDNA express significantly higher levels of CRIPT compared to control neurons transfected with GFP (Supplementary Fig. 2). CRIPT overexpression resulted in decreased dendritic branching proximal to the soma (Fig. 1a, b, c), and overexpression of a truncated version of CRIPT, CRIPTΔPDZ, further decreased dendritic branching proximally and distally to the soma (Fig. 1a, b, c). More specifically, neurons overexpressing CRIPTΔPDZ showed significant decreases in number of secondary dendrites (Fig. 1e), number of tertiary and higher order dendrites (Fig. 1f), total number of segments (Fig. 1g), total number of branch points (Fig. 1h) and total number terminal points (Fig. 1i) when compared to control neurons. Overexpression of CRIPT or CRIPTΔPDZ did not affect number of primary dendrites (Fig. 1d), the length of primary (Fig. 1j), secondary (Fig. 1k) or higher order dendrites (Fig. 1l), nor total dendrite length (Fig. 1m). Taken together, these data support the hypothesis that CRIPT regulates dendritic arborization in hippocampal neurons and that the PDZ-binding domain of CRIPT plays a significant role in modulating dendrite branching.
Fig. 1. CRIPT and CRIPTΔPDZ overexpression decreases dendritic arborization.
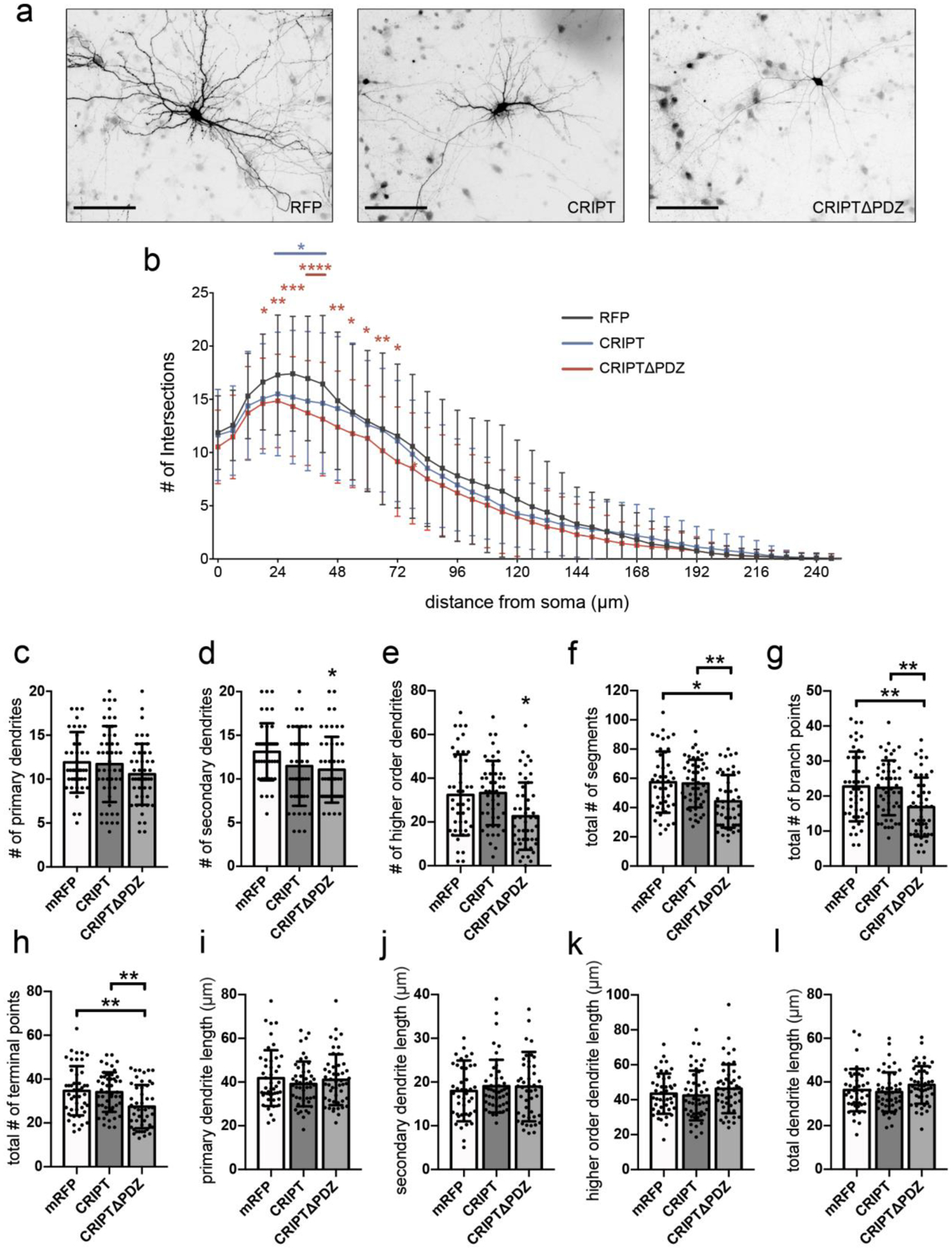
Rat hippocampal neurons were cultured from gestation day 18 embryos and were co-transfected with cDNA encoding eGFP and 1) RFP (control) or 2) RFP-tagged CRIPT or 3) CRIPTΔPDZ on DIV7 and fixed on DIV12. a Representative micrographs of transfected neurons. b Sholl analysis of dendritic arborization. Asterisks at single data points indicate significant difference from control. Asterisks above lines indicate significant difference from control for data points beneath the line. c CRIPT and CRIPTΔPDZ overexpression do not affect number of primary dendrites. d-h CRIPTΔPDZ, but not CRIPT, overexpression decreases number of (d) secondary dendrites, (e) higher order dendrites, (f) total number of segments, (g) total number of branch points, and (h) total number of terminal points. i-l CRIPT and CRIPTΔPDZ overexpression do not affect length of primary, secondary, higher order dendrites, or total dendrite length. Data obtained from five biological replicates. *p < 0.05, **p < 0.01, ***p < 0.001 ****p < 0.0001 as determined by two-way ANOVA followed by Bonferroni multiple comparisons test (Sholl analysis), and one-way ANOVA followed by Tukey multiple comparisons test (all other analyses). n(RFP) = 45; n(CRIPT) = 49; n(CRIPTΔPDZ) = 49 neurons from three independent cultures. Scale bars, 100μm. Error bars are ± s.d.
CRIPT overexpression increases spine density in a PDZ-binding domain-dependent manner
CRIPT directly binds to PSD-95 and microtubules and is abundant in synaptic spines [25]. As such, we hypothesized that CRIPT regulates spine morphogenesis during development. To test this, we transfected rat hippocampal neurons with cDNA encoding CRIPT or CRIPTΔPDZ on DIV 14 and assessed spine density and morphology on DIV17 using confocal microscopy. Interestingly, overexpression of CRIPT resulted in increased number of dendritic spines (Fig. 2a, b). CRIPT, but not CRIPTΔPDZ, overexpression increased the number of mushroom (Fig. 2c) and immature spines (Fig. 2d) but not filopodia (Fig. 2e). Expression of CRIPT lacking the PDZ-domain binding motif did not affect dendritic spine density and morphology, compared to control, suggesting that the PDZ-domain binding motif is crucial for CRIPT-mediated changes to spines or that CRIPTΔPDZ acts as a dominant-negative mutant by attenuating the interaction of PSD-95 with microtubules. In addition, overexpression of CRIPT or CRIPTΔPDZ did not affect spine length (Fig. 2f), spine head width (Fig. 2g), spine neck width (Fig. 2h) and head to neck length ratio (Fig. 2i).
Fig. 2. Overexpression of CRIPT, but not CRIPTΔPDZ, results in increased spine density.

a Representative micrographs of dendritic segments from DIV17 hippocampal rat neurons co-transfected with cDNA encoding eGFP and 1) RFP (control) or 2) CRIPT or 3) CRIPTΔPDZ. Filled arrowheads point to mature spines, and open arrowheads point to immature spines and filopodia. b–d CRIPT, but not CRIPTΔPDZ, overexpression increases number of (b) dendritic spines, (c) mushroom spines, and (d) immature spines. e Number of filopodia is not altered by CRIPT or CRIPTΔPDZ overexpression. f-i CRIPT and CRIPTΔPDZ overexpression do not affect (f) spine length, (g) spine head width, (h) spine neck width, or (i) spine head to neck ratio. n = 20–25 neurons per condition from three three independent cultures. *p < 0.05, ***p < 0.001 ****p < 0.0001 as determined by one-way ANOVA followed by Tukey’s multiple comparisons test. Scale bars, 5μm. Error bars are ± s.d. Mushroom spines are defined as having a head width/neck width > 1.5μm and total length <5μm and are considered mature spines. Thin spines are defined as having head width/neck width between 1 and 1.5μm and total length <5μm and are considered immature spines. Stubby spines have head width/neck width ≤ 1μm and total length ≤ 1μm and are considered immature spines. Filopodia have head width/neck width ≤ 1μm and total length > 1μm.
PSD-95-binding peptides reduce the interaction between CRIPT and PSD-95
To further investigate the role of CRIPT in dendritic arborization during development, we performed an additional set of experiments that do not rely on overexpression but instead use peptides to uncouple PSD-95 interactions with other proteins. Specifically, we used a series of previously described PSD-95-binding peptides based on either the CRIPT C-terminus (YKQTSV) or a PDZ3-binding consensus sequence generated from library data (KKETEV) [34]. Peptides were further modified to either include an Fmoc group at the amino terminus to increase the binding affinity for PDZ3 (data unpublished) or amino terminal myristoylation (N-Myr) to facilitate cell permeabilization [35] (Fig. 3a, b, c, d). As these peptide sequences have an affinity for PDZ3, we hypothesized that they would compete with CRIPT for binding to PSD-95, and thus, could be used to assess how the competitive inhibition of CRIPT and PSD-95 interaction affects dendritic arborization. Nomenclature for the peptides follows: HMC2058 (Fmoc CRIPT C-terminus), HMC2044 (myristoylated CRIPT C-terminus), HMC2049 (Fmoc PDZ3-binding consensus sequence), and TW1036 (myristoylated PDZ3-binding consensus sequence). To examine whether PSD-95-binding peptides affect the interaction between CRIPT and PSD-95, we performed a co-immunoprecipitation assay in which PSD-95 was immunoprecipitated in the presence of different amounts of peptides and assessed the amount of CRIPT that co-immunoprecipitated with PSD-95 using Western blot analysis. Incubation of brain lysates with peptides bearing the Fmoc group, HMC2058 and HMC2049, at 1μM, 10μM, and 100μM during immunoprecipitation resulted in a robust decrease in the amount of co-immunoprecipitated CRIPT (Fig. 3e; Supplementary Fig. 3–5). In contrast, of the two myristoylated peptides, only incubation with 100μM HMC2044 resulted in a significant decrease in the amount of co-immunoprecipitated CRIPT, while incubation with HMC2044 at lower concentrations and with TW1036 at all concentrations tested did not affect the amount of co-immunoprecipitated CRIPT (Fig. 3f), consistent with the idea that these peptides bind with lower affinity to PDZ3 than does CRIPT.
Fig. 3. PSD-95 PDZ-domain binding peptides reduce the interaction of CRIPT and PSD-95.

The YKQTSV sequence is from the CRIPT C-terminus, while KKETEV was generated as a PDZ3-binding consensus sequence based on library data [34]. Amino terminal myristoylation (N-Myr) was used to aid in cell permeabilization, while an added Fmoc group at the amino terminus was used to increase the binding affinity for PDZ3 (Spaller, unpublished data). a Structure of HMC2058 (Fmoc-YKQSTV), based on the CRIPT carboxy terminal sequence with added Fmoc group. b Structure of HMC2044 (N-Myr-YKQTSV), based on CRIPT carboxy terminal sequence with added amino terminal myristoylation. c Structure of HMC2049 (Fmoc-KKETEV), based on a PDZ3-binding consensus sequence with added Fmoc group. d Structure of TW1036 (N-Myr-KKETEV), based on a PDZ3-binding consensus sequence with added amino terminal myristoylation. e-f Representative immunoblots and densitometric quantification of relative amount of CRIPT co-immunoprecipitated with PSD-95 from rat brain extracts in the presence of different concentrations of PSD-95-binding peptides. Western blot densitometric results were normalized to the amount of CRIPT co-immunoprecipitated with PSD-95 in vehicle (DMSO) condition, represented by the dotted line. e HMC2058 and HMC2049 significantly decrease the amount of CRIPT that co-immunoprecipitates with PSD-95 at concentrations of 1μM,10μM, and 100μM. f HMC2044 at a concentration of 100μM significantly decreases the amount of CRIPT that co-immunoprecipitates with PSD-95. Lower concentrations (1μM and 10μM) of HMC2044 and all concentrations tested of TW1036 do not affect the amount of CRIPT that co-immunoprecipitates with PSD-95. Data obtained from seven independent experiments. The controls for these experiments include the KKETEV PDZ3-binding consensus sequence based on library data, vehicle (0.1% DMSO), and addition of the Fmoc group. Fmoc acts as control for amino terminal myristoylation (N-Myr) to control for the lipid moiety of the lipopeptide. **p < 0.01, ***p < 0.001 ****p < 0.0001 vs. co-immunoprecipitation in presence of vehicle (DMSO) as determined by one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars are ± s.d.
PSD-95-binding peptides based on the CRIPT C-terminus decrease dendritic arborization
To assess whether competitive inhibition of the interaction between PSD-95 and CRIPT affects dendritic arborization, we transfected rat primary hippocampal neurons with cDNA encoding mRFP (to visualize dendrites) on DIV9, treated with 100μM of respective PSD-95-binding peptides, or vehicle (0.1% DMSO) on DIV10 for 48 h, and analyzed dendritic branching on DIV12 with Sholl analysis. The timing of this treatment corresponds to the period of active dendritic branching in our cultures [20]. Thus, any changes observed are most likely due to inhibition of branching rather than pruning, which occurs a bit later in culture [20,36].Treatment of hippocampal neurons with PSD-95-binding peptides based on the C-terminus of CRIPT, HMC2058 and HMC2044, significantly reduced dendritic arborization compared to treatment with DMSO vehicle (Fig. 4a,c). HMC2049, based on the PDZ3-binding consensus sequence with an added Fmoc group, did not alter dendritic arborization as determined by Sholl analysis (Fig. 4b). Surprisingly, TW1036, which is also based on the PDZ3-binding consensus sequence except with amino terminal myristoylation instead of Fmoc, reduced dendritic branching at distal regions from the soma at 50μm to 150μm (Fig. 4c). Treatment with peptides based on the C-terminus of CRIPT, HMC2058 and HMC2044, significantly reduced the number of secondary dendrites (Fig. 4e), higher order dendrites (Fig. 4f), segments (Fig. 4g), branch points (Fig. 4h), and terminal points (Fig. 4i). Treatment with HMC2044, but not HMC2058, significantly reduced the number of primary dendrites (Fig. 4d). Treatment with HMC2058 or HMC2044 did not affect primary (Fig. 4j), secondary (Fig. 4k), higher order (Fig. 4l) dendrites, or total dendrite length (Fig. 4m). Notably, of the two peptides based on the PDZ3-binding consensus sequence, treatment with HMC2049 (bearing the Fmoc group) resulted in significantly increased number of primary dendrites (Fig. 4d), decreased number of secondary dendrites (Fig. 4e) and higher order dendrites (Fig. 4f), and did not affect number of segments (Fig. 4g), branch points (Fig. 4h), or terminal points (Fig. 4i). Moreover, HMC2049 treatment significantly increased secondary (Fig. 4k) and total dendrite length (Fig. 4m), but did not affect primary (Fig. 4j) and higher order dendrite length (Fig. 4l). In contrast, the other peptide based on the PDZ3-binding consensus sequence, treatment with TW1036 (containing the myristoyl group) did not affect the number and length of dendrites (Fig. 4d–m). Taken together, these results suggest that the interaction of PSD-95 with CRIPT shapes the dendritic arbor distinctly from the interaction of PSD-95 with other PDZ3-binding proteins.
Fig. 4. Dendritic arborization significantly decreases by treatment with PSD-95-binding peptides based on the CRIPT carboxy terminal sequence.

Primary rat hippocampal neurons were transfected with cDNA encoding mRFP on DIV9, treated with 100μM of indicated PSD-95-binding peptides on DIV10 for 48 h and fixed on DIV12. a Representative micrographs of neurons treated with PSD-95 binding peptides. b Sholl analysis of dendritic arborization. Asterisks above lines indicate significant difference from control. PSD-95-binding peptides based on the carboxy terminal sequence of CRIPT (HMC2058 and HMC2044) significantly reduce dendritic arborization compared with DMSO-treated control neurons. TW1036, which is based on the PDZ3-binding consensus sequence, differentially decreases dendritic branching in regions distal to the neuronal soma. HMC2049, also based on the PDZ3-binding consensus sequence, does not significantly alter the dendritic arbor. c Treatment with HMC2049 significantly increases number of primary dendrites, while HMC2044 significantly decreases number of primary dendrites compared to vehicle control. d–e Treatment with HMC2058, HMC2049, and HMC2044, significantly decreases number of (d) secondary dendrites and (e) number of higher order dendrites. f-h Treatment with HMC2058 and HMC2044 significantly reduces (f) total number of segments, (g) total number of branch points, and (h) total number of terminal points. i PSD-95-binding peptides do not affect primary dendrite length. j Treatment with HMC2049 significantly increases secondary dendrite length compared to vehicle control. k Higher order dendrite length is not affected by PSD-95-binding peptide treatment. l Treatment with HMC2049 significantly increases total dendrite length compared to vehicle control. Data obtained from five independent experiments. **p < 0.01, ***p < 0.001 ****p < 0.0001 by two-way ANOVA followed by Bonferroni multiple comparisons test, and one-way ANOVA followed by Bonferroni multiple comparisons test. Scale bars, 100μm. Error bars are ± s.d.
CRIPT knockdown increases dendritic arborization and proximal dendritic length
To investigate whether decreased CRIPT expression results in changes to dendritic arborization, we transfected rat hippocampal neurons with cDNA encoding eGFP (to trace dendrites) and either negative control siRNA or CRIPT siRNA on DIV7 and assessed dendrite branching on DIV12. Transfection with CRIPT siRNA resulted in approximately 65% decrease in CRIPT expression (Fig. 5a). Decreased CRIPT expression resulted in increased dendritic arborization as determined by Sholl analysis (Fig. 5b,c). Transfection with CRIPT siRNA increased higher order dendrites (Fig. 5f), segments (Fig. 5g), primary dendrite length (Fig. 5j), and secondary dendrite length (Fig. 5k) but had no effect on numbers of primary and secondary dendrites (Fig. 5 d,e), branch and terminal points (Fig. 5h,i), or higher order and total dendrite length (Fig. 5 l,m). Since knockdown of CRIPT resulted in opposite effects on the dendritic arbor than does overexpression of CRIPT, our data suggest that CRIPT regulates dendrite growth and branching.
Fig. 5. CRIPT knockdown increases dendritic arborization and proximal dendrite length.

Rat hippocampal neurons were cultured from embryos at gestation day 18 and were co-transfected with cDNA encoding eGFP and 1) negative control siRNA or 2) CRIPT siRNA on DIV7 and fixed on DIV12. a Representative immunoblots and quantification of CRIPT expression in lysates prepared from cultures transfected with either negative control siRNA or CRIPT siRNA on DIV7. Cultures were lysed on DIV12, and proteins were resolved using Western blot analysis. b Representative images of siRNA transfected neurons. c Sholl analysis of dendritic arborization. Asterisks above lines indicate significant difference from control for data points beneath the line. d-e CRIPT knockdown does not affect number of (d) primary dendrites or (e) secondary dendrites. f-h CRIPT knockdown increases number of (f) higher order dendrites and (g) total number of segments. h-i CRIPT knockdown does not affect (h) total number of branch points or (i) total number of terminal points. j-k CRIPT knockdown increases length of (j) primary and (k) secondary dendrites. l-m CRIPT knockdown does not affect (l) higher order dendrites, or (m) total dendrite length. Data obtained from two independent cultures replicates. *p < 0.05, **p < 0.01, ***p < 0.001 as determined by two-way ANOVA followed by Bonferroni multiple comparisons test (Sholl analysis), and two-tailed Student’s t-test (all other analyses). n(negative control siRNA) = 38; n(CRIPT siRNA) = 57; neurons from two independent experiments. Scale bars, 100μm. Error bars are ± s.d.
Discussion
Proper regulation of the processes underlying dendritogenesis is essential for appropriate synaptic and neuronal circuit function. Aberrations in dendritic arborization and spine formation during development result in a multitude of neuropsychiatric and developmental disorders, which significantly impair neurocognition. Here, for the first time, we show distinct effects of CRIPT and CRIPTΔPDZ overexpression on dendritic arborization and spinogenesis in hippocampal neurons. CRIPT overexpression results in a modest decrease in dendritic branching proximal to the soma and a significant increase in number of immature and mature synaptic spines (Fig. 6b). Truncation of the PDZ-binding domain of CRIPT resulted in significant decreases in dendritic branching proximal and distal to the soma, significant decreases in number of secondary, higher order, and total dendrites, and elimination of increased spinogenesis observed with overexpression of full length CRIPT (Fig. 6c). We also show that PSD-95-binding peptides based on the CRIPT carboxyl terminus compete with CRIPT for PSD-95 binding and that treatment of neurons with these peptides results in significant decreases in proximal and distal dendritic branching, which mimic those observed with CRIPTΔPDZ overexpression. Our results highlight the significance of the CRIPT PDZ-domain binding motif in dendritic arborization and spine formation during development.
Fig. 6. Proposed model for role of CRIPT-PSD-95 interaction in neuronal development.

a Under baseline conditions, CRIPT binds to third PDZ domain of PSD-95. This interaction allows for attachment of PSD-95 to the microtubule cytoskeleton. Binding of PDZ-3 ligands to PSD-95 is necessary for PSD-95 multimerization and can induce conformational changes in PSD-95 SH3 and GK domains. [37,43] b Overexpression of full length CRIPT results in modest decreases in dendritic branching proximal to the neuronal soma and a substantial increase in the number of immature and mature dendritic spines. Treatment with PSD-95 PDZ3-binding peptide ligands does not recreate diminished dendritic branching. c Overexpression of CRIPT lacking the PDZ-domain binding motif results in extensive decreases in dendritic arborization proximal and distal to the soma but does not affect dendritic spine number. Treatment with PSD-95 peptide ligands based on the CRIPT carboxyl terminus recapitulates the significant decreases in dendritic branching observed with CRIPTΔPDZ overexpression.
The data reported in this study complement previous investigations of CRIPT and its role in dendritic arborization and in anchoring PSD-95 to the synaptic cytoskeleton during development [25,26,30,37]. PSD-95 family scaffolding proteins are essential components of the PSD as they organize and regulate postsynaptic neurotransmitter receptor trafficking, coordinate downstream postsynaptic signaling enzymes, and promote the structural arrangement of the postsynaptic cytoskeleton [38]. Early stage excitatory synaptic contacts contain a greater amount of SAP-102 than other PSD-95 family members, and through cell adhesion signaling, SAP-102 initiates recruitment of pre- and postsynaptic machinery, which travel to synaptic sites via microtubules in distinct complexes [39]. PSD-95 is one of the first proteins found in the PSD and is an essential scaffolding backbone of the PSD, as PSD-95 remains stabilized in position while a dynamic matrix of PSD machinery undergoes rapid and extensive changes in structure in response to synaptic activity [8,17].
CRIPT, one of the few identified PDZ3 ligands, induces filamentous redistribution of PSD-95 to microtubules through its carboxyl terminal PDZ3-binding motif [25]. We previously demonstrated that overexpression of PSD-95 prior to DIV12 in cultured hippocampal neurons decreases dendritic arborization up to 100μm away from the soma by disrupting microtubule organization at dendritic branch points [21]. The decrease in dendrites we observed here when CRIPTΔPDZ is overexpressed is similar to the decrease in dendrites we observed when PSD-95 is overexpressed. This notion is further supported by the similarity in deficits in dendrite branching observed when we treated rat hippocampal neurons with PSD-95-binding peptides based on the carboxyl terminus of CRIPT. Treatment with either of the two peptides based on the CRIPT carboxyl terminus decreased dendritic arborization in a similar manner to CRIPTΔPDZ and PSD-95 overexpression, with greater proximal deficits by treatment with HMC2044, which we attribute to greater cell permeabilization, due to the myristoylation.
We were surprised to observe marginal decreases in dendrites proximal to the soma when we overexpressed full length CRIPT and attribute this to saturation of CRIPT-mediated regulation of dendritogenesis at baseline. Recently, it was shown that CRIPT knockdown in rat spinal cord neurons decreases total number of dendritic branches and total length of the dendritic arbor [30]. This observed effect is similar to that seen with CRIPTΔPDZ overexpression in the present study, and taken together, these results suggest that full length CRIPT is required for proper dendritogenesis during development. Moreover, the same study found no effect on dendritic arborization in spinal cord neurons when CRIPT or a palmitoylated version of CRIPT was overexpressed [30]. Although we also found no effect on number or length of dendritic branches when CRIPT was overexpressed, the use of Sholl analysis revealed a marginal decrease in the number of dendritic intersections proximal to the cell body. Together, these results indicate that CRIPT alone is not sufficient to promote dendritogenesis and that expression levels of CRIPT must be tightly regulated in for proper dendrite patterning in regions near the soma.
Our data also show that increased spinogenesis occurs when CRIPT is overexpressed and that this effect requires the PDZ-domain binding motif. Although not examined, we would predict that treatment with peptides that disrupt CRIPT-PSD-95 interactions would decrease spines. This observed result may be due to an increase in the creation of new spines or an increase in the size of spines. PSD-95 overexpression increases the number and size of dendritic spines as well as synaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor clustering [40]. Formation of dendritic segments and dendritic spines is independent of presynaptic glutamatergic signaling and is likely to be a combination of cell-autonomous processes, which regulate the targeting and transport of a host of scaffold, cell adhesion, and receptor proteins, as well as necessary organelles [41]. Dendritic spines may be persistent and stable or transient and rapidly pruned [42], and PSD-95 clustering affects spine stability with persistent spines containing a distinct PSD [42]. Dendritic spines without a distinct PSD, or those that lose the PSD, are more likely to be pruned [42]. As CRIPT induces PSD-95 redistribution onto microtubules and co-localizes with PSD-95 at the PSD, we hypothesize that CRIPT overexpression-mediated increases in spine density are likely due to increased PSD-95 microtubule distribution. Moreover, binding of CRIPT-derived PDZ3 ligands to PSD-95 induces conformational alterations in the PSD-95 SH3-GK domain, and consequently, affects differential interactions between PSD-95 binding partners and PSD-95 complex formation [37]. This suggests different affinities of binding partners and stabilities of PSD-95 protein complexes in the PSD, and increased CRIPT levels may affect PSD-95 protein complex formation. Specifically, ligand binding to the PDZ3 domain of PSD-95 affects how PSD-95 monomers interact and influences the formation of PSD-95 multimer complexes [43]. Thus, CRIPT overexpression may also affect PSD-95 multimer complex formation, and subsequently, influence the synaptic function of PSD-95 and PSD formation. The binding of SynGAP, a Ras/Rap GTPase activator abundant in the PSD, to the PDZ3 domain of PSD-95 requires an α-helix extension adjacent to the PDZ3 domain, and excessive CRIPT binding to PSD-95 may affect this interaction [44]. Effects of ligand binding to PDZ3 are evident in the present study, as differential effects on dendritic arborization were observed with treatment with PSD-95-binding peptides based on the carboxyl terminus of CRIPT and a general consensus sequence.
This study contributes to the current understanding of the functional significance of the interaction between CRIPT and PSD-95 in dendritogenesis and spinogenesis during development. Our data showing distinct effects of PSD-95 PDZ3-binding peptides based on CRIPT versus a general sequence demonstrate the complexity of the processes regulating neuronal development. Our investigation of CRIPT and CRIPTΔPDZ overexpression in dendritic spine number, morphology, and maturity highlights a multifunctional role for CRIPT in regulation of morphological changes in neurons.
Materials and Methods
CRIPT and CRIPTΔPDZ Overexpression Plasmids
CRIPT open reading frame (ORF) was amplified from pGW1-CRIPT (gift from Dr. David Bredt) using the forward primer 5’-GGACAAGAATTCGCCACCAATGGTATGTGAAAAGTGTGAAAAG-3’ and reverse primer 5’-CCTGTTGGTACCCTAGACGGAAGTTTGCTTGTAG-3’. The resulting ORF was ligated at EcoRI and KpnI sites into a pmRFP vector (generated by subcloning mRFP in place of EGFP in pEGFP-C1 purchased from Clontech, at NheI and XhoI sites) using T4 DNA ligase (all enzymes from New England Biolabs). DH5α E. coli were transformed with ligation mixes, and colonies that grew in the presence of kanamycin were screened for the CRIPT insert via restriction digestion. Constructs were confirmed by sequencing (Genscript). For amplification of CRIPTΔPDZ ORF, all steps were followed except the reverse primer used was 5’-CCTTCGGGTACCCTACTTGTAGTTTTTAGTATCCAAAAC-3’.
Dendritic Arborization Analysis
All animal studies were performed in accordance with US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and were approved by the Rutgers University Institutional Animal Care and Use Committee. Hippocampi were dissected from embryonic rats at gestation day 18 (E18) as previously reported [45–47]. Hippocampi were dissociated by trituration and plated at a density of 1.05 × 105 cells per cm2 in a 24 well plate on 12mm glass coverslips coated with poly-D-lysine (100μg/ml, Sigma-Aldrich). Cultures were maintained in Neurobasal medium supplemented with GlutaMAX and B27 (all from ThermoFisher) in a humidified incubator at 37°C and 5% CO2. Neurons were co-transfected with pEGFP-C1 and 1) pmRFP or 2) pmRFP-CRIPT or 3) pmRFP-CRIPTΔPDZ using Lipofectamine-LTX on day in vitro (DIV) 7 as per the manufacturer’s protocol. Cells were fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences; cat. #15714) in phosphate-buffered saline (PBS) followed by blocking for 1h in PBS containing 2% normal goat serum, 0.1% Triton-X, and 0.04% sodium azide. Neurons were immunostained with anti-MAP2 (1:500; BD Pharmingen, cat. #556320) and anti-GFP (1:500; ThermoFisher, cat. #PA1–9533) overnight at 4°C. Secondary antibodies used were goat anti-mouse Alexa Fluor® 647 and goat anti-chicken Alexa Fluor® 488 (both from ThermoFisher) at 1:200 dilution. Coverslips were mounted onto slides using Fluormount-G (Southern Biotech) and left to dry overnight at room temperature (RT). Micrographs of transfected neurons were captured using an Olympus Optical IX50 (Tokyo, Japan) microscope equipped with a Cooke SensiCam camera and Image Pro software (Media Cybernetics). All images were captured under 20X objective. NeuronJ plugin on ImageJ was used to trace dendrites with the experimenter blinded to the condition. Semi-automated Sholl analysis was performed as previously described by our laboratory using the Bonfire program [33]. Briefly, Bonfire sums the dendrites that cross concentric circles spaced 6μm apart, starting from the cell body. The dendrites are identified as GFP-positive and MAP2-positive, and we have observed that MAP2+ immunostaining is the same as GFP expression except for the axon, which is MAP2-negative. Dendrites are traced, and the program computes Sholl curves and characteristics, such as primary, secondary, and higher order dendrite numbers, dendrite lengths, tips, and branch points. Data at each distance are averaged across neurons and plotted in Excel or GraphPad Prism. All experiments were performed using cells from three mothers and cultured on different days except for siRNA studies where neurons were from two mothers. Cells from multiple coverslips per culture were used. Statistical analysis was performed using two-way ANOVA followed by Bonferroni multiple comparisons test.
CRIPT knockdown using siRNA
Neurons were transfected with the following siRNAs: Silencer Select Pre-Designed siRNA (siRNA ID: s132723, cat. #4390771), Silencer Select Pre-Designed siRNA (siRNA ID: s132724, cat. #4390771), and Silencer Select Negative Control No. 1 siRNA cat. #4390843). Rat hippocampal neurons were transfected on DIV7 with cDNA encoding eGFP (for dendrite tracing) and either 1) negative control siRNA or 2) pooled siRNA (from two pre-designed siRNA constructs) targeting CRIPT using the RNAiMAX kit as per manufacturer’s instructions. Neurons were fixed on DIV12 for Sholl analysis dendrite branching analysis as described above.
Dendritic Spine Analysis
Hippocampal neurons were dissected and plated at a density of 1.05 × 105 cells per cm2 on 12mm glass coverslips coated with poly-D-lysine (100μg/ml) as described above. Neurons were transfected on DIV14 using the calcium phosphate transfection method [48] with 4μg each of pEGFP-C1 and 1) pmRFP or 2) pmRFP-CRIPT or 3) pmRFP-CRIPTΔPDZ. On DIV17, neurons were fixed with 4% PFA in PBS followed by blocking for 1h and immunostaining with anti-MAP2 and anti-GFP as described above. Micrographs were captured using a Zeiss LSM800 confocal microscope under a 40X oil objective. Dendritic spine morphology was analyzed using ImageJ as previously described [49]. Mushroom spines are defined as having a head width/neck width > 1.5μm and total length <5μm and are considered mature spines. Thin spines are defined as having head width/neck width between 1 and 1.5μm and total length <5μm and are considered immature spines. Stubby spines have head width/neck width ≤ 1μm and total length ≤ 1μm and are considered immature spines. Filopodia have head width/neck width ≤ 1μm and total length > 1μm. Spine morphology was assessed in two dendritic segments (10–30μm in length; 20–80μm away from the soma) per neuron. A minimum of 15 neurons per experimental condition was analyzed across three independent experiments with the experimenter blinded to the condition. All experiments were performed using cells from three mothers and cultured on different days. One-way ANOVA followed by Bonferroni multiple comparisons test was used for statistical analysis.
PSD-95-binding Peptide Treatment
Primary rat hippocampal neurons were dissected and plated at a density of 1.05 × 105 cells per cm2 as described above. Neurons were transfected with pmRFP on DIV9 using Lipofectamine 2000 following the manufacturer’s protocol and treated with 100μM of respective PSD-95-binding peptide (in DMSO), or vehicle (0.1% DMSO) on DIV10 for 48 hours. On DIV12, cells were fixed with 4% PFA in PBS followed by blocking for 1h in PBS containing 2% normal goat serum, 0.1% Triton-X, and 0.04% sodium azide. Neurons were immunostained against MAP2 as described above and mounted onto slides using Fluormount-G. Micrographs of transfected neurons were captured using an Olympus Optical IX50 microscope and dendrites were analyzed by semi-automated Sholl analysis as previously described [33]. Statistical analysis was performed using two-way ANOVA followed by Bonferroni multiple comparisons test.
Co-immunoprecipitation and Western blot Analysis
Rat whole brain lysate was prepared by homogenizing the tissue in lysis buffer consisting of 1x protease inhibitor (Sigma, cat. #11836170001) in 10ml TEE (20mM Tris pH7.4, 1mM EDTA and 1mM EGTA). Homogenized brain sample was rotated at 4°C for 1hr in 10mL of 2x RIPA buffer (100mM Tris pH7.4, 300mM NaCl, 2mM EDTA, 1% sodium deoxycholate, 2% NP-40 and 0.2% SDS) containing 100mM PMSF. The lysate was then spun at 18000 × g at 4°C for 15 min. Protein concentration in the resulting supernatant was assessed with BCA assay (ThermoFisher), and lysates were either used for co-immunoprecipitation assay immediately or frozen at −20°C. Whole rat brain lysate (1mL; 7mg of protein) was precleared by incubating with 50μL of Protein A beads (ThermoFisher cat. #20333) at 4°C for 1h. After preclearing, the lysate was incubated with differing concentrations (1, 10 and 100μM) of respective PSD-95 binding peptides or vehicle (0.1% DMSO) for 24h at 4°C under rotation. After 24h, either control normal rat IgG or PSD-95 antibody (5μg; UC Davis/NIH Neuromab Facility clone K28/43) was added to the lysate for co-immunoprecipitation analysis. Precipitated protein was subjected to SDS PAGE, transferred to polyvinylidene difluoride (PVDF) membrane, and analyzed by Western blot analysis as previously described [47]. Briefly, PVDF membrane was blocked in TBST containing 2% BSA for 1h at RT followed by incubation with indicated primary antibody diluted in TBST containing 2% BSA: monoclonal mouse anti-PSD95 (1:1000, K28/43), polyclonal rabbit anti-cypin (1:500, BF6) or polyclonal rabbit anti-CRIPT (1:500; Proteintech cat. #11211–1-AP). Relevant bands on scanned blots were analyzed using ImagePro software (Media Cybernetics). Five independent experiments were performed.
PSD-95-binding Peptide Synthesis and Characterization
Peptides were synthesized using a standard Fmoc-based solid phase synthesis protocol. All syntheses utilized preloaded Fmoc-Val wang resin loaded into a peptide synthesis reaction vessel mounted on a wrist-action shaker. Resin equivalent to 0.2 mmol was used for each reaction. The initial suspension and swelling of the resin (30 min) took place with shaking using dimethylformamide (DMF) followed by washing. Thereafter, the procedure consisted of iterative deprotection and coupling steps with interspersed washing steps. Fmoc deprotection was effected by piperidine-DMF (20%, v/v; 10 × the resin volume) treatment for 1 min, washing, and a repeat with fresh deprotection solvents for 1 min, followed by DMF washing (10 × the resin volume). Sequential coupling of residues involved mixing of Fmoc amino acid (1 mmol), DIPEA (2 mmol), HCTU (2 mmol), and DMF (5 × the resin volume) with gentle shaking for 3 min. Coupling and deprotection steps were repeated for each added residue, with intervening washing steps (DMF, 10 × the resin volume). After the final amino acid coupling the N-terminal modifications were completed. HMC2058 and HMC2049 retained their N-terminal Fmoc group by skipping the deprotection steps after the final amino acid addition. N-myristoyation of HMC2044 and TW1036 occurred after the final amino acid addition and subsequent deprotection. Deprotection was followed by DMF washing (10 × the resin volume) then addition of myristic anhydride (2 mmol) in 9 mL of DMF for 30 minutes at 50°C using microwave heating. After N-terminal modifications were complete the resin was washed with dichloromethane (DCM) three times (10 × the resin volume). Finally, the resin cleavage solution [5 × the resin volume, TFA/TIS (triisopropylsilane)/thioanisole/anisole, 92: 4:2:2, v/v] was added with shaking for 2 h. The solution was collected in a 50 mL conical tube and 20 mL of cold ethyl ether (− 80 °C) was added. The solution was mixed, and the peptide precipitate formed immediately. After centrifugation (10 min at 8000 rpm), the supernatant was decanted, and the peptide was dissolved in distilled water and methanol (5 mL of methanol and 35 mL of water), frozen at −80 °C, and lyophilized for 24–48 h until a white powder was obtained. Peptides were purified by RP-HPLC, and molecular masses were confirmed by LCMS.
Statistics:
GraphPad Prism 7 software was used to perform all statistical analyses. For Sholl data, statistical analysis was performed using two-way ANOVA followed by Bonferroni multiple comparisons test. All other dendritic branching data from Fig.1 and Fig. 4 were analyzed using one-way ANOVA followed by Tukey multiple comparisons test. Dendrite spine data were analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test. Co-immunoprecipitation data were analyzed using one-way ANOVA followed by Dunnett’s multiple comparisons test. Two-tailed Student’s t-test was used for all for all comparisons of dendrite number, length, and branch points in Fig. 5. Outliers were removed using ROUT (Q = 1%) method in GraphPad Prism 7. Extended statistical results are in Supplementary Tables.
Supplementary Material
Acknowledgements
This work was funded by National Science Foundation grant IOS-1353724 and New Jersey Commission on Brain Injury Research grant # CBIR14IRG019 to B.L.F. A.O. was supported by National Institutes of Health Biotechnology Training Grant T32 GM008339-20 and a Predoctoral Fellowship from the New Jersey Commission on Brain Injury Research # CBIR19FEL018. HM was supported by National Institutes of Health IRACDA (Institutional Research and Career Development Award) INSPIRE - IRACDA2K12GM093854–07A1. MS received funding from Norris Cotton Cancer Center, Geisel School of Medicine at Dartmouth.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Eilers J, Konnerth A (1997) Dendritic signal integration. Current Opinion in Neurobiology 7 (3):385–390. doi: 10.1016/S0959-4388(97)80067-0 [DOI] [PubMed] [Google Scholar]
- 2.Hausser M, Spruston N, Stuart GJ (2000) Diversity and dynamics of dendritic signaling. Science (New York, NY) 290 (5492):739–744 [DOI] [PubMed] [Google Scholar]
- 3.Vetter P, Roth A, Hausser M (2001) Propagation of action potentials in dendrites depends on dendritic morphology. Journal of neurophysiology 85 (2):926–937. doi: 10.1152/jn.2001.85.2.926 [DOI] [PubMed] [Google Scholar]
- 4.Kulkarni VA, Firestein BL (2012) The dendritic tree and brain disorders. Molecular and Cellular Neuroscience 50 (1):10–20. doi: 10.1016/j.mcn.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann WE, Moser HW (2000) Dendritic anomalies in disorders associated with mental retardation. Cerebral cortex (New York, NY : 1991) 10 (10):981–991 [DOI] [PubMed] [Google Scholar]
- 6.Stephan KE, Baldeweg T, Friston KJ (2006) Synaptic plasticity and dysconnection in schizophrenia. Biological psychiatry 59 (10):929–939. doi: 10.1016/j.biopsych.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 7.Calabresi P, Picconi B, Parnetti L, Di Filippo M (2006) A convergent model for cognitive dysfunctions in Parkinson’s disease: the critical dopamine-acetylcholine synaptic balance. The Lancet Neurology 5 (11):974–983. doi: 10.1016/s1474-4422(06)70600-7 [DOI] [PubMed] [Google Scholar]
- 8.Blanpied TA, Ehlers MD (2004) Microanatomy of dendritic spines: emerging principles of synaptic pathology in psychiatric and neurological disease. Biological psychiatry 55 (12):1121–1127. doi: 10.1016/j.biopsych.2003.10.006 [DOI] [PubMed] [Google Scholar]
- 9.Südhof TC (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455 (7215):903–911. doi: 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo G (2011) The cytoskeletal and signaling mechanisms of axon collateral branching. Developmental neurobiology 71 (3):201–220. doi: 10.1002/dneu.20852 [DOI] [PubMed] [Google Scholar]
- 11.Stiess M, Bradke F (2011) Neuronal polarization: The cytoskeleton leads the way. Developmental neurobiology 71 (6):430–444. doi: 10.1002/dneu.20849 [DOI] [PubMed] [Google Scholar]
- 12.Jan YN, Jan LY (2010) Branching out: mechanisms of dendritic arborization. Nature reviews Neuroscience 11 (5):316–328. doi: 10.1038/nrn2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conde C, Cáceres A (2009) Microtubule assembly, organization and dynamics in axons and dendrites. Nature Reviews Neuroscience 10 (5):319–332. doi: 10.1038/nrn2631 [DOI] [PubMed] [Google Scholar]
- 14.Parrish JZ, Emoto K, Kim MD, Jan YN (2007) Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annual review of neuroscience 30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907 [DOI] [PubMed] [Google Scholar]
- 15.Georges PC, Hadzimichalis NM, Sweet ES, Firestein BL (2008) The yin-yang of dendrite morphology: unity of actin and microtubules. Molecular neurobiology 38 (3):270–284. doi: 10.1007/s12035-008-8046-8 [DOI] [PubMed] [Google Scholar]
- 16.Sainath R, Gallo G (2015) Cytoskeletal and signaling mechanisms of neurite formation. Cell Tissue Res 359 (1):267–278. doi: 10.1007/s00441-014-1955-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim E, Sheng M (2004) PDZ domain proteins of synapses. Nature Reviews Neuroscience 5 (10):771–781. doi: 10.1038/nrn1517 [DOI] [PubMed] [Google Scholar]
- 18.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science (New York, NY) 290 (5495):1364–1368 [PubMed] [Google Scholar]
- 19.Kilinc D (2018) The Emerging Role of Mechanics in Synapse Formation and Plasticity. Frontiers in Cellular Neuroscience 12 (483). doi: 10.3389/fncel.2018.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, Firestein BL (2006) Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci 26 (40):10164–10176. doi: 10.1523/JNEUROSCI.2379-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sweet ES, Previtera ML, Fernandez JR, Charych EI, Tseng CY, Kwon M, Starovoytov V, Zheng JQ, Firestein BL (2011) PSD-95 alters microtubule dynamics via an association with EB3. The Journal of neuroscience : the official journal of the Society for Neuroscience 31 (3):1038–1047. doi: 10.1523/jneurosci.1205-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yanai H, Satoh K, Matsumine A, Akiyama T (2000) The colorectal tumour suppressor APC is present in the NMDA-receptor-PSD-95 complex in the brain. Genes to cells : devoted to molecular & cellular mechanisms 5 (10):815–822 [DOI] [PubMed] [Google Scholar]
- 23.Munemitsu S, Souza B, Muller O, Albert I, Rubinfeld B, Polakis P (1994) The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer research 54 (14):3676–3681 [PubMed] [Google Scholar]
- 24.Takamori N, Shimomura A, Senda T (2006) Microtubule-bundling activity of APC is stimulated by interaction with PSD-95. Neuroscience letters 403 (1–2):68–72. doi: 10.1016/j.neulet.2006.04.045 [DOI] [PubMed] [Google Scholar]
- 25.Niethammer M, Valtschanoff JG, Kapoor TM, Allison DW, Weinberg RJ, Craig AM, Sheng M (1998) CRIPT, a Novel Postsynaptic Protein that Binds to the Third PDZ Domain of PSD-95/SAP90. 20 (4):693–707. doi: 10.1016/s0896-6273(00)81009-0 [DOI] [PubMed] [Google Scholar]
- 26.Passafaro M, Sala C, Niethammer M, Sheng M (1999) Microtubule binding by CRIPT and its potential role in the synaptic clustering of PSD-95. Nature Neuroscience 2 (12):1063–1069. doi: 10.1038/15990 [DOI] [PubMed] [Google Scholar]
- 27.Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE (2019) Mouse Genome Database (MGD) 2019. Nucleic Acids Res 47 (D1):D801–d806. doi: 10.1093/nar/gky1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaheen R, Faqeih E, Ansari S, Abdel-Salam G, Al-Hassnan ZN, Al-Shidi T, Alomar R, Sogaty S, Alkuraya FS (2014) Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res 24 (2):291–299. doi: 10.1101/gr.160572.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leduc MS, Niu Z, Bi W, Zhu W, Miloslavskaya I, Chiang T, Streff H, Seavitt JR, Murray SA, Eng C, Chan A, Yang Y, Lalani SR (2016) CRIPT exonic deletion and a novel missense mutation in a female with short stature, dysmorphic features, microcephaly, and pigmentary abnormalities. Am J Med Genet A 170 (8):2206–2211. doi: 10.1002/ajmg.a.37780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Jablonski AM, Mojsilovic-Petrovic J, Ding H, Seeholzer S, Newton IP, Nathke I, Neve R, Zhai J, Shang Y, Zhang M, Kalb RG (2017) SAP97 Binding Partner CRIPT Promotes Dendrite Growth In Vitro and In Vivo. eNeuro 4 (6). doi: 10.1523/ENEURO.0175-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kutzing MK, Langhammer CG, Luo V, Lakdawala H, Firestein BL (2010) Automated Sholl analysis of digitized neuronal morphology at multiple scales. J Vis Exp (45). doi: 10.3791/2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweet ES, Langhammer CL, Kutzing MK, Firestein BL (2013) Semiautomated analysis of dendrite morphology in cell culture. Methods Mol Biol 1018:261–268. doi: 10.1007/978-1-62703-444-9_24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langhammer CG, Previtera ML, Sweet ES, Sran SS, Chen M, Firestein BL (2010) Automated Sholl analysis of digitized neuronal morphology at multiple scales: Whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytometry A 77 (12):1160–1168. doi: 10.1002/cyto.a.20954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saro D, Li T, Rupasinghe C, Paredes A, Caspers N, Spaller MR (2007) A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry 46 (21):6340–6352. doi: 10.1021/bi062088k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patra CR, Rupasinghe CN, Dutta SK, Bhattacharya S, Wang E, Spaller MR, Mukhopadhyay D (2012) Chemically modified peptides targeting the PDZ domain of GIPC as a therapeutic approach for cancer. ACS Chem Biol 7 (4):770–779. doi: 10.1021/cb200536r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dotti CG, Sullivan CA, Banker GA (1988) The establishment of polarity by hippocampal neurons in culture. The Journal of Neuroscience 8 (4):1454. doi: 10.1523/JNEUROSCI.08-04-01454.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rademacher N, Kuropka B, Kunde S-A, Wahl MC, Freund C, Shoichet SA (2019) Intramolecular domain dynamics regulate synaptic MAGUK protein interactions. eLife 8:e41299. doi: 10.7554/eLife.41299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Shang Y, Zhang M (2016) Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nature Reviews Neuroscience 17 (4):209–223. doi: 10.1038/nrn.2016.18 [DOI] [PubMed] [Google Scholar]
- 39.Gerrow K, Romorini S, Nabi SM, Colicos MA, Sala C, El-Husseini A (2006) A Preformed Complex of Postsynaptic Proteins Is Involved in Excitatory Synapse Development. Neuron 49 (4):547–562. doi: 10.1016/j.neuron.2006.01.015 [DOI] [PubMed] [Google Scholar]
- 40.El-Husseini AE-D, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 Involvement in Maturation of Excitatory Synapses. Science (New York, NY) 290 (5495):1364. doi: 10.1126/science.290.5495.1364 [DOI] [PubMed] [Google Scholar]
- 41.Sigler A, Oh WC, Imig C, Altas B, Kawabe H, Cooper BH, Kwon H-B, Rhee J-S, Brose N (2017) Formation and Maintenance of Functional Spines in the Absence of Presynaptic Glutamate Release. Neuron 94 (2):304–311.e304. doi: 10.1016/j.neuron.2017.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cane M, Maco B, Knott G, Holtmaat A (2014) The Relationship between PSD-95 Clustering and Spine Stability In Vivo. The Journal of Neuroscience 34 (6):2075. doi: 10.1523/JNEUROSCI.3353-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rademacher N, Kunde S-A, Vera, Sarah (2013) Synaptic MAGUK Multimer Formation Is Mediated by PDZ Domains and Promoted by Ligand Binding. Chemistry & Biology 20 (8):1044–1054. doi: 10.1016/j.chembiol.2013.06.016 [DOI] [PubMed] [Google Scholar]
- 44.Zeng M, Shang Y, Araki Y, Guo T, Huganir RL, Zhang M (2016) Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 166 (5):1163–1175.e1112. doi: 10.1016/j.cell.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akum BF, Chen M, Gunderson SI, Riefler GM, Scerri-Hansen MM, Firestein BL (2004) Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat Neurosci 7 (2):145–152. doi: 10.1038/nn1179 [DOI] [PubMed] [Google Scholar]
- 46.Firestein BL, Brenman JE, Aoki C, Sanchez-Perez AM, El-Husseini AE, Bredt DS (1999) Cypin: a cytosolic regulator of PSD-95 postsynaptic targeting. Neuron 24 (3):659–672 [DOI] [PubMed] [Google Scholar]
- 47.O’Neill KM, Donohue KE, Omelchenko A, Firestein BL (2018) The 3’ UTRs of Brain-Derived Neurotrophic Factor Transcripts Differentially Regulate the Dendritic Arbor. Front Cell Neurosci 12:60. doi: 10.3389/fncel.2018.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon M, Firestein BL (2013) DNA transfection: calcium phosphate method. Methods Mol Biol 1018:107–110. doi: 10.1007/978-1-62703-444-9_10 [DOI] [PubMed] [Google Scholar]
- 49.Patel MV, Sewell E, Dickson S, Kim H, Meaney DF, Firestein BL (2019) A Role for Postsynaptic Density 95 and Its Binding Partners in Models of Traumatic Brain Injury. Journal of Neurotrauma 36 (13):2129–2138. doi: 10.1089/neu.2018.6291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


