Abstract
N-nitroso-containing natural products are bioactive metabolites with antibacterial and anticancer properties. In particular, compounds containing the diazeniumdiolate (N-nitrosohydroxylamine) group display a wide range of bioactivities ranging from cytotoxicity to metal chelation. Despite the importance of this structural motif, knowledge of its biosynthesis is limited. Here, we describe the discovery of a biosynthetic gene cluster in Streptomyces alanosinicus ATCC 15710 responsible for producing the diazeniumdiolate natural product L-alanosine. Gene disruption and stable isotope feeding experiments identified essential biosynthetic genes and revealed the source of the N-nitroso group. Additional biochemical characterization of the biosynthetic enzymes revealed that the non-proteinogenic amino acid L-2,3-diaminopropionic acid (L-Dap) is synthesized and loaded onto a free-standing peptidyl carrier protein (PCP) domain in L-alanosine biosynthesis, which we propose may be a mechanism of handling unstable intermediates generated en route to the diazeniumdiolate. These discoveries will facilitate efforts to determine the biochemistry of diazeniumdiolate formation.
Keywords: biosynthesis, enzymes, natural products, N-nitroso compounds, N-N bond
Graphical Abstract
Diazeniumdiolate biosynthesis: L-alanosine is an unusual diazeniumdiolate-containing natural product. The gene cluster that encodes L-alanosine production has been identified and the origin of the N-nitroso group has been characterized. This discovery advances understanding of the biological origins of this functional group.
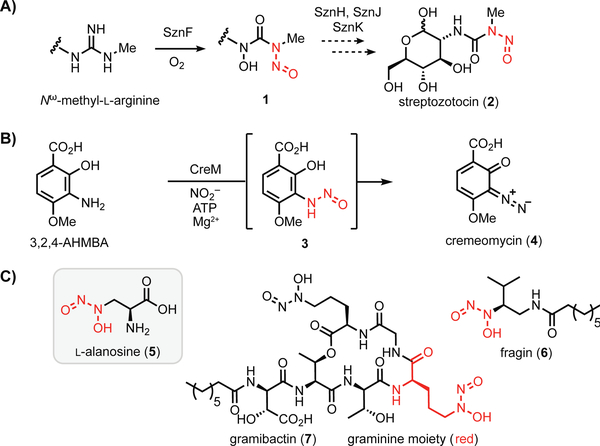
N-nitroso-containing small molecules play prominent roles in human health, disease, and therapeutics development.[1,2] In biological systems, nitric oxide synthases (NOSs) convert L-arginine into L-citrulline and nitric oxide (NO), which can undergo further oxidation to nitrite (NO2−) in aqueous systems. Both NO2− and NO can react with secondary amines to afford N-nitrosamines, which are notorious environmental toxins and carcinogens, yet have been exploited as a bioactive pharmacophore in several N-nitrosourea-containing chemotherapeutic drugs.[1,3] While many of these compounds are formed abiotically, the N-nitrosourea in streptozotocin (2) was recently reported to arise from the action of a metalloenzyme[4] (SznF, Figure 1a), and the diazo functional group in cremeomycin (4)[5] is thought to be produced via a transient N-nitrosamine intermediate (3) generated by an ATP-dependent enzyme (Figure 1b). Recent studies have linked bacterial diazeniumdiolate-containing metabolites to quorum sensing (6) and metal-acquisition (7), revealing an emerging ecological role for this group of natural products (Figure 1c).[6,7] However, the genetic and biochemical basis for diazeniumdiolate biosynthesis remains poorly understood.
Figure 1.
Oxidized nitrogen species in the biosynthesis of N-N bond-containing natural products. A) Biosynthesis of the N-nitrosourea group in streptozotocin. B) Biosynthesis of cremeomycin may proceed through an N-nitrosamine intermediate. C) Selected diazeniumdiolate-containing natural products.
L-alanosine (5, Figure 1c) is a naturally occurring diazeniumdiolate-containing amino acid produced by the bacterium Streptomyces alanosinicus ATCC 15710 that was discovered in the 1960s from a Brazilian soil sample.[8] 5 has since been studied for its antibiotic, antiviral, and antitumor activities. The unusual diazeniumdiolate group confers metal chelating properties to 5.[9] This moiety also has been demonstrated to release NO upon metabolism of 5 by L-amino acid oxidases, generating reactive nitrogen species.[10] In the decades following its isolation, 5 has been demonstrated to act as an antimetabolite targeting de novo purine biosynthesis and has been investigated in clinical trials to treat various tumors (SDX-102).[11]
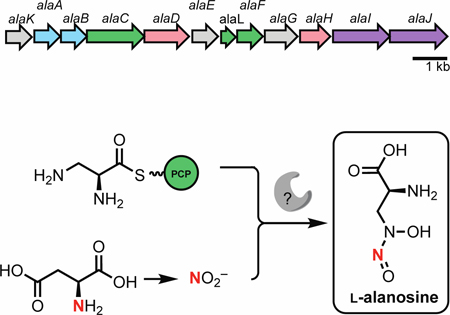
Despite numerous studies exploring 5 as a potential therapeutic agent, the biosynthetic gene cluster responsible for its production has not been identified. Recently, the Eberl and Hertweck groups reported the biosynthetic gene clusters for the N-nitrosamines fragin (6)[6] and gramibactin (7),[7] revealing putative enzymes that could install the diazeniumdiolate moiety. HamC, which has been demonstrated to oxidize p-aminobenzoic acid to p-nitrobenzoic acid in vitro, is proposed to mediate N-N bond formation in fragin biosynthesis, but this putative reaction has yet to be verified in vivo and in vitro. Similarly, the SznF homolog GrbD from the gramibactin pathway is proposed to catalyze N-N bond formation, but its activity has not been demonstrated.[6,12] Elucidating the biosynthesis of 5 would therefore improve our limited insights into enzymatic installation of the diazeniumdiolate, and more broadly the N-nitroso group. Here, we report the discovery of the L-alanosine (ala) biosynthetic gene cluster (Figure 2a). Using a combination of feeding studies, in vivo gene inactivation experiments, and in vitro biochemistry, we have revealed a plausible biosynthetic pathway, paving the way for further understanding of N-nitroso assembly in living organisms.
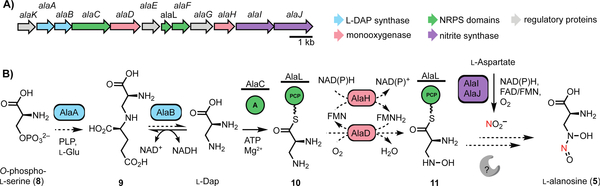
Figure 2.
The L-alanosine gene cluster encodes a pathway for diazeniumdiolate biosynthesis. A) The putative L-alanosine (ala) gene cluster and B) biosynthetic hypothesis for the production of 5 from O-phospho-L-serine (8). Solid arrows represent enzyme reactions confirmed in vitro, dashed arrows represent proposed transformations.
We initially hypothesized that 5 could be derived from N-hydroxylation and N-nitrosation of the putative biosynthetic precursor L-2,3-diaminopropionic acid (L-Dap, Figure 2b), a nonproteinogenic amino acid involved in several siderophore biosynthetic pathways.[13–15] In staphyloferrin B biosynthesis, SbnA uses pyridoxal 5’-phosphate (PLP) as a cofactor to ligate O-phospho-L-serine (8) and L-glutamate to form a dipeptide N-(1-amino-1-carboxyl-2-ethyl)-glutamic acid (9) that is cleaved by deaminase SbnB to generate L-Dap (Figure S1).[16] To identify the ala gene cluster, we sequenced the genome of S. alanosinicus ATCC 15710 and searched for homologs of the L-Dap biosynthetic genes. This strategy revealed a 13.5 kb genomic region that encodes homologs of SbnAB (AlaAB), an amino acid adenylation domain-containing protein (AlaC), a free-standing peptidyl carrier protein (PCP) domain (AlaL), and a thioesterase (TE) enzyme (AlaF) (Figure 2a, Table 1). These biosynthetic enzymes are encoded alongside homologs of the enzymes CreD (AlaJ) and CreE (AlaI), which are known to produce NO2− from L-aspartic acid in cremeomycin biosynthesis,[5] a predicted flavin-dependent N-hydroxylase (AlaD), and a potential flavin reductase (AlaH) (Figure 2a). The putative ala gene cluster also contains a transcriptional regulator (AlaK), a GAF-domain containing protein (AlaE) and a PAS-domain containing protein (AlaG) that likely regulate the transcription of this gene cluster. The ala gene cluster boundaries were initially determined using antiSMASH v 5.0.0 with ClusterBlast and the genome neighborhood feature of IMG/JGI (Figure S8). This analysis revealed highly similar gene clusters in a number of additional Streptomyces species.
Table 1.
Annotations of the alanosine (ala) biosynthetic gene cluster. The nucleotide sequences have been deposited into the NCBI database (Genbank accession numbers for AlaA-L are MN603934-MN603945).
| Protein | Size (aa) | Predicted Function | Closest Homolog | Accession # | %Identity/ %Similarity |
|---|---|---|---|---|---|
| AlaA | 324 | 2,3-diaminopropionate biosynthesis protein | SbnA, Streptomyces hirsutus | WP_055594931.1 | 87/91 |
| AlaB | 353 | 2,3-diaminopropionate biosynthesis protein | SbnB, Streptomyces hirsutus | WP_055594930.1 | 84/89 |
| AlaC | 557 | amino acid adenylation domain-containing protein | Streptomyces hirsutus | WP_055594929.1 | 79/84 |
| AlaD | 385 | acyl-CoA dehydrogenase | Streptomyces hirsutus | WP_055594947.1 | 89/94 |
| AlaE | 190 | hypothetical protein | Streptomyces hirsutus | WP_055594928.1 | 80/88 |
| AlaF | 237 | thioesterase | hypothetical protein, Streptomyces hirsutus | WP_079034952.1 | 76/83 |
| AlaG | 286 | hypothetical protein | Streptomyces hirsutus | WP_055594925.1 | 86/92 |
| AlaH | 177 | NADPH-dependent FMN reductase | Streptomyces hirsutus | WP_055594946.1 | 87/92 |
| AlaI | 657 | FAD/NAD(P)-binding protein | FAD/NAD(P)-binding protein [Streptomyces kanamyceticus] | WP_055544225.1 | 76/82 |
| AlaJ | 470 | nitro-succinate lyase | 3-carboxy-cis,cis-muconate cycloisomerase [Streptomyces sp. BK161] | TDT22211.1 | 75/80 |
| AlaK | 273 | SARP family regulator | CreF [Streptomyces cremeus] | ALA99205.1 | 57/69 |
| AlaL | 79 | carrier protein | MULTISPECIES: [Streptomyces] | WP_030834819.1 | 91/79 |
Based on the predicted functions of the enzymes encoded in this cluster, we proposed a biosynthetic hypothesis for the assembly of 5 (Figure 2b). AlaAB would generate L-Dap, which could be loaded onto the phosphopantethienyl (ppant) arm of the free-standing PCP AlaL by A domain AlaC. This biosynthetic logic parallels the proposed pathway for fragin construction in which diazeniumdiolate installation could occur on enzyme-tethered intermediates.[6] The predicted flavin-binding enzyme AlaD and the flavin reductase AlaH could be a two-component flavin N-monooxygenase involved in forming N-hydroxy-L-Dap (11), and an unknown enzyme (potentially one of the remaining proteins encoded by the ala gene cluster) could install the N-nitroso group using NO2− generated by AlaIJ. Notably, the S. alanosinicus genome does not encode homologs of any biosynthetic enzymes from the fragin and L-graminine pathways. Furthermore, the genome lacks homologs of SznF and KtzT, recently reported N-N bond-forming enzymes in streptozotocin and piperazate biosynthesis, respectively.[4,17] Moreover, homologs of other suspected N-N bond-forming enzymes, Spb40/Tri28 from the s56-p1 and triacsin biosynthetic pathways[18,19] and FzmP/KinJ from fosfazinomycin and kinamycin biosynthetic pathways, are also absent.[20] Our bioinformatics analysis therefore suggests that biosynthesis of 5 either employs a novel N-N bond-forming enzyme or generates the diazeniumdiolate functional group non-enzymatically.
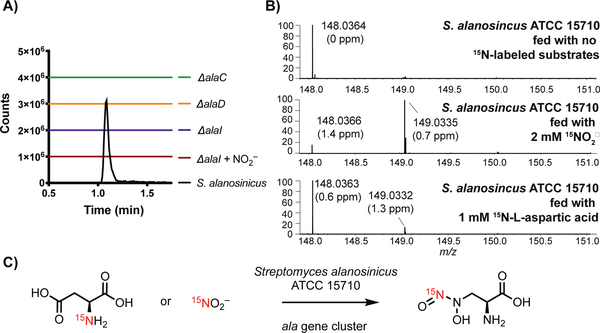
To establish the link between the ala gene cluster and biosynthesis of 5, we performed several gene inactivation experiments in S. alanosinicus. Production of 5 was abolished when alaC, alaD, and alaI were deleted via the well-established PCR-targeting and λ-red-mediated recombination platform (Figure 3a). This confirms that the activity of the A domain (AlaC), redox chemistry (via AlaD) and NO2− generation (via AlaI) are all essential. When the 𝛥alaI mutant was supplemented with NO2−, 5 was not detected (Figure 3a, maroon line). This suggests that AlaI may be important for NO2− incorporation, or that the deletion of alaI has introduced a polar mutation that has impacted a previously undetected regulatory mechanism in the gene cluster. Lastly, the deletion of alaC and concomitant loss of 5 imply that the non-ribosomal peptide synthetase (NRPS) machinery is necessary for biosynthesis and may facilitate the generation of an unstable, enzyme-bound intermediate during diazeniumdiolate assembly. Taken together, these gene-inactivation experiments confirmed the role of the ala gene cluster in the biosynthesis of 5 and identified several indispensable enzymes for further in vitro characterization.
Figure 3.
Gene inactivation and feeding studies link ala biosynthetic genes to L-alanosine (5) production A) Extracted ion chromatograms (EICs) of 5 ([M-H]− = 148.0364) in fermentation extracts of gene deletion mutants and wild type S. alanosinicus. The EICs are generated within a 5 ppm window. B) Feeding studies using 15N sources with wild type S. alanosinicus to determine the source of the distal N-nitroso nitrogen atom of 5. LC-MS/MS analysis confirmed the distal N-nitroso nitrogen was labeled when 15N-nitrite was fed (Figure S2 and S3). Incorporation of 15N into 5 was also observed when 3 mM 15NO2− was fed, although the cell viability was lower due to toxicity (Figure S3) C) Summary of the results of the feeding studies.
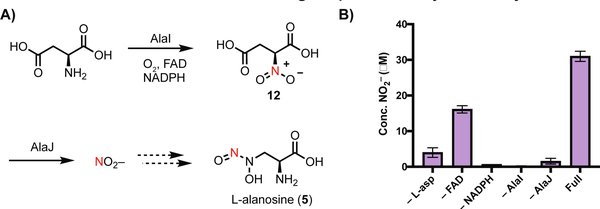
Because the putative ala gene cluster encodes homologs of the nitrite-generating enzymes CreD and CreE (AlaJ and AlaI, respectively, Figure 4a), we proposed that the precursor of the distal nitrogen of the N-nitroso group is NO2−. In addition to its role in non-enzymatic N-nitrosation reactions, NO2− is used biosynthetically as a source of nitrogen in the formation of other N-N bond linkages, including diazo and hydrazide groups.[20,21] To test this hypothesis, we overexpressed and purified N-His6-AlaI and N-Strep-AlaJ, and we demonstrated their ability to generate NO2− from L-aspartic acid with the necessary cofactors (Figure 4b). We next fed 15NO2− to cultures of S. alanosinicus and observed incorporation of 15N into 5 (~ 80%, Figure 3b). Tandem high resolution-mass spectrometry (HR-LCMS/MS) revealed the distal N-nitroso nitrogen is labeled (Figure S2 and S3). Feeding 15NO3− also resulted in lower enrichment of 15N-labeled 5 (Figure S5); this could potentially arise from conversion of NO3− to NO2− by nitrate reductases encoded by S. alanosinicus. Finally, we also fed 15N-L-aspartic acid, the presumed precursor of 15NO2− and observed enrichment of 15N-5 (~ 10%, Figure 3b). Together, these results suggest that the biosynthesis of 5 employs NO2− generated from L-aspartic acid, and the distal nitrogen of the diazeniumdiolate is derived from NO2−.
Figure 4.
AlaI and AlaJ are a nitrite synthase. A) Proposed mechanism of NO2− generation from L-aspartic acid by flavin dependent monooxygenase AlaI and nitrosuccinate lyase AlaJ B) AlaI and AlaJ produce NO2− from L-aspartic acid in vitro. NO2- production was detected by the Griess assay. Assay mixtures contained FAD (10 μM), NADPH (5 mM), N-Strep-AlaJ (5 μM), N-His6-AlaI (5 μM) and L-aspartic acid (1 mM) in reaction buffer (50 mM HEPES, 10 mM MgCl2, pH = 8.0) and were analyzed after a 1 h incubation at room temperature. AlaI purified with FAD, hence we still observed activity without exogenous FAD added. Data are mean ± standard deviation (s.d.) of two biological replicates.
Having established the origin of the N-nitroso group in 5, we next sought to confirm the role of L-Dap in this biosynthetic pathway. We first overexpressed and purified N-His6-AlaB for in vitro biochemical assays. SbnB, a homolog of AlaB, was previously characterized by performing the L-Dap synthase reaction in reverse to generate 9 (Figure S1).[16] We employed the same strategy to characterize AlaB by incubating the enzyme with L-Dap and NADH. We observed rapid consumption of NADH and production of 9 (Figure 2b, Figure S6 and S7). Therefore, AlaB is a functional homolog of the previously characterized L-Dap synthase component SbnB.[16]
The involvement of a free-standing A domain in the biosynthesis of 5 and its putative loading of L-Dap are unusual, as there are only three examples of biochemically characterized A domains that utilize this amino acid.[22–24] A domain active sites contain 8–10 binding-pocket residues that dictate substrate specificity.[25] Several bioinformatic methods that predict A domain substrate specificity from these amino acid residues have been developed.[26] However, these tools[27] failed to predict L-Dap as the substrate of AlaC. The same is true of other A domains that have been experimentally demonstrated to activate L-Dap (Table S1).
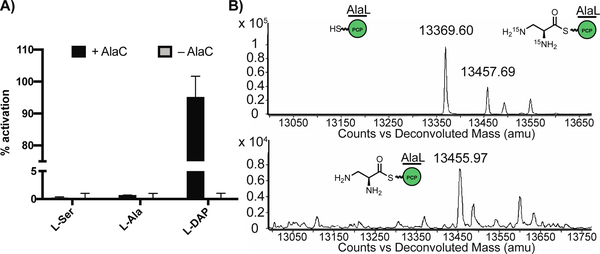
To test if AlaC can recognize and load L-Dap, we overexpressed and purified N-His6-tagged AlaC and N-His6-C-His6-tagged AlaL for in vitro biochemical assays. The preferred substrate of AlaC was confirmed to be L-Dap using the ATP-[32P]Pi exchange assay (Figure 5a). The ability of purified apo-AlaL to undergo successful posttranslational modification was determined via incubation with the promiscuous ppant transferase Sfp and the fluorescent coenzyme A (CoA) analog BODIPY-CoA.[28] Finally, we demonstrated that isotopically labeled L-15N2-Dap was activated by AlaC and loaded onto the ppant arm of holo-AlaL using whole-protein mass spectrometry (Figure 5b). Thus, the NRPS machinery encoded in the ala gene cluster is capable of activating L-Dap and loading it onto the PCP AlaL. Given that alaC was determined to be necessary for the production of 5, the AlaL-tethered L-Dap aminoacyl thioester 10 (Figure 2b) is likely a biosynthetic intermediate.
Figure 5.
AlaC selectively activates L-Dap for loading onto AlaL. A) ATP-32PPi exchange assay shows that AlaC preferentially activates L-Dap over other amino acids from 100 μL incubations of N-His6-AlaC (1 μM) with 5 mM dithiothreitol, 5 mM ATP, 1 mM of amino acid substrate, and 4 mM Na4PPi/[32P]PPi in reaction buffer (50 mM HEPES, 200 mM NaCl, 10 mM MgCl2, pH = 8) at room temperature for 30 min. Data are mean ± s.d. of two biological replicates. B) Deconvoluted whole protein mass spectra (ESI+) showing holo-N-His6-AlaL-C-His6 (13369.60) is loaded with 15N2-L-Dap (13457.69) or L-Dap (13455.97). 50 μL reaction mixtures were set up with coenzyme A (1 mM), Sfp (5 μM), and N-His6-AlaL-C-His6 (20 μM) in a solution of reaction buffer (50 mM HEPES, 200 mM NaCl, 10 mM MgCl2, pH = 8). After incubation at room temperature for 2 h, AlaC (20 μM) and L-Dap or L-15N2-Dap (250 μM) were added, followed by ATP (5 mM) to initiate the reaction. Incubated for 1 h at room temperature.
In summary, we have identified a set of genes that are required for the biosynthesis of 5 in S. alanosinicus. While genetic deletion has confirmed these genes are essential, we cannot currently conclude they are minimally required, as all attempts to heterologous express the ala gene cluster in Streptomyces lividans TK64 have been unsuccessful. The ala gene cluster we have introduced into this heterologous host is flanked by ~9 kb of native genomic DNA. This negative result could suggest that the ala gene cluster is not expressed in this system, that an additional enzyme(s) outside of the gene cluster might be required for L-alanosine production, or that S. alanosinicus produces cofactors and chaperones that are necessary for L-alanosine production that are not present in S. lividans TK64.
We have also demonstrated the importance of the free-standing NRPS biosynthetic machinery to this pathway, both by generating genetic knockouts of S. alanosinicus and through in vitro biochemical assays. We have confirmed the role of L-Dap as a biosynthetic precursor by showing that AlaB is a functional homolog of the L-Dap synthase SbnB and characterizing a new L-Dap specific A domain. The previously characterized biosynthetic uses of L-Dap have been as a building block for NRPS assembly lines, so its utilization as the core of a secondary metabolite is unusual. This may suggest that as yet undiscovered non-ribosomal peptide natural products contain L-alanosine building blocks in the same manner as gramibactin contains L-graminine building blocks.[7] We have also demonstrated a critical role for NO2− synthesis in this pathway, having confirmed through stable-isotope feeding studies that L-aspartic acid is the source of the N-nitroso group and shown that AlaI and AlaJ generate NO2− from L-aspartic acid in vitro. The presence of known NO2− generation enzymes in the ala gene cluster, and confirmation of their necessity for the production of 5, provides preliminary insights into diazeniumdiolate formation.
However, the mechanism of diazeniumdiolate formation in L-alanosine biosynthesis remains to be determined, and the remaining proteins encoded by the ala gene cluster may prove to be important in this process. The hypothetical proteins AlaE and AlaG contain a GAF and PAS domain, respectively, and may be part of a nitric oxide sensing system. It is possible that the PAS domain, which contains residues for heme-binding, may be acting to bind nitric oxide (or nitrite) and promote N-N bond formation in L-alanosine biosynthesis. As highlighted earlier, it is also possible that an N-N bond forming enzyme(s) is encoded elsewhere in the S. alanosinicus genome. We have found a potential homolog of the diazotizing enzyme from cremeomycin biosynthesis[5] (CreM 53.9% identity, 67.0% similarity) in the S. alanosinicus genome, and while it appears to be part of a separate biosynthetic gene cluster, we have not yet ruled out its invovlement in L-alanosine biosynthesis. If this enzyme is not involved, the lack of known homologs of N–nitrosating enzymes in the ala gene cluster and the S. alanosinicus genome hints at a potentially distinct mechanism of N-N bond-formation in L-alanosine biosynthesis.
Supplementary Material
Acknowledgements
We thank Dr. L. Zha for assistance with the ATP-PPi exchange assay, Dr. B. A. Schneider for assistance with the BODIPY-CoA loading assay, and Dr. A. J. Waldman for guidance with the nitrite production assay. M.E.M. acknowledges the Damon Runyon Foundation for a Postdoctoral Fellowship (DRG 2307-17). We acknowledge financial support from the NIH (DP2 GM105434), a Cottrell Scholar Award (to E.P.B.), a Camille Dreyfus Teacher-Scholar Award (to E.P.B.), and Harvard University.
Footnotes
Experimental Section
Complete experimental details are provided in the Supporting Information.
References
- [1].Lundberg JO, Weitzberg E, Gladwin MT, Nat. Rev. Drug Discov. 2008, 7, 156–167. [DOI] [PubMed] [Google Scholar]
- [2].Lundberg JO, Weitzberg E, Cole JA, Benjamin N, Nat. Rev. Microbiol.2004, 2, 593–602. [DOI] [PubMed] [Google Scholar]
- [3].Weiss RB, Issell BF, Cancer Treat. Rev. 1982, 9, 313–330. [DOI] [PubMed] [Google Scholar]
- [4].(a) Ng TL, Rohac R, Mitchell AJ, Boal AK, Balskus EP, Nature 2019, 566, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) He HY, Henderson AC, Du YL, Ryan KS, J. Am. Chem. Soc. 2019, 141, 4026–4033. [DOI] [PubMed] [Google Scholar]
- [5].(a) Waldman AJ, Pechersky Y, Wang P, Wang JX, Balskus EP, ChemBioChem 2015, 16, 2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sugai Y, Katsuyama Y, Ohnishi Y, Nat. Chem. Biol. 2015, 12, 73–77. [DOI] [PubMed] [Google Scholar]
- [6].Jenul C, Sieber S, Daeppen C, Mathew A, Lardi M, Pessi G, Hoepfner D, Neuburger M, Linden A, Gademann K, Eberl L, Nat. Commun. 2018, 9, 1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hermenau R, Ishida K, Gama S, Hoffman B, Pfeifer-Leeg M, Plass W, Mohr JF, Wichard T, Saluz H-P, Hertweck C, Nat. Chem. Biol. 2018, 14, 841–843. [DOI] [PubMed] [Google Scholar]
- [8].Murthy YKS, Thiemann JE, Coronelli C, Sensi P, Nature 1966, 211, 1198–1199. [DOI] [PubMed] [Google Scholar]
- [9].Powis G, Kovach JS, Biochemical Pharmacology 1981, 30, 771–776. [DOI] [PubMed] [Google Scholar]
- [10].Alston TA, Bright HJ, Biochem. Biophys. Res. Commun. 1982, 105, 560–566. [DOI] [PubMed] [Google Scholar]
- [11].Kindler HL, Burris HA 3rd, Sandler AB, Oliff IA, Invest. New Drugs 2009, 27, 75–81. [DOI] [PubMed] [Google Scholar]
- [12].Hermenau R, Mehl JL, Ishida K, Dose B, Pidot SJ, Stinear TP, Hertweck C, Angew. Chem. Int. Ed. Engl 2019, 58, 13024–13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Felnagle EA, Rondon MR, Berti AD, Crosby HA, Thomas MG, Appl. Environ. Microbiol. 2007, 73, 4162–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Thomas MG, Chan YA, Ozanic SG, Antimicrob. Agents Chemother. 2003, 47, 2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dale SE, Doherty-Kirby A, Lajoie G, Heinrichs DE, Infect. Immun. 2004, 72, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kobylarz MJ, Grigg JC, Takayama SJ, Rai DK, Heinrichs DE, Murphy ME, Chem Biol. 2014, 21, 379–388. [DOI] [PubMed] [Google Scholar]
- [17].Du Y-L, He H-Y, Higgins MA, Ryan KS, Nat. Chem. Biol 2017, 13, 836–838. [DOI] [PubMed] [Google Scholar]
- [18].(a) Matsuda K, Hasebe F, Shiwa Y, Kanesaki Y, Tomita T, Yoshikawa H, Shin-ya K, Kuzuyama T, Nishiyama M, ACS Chem. Biol. 2017, 12, 124–131. [DOI] [PubMed] [Google Scholar]; (b) Matsuda K, Tomita T, Shin-ya K, Wakimoto T, Kuzuyama T, Nishiyama M, J. Am. Chem. Soc. 2018, 140, 9083–9086. [DOI] [PubMed] [Google Scholar]
- [19].Twigg FF, Cai W, Huang W, Liu J, Sato M, Perez TJ, Geng J, Dror MJ, Montanez I, Tong TL, Lee H, Zhang W, ChemBioChem 2019, 20, 1145–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang KKA, Ng TL, Wang P, Huang ZD, Balskus EP, van der Donk WA, Nat. Commun. 2018, 9, 3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Graham DE, Spain JC, Parry RJ, Hettich RL, Mahan KM, Klingeman DM, Giannone RJ, Gulvick CA, Fida TT. Nitration Enzyme Toolkit for the Biosynthesis of Energetic Materials. 2018, Oak Ridge National Lab, Oak Ridge, TN. [Google Scholar]
- [22].Xu Z, Sun Z, Li S, Xu Z, Cao C, Xu Z, Feng X, Xu H, Scientific Reports 2015, 5, 17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].(a) Zhao C, Song C, Luo Y, Yu Z, Sun M, FEBS Letters 2008, 582, 3125–3131. [DOI] [PubMed] [Google Scholar]; (b) Kevany BM, Rasko DA, Thomas MG, Appl. Environ. Microbiol. 2008, 75, 1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Felnagle EA, Rondon MR, Berti AD, Crosby HA, Thomas MG, Appl. Environ. Microbiol. 2007, 73, 4162–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hur GH, Vickery CR, Burkart MD, Nat. Prod. Rep. 2012, 29, 1074–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chevrette MG, Aicheler F, Kohlbacher O, Currie CR, Medema MH, Bioinformatics 2017, 33, 3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bachman BO, Ravel J, 2009. Methods in enzymology, 458, 181–217. [DOI] [PubMed] [Google Scholar]
- [28].Nakamura H, Wang JX, Balskus EP, Chem. Sci. 2015, 6, 3816–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.