Abstract
Members of the Bacteroidetes phylum, represented by Alistipes finegoldii, are prominent anaerobic, Gram-negative inhabitants of the gut microbiome. The lipid biosynthetic pathways were analyzed using bioinformatic analyses, lipidomics, metabolic labeling and biochemistry to characterize exogenous fatty acid metabolism. A. finegoldii only produced saturated fatty acids. The most abundant lipids were phosphatidylethanolamine (PE) and sulfonolipid (SL). Neither phosphatidylglycerol nor cardiolipin are present. PE synthesis is initiated by the PlsX/PlsY/PlsC pathway, whereas the SL pathway is related to sphingolipid biosynthesis. A. finegoldii incorporated medium-chain fatty acids (≤14 carbons) into PE and SL after their elongation, whereas long-chain fatty acids (≥16 carbons) were not elongated. Fatty acids >16 carbons were only incorporated into the 2-position at the PlsC step, the only biosynthetic enzyme that utilizes long-chain acyl-ACP. The ability to assimilate a broad-spectrum of fatty acid chain lengths present in the gut environment is due to the expression of two acyl-acyl carrier protein (ACP) synthetases. Acyl-ACP synthetase 1 had a substrate preference for medium-chain fatty acids and synthetase 2 had a substrate preference for long-chain fatty acids. This unique combination of synthetases allows A. finegoldii to utilize both medium- and long-chain fatty acid nutrients available in the gut environment to assemble its membrane lipids.
Keywords: fatty acid synthesis, bacteria, acyl-CoA synthetase, acyl-ACP synthetase, phospholipid, sulfonolipid, Bacteroidetes, microbiome
Graphical abstract
Members of the Bacteroidetes phylum are major contributors to the human gut microbiome. Here, we show that Alistipes finegoldii, an anaerobic commensal, acquires the fatty acid nutrients available in the gut environment to construct its membrane lipids utilizing a unique combination of acyl-acyl carrier protein synthetases with different substrate selectivities.
Introduction
The human gut microbiome is a focus of active research because it constitutes the greatest biomass of commensal microorganisms in humans and has been implicated in obesity, non-alcoholic fatty liver disease, colorectal cancer, inflammatory bowel disease, and other conditions (Schmidt et al., 2018; Kau et al., 2011). Despite their importance to human physiology, details of the biochemistry and physiology of bacteria that constitute the gut microbiome are just beginning to emerge. Most gut bacteria are strict anaerobes and initially species were known only by their 16s RNA sequences (Caporaso et al., 2011). These species were initially unculturable, but recently methods for the systematic isolation of hundreds of species have been developed (Browne et al., 2016; Lagier et al., 2018). The phylum Bacteroidetes contains rod-shaped Gram-negative bacteria that are widely distributed in the environment as well as in the guts and on the skin of animals (Gupta and Lorenzini, 2007). Members of class Bacteroidia are best known as major contributors to the 1.5 kg of bacteria in the healthy human gut microbiome (Human Microbiome Project, 2012; Johnson et al., 2017). Perturbations in the abundance of various Bacteroidia species in the microbiome, such as by diet or antibiotic treatment, have been associated with metabolic diseases such as diabetes and obesity (Finucane et al., 2014; Walters et al., 2014). Although Bacteroidetes are Gram-negative, they are phylogenetically distant from the well-characterized Proteobacteria phylum containing common Gram-negative bacteria (Johnson et al., 2017). These Gram-negative bacteria are thought to have lipid biosynthetic systems like those described in Escherichia coli, but the metabolic pathways have not been investigated in detail and they do contain unusual lipids. One unique feature of the environmental Bacteroidetes taxa is the presence of sphingolipids and sphingolipid-related lipids, like sulfonolipids (SL) (Geiger et al., 2010; Sohlenkamp and Geiger, 2016; Heaver et al., 2018). SL are an unusual class of sphingolipids containing sulfonic acid in the sphingoid base, which is derived from cysteic acid and saturated 15-carbon fatty acids (White, 1984; Walker et al., 2017; Abbanat et al., 1986; Godchaux and Leadbetter, 1988). SL are found in the outer membrane (Godchaux and Leadbetter, 1988) and are required for gliding activity in Cytophaga species (Abbanat et al., 1986). Other unusual lipids found in Bacteroidetes include acylated amino acids and dipeptides and various ceramide derivatives (Farrokhi et al., 2013; Olsen and Nichols, 2018; Nemati et al., 2017; Rizza et al., 1970).
Despite the association of gut microbiome Bacteroidetes taxa with human health and disease, little is known about their biology in general, or their fatty acid metabolism. This study investigates the pathways and enzymes responsible for the utilization of exogenous fatty acids in Alistipes finegoldii using a combination of bioinformatics, metabolic labeling and biochemistry. A. finegoldii is a fully-sequenced representative of the commensal gut Bacteroidetes that can be cultured in the laboratory. A. finegoldii encodes a bacterial type II fatty acid biosynthesis system (FASII) that primarily produces saturated fatty acids ≤16 carbons. The major lipid species in A. finegoldii are phosphatidylethanolamine (PE) and sulfonolipid (SL), and they lack phosphatidylglycerol. A. finegoldii efficiently incorporates extracellular medium-chain fatty acids (≤14 carbons) into PE and SL after their elongation by FASII. Long-chain fatty acids (≥16 carbons) are incorporated, but not elongated. The incorporation of the common dietary unsaturated fatty acids (18:1 and 18:2) is restricted to the 2-position of PE and excluded from SL. These results show that A. finegoldii encodes a unique combination of acyl-ACP synthetases with different substrate specificities to exploit the fatty acid nutrients provided in the gut environment to manufacture its membrane lipids.
Results
Roadmap for Alistipes lipid synthesis
A. finegoldii is a fully-sequenced Bacteroidetes resident of the human gut that can be propagated in an oxygen-free environmental chamber. We began with a bioinformatics analysis of the enzymes in bacterial fatty acid and phospholipid synthesis to provide a roadmap for the investigation of fatty acid metabolism in this organism (Table 1). Phospholipid synthesis is composed of 4 modules. First, acetyl-CoA generated from central metabolism is activated into malonyl-CoA in the initiation module. A. finegoldii is not predicted to encode homologs of known acetyl-CoA carboxylase complex genes (accABCD) but does encode homologs of the malonyl-CoA-ACP transacylase (FabD) and 3-ketoacyl-ACP synthase III (FabH) (Table 1). Second, a four-enzyme repetitive cycle extends the chain length of the acyl-ACP in the elongation module. There are two enzymes predicted to carry out the enoyl-ACP reductase step, FabI and FabK (Table 1). There is no evidence for genes (fabA, fabB, fabM, etc.) required to introduce double bonds into the growing acyl chain leading to the clear prediction that A. finegoldii does not synthesize unsaturated fatty acids. A. finegoldii is predicted to encode for the PlsX/Y/C pathway for phosphatidic acid synthesis in the acyltransfer module (Table 1), which is found in all characterized bacterial taxa except Gammaproteobacteria (Yao and Rock, 2013). The only known phospholipid A. finegoldii is predicted to synthesize is PE (Table 1). Notably absent is the pgsA gene.
Table 1. Predicted lipid metabolism genes in A. finegoldii.
Gene identifications were made using tBLASTn to query Uniprot sequences against A. finegoldii DSM 17242 taxid: 679935.
| FASII | ALFI_RS ID | Alfi ID | E value | Query (Uniprot) |
|---|---|---|---|---|
| FabD | ALFI_RS12490 | Alfi_2558 | 1.00E-53 | P0AAI9 |
| FabH | ALFI_RS02965 | Alfi_0603 | 2.00E-69 | P0A6R0 |
| FabF | ALFI_RS01295 | Alfi_0254 | 1.00E-104 | P0AAI5 |
| FabG | ALFI_RS14995 | Alfi_3068 | 5.00E-63 | P0AEK2 |
| FabZ | ALFI_RS00450 | Alfi_0083 | 2.00E-32 | P0A6Q6 |
| FabI | ALFI_RS10055 | Alfi_2059 | 8.00E-27 | A0A1D3INF0 |
| FabK | ALFI_RS01405 | Alfi_0277 | 1.00E-51 | Q9FBC5 |
| Phospholipid synthesis | ||||
| PlsX | ALFI_RS02970 | Alfi_0604 | 2.00E-53 | P65739 |
| PlsY | ALFI_RS12850 | Alfi_2636 | 7.00E-27 | P67164 |
| PlsC | ALFI_RS14035 | Alfi_2876 | 4.00E-11 | P26647 |
| CdsA | ALFI_RS12060 | Alfi_2470 | 1.00E-28 | P0ABG1 |
| PssA | ALFI_RS11535 | Alfi_2358 | 3.00E-16 | P39823 |
| Psd | ALFI_RS12065 | Alfi_2471 | 1.00E-22 | A0A1P8Y2N7 |
| Sulfonolipid synthesis | ||||
| Condensing enzyme | ALFI_RS07180 | Alfi_1465 | 2.00E-59 | P22557 |
| Condensing enzyme | ALFI_RS05985 | Alfi_1224 | 4.00E-37 | P22557 |
| O-Acyltransferase | ALFI_RS07510 | Alfi_1534 | 3.00E-06 | P26647 |
| Synthetases | ||||
| Aas1 | ALFI_RS12845 | Alfi_2635 | 1.00E-39 | P69451 |
| Aas2 | ALFI_RS01860 | Alfi_0371 | 2.00E-41 | Q5F969 |
| Alfi_3181 | ALFI_RS15545 | Alfi_3181 | 1.00E-34 | P69451 |
The Bacteroidetes phylum has been classified into environmental-associated Bacteroidetes and human/animal-associated Bacteroidetes groups (Johnson et al., 2017). Both environmental Bacteroidetes and human Bacteroidetes lack a pgsA gene. Human-associated Bacteroidetes lack genes encoding the AccABCD complex, but environmental-associated Bacteroidetes encode for accABCD gene homologs, suggesting that the commensal bacteria may use another mechanism to produce malonyl-CoA. Chlorobi is closely related to Bacteroidetes (Johnson et al., 2017) and they encode for accABCD and pgsA gene homologs. Together, the bioinformatics analysis suggests that phosphatidylglycerol is missing in the Bacteroidetes phylum, but present in the Chlorobi. Sulfonolipids are sphingolipid-related lipids known to be produced by Bacteroidetes taxa (Walker et al., 2017) that are absent in bacterial models such as E. coli or S. aureus. Genes homologous to 2-amino-3-ketobutyrate coenzyme A ligase and 8-amino-7-oxononanoate synthase were identified that correlate with the presence of SL in Bacteroidetes (Walker et al., 2017) (Table 1). These enzymes are members of the α-oxoamine synthase family that includes serine palmitoyl transferase, the first step in sphingolipid synthesis. However, the biochemical function(s) of these genes in SL biosynthesis has not been experimentally validated. The acyltransferase gene (Alfi_1534) predicted in the SL pathway (Table 1) has a PlsC-like domain and may contain a flippase/acyltransferase domain. The genes predicted to be involved in the synthesis of sulfonolipids are found in both environmental- and human-associated Bacteroidetes.
Lipid composition of A. finegoldii
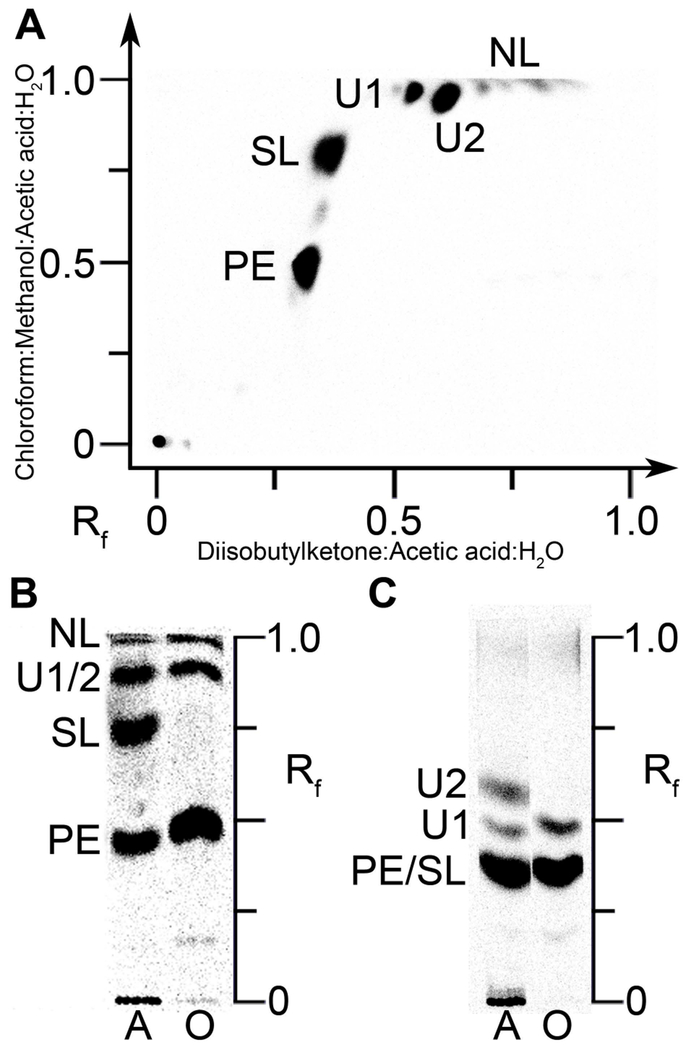
The lipid metabolism of A. finegoldii was first investigated by labeling an A. finegoldii cell culture with [14C]acetate and separating the lipid species by 2-dimensional thin-layer chromatography (Fig. 1A). Four major lipid classes were resolved, and quantitation of the chromatograms showed that the label was distributed between PE (37%), SL (28%), and two unknown lipids referred to as U1 (9%) and U2 (25%). PE is widely distributed in bacteria, but SL is more restricted in its distribution (Walker et al., 2017; Baronio et al., 2010). There were also two minor, unidentified neutral lipids (NL) (Fig. 1A). We were unable to detect phosphatidylglycerol or cardiolipin by metabolic labeling or mass spectrometry in A. finegoldii lipid extracts. We were able to easily detect these two phospholipids in S. aureus extracts but there was no indication that they were present in A. finegoldii. These analytical data support the conclusion from the bioinformatic analysis that A. finegoldii lacks two acidic phospholipids that are major membrane phospholipids in most bacteria.
Figure 1. Polar lipids of A. finegoldii.
PE, phosphatidylethanolamine; SL, sulfonolipid; U1 and U2 were not identified; NL, neutral lipids. PE and SL were identified by mass spectrometry analysis as shown in subsequent figures.
A. A. finegoldii was labeled with [14C]acetate and the lipid extract was separated by two-dimensional chromatography on Silica Gel H thin layers using chloroform:methanol:acetic acid:water (80/25/10/2, v/v) in the first dimension followed by diisobutylketone:acetic acid:water (80/55/15, v/v) in the second dimension. The labeled lipids were visualized with a PhosphorImager system.
B. Lipid extracts from cells labeled with either [14C]acetate (A) or [14C]oleate (O) were separated using the first-dimension solvent system.
C. Separation of [14C]acetate (A) or [14C]oleate (O) labeled lipids using the second-dimension solvent system.
The second experiment labeled the growing cells with either [14C]acetate or [14C]oleate followed by the analysis of the samples in two, 1-dimensional chromatography systems (Figs. 1B and 1C). PE was labeled with either [14C]acetate or [14C]oleate; however, SL was only labeled with [14C]acetate (Fig. 1B). U1 was labeled with either [14C]acetate or [14C]oleate, but U2 was only labeled with [14C]acetate (Fig. 1C). These data showed that A. finegoldii was able to incorporate exogenous oleate into PE, but fatty acids derived from de novo FASII were the only acyl chains incorporated into SL. Thus, A. finegoldii can activate and incorporate exogenous fatty acids into its glycerolipids.
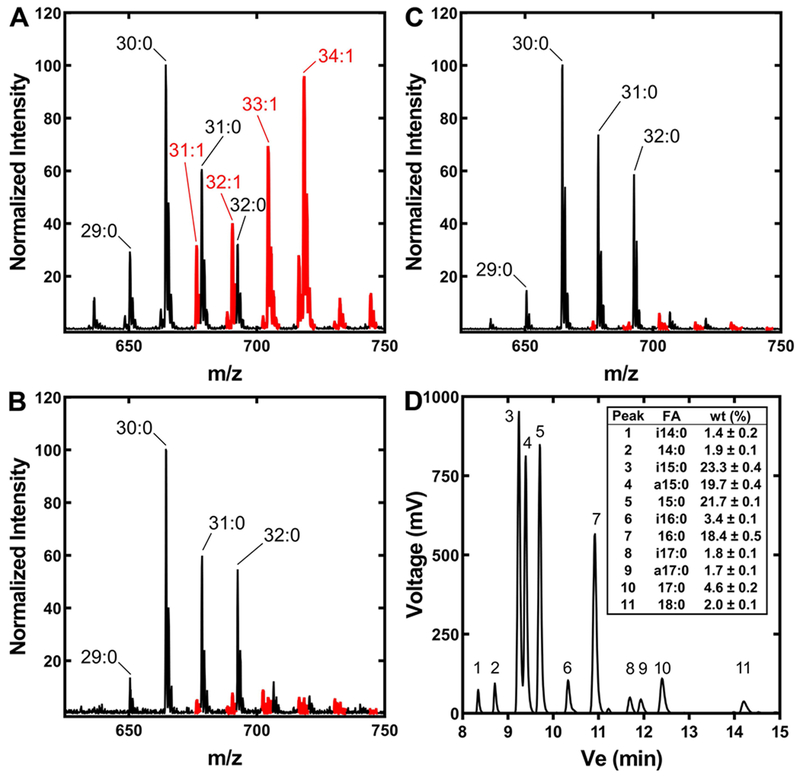
The PE mass spectrum of A. finegoldii grown in the recommended DSM Medium 104 showed the presence of molecular species with unsaturated fatty acids (Fig. 2A). The fatty acid was identified as oleate (18:1Δ9) by gas chromatography, with no other unsaturated fatty acid present in a significant amount (not shown). This result was at odds with the bioinformatic pathway analysis that indicated that A. finegoldii does not make unsaturated fatty acids (Table 1). DSM Medium 104 for the growth of A. finegoldii contains 0.1% Tween 80 (polyethylenesorbitan monooleate), a nonionic detergent containing esterified oleate. Removal of Tween 80 from the growth medium almost completely eliminated the presence of oleate in the mass spectrum of PE molecular species (Fig. 2B). These data showed that Tween 80 was the source of the oleate. There was no difference in the growth rate of A. finegoldii in the presence or absence of Tween 80 (Fig. 3A), illustrating that this additive is not needed for optimal growth of the organism. However, the mass spectrum in the medium lacking Tween 80 also showed several minor molecular species that likely contained unsaturated fatty acids (Fig. 2B). Removal of the beef extract from the growth medium by culturing on WCAB defined minimal medium lowered unsaturated PE molecular species even further (Fig. 2C). The fatty acid composition of A. finegoldii grown on WCAB minimal medium was determined by gas chromatography (Fig. 2D). Fifteen carbon fatty acids were the most abundant (65%) and the bacterium synthesized straight chain and branched-chain iso and anteiso pentadecanoic acid. The prevalence of branched-chain fatty acids meant that FabH uses branched-chain acyl-CoAs derived from branched-chain amino acid degradation precursors to generate the corresponding iso and anteiso fatty acids (Choi et al., 2000). The abundance of 15:0 suggests propionyl-CoA is used for initiation, and the 16:0 fatty acids suggests initiation from either acetyl- or butyryl-CoA, all of which are fermentation products of the Bacteroidetes. Stearate was only 2% of the total, setting the upper limit for the elongation of acyl-ACP by A. finegoldii FASII. Although A. finegoldii grew more slowly and to a lower density in the WCAB medium than in DSM Medium 104 (Fig. 3A), these data confirm that the bacterium lacks the capacity for de novo unsaturated fatty acid biosynthesis.
Figure 2. A. finegoldii PE and fatty acid composition.
A. finegoldii was grown in three different media, the lipids extracted and the PE molecular species profiled by mass spectrometry. PE molecular species containing an unsaturated fatty acid are indicated in red.
A. A representative mass spectrum of PE molecular species from A. finegoldii grown in DSM Medium 104.
B. A representative mass spectrum of PE molecular species from A. finegoldii grown in DSM Medium 104 without Tween 80.
C. A representative mass spectrum of PE molecular species from A. finegoldii grown in a defined minimal medium (Wilkins-Chalgren Anaerobe Broth, WCAB).
D. Fatty acid composition of A. finegoldii grown in WCAB medium. Methyl esters were prepared from lipid extracts and a representative gas chromatogram is shown. Inset, average fatty acid composition from three biological replicates. Only fatty acids >1% of the total were quantified. Mean ± S.E.M. i, iso; a, anteiso.
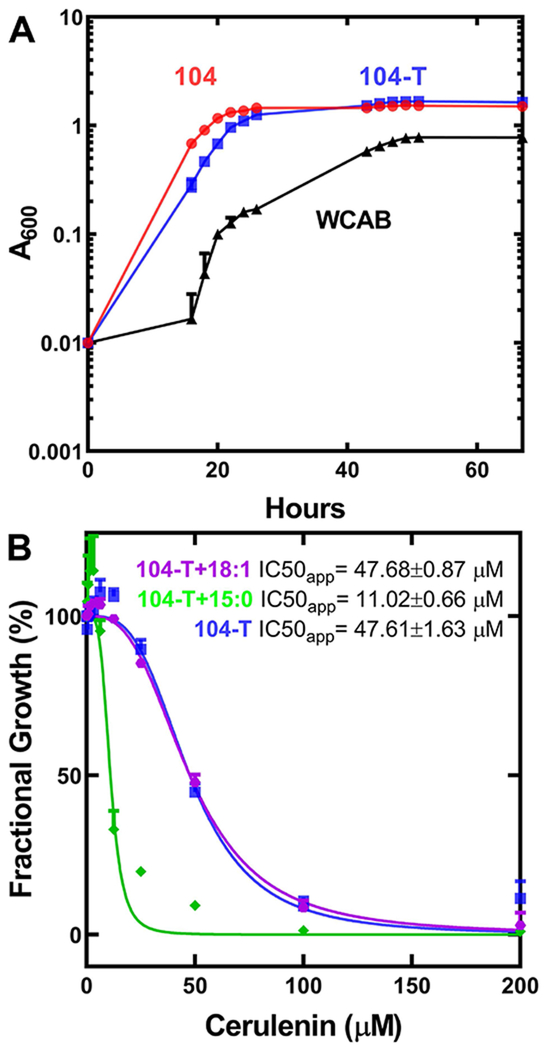
Figure 3. A. finegoldii growth and FASII essentiality.
A. The growth of A. finegoldii was monitored in either DSM Medium 104 (104, red), DSM Medium 104 excluding Tween 80 (104-T, blue), or lipid-free WCAB defined medium (WCAB, black). Growth was monitored over three days, and data is presented as the mean A600 ± S.E.M. of three independent cultures.
B. Concentration-dependent growth inhibition of A. finegoldii by the FASII inhibitor cerulenin in 104-T medium (blue), in 104-T medium supplemented with 50 μM anteiso-15:0 (green), or 104-T medium supplemented with 50 μM 18:1Δ9 (purple). A. finegoldii was back-diluted 1:100 from a stationary phase stock culture grown in 104-T medium into 104-T medium containing 1% v/v DMSO, and the indicated concentrations of cerulenin and the A600 of the cultures was measured at 48 hours. Growth data is the mean A600 ± S.E.M. of three biological replicates.
The essentiality of de novo FASII in A. finegoldii was addressed using the covalent condensing enzyme inhibitor, cerulenin (Vance et al., 1972; D’Agnolo et al., 1973) (Fig. 3B). The minimal inhibitory concentration for cerulenin was 100 μM, and either the addition of exogenous oleate or anteiso-pentadecanoic acid did not overcome cerulenin growth inhibition. These data show that FASII is required for A. finegoldii growth, and although the bacterium can incorporate exogenous fatty acids into PE, it cannot survive solely on exogenous fatty acids.
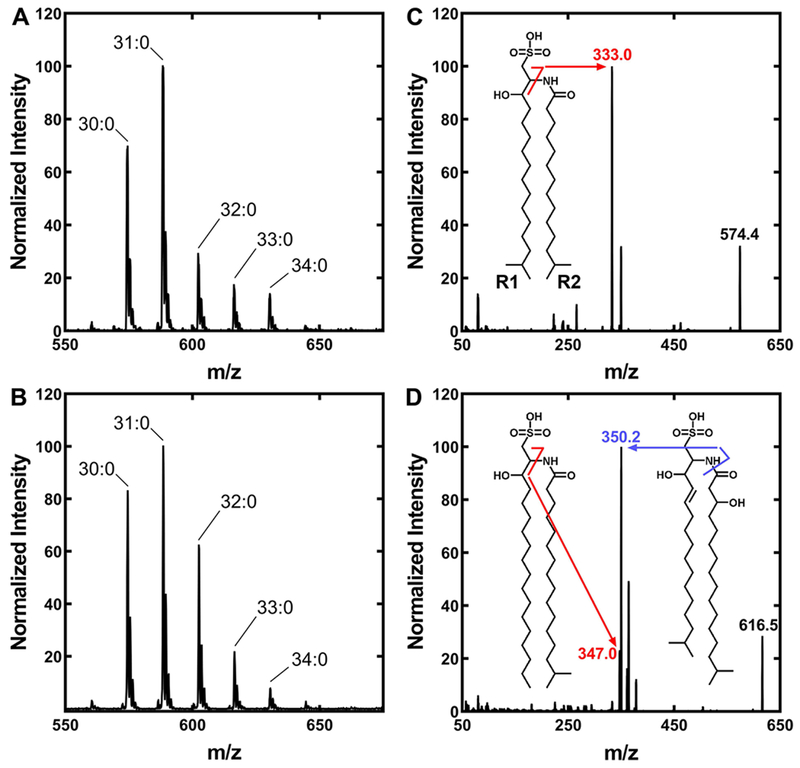
A. finegoldii also contains SL (Fig. 1), and the structure of this lipid was investigated by mass spectrometry (Fig. 4). There was no indication of oleate in the spectrum of SL (Fig. 4A) even when the strain was grown in Medium 104 containing Tween 80 (Fig. 4A), consistent with the lack of [14C]oleate incorporation into this lipid (Fig. 1B & 1C). The SL molecular species were not altered by growth in the minimal WCAB medium (Fig. 4B). The SL mass spectra appear to be a single series with different fatty acid chain lengths, but there were two different SL species detected. The peak labeled 30:0 was fragmented and consisted entirely of an SL called sulfobacin B (Walker et al., 2017) that is constructed from two 15-carbon fatty acids (Fig. 4C). However, the molecular species labeled 33:0 in Fig. 4B is a mixture of two SL species (Fig. 4D). This peak does contain the SL described above constructed from 16:0 and 17:0 fatty acids (Fig. 4D, inset). However, the major species in this peak is an SL constructed from a 3-hydroxy-17:0 and a trans-2-15:0 fatty acid (Fig. 4D, inset). This SL structure is called flavocristamide A (Walker et al., 2017). The 3-hydroxy- and trans-2 acyl chains correspond to acyl-ACP intermediates that were extracted from the FASII elongation cycle to construct flavocristamide A.
Figure 4. Molecular species of A. finegoldii SL.
The total apparent number of carbons in the SL that are derived from fatty acids are indicated.
A. A representative mass spectrum of the SL fraction of A. finegoldii grown in DSM Medium 104.
B. A representative mass spectrum of SL from A. finegoldii grown in WCAB minimal medium.
C. A representative MS/MS spectrum of the 30:0 SL molecular species. Inset, the structure of the single 30:0 SL species present that was derived from two 15:0 fatty acids (sulfobacin B). The diagnostic peak is indicated along with the fragmentation pattern. R1 of SL is the fatty acid condensed with cysteic acid, and R2 is the fatty acid attached to the amine.
D. A representative MS/MS spectrum of the 33:0 SL molecular species. Inset, a representative MS/MS spectrum showing the structures of the two SL molecules that comprise the 33:0 SL molecular species. The minor species is SL derived from 16:0 and 17:0 fatty acids. The major species is derived from a trans-2–15:0 and a 3-hydroxy-17:0 fatty acids (flavocristamide A). The 32:0 SL molecular species is also a mixture of two SL species (not shown).
Exogenous fatty acid metabolism
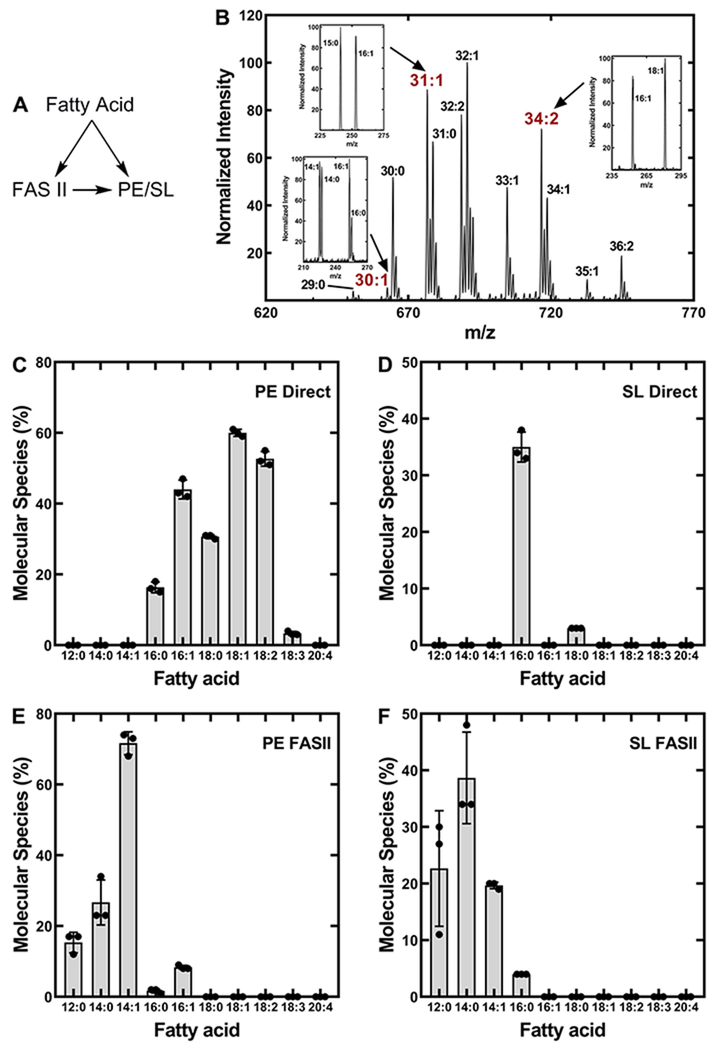
A. finegoldii was grown in the presence of different fatty acids followed by the analysis of the PE and SL by mass spectrometry to assess the ability of the bacterium to incorporate extracellular fatty acids into membrane lipids (Fig. 5). The saturated fatty acids were labeled with deuterium so that molecular species containing these fatty acids could be distinguished from those generated from de novo biosynthesis. Because unsaturated fatty acids were not produced by Alistipes, they could be recognized without a heavy label. Upon entering the cell, the exogenous fatty acids had two possible fates. They could be incorporated into PE/SL without alteration, or they could be funneled into FASII where they would be elongated prior to their incorporation (Fig. 5A). In each labeling experiment, the proportion of the total molecular species derived from direct incorporation and the proportion that was elongated prior to incorporation was estimated by quantifying the mass spectra of PE and SL. For example, A. finegoldii was labeled with 14:1 fatty acid and the PE mass spectrum was obtained (Fig. 5B). An estimate for the percent of the total PE molecular species that contained a 14:1-derived fatty acid was calculated by integrating the total ion current for each molecular species with an unsaturated fatty acid, and comparing this value to the total area of all molecular species. In this example, the PE molecular species that did not contain an unsaturated fatty acid (30:0 and 31:0) were in the minority (Fig. 5B), illustrating the efficiency of exogenous fatty acid utilization for PE synthesis. The proportion of the fatty acid that was incorporated intact compared to the amount of the fatty acid that was elongated by FASII prior to incorporation was analyzed by verifying the composition of each peak by fragmentation. The 31:1 peak contained 15:0 and 16:1, and the 34:2 peak consisted of 16:1 and 18:1 (Fig. 5B, inset). The two peaks containing intact 14:1 (29:1 and 30:1) were minor elements in the spectrum (Fig. 5B, inset). This analysis was performed on each PE molecular species in the spectrum. In this example, 70% of the PE species were derived from exogenous 14:1, and >99% of the 14:1 was elongated to 16:1 or 18:1 prior to incorporation into both positions of PE. This analysis was repeated in triplicate for each fatty acid labeling experiment to map the utilization of extracellular medium- and long-chain fatty acids by A. finegoldii for PE and SL synthesis.
Figure 5. Metabolism of exogenous fatty acids by A. finegoldii.
A, A. finegoldii was grown in the presence of a series of heavy-labeled saturated or unsaturated fatty acids. The incorporation of each of these fatty acids into PE and SL was determined by mass spectrometry. There are two possible fates for the fatty acids after their entry into the cell. The exogenous fatty acid may be directly incorporated into PE or SL, or it may enter the FASII elongation cycle and its elongation product is incorporated into PE or SL.
B. An example of how the proportion of an exogenous fatty acid directly incorporated into PE or elongated by FASII prior to incorporation was determined by mass spectrometry. A. finegoldii was grown in the presence of 14:1 and the PE mass spectrum was obtained. Peaks with an unsaturated fatty acid were fragmented to determine their fatty acid composition. Insets, the 30:1 peak contained 16:1/14:0 and 16:0/14:1, the 31:1 peak was composed of 16:1/15:0, and the 34:1 peak was composed of 18:1/16:0. The percent contribution of each molecular species to the total was determined by integrating the ion current in each peak.
C. The percent of molecular species formed from the direct incorporation of fatty acids into PE without elongation.
D. The percent of molecular species with fatty acids that were directly incorporated into SL without elongation.
E. The percent of molecular species that contained a fatty acid that was elongated prior to incorporation into PE.
F. The percent of molecular species containing a fatty acid that was elongated prior to incorporation into SL. There were three biological replicates and the mean ± S.E.M. is plotted.
The fatty acids had distinctly different fates depending on their chain length. Long-chain fatty acids (16:0, 16:1, 18:0, 18:1, 18:2 & 18:3) were incorporated into PE without elongation (Fig. 5C). Only minor amounts of 18 carbon fatty acid are produced by the de novo FASII pathway (Fig. 2D, inset), therefore the inability of the cells to elongate these fatty acids reflects the substrate selectivity of the FabF elongation condensing enzyme of FASII. Palmitic acid (16:0) was incorporated into both PE and SL, and 16:0 predominately entered these lipids without first being elongated by FASII (Fig. 5C and 5D). Long-chain unsaturated fatty acids were not incorporated into SL, suggesting that the substrate selectivities of both enzymes that introduce fatty acids into SL were not capable of utilizing these substrates (Fig. 5D). Acyl chains of 12:0, 14:0 and 14:1 were incorporated into both PE and SL, and in each case entered FASII and were elongated prior to their incorporation into a membrane lipid (Fig. 5E & 5F). Because acyl-ACP are the substrates for FASII elongation, these data provide clear evidence that these shorter-chain fatty acids were being ligated to ACP. Palmitoleic acid (16:1) was elongated to 18:1 as well as directly incorporated into PE, but neither 16:1 nor 18:1 were incorporated into SL (Fig. 5D & 5F) reflecting the substrate specificities of SL biosynthetic enzymes. These data reveal that A. finegoldii channels shorter-chain fatty acids (<16:0) into FASII, and the elongation products were used for both PE and SL synthesis depending on the substrate preferences of the biosynthetic enzymes.
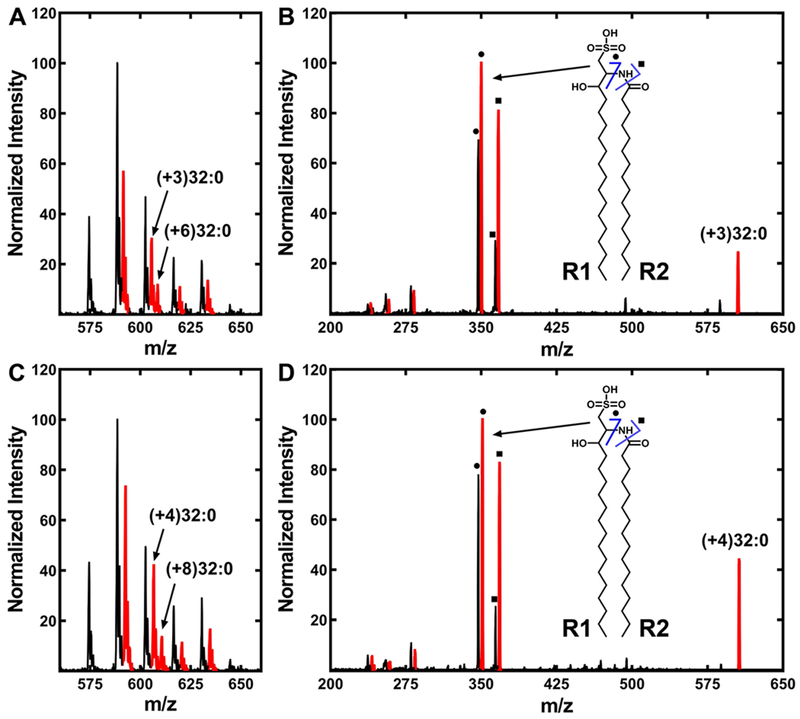
There are two fatty acids used in SL synthesis, and mass fragmentation experiments of the heavy-labeled SL were used to determine if the exogenous fatty acids were incorporated into both R1 and R2 positions of SL, or only into one of them (Fig. 6). SL incorporated [d3]14:0 after elongation (Fig. 5F) and [d4]16:0 without elongation (Fig. 5D). Growth with [d3]14:0 resulted in both (+3)32:0-SL and (+6)32:0-SL molecular species showing that [d3]14:0 was incorporated into both SL positions (Fig. 6A). Fragmentation of the (+3)32:0-SL peak (m/z = 605) confirmed that [d3]14:0 was incorporated into both the R1 and R2 positions (Fig. 6B, inset). The direct incorporation of [d3]14:0 into SL was too low to be analyzed meaning that the elongation of [d3]14:0 to [d3]16:0 was required prior to its incorporation into SL. The SL mass spectrum of cells labeled with [d4]16:0 showed a prominent molecular species (m/z = 592.7) containing one heavy fatty acid (+4)31:0-SL, but there were no (+8)31:0-SL species detected (Fig. 6C). This result was expected because this SL molecular species is constructed from a 16:0 and a 15:0 fatty acid. The 32:0-SL species arises from two 16:0 fatty acids, and both (+4)32:0-SL and (+8)32:0-SL species were detected (Fig. 6C), meaning that 16:0 could be incorporated into both the R1 and R2 positions. Fragmentation of the (+4)32:0-SL peak (m/z = 606) containing a single [d4]16:0 verified that the [d4]16:0 was incorporated into both the R1 and R2 positions of SL (Fig. 6D). The diagnostic peak pairs at m/z = 347 and 351, and m/z = 364 and 368 show that R1 was a mixture of light and heavy 16:0 (Fig. 6D, inset). The R2 fatty acid was also a mixture based on the less abundant m/z = 237 and 241 pair of peaks derived from a cleavage event that released R2 (not highlighted in the spectrum).
Figure 6. Incorporation of extracellular myristate and palmitate into SL.
A. finegoldii was labeled with either [d3]14:0 or [d4]16:0, and the lipids were extracted as described under “Experimental Procedures.” Mass spectra of the SL were obtained in both cases and fragmented to determine the positional distribution of the incorporated fatty acids into the SL molecule. R1 of SL is the fatty acid condensed with cysteic acid, and R2 is the fatty acid attached to the amine.
A. SL from cells labeled with [d3]14:0.
B. Fragmentation of the (+3)32:0-SL molecular species. Inset, ● and ■ indicate the two diagnostic fragments arising from (+3)32:0-SL as shown in the structure diagram.
C. SL molecular species from cells labeled with [d4]16:0.
D. Fragmentation of the (+4)32:0-SL molecular species. Inset, ● and ■ indicate the two diagnostic fragments arising from (+4)32:0-SL as shown in the structure diagram. SL molecular species (A, C) or fragments (B, D) containing a deuterium label are indicated in red.
Characterization of AfPlsC
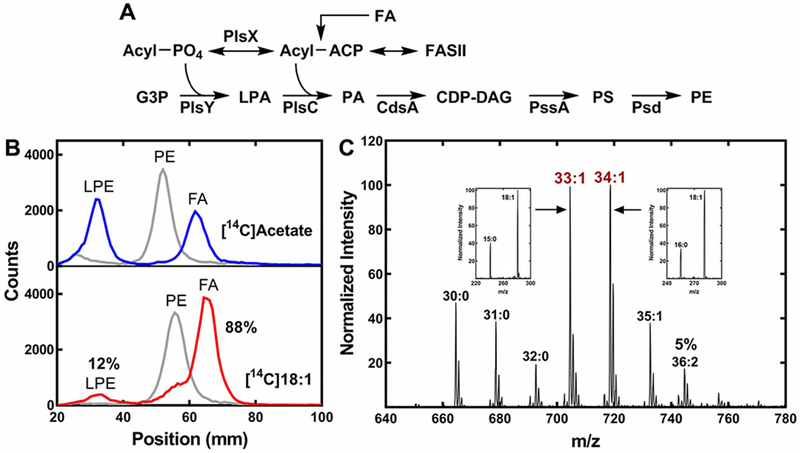
The mass spectrum of PE from oleate-labeled cells showed that 18:1 was always paired with a saturated fatty acid (Fig. 2A). This pattern of incorporation would be explained if exogenous oleate was predominately incorporated into one position of the glycerol backbone in the pathway leading to PE (Fig. 7A). Therefore, we isolated [14C]PE from A. finegoldii labeled with either [14C]acetate or [14C]oleate, and digested the lipid with phospholipase A2 to determine the positional distribution of the label (Fig. 7B). There was an equal distribution of label between the 1- and 2-positions when the PE was labeled with [14C]acetate, reflecting the incorporation of acyl-ACP from FASII into both positions. However, in the cells labeled with [14C]oleate, the radioactivity was found primarily in the 2-position (88%) (Fig. 7B). This result was consistent with the fragmentation of the 33:1 and 34:1 PE molecular species (Fig. 7C, insets). In this experiment, the intensity of the 18:1 was >2.2 times higher than that of the paired 15:0 or 16:0 signals, and is consistent with the conclusion that 18:1 occupies the 2-position in these PE molecular species (Han and Gross, 1995; Hsu and Turk, 2001; Mazzella et al., 2004). The [14C]18:1 incorporated into the 1-position was due to its utilization by PlsX/PlsY. This data showed that exogenous oleate was channeled predominately into the 2-position, and explains the pattern of PE molecular species produced from growth with exogenous oleate (Fig. 2A & 7C).
Figure 7. Exogenous oleate incorporation by A. finegoldii.
A. Pathway for the synthesis of PE in A. finegoldii. Acyl-ACP is formed from fatty acid synthesis (FASII) or by the activation of an exogenous fatty acid (FA). The acyl-ACP is converted to acyl-PO4 by PlsX, and the 1-position of glycerol-phosphate (G3P) is acylated by PlsY to form 1-acyl-sn-glycero-3-phosphate (LPA). PlsC acylates the 2-position with an acyl-ACP to form phosphatidic acid (PA) that is converted to CDP-dacylglycerol (CDP-DAG) by CdsA. PssA converts PA to phosphatidylserine (PS), which is then decarboxylated by Psd to form PE.
B. A. finegoldii was metabolically labeled with either [14C]acetate or [14C]oleate and the [14C]PE was isolated by thin-layer chromatography, then digested with phospholipase A2 to liberate the fatty acid at the 2-position. The products, fatty acid (FA) and 1-acyl-glycerophosphoethanolamine (LPE), were separated by thin-layer chromatography and the radioactive products were localized and quantified using a Bioscan Counter. Gray trace, [14C]PE starting material; blue trace, digestion of [14C]PE from [14C]acetate-labeled cells; red trace, digestion of [14C]PE derived from [14C]oleate-labeled cells (18:1). 88% of the [14C]oleate label was incorporated in the 2-position, demonstrating the preference for this position.
C. Representative mass spectrum of PE molecular species in cells grown with 20 μM 18:1. Insets, the fatty acids present in the 33:1 and 34:1 molecular species was determined by MS/MS. The 36:2 molecular species was 5% of the total ion current.
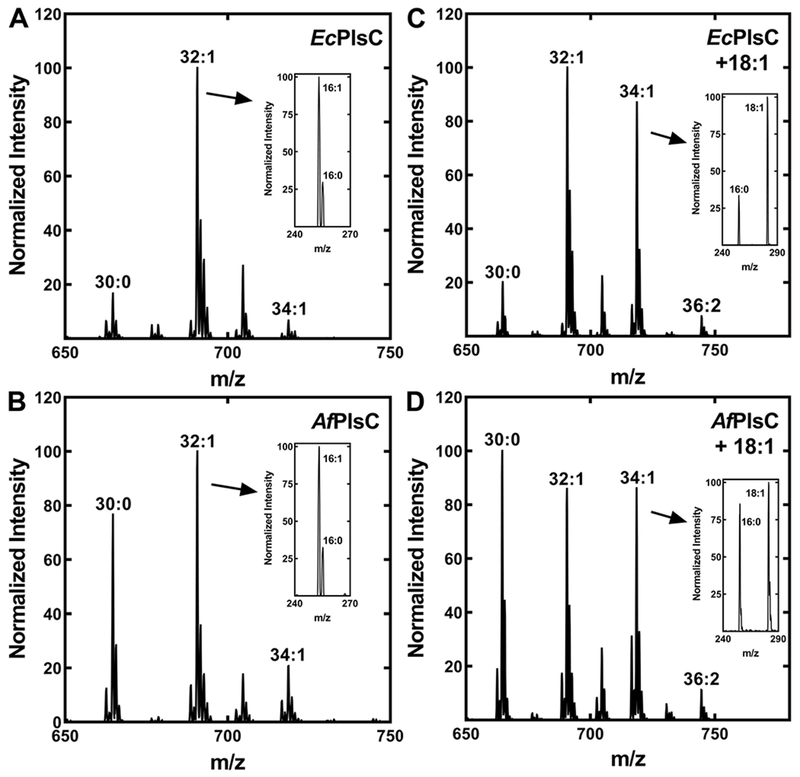
PlsC is responsible for acylation of the 2-position (Fig. 7A). EcPlsC utilizes both acyl-CoA and acyl-ACP acyl donors (Parsons et al., 2013; Yao and Rock, 2013), whereas SaPlsC uses only acyl-ACP (Lu et al., 2006). The AfPlsC gene (Alfi_2876) was identified based on its 32% identify with E. coli plsC, and we examined the properties of AfPlsC by expressing the protein in E. coli strain SM2-1 (plsC(Ts)). The expression of AfPlsC complemented the growth of strain SM2-1 at 42°C demonstrating that it possessed acyl-ACP-dependent PlsC activity (not shown). EcPlsC prefers to place 16:1 into the 2-position, and the complementation of strain SM2-1 (plsC(Ts)) with a plasmid expressing EcPlsC (pEcPlsC) led to the predominant synthesis of the 32:1 PE molecular species consisting of 16:0 and 16:1 (Fig. 8A). Fragmentation of this peak identified 16:1 and 16:0 fatty acids with an intensity ratio consistent with 16:1 occupying the 2-position (Han and Gross, 1995; Hsu and Turk, 2001; Mazzella et al., 2004) as expected (Yao and Rock, 2013) (Fig. 8A, inset). Two minor PE molecular species were 30:0 and 34:1. The former arises from incorporation of 14:0-ACP into the 2-position, and the 34:1 species corresponds to the incorporation of 18:1-ACP. The 14:0-ACP would be available from FASII as an intermediate in the formation of 16:0-ACP, but E. coli does not significantly elongate 16:1-ACP to 18:1-ACP at 42°C (Garwin et al., 1980a; Garwin et al., 1980b), accounting for the low abundance of the 34:1 PE molecular species. Strain SM2-1 complemented with AfPlsC also produced the 32:1 PE molecular species, but the 30:0 PE molecular species was substantially higher than in the EcPlsC complemented strain (Fig. 8B). Fragmentation of the 32:1 peak showed that 16:1 also occupied the 2-position in this case (Fig. 8B, inset). The 14:0 acyl chains were located in the 2-position based on the fragmentation pattern of the 30:0 PE molecular species (not shown). These data indicate that AfPlsC utilized both unsaturated 16:1-ACP and the shorter-chain 14:0-ACP as substrates.
Figure 8. Acyl carrier and chain length selectivity of AfPlsC.
E. coli strain SM2–1 (plsC(Ts)) was complemented with plasmids expressing either EcPlsC or AfPlsC and grown at the non-permissive temperature (42°C). The PE molecular species were determined to assess AfPlsC acyl carrier and acyl chain selectivity. The insets show the fragmentation of a relevant peak in the spectrum to confirm the identity and positional distribution of the fatty acids. The spectra shown are representative of three independent experiments.
A. PE molecular species profile of E. coli strain SM2-1 complemented with EcPlsC.
B. PE molecular species profile of E. coli strain SM2-1 complemented with AfPlsC.
C. PE molecular species profile of E. coli strain SM2-1 complemented with EcPlsC grown in the presence of exogenous oleate.
D. PE molecular species profile of E. coli strain SM2-1 complemented with AfPlsC grown in the presence of exogenous oleate.
Exogenous fatty acids are activated by acyl-CoA synthetase in E. coli and utilized by EcPlsC (Parsons and Rock, 2013; Yao and Rock, 2013). Therefore, when strain SM2–1 (plsC(Ts))/pEcPlsC was grown in the presence of an exogenous 18:1 supplement, there was a significant increase in the abundance of the 34:1 (16:0/18:1) PE molecular species (Fig. 8C). Fragmentation of the new 34:1 peak showed that 18:1 occupied the 2-position (Fig. 8C, inset). Growth of strain SM2-1/pAfPlsC in the presence of exogenous 18:1 also led to the appearance of 34:1 PE (Fig. 8D). The fragmentation pattern of the 34:1 PE molecular species indicated that 16:0 and 18:1 were equally distributed between the 1- and 2-positions (Fig. 8D, inset). Also, the appearance of a 36:2 molecular species showed that AfPlsC can use acyl-CoA. This is anticipated because acyl-CoAs are used as substrates analogs to assay acyl-ACP-dependent enzymes. However, the lower incorporation of 18:1 and higher levels of 30:0 PE molecular species in AfPlsC-complemented cells compared to EcPlsC suggests that 14:0-ACP is the preferred substrate for AfPlsC in E. coli.
Synthetases of A. finegoldii
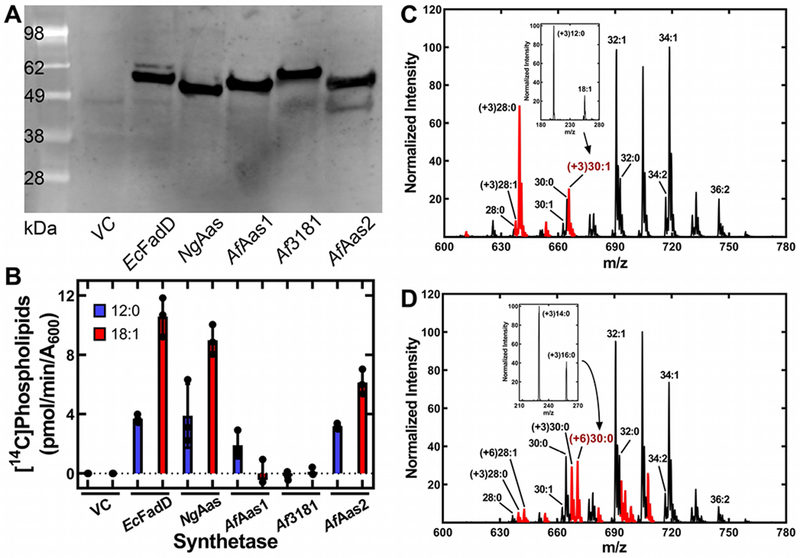
Acyl-CoA and acyl-ACP synthetases are two enzymes that have been described in Gram-negative bacteria for the activation of exogenous fatty acids (Black et al., 1992; Yao and Rock, 2015; Jiang et al., 2006; Yao et al., 2015b; Yao et al., 2016). A bioinformatic search of the A. finegoldii genome using either E. coil acyl-CoA synthetase (EcFadD) or Neisseria acyl-ACP synthetase (NgAas) identified three candidate genes that possessed the acyl-adenylate (AMP) binding sequence motif (Jackowski et al., 1994; Black and DiRusso, 2003; Black et al., 1997). The three genes (locus tags: Alfi_2635, Alfi_0371 and Alfi_3181) were equally related to both EcFadD and NgAas, consistent with the difficulty in determining either the acyl carrier or the acyl chain specificity from a bioinformatic analysis. Therefore, we cloned the three genes into E. coli expression vectors and introduced them into E. coli strain LCH30 (aas fadD), which is incapable of activating exogenous fatty acids for incorporation into phospholipid (Hsu et al., 1991). Strain LCH30 was transformed with each of the putative A. finegoldii synthetases, along with NgAas, EcFadD and empty vector controls. The expressed proteins were 6x-His-tagged and were visualized by immunoblotting of the soluble cell extracts with an anti-His-tag antibody to illustrate that they were all expressed at similar levels (Fig. 9A). The strains were next labeled with either [14C]laurate or [14C]oleate to determine which of the genes supported medium- and long-chain fatty acid activation. The vector control did not incorporate either labeled fatty acid and the NgAas and EcFadD controls exhibited robust laurate and oleate activation and incorporation (Fig. 9B). The expression of either Alfi_2635 (AfAas1) or Alfi_0371 (AfAas2) supported the incorporation of laurate into phospholipid. AfAas2 supported oleate incorporation, but AfAas1 expression did not. Alfi_3181 did not support the activation of either fatty acid (Fig. 9B). These data indicated that AfAas1 and AfAas2 were either acyl-CoA or acyl-ACP synthetases.
Figure 9. Analysis of the acyl-ACP synthetases of A. finegoldii.
E. coli strain LCH30 (aas fadD) was transformed with a series of expression vectors directing the synthesis of 6xHis-tagged versions of the three Alistipes genes encoding putative synthetases, along with two controls expressing Neisseria acyl-ACP synthetase (NgAas) or E. coli acyl-CoA synthetase (EcFadD). Cells were grown to mid-log phase and either metabolically labeled or extracted to biochemically characterize the expressed enzymes.
A. Western blot of the expressed proteins using anti-His-tag antibody for detection to validate expression of the synthetases.
B. Metabolic labeling of the strain set with either [14C]12:0 or [14C]18:1 fatty acids. Lipids were extracted, and the amount of radioactivity incorporated into phospholipids determined by scintillation counting.
C. Mass spectrum of PE synthesized by strain LCH30 complemented with EcFadD and labeled with [d3]12:0. Inset, fragmentation of the (+3)30:1 molecular species showing [d3]12:0 was not elongated.
D. Mass spectrum of strain LCH30 complemented with AfAas1 and labeled with [d3]12:0. Inset, fragmentation of the (+6)30:0 PE molecular species showing the presence of [d3]14:0 and [d3]16:0, demonstrating [d3]12:0 was elongated. This spectrum is representative of those obtained from cells complemented with either NgAas or AfAas2 and labeled with [d3]12:0. PE molecular species (C, D) containing a deuterium label are indicated in red.
The acyl-CoA synthetases were distinguished from the acyl-ACP synthetases by labeling with exogenous [d3]12:0. Acyl-CoA cannot be elongated by E. coli, and only [d3]12:0 was incorporated into PE in cells expressing EcFadD (Fig. 9C). The (+3)28:0 PE species (16:0/[d3]12:0) becomes a prominent feature of the spectrum, and there are no (+3)PE molecular species that contain a fatty acid longer than [d3]12:0. The fragmentation of the (+3)30:1 molecular species showed that it contained [d3]12:0 and 18:1 (Fig. 9C, inset). In strain LCH30 complemented with either NgAas, AfAas1 or AfAas2, the majority of the [d3]12:0 was elongated prior to incorporation into phospholipid. An example spectrum from cells expressing AfAas1 illustrates the incorporation pattern that was similar for all three acyl-ACP synthetases (Fig. 9D). The fragmentation pattern of the (+6)30:0 PE molecular species showed the presence of [d3]16:0 and [d3]14:0 (Fig. 9D, inset), confirming the elongation of [d3]12:0. Also, the (+6)32:0 peak composed of two 16:0 fatty acids can only arise from the elongation of [d3]12:0. These data established the existence of two acyl-ACP synthetases in A. finegoldii.
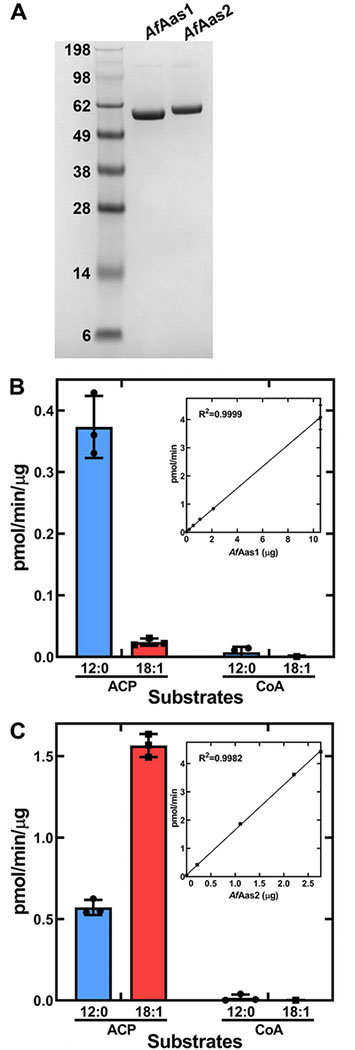
The identity of these enzymes as acyl-ACP synthetases was verified by biochemical analysis of the purified proteins (Fig. 10). The amino-terminal His-tagged versions of AfAas1 and AfAas2 were cloned into the pET expression vector, expressed in E. coli and purified by Ni2+ affinity chromatography. The purified proteins were soluble, homogeneous protein preparations (Fig. 10A). They were both tested for activity using either CoA or E. coli ACP as acyl acceptors, and either [14C]12:0 or [14C]18:1 as acyl donors. AfAas1 selectively activated 12:0 and did not activate 18:1 (Fig. 10B). AfAas2 was most active with 18:1, but also activated 12:0 at approximately one third the rate of 18:1 (Fig. 10C). These data confirm the substrate specificities of the two synthetases deduced from the expression experiments.
Figure 10. Biochemical analysis of purified AfAas1 and AfAas2.
A. A Coomassie-stained SDS gel electrophoresis illustrating the purity of AfAas1 and AfAas2 used in the biochemical assays to determine the acyl acceptor (ACP or CoA) and acyl donor (12:0 or 18:1) specificities of the two enzymes.
B. Biochemical activity of AfAas1. AfAas1 activity using [14C]12:0 as substrate was linear with protein and had a specific activity of 0.38 ± 0.002 pmol/min/μg protein (Inset).
C. Biochemical activity of AfAas2. AfAas2 activity using [14C]18:1 as substrate was linear with protein and had a specific activity of 1.59 ± 0.02 pmol/min/μg protein (Inset). AfAas2 activity using [14C]12:0 as substrate was 0.57 ± 0.03 pmol/min/μg protein.
Discussion
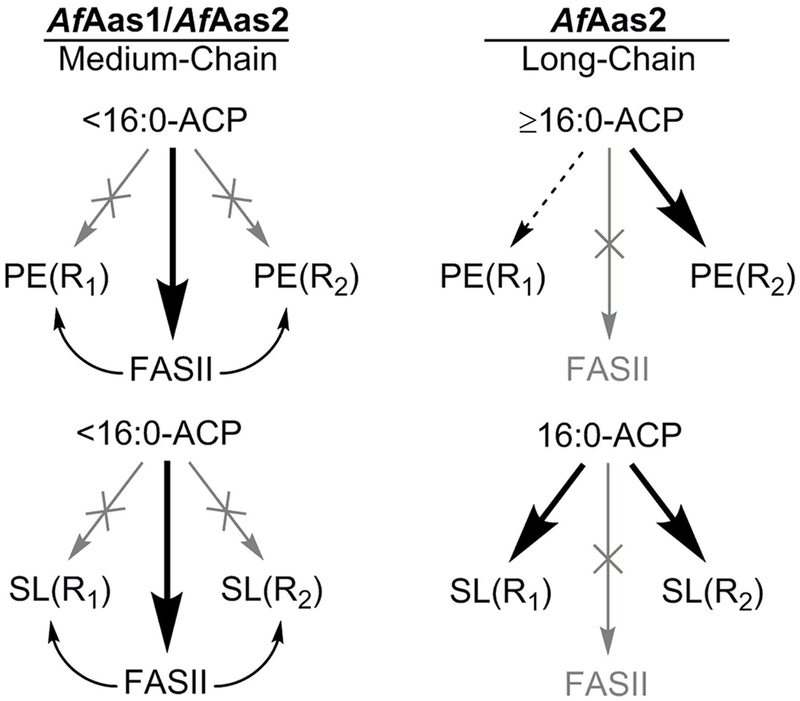
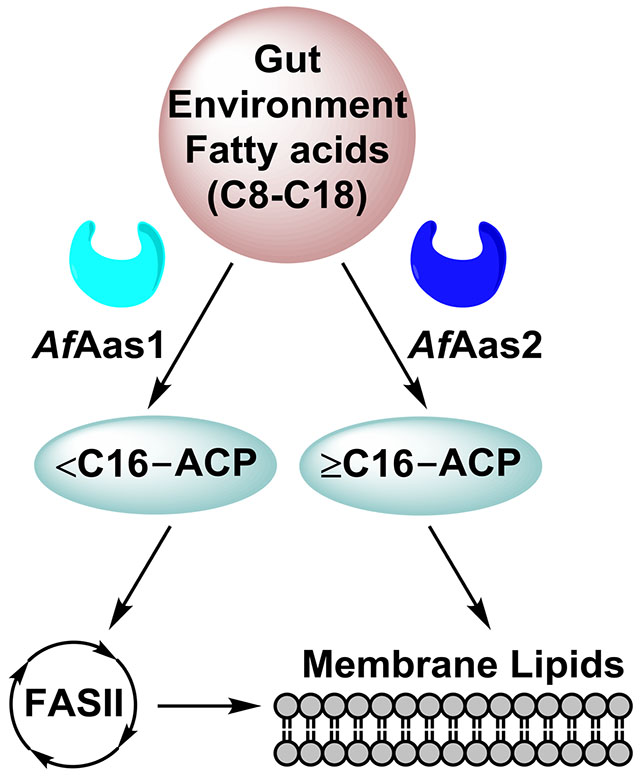
A. finegoldii uses a unique combination of acyl-ACP synthetases to activate a spectrum of exogenous fatty acids for membrane lipid synthesis (Fig. 11). AfAas1 prefers medium-chain fatty acid substrates and has little to no activity with chain-lengths ≥16 carbons. Thus, the substrate binding site of AfAas1 excludes long-chain fatty acids. AfAas1 substrate selectivity is most closely related to the acyl-ACP synthetases from Vibrio harveyi (Byers and Holmes, 1990; Jiang et al., 2006; Jiang et al., 2010) and Chlamydia trachomatis (Yao et al., 2015b). These synthetases prefer saturated fatty acids, and function best with acyl chains ≤16 carbons. Acyl-ACP arising from AfAas1 action are too short to be the best substrates for the enzymes of either PE or SL synthesis, and are elongated by FASII prior to their incorporation. This utilization pathway also results in the elongated medium-chain fatty acids being found in both positions of the PE and SL backbones. AfAas2 prefers long-chain saturated and unsaturated fatty acids, although its larger substrate pocket cannot exclude medium-chain fatty acids (Fig. 11). AfAas2 is most similar in its substrate selectivity to NgAas that can activate long-chain saturated and unsaturated fatty acids. Acyl-ACP chain lengths >16 carbons are not elongated by the FabF condensing enzyme of FASII, and are also excluded from incorporation into the 1-position of PE. This indicates that the A. finegoldii PlsX/PlsY system discriminates against these chain lengths, leading to their incorporation only into the 2-position of PE by PlsC. The only long-chain fatty acid used directly for SL was 16:0, and this fatty acid is incorporated into both positions of SL without elongation. Unsaturated, long-chain acyl-ACP produced by AfAas2 are not used to synthesize SL. The exclusion of these acyl chains from the SL pathway is attributed to the substrate selectivity of the enzymes in the SL biosynthetic pathway that exclude the long, kinked acyl chains of 18-carbon unsaturated fatty acids.
Figure 11. The different metabolic fates for medium- and long-chain fatty acids in A. finegoldii.
A. finegoldii uses two acyl-ACP synthetases with different substrate specificities to activate a spectrum of exogenous fatty acid chain lengths found in the gut environment. R1 and R2 refer to the 1- and 2-positions of PE, respectively. R1 of SL is the fatty acid condensed with cysteic acid, and R2 is the fatty acid attached to the amine (see Fig. 4C for structure). Medium-chain fatty acids <16 carbons are ligated to ACP by AfAas1. These medium-chain acyl-ACPs are elongated by FASII prior to the incorporation of the elongated acyl-ACP into both the R1 and R2 positions of both PE and SL. Long-chain fatty acids ≥16 carbons are activated by AfAas2. Acyl-ACP 16 carbons or longer cannot enter FASII and are funneled into the R2 position of PE at the PlsC step. AfAas2 can also use medium-chain fatty acids as substrates, and these acyl-ACP are handled in the same manner as medium-chain acyl-ACP derived from AfAas1. The only long-chain acyl-ACP used for SL synthesis is 16:0-ACP, and it is incorporated into both the R1 and R2 positions of SL.
The utilization of acyl-ACP synthetases for the activation of fatty acids is consistent with the overall lipid metabolic program in A. finegoldii. Gram-negative bacteria like E. coli employ the PlsB/C pathway to phosphatidic acid and utilize acyl-CoA synthetase to activate exogenous fatty acids (Yao and Rock, 2015). The acyltransferases of these bacteria use either acyl-CoA or acyl-ACP substrates (Yao and Rock, 2013), and many possess a suite of fatty acid β-oxidation genes and acyl-CoAs derived from exogenous fatty acid that are also used as a carbon source (Iram and Cronan, 2006). This metabolism is characteristic of the Gammaproteobacteria, but A. finegoldii is like many other Gram-negative bacteria in that they use the more widely-distributed PlsX/Y/C pathway for phosphatidic acid synthesis (Yao and Rock, 2013). Gram-positive bacteria also use the PlsX/Y/C pathway and activate exogenous fatty acids using a fatty acid kinase system (Parsons et al., 2014a; Parsons et al., 2014c). Fatty acid kinase is not found in A. finegoldii or other Gram-negative bacteria. We did not detect an acyl-CoA synthetase, although the complementation experiments indicate that AfPlsC can utilize acyl-CoA substrates to some extent. Acyl-ACP synthetases are the most flexible activation systems for Gram-negative PlsX/Y/C bacteria because the acyl-ACP can be converted to the preferred chain lengths by FASII, or if suitable, be directly utilized by PlsX/Y/C for membrane glycerolipid synthesis. SL are an unusual and abundant membrane lipid in A. finegoldii that contain two fatty acids derived from FASII. Our data are consistent with acyl-ACP as the acyl donors for these important lipids. The acyl-ACP synthetases direct environmental medium-chain fatty acids to the SL pathway. The acyl-ACP that enters FASII is also used to form the 3-hydroxy- and trans-2-fatty acids in the flavocristamide version of SL (Walker et al., 2017) and the 3-hydroxy-fatty acids of LPS (d’Hennezel et al., 2017).
A. finegoldii provides a blueprint for understanding fatty acid acquisition by anaerobic Bacteroidetes, a major contributor to the commensal gut microbiome. The gut environment is a rich source of fatty acids arising from food digestion. Long-chain saturated and unsaturated fatty acids are derived from animal and plant glycerolipids and triglycerides, and medium-chain saturated fatty acids are found in dairy products, and coconut and palm kernel oils (Marten et al., 2006). These medium-chain fatty acids are not used for lipid synthesis by mammals, but rather are excellent substrates for energy and ketone production via mammalian β-oxidation (Schonfeld and Wojtczak, 2016). AfAas1 provides A. finegoldii a mechanism to utilize medium-chain fatty acids in the gut as building blocks for the energy efficient construction of its membrane lipids. AfAas2 prefers the saturated and unsaturated long-chain fatty acids that are the digestion products of common glycerolipids. Phylogenetic analysis of the 16S rRNA sequence of the Bacteroidetes phylum shows that the human/mammal-associated commensal taxa share a recent common ancestor that branches from the environmental taxa (Johnson et al., 2017). The gene encoding AfAas1 is only found in the commensal Bacteroidetes where medium-chain fatty acids would be a potential source of acyl chains. All members of the Bacteroidetes phylum express AfAas2, an acyl-ACP synthetase with a substrate preference for the most common acyl chains in the environment.
Experimental Procedures
Materials
Sources of supplies were: trypticase peptone, tryptone, peptone, glucose, yeast extract, and beef extract, BD Medical Technologies (Franklin Lakes, NJ); haemin, arginine, cysteine, DMSO, CoA and Tween 80, Naja mossambica mossambica snake venom phospholipase A2, monoclonal anti-polyhistidine-alkaline phosphatase antibody, Sigma-Aldrich (St. Louis, MO); vitamin K1 and media salts, Fisher Scientific (Hampton, NH); Brij-58, G Biosciences (St. Louis, MO); antibiotics, GoldBio (St. Louis, MO); [1-14C]acetic acid (50.5 mCi/mmol, 1 mCi/ml) and [1-14C]oleic acid (59 mCi/mmol, 0.1 mCi/ml), PerkinElmer (Waltham, MA); [1-14C]lauric acid (59 mCi/mmol, 0.1 mCi/ml), American Radiolabeled Chemicals, Inc (St. Louis, MO); (methyl-d3)12:0, (methyl-d3)14:0, (7,7,8,8-d4)16:0, and (methyl-d3)18:0, Cambridge Isotope Laboratories, Inc (Tewksbury, MA); and methyl ester standards, 14:1(Δ9), 16:1(Δ9), 18:1(Δ9), 18:2(Δ9,12), 18:3(Δ9,12,15), 20:4(Δ5,8,11,14), Matreya, LLC (State College, PA). All solvents were chromatography grade. E. coli ACP was prepared as described (Yao et al., 2013).
Bacteriology
Bacterial strains and plasmids used in this study are listed in Table 2. A. finegoldii were grown anaerobically as 5 ml cultures in 16 × 100 mm borosilicate glass tubes at 37°C in a Whitley M45 HEPA Variable Atmosphere Workstation (10% CO2 and 5% H2; 70% humidity). A. finegoldii cells were cultured in three broths: DSM Medium 104 (with vitamin solution) (Leibniz-Institut DSMZ, Germany), DSM Medium 104 excluding Tween-80, and Wilkins-Chalgren anaerobe broth (WCAB) (Leibniz-Institut DSMZ, Germany) with vitamin K used instead of menadione (a vitamin K analog), and the sodium pyruvate was also omitted because it is not required for A. finegoldii to grow. Growth rate experiments were performed by 1:100 back-dilution of a stationary phase culture grown in the medium of investigation. The A600 in the 16×100 mm borosilicate glass tubes were measured using a Biochrom WPA CO8000 cell density meter.
Table 2.
Bacterial strains and plasmids
| Strain | Relevant Genotype | Source |
|---|---|---|
| A. finegoldii | Strain DSM 17242 (DSMZ Collection) | (Rautio et al., 2003) |
| E. coli SM2-1 | plsC101(Ts) metCl6S::Tn10 | (Coleman, 1990) |
| E. coli LCH30 | aas-1 fadD88 metB1 relA1 spoT1 | (Hsu et al., 1991) |
| Plasmid | Description | Source |
| pPJ131 | E. coli expression vector | (Lu et al., 2006) |
| pEcPlsC | E. coli plsC in pPJ131 | (Robertson et al., 2017) |
| pCtPlsC | Chlamydia trachomatis plsC in pPJ131 | (Robertson et al., 2017) |
| pAfPlsC | A. finegoldii plsC (Alfi_2876) in pPJ131 | This study |
| pEcFadD | E. coli fadD in pPJ131 | (Yao et al., 2016) |
| pNgAas | Neisseria gonorrhoeae aas in pPJ131 | (Yao et al., 2016) |
| pAfAas1 | A. finegoldii aas1 (Alfi_2635) in pPJ131 | This study |
| pAf3181 | A. finegoldii Alfi_3181 in pPJ131 | This study |
| pAfAas2 | A. finegoldii aas2 (Alfi_0371) in pPJ131 | This study |
| pET_AfAas1 | A. finegoldii aas1 (Alfi_2635) in pET | This study |
| pET_AfAas2 | A. finegoldii aas2 (Alfi_0371) in pET | This study |
The minimum inhibitory concentration for cerulenin with or without exogenous fatty acid supplementation against A. finegoldii was determined using a modified broth microdilution method (Parsons et al., 2011). Fatty acids and cerulenin were added in DMSO to a final concentration of 1%, and the control media also had 1% DMSO. A stock culture of A. finegoldii grown in DSM Medium 104 excluding Tween 80 was diluted 1:100 in the same medium with no fatty acid, 50 μM 15:0, or 50 μM 18:1. Briefly, 100 μl of diluted cells was added to each well of a U-bottom 96-well plate except the first column of wells. A 200 μl aliquot of diluted cells mixed with 400 μM cerulenin was added to the first column of wells, and 100 μl was serially diluted through the plate excluding the last column of wells leaving 100 μl of cells in each well with the appropriate concentration of cerulenin. The plate was incubated at 37°C as described above for 48 hours and the A600 determined using a SPECTRAmax 340PC Microplate Reader. Cells grown with 0 μM cerulenin were used as a reference (i.e., 100% growth).
Molecular Biology
DNA sequences encoding Alfi_2876 (AfPlsC), Alfi_2635 (AfAas1), Alfi_0371 (AfAas2), and Alfi_3181 were codon optimized and purchased as GeneArt DNA sequences from ThermoFisher Scientific (Waltham, MA). The DNA sequences were designed with 5′-NdeI and 3′-EcoRI restriction sites for cloning into the pPJ131 vector (Lu et al., 2006). The pPJ131 vector contains an amino-terminal His6-tag that was cloned in frame with each gene using the NdeI restriction site. The pEcPlsC vector (Robertson et al., 2017), pEcFadD vector (Yao et al., 2016), and pNgAas vector (Yao et al., 2016) were described previously. AfAas1 and AfAas2 were cloned into the pET28b vector (Novagen) using the 5′-NdeI and 3′-EcoRI restriction sites to introduce an amino-terminal His6-tag.
Bioinformatics analysis
Homologs to known lipid synthesis genes were searched against the A. finegoldii DSM 17242 genome using tBLASTn with FabK, Streptococcus pneumoniae; PssA, Bacillus subtilis; Staphylococcus aureus; PlsX and PlsY; Streptomyces autolyticus, Psd; Neisseria gonorrhoeae, FabI and Aas; Homo sapiens, sulfonoliid condensing enzymes; and E. coli for all other genes. Hits with an Expect value < e−30 were searched back against the query genome to ensure the hit is not more homologous against another protein in the query genome. Interesting gene features were searched against environmental associated Bacteroidetes group (Johnson et al., 2017), human associated Bacteroidetes group (Johnson et al., 2017), Ignavibacteriaceae family, and Chlorobiaceae family to understand the distribution of the features.
Analysis of AfPlsC
The E. coli PlsC amino acid sequence (Uniprot Accession: P26647) were used as a query sequence in a tBLASTn search against the A. finegoldii DSM 17242 genome (taxid:679935). The search returned the gene predicted to encode AfPlsC, Alfi_2876, with an Expect value of 4e−11. The function of the predicted A. finegoldii PlsC was confirmed by complementation of E. coli strain SM2-1 (plsC(Ts)) (Yao et al., 2015a). Strain SM2-1 is not viable at 42°C unless complemented with a functional PlsC. Plasmids expressing E. coli PlsC (pEcPlsC) or A. finegoldii PlsC (pAfPlsC) were transformed into strain SM2-1 (plsC(Ts)) and recovered on Luria plates containing 100 μg/ml carbenicillin at 30°C. Individual transformants were then tested for growth on Luria broth plates at 30°C and 42°C. Overnight cultures of strain SM2-1 (plsC(Ts)) harboring the PlsC expression plasmids were grown at 30°C, subcultured into Luria broth with and without exogenous 18:1(Δ9), and grown at 42°C to A600=0.8. Lipids were extracted via the Bligh and Dyer method (Bligh and Dyer, 1959) from each experiment, and PE molecular species composition was determined (Robertson et al., 2017).
Metabolic labeling
Cultures of A. finegoldii (5 ml) were grown for two days in the medium of interest. Fatty acids (20 μM) used in the metabolic labeling experiments were; (methyl-d3)12:0, (methyl-d3)14:0, (7,7,8,8-d4)16:0, (methyl-d3)18:0, 14:1(Δ9), 16:1(Δ9), 18:1(Δ9), 18:2(Δ9,12), 18:3(Δ9,12,15), 20:4(Δ5,8,11,14), 1 μCi/ml [14C]acetate, or 0.1 μCi/ml [14C]18:1Δ9 added to the medium prior to cell growth, and Brij-58 added to a final concentration of 0.1% v/v. Cells were harvested by centrifugation and washed twice in 1 ml of 10 mg/ml fatty acid free bovine serum albumin in phosphate-buffered saline. The cell pellet was resuspended in 1 ml water and 2.4 ml of 2% acetic acid in methanol. Chloroform (1 ml) was added and the suspension incubated at room temperature for 30 min. An additional 1.5 ml chloroform and 1.2 ml water were added and the suspension was centrifugalized at 30 x g for 10 min to separate the organic and aqueous phases. The organic phase was collected and dried under nitrogen for storage at −20°C.
Lipidomics
Fatty acid methyl esters were prepared by suspending the dry extract in 2 ml MeOH containing 4 drops of acetyl chloride and incubating overnight at room temperature. The reaction was dried under nitrogen, resuspended in 1 ml hexane, and extracted with 2 ml water. The organic phase was collected, dried, and stored under nitrogen at −20°C. Methyl esters were quantified using a Hewlett-Packard model 5809A gas chromatograph equipped with a flame ionization detector. Separations were achieved using a 30 m x 0.53 mm x 0.50 μm Agilent J&W DB-225 GC Column as described (Parsons et al., 2014b). The retention times of methyl ester standards (Sigma-Aldrich) were used to identify the methyl esters.
Polar lipids were analyzed on Silica Gel H plates (Analtech, Inc.) developed with chloroform:methanol:acetic acid:water (80/25/10/2, v/v) or diisobutylketone/acetic acid/water (80/55/15, v/v). 2-Dimensional thin layer chromatography used Silica Gel H plates (Analtech, Inc.) developed in the first dimension with chloroform:methanol:acetic acid:water (80/25/10/2, v/v), and in the second dimension with diisobutylketone/acetic acid/water (80/55/15, v/v).
The distribution of fatty acids between the 1- and 2-positions of A. finegoldii PE was determined by radiolabeling cultures with [14C]acetate or [14C]oleate, and purifying labeled PE from the lipid extracts by thin-layer chromatography on Silica Gel H plates. Silica-bound [14C]PE was digested with Naja mossambica mossambica snake venom phospholipase A2 to deacylate the 2-position fatty acid as described (Robertson et al., 2017). After 3 h at room temperature, lipids were extracted via the Bligh and Dyer method (Bligh and Dyer, 1959) and separated on a Silica Gel H plate (Analtech, Inc.) developed with chloroform:methanol:ammonium hydroxide (80/25/4, v/v), and the distribution of radioactivity on the plate was determined using a Bioscan Imaging detector.
The dried lipid extracts were resuspended in chloroform:methanol (1:1). PE and SL were analyzed using a Shimadzu Prominence UFLC attached to a QTrap 4500 equipped with a Turbo V ion source (Sciex). Samples were injected onto an Acquity UPLC BEH HILIC, 1.7 μM, 2.1 x 150 mm column (Waters) at 45°C a flow rate of 0.2 ml/min. Solvent A was acetonitrile, and solvent B is 15 mM ammonium formate, pH 3. The HPLC program was the following: starting solvent mixture of 96% A / 4% B, 0 to 2 min isocratic with 4% B; 2 to 20 min linear gradient to 80% B; 20 to 23 min isocratic with 80% B; 23 to 25 min linear gradient to 4% B; 25 to 30 min isocratic with 4% B. The system was controlled by the Analyst® software (Sciex).
The QTrap 4500 was operated in the Q1 negative mode or Precursor negative mode to analyze SL. The ion source parameters for Q1 were: ion spray voltage, −4500 V; curtain gas, 25 psi; temperature, 350°C; ion source gas 1, 40 psi; ion source gas 2, 60 psi; and declustering potential, −40 V. In precursor mode, the ion source parameters were the same as above along with collision energy, −60 V; collision gas, medium; and the precursor ion was 79.8 Da. The QTrap 4500 was operated in the Q1 positive mode or neutral loss (NL) positive mode to analyze PE. For Q1, the ion source parameters were: ion spray voltage, 5500 V; curtain gas, 25 psi; temperature, 350 °C; ion source gas 1, 40 psi; ion source gas 2, 60 psi; and declustering potential, 40 V. The ion source parameters for neutral loss were the same as above along with collision energy, 30 V; collision gas, medium, and neutral loss ion was 141.0 Da.
Product Ion scans of PE and SL were obtained following separation using a Discovery DSC-NH2 solid-phase extraction column (Supelco, Bellefonte, PA) (Alvarez and Touchstone, 1992). The column was conditioned with 8 ml of hexane, and lipid extract was added. Non-polar lipids were eluted with 6 ml of 2:1 (v/v) choloform:isopropanol; fatty acids were eluted with 6 ml of ether + 2% acetic acid; PE was eluted with 6 ml of methanol; and SL was eluted with 6 ml of choloform:methanol:0.8 M sodium acetate 60:30:4.5 (v/v). The ion source parameters for product scan in the negative mode were: ion spray voltage, −4500 V; curtain gas, 15 psi; collision gas, medium; temperature, 270°C; ion source gas 1, 15 psi; ion source gas 2, 20 psi; and declustering potential, −40 V. For PE species, the collision energy ranged from −30 V to −38 V. For SL species the collision energy ranged from −75 V to −88 V. The sample was introduced to the QTrap 4500 by direct injection.
Characterization of the synthetases
The Neisseria gonorrhoeae acyl-ACP synthetase amino acid sequence (Uniprot Accession: Q5F969) and the E. coli acyl-CoA synthetase amino acid sequence (Uniprot Accession: P69451) were used for query sequences in a tBLASTn search against the Alistipes finegoldii DSM 17242 genome (taxid:679935). Both searches independently found Alfi_2635 (AfAas1), Alfi_0371 (AfAas2), and Alfi_3181 with the highest E values of 1e−34 to 2e−41. All three genes contain an AMP-binding sequence (Jackowski et al., 1994; Black and DiRusso, 2003) and a fatty acyl-CoA synthetase signature motif (Black and DiRusso, 2003; Black et al., 1997) characteristic of a synthetase. Both searches found a pseudogene with an E value of 2e−26 to 5e−42. This gene is annotated as a pseudogene (Liu et al., 2004) due to a premature stop codon located between the predicted AMP-binding sequence and acyl-CoA synthetase signature motif.
The function of the predicted A. finegoldii synthetases was confirmed by complementation of E. coli strain LCH30 (aas fadD) (Hsu et al., 1991). Strain LCH30 is not capable of utilizing exogenous fatty acids unless complemented with a functional synthetase. Plasmids expressing nothing (pPJ131), E. coli FadD (pEcFadD), N. gonorrhoeae Aas (pNgAas), A. finegoldii Aas1 (pAfAas1), A. finegoldii Aas2 (pAfAas2), or A. finegoldii Alfi_3181 (pAf3181) were transformed into strain LCH30 and recovered on Luria plates containing 100 μg/ml carbenicillin. Individual transformants were then tested for [14C]oleate or [14C]laurate incorporation into phospholipids. Overnight cultures of strain LCH30 harboring the synthetase expression plasmids were back diluted into Luria broth containing 1% v/v DMSO and 0.1 μCi/ml [14C]laurate or [14C]oleate, and labeled at 37°C for one hour after reaching mid-log phase. Lipids were extracted via the Bligh and Dyer method (Bligh and Dyer, 1959) from each experiment and the total radioactive incorporation was measured via liquid scintillation counting. Synthetase expression was visualized by immunoblot analysis of each lysate supernatant using a monoclonal anti-polyhistidine-alkaline phosphatase antibody (Sigma-Aldrich), and the ECF substrate (GE Healthcare) on a Typhoon FLA 9500 imager.
The pET-AfAas1 and pET-AfAas2 expression vectors were transformed into competent cells of E. coli strain BL21(DE3) (Thermo Scientific). Expression of the amino-terminal His6-tagged synthetases was induced with 1 mM IPTG overnight at 16°C, and cells were harvested by centrifugation at 6,000 x g for 20 min. Cell pellets were resuspended in binding buffer (10 mM imidazole, 20 mM Tris-HCl, pH 7.5, 500 mM NaCl), and lysed using a microfluidizer. Cell debris were removed by centrifugation at 40,000 × g for 20 min, and the supernatant was loaded onto a Ni2+-NTA column equilibrated in binding buffer. The column was washed with 5 column volumes each of binding buffer containing increasing concentrations of imidazole (20 mM, 40 mM), and then eluted with 250 mM imidazole, 20 mM Tris-HCl pH 7.5, 500 mM NaCl. The proteins were further purified by size exclusion on a Superdex 200 16/600 column (GE Healthcare) using 20 mM Tris-HCl, pH 7.5, 200 mM NaCl as the running buffer.
The acyl carrier and chain length selectivity for each synthetase was determined by incubating 0, 100, 250, 500, 1000, and 5000 μM AfAas1 or 0, 100, 250, 500, 1000, 1250 μM AfAas2 for 30 minutes at 37°C with E. coli ACP (50 μM) or CoA (50 μM); 100 mM Tris-HCl, pH 7.5; 10 mM MgCl2; 1% Triton X-100; 10 μM DTT; 25 μM EDTA; 5 mM ATP; and 80 μM [1-14C]lauric acid (59 mCi/mmol, 0.1 mCi/ml) or [1-14C]oleic acid (59 mCi/mmol, 0.1 mCi/ml) in a 40 μl reaction. After incubation, the 40 μl were spotted on a Whatman 3MM paper disc and dried to stop the reaction. The discs were washed twice in chloroform:methanol:acetic acid (3:6:1 v/v) and measured in a scintillation counter.
Acknowledgments
We thank Pam Jackson for strain construction, Ellie Margolis for training and access to the Whitley M45 HEPA Variable Atmosphere Workstation, and the Hartwell Center DNA Sequencing Shared Resource for DNA sequencing.
This work was supported by National Institutes of Health grant GM034496 (C.O.R.), Cancer Center Support Grant CA21765 and the American Lebanese Syrian Associated Charities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest
The authors declare that they have no conflicts of interest with the contents of this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Abbanat DR, Leadbetter ER, Godchaux W, and Escher A (1986) Sulfonolipids are molecular determinants of gliding motility. Nature 324, 367–369. [Google Scholar]
- Alvarez JG, and Touchstone JC (1992) Separation of acidic and neutral lipids by aminopropyl-bonded silica gel column chromatography. J. Chromatogr. A 577, 142–145. [DOI] [PubMed] [Google Scholar]
- Baronio M, Lattanzio VM, Vaisman N, Oren A, and Corcelli A (2010) The acylhalocapnines of halophilic bacteria: structural details of unusual sulfonate sphingoids. J. Lipid Res 51, 1878–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN, and DiRusso CC (2003) Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol. Mol. Biol. Rev 67, 454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN, DiRusso CC, Metzger AK, and Heimert TL (1992) Cloning, sequencing, and expression of the fadD gene of Escherichia coli encoding acyl coenzyme A synthetase. J. Biol. Chem 267, 25513–25520. [PubMed] [Google Scholar]
- Black PN, Zhang Q, Weimar JD, and DiRusso CC (1997) Mutational analysis of a fatty acyl-coenzyme A synthetase signature motif identifies seven amino acid residues that modulate fatty acid substrate specificity. J. Biol. Chem 272, 4896–4903. [DOI] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, and Lawley TD (2016) Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers DM, and Holmes CG (1990) A soluble fatty acyl-acyl carrier protein synthetase from the bioluminescent bacterium Vibrio harveyi. Biochem. Cell Biol 68, 1045–1051. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A 108, 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-H, Heath RJ, and Rock CO (2000) β-Ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol 182, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J (1990) Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J. Biol. Chem 265, 17215–17221. [PubMed] [Google Scholar]
- D’Agnolo G, Rosenfeld IS, Awaya J, Omura S, and Vagelos PR (1973) Inhibition of fatty acid biosynthesis by the antibiotic cerulenin. Specific inactivation of β-ketoacyl-acyl carrier protein synthetase. Biochim. Biophys. Acta 326, 155–166. [DOI] [PubMed] [Google Scholar]
- d’Hennezel E, Abubucker S, Murphy LO, and Cullen TW (2017) Total lipopolysaccharide from the human gut microbiome silences Toll-like receptor signaling. mSystems 2, e00046–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokhi V, Nemati R, Nichols FC, Yao X, Anstadt E, Fujiwara M, Grady J, Wakefield D, Castro W, Donaldson J, and Clark RB (2013) Bacterial lipodipeptide, Lipid 654, is a microbiome-associated biomarker for multiple sclerosis. Clin. Transl. Immunol 2, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane MM, Sharpton TJ, Laurent TJ, and Pollard KS (2014) A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS ONE 9, e84689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, and Sonnenburg JL (2011) Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwin JL, Klages AL, and Cronan JE Jr. (1980a) β-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J. Biol. Chem 255, 3263–3265. [PubMed] [Google Scholar]
- Garwin JL, Klages AL, and Cronan JE Jr. (1980b) Structural, enzymatic, and genetic studies of β-ketoacyl-acyl carrier protein synthases I and II of Escherichia coli. J. Biol. Chem 255, 11949–11956. [PubMed] [Google Scholar]
- Geiger O, Gonzalez-Silva N, Lopez-Lara IM, and Sohlenkamp C (2010) Amino acid-containing membrane lipids in bacteria. Prog. Lipid Res. 49, 46–60. [DOI] [PubMed] [Google Scholar]
- Godchaux W, and Leadbetter ER (1988) Sulfonolipids re localized in the outer membrane of the gliding bacterium Cytophaga johnsonae. Arch. Microbiol 150, 42–47. [Google Scholar]
- Gupta RS, and Lorenzini E (2007) Phylogeny and molecular signatures (conserved proteins and indels) that are specific for the Bacteroidetes and Chlorobi species. BMC Evol. Biol. 7, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, and Gross RW (1995) Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom 6, 1202–1210. [DOI] [PubMed] [Google Scholar]
- Heaver SL, Johnson EL, and Ley RE (2018) Sphingolipids in host-microbial interactions. Curr. Opin. Microbiol 43, 92–99. [DOI] [PubMed] [Google Scholar]
- Hsu FF, and Turk J (2001) Studies on phosphatidylglycerol with triple quadrupole tandem mass spectrometry with electrospray ionization: Fragmentation processes and structural characterization. J. Am. Soc. Mass Spectrom. 12, 1036–1043. [Google Scholar]
- Hsu L, Jackowski S, and Rock CO (1991) Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. J. Biol. Chem 266, 13783–13788. [PubMed] [Google Scholar]
- Human Microbiome Project, C. (2012) Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iram SH, and Cronan JE (2006) The β-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J. Bacteriol 188, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S, Jackson PD, and Rock CO (1994) Sequence and function of the aas gene in Escherichia coli. J. Biol. Chem 269, 2921–2928. [PubMed] [Google Scholar]
- Jiang Y, Chan CH, and Cronan JE (2006) The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry 45, 10008–10019. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Morgan-Kiss RM, Campbell JW, Chan CH, and Cronan JE (2010) Expression of Vibrio harveyi acyl-ACP synthetase allows efficient entry of exogenous fatty acids into the Escherichia coli fatty acid and lipid A synthetic pathways. Biochemistry 49, 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EL, Heaver SL, Walters WA, and Ley RE (2017) Microbiome and metabolic disease: revisiting the bacterial phylum Bacteroidetes. J. Mol. Med. (Berl) 95, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, and Gordon JI (2011) Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain JM, Fournier PE, and Raoult D (2018) Culturing the human microbiota and culturomics. Nat. Rev. Microbiol 16, 540–550. [DOI] [PubMed] [Google Scholar]
- Liu Y, Harrison PM, Kunin V, and Gerstein M (2004) Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-J, Zhang Y-M, Grimes KD, Qi J, Lee RE, and Rock CO (2006) Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Molec. Cell 23, 765–772. [DOI] [PubMed] [Google Scholar]
- Marten B, Pfeuffer M, and Schrezenmeir J (2006) Medium-chain triglycerides. Int. Dairy J. 16, 1374–1382. [Google Scholar]
- Mazzella N, Molinet J, Syakti AD, Dodi A, Doumenq P, Artaud J, and Bertrand JC (2004) Bacterial phospholipid molecular species analysis by ion-pair reversed-phase HPLC/ESI/MS. J. Lipid Res. 45, 1355–1363. [DOI] [PubMed] [Google Scholar]
- Nemati R, Dietz C, Anstadt EJ, Cervantes J, Liu Y, Dewhirst FE, Clark RB, Finegold S, Gallagher JJ, Smith MB, Yao X, and Nichols FC (2017) Deposition and hydrolysis of serine dipeptide lipids of Bacteroidetes bacteria in human arteries: relationship to atherosclerosis. J. Lipid Res. 58, 1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen I, and Nichols FC (2018) Are sphingolipids and serine dipeptide lipids underestimated virulence factors of Porphyromonas gingivalis? Infect. Immun 86, e00035–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, and Rock CO (2014a) Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A 111, 10532–10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Frank MW, Eleveld MJ, Schalkwijk J, Broussard TC, de Jonge MI, and Rock CO (2014b) A thioesterase bypasses the requirement for exogenous fatty acids in the plsX deletion of Streptococcus pneumoniae. Mol. Microbiol 96, 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Frank MW, Jackson P, Subramanian C, and Rock CO (2014c) Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol. Microbiol 92, 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Frank MW, Subramanian C, Saenkham P, and Rock CO (2011) Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U. S. A 108, 15378–15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, and Rock CO (2013) Bacterial lipids: Metabolism and membrane homeostasis. Prog. Lipid Res. 52, 249–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JB, Yao J, Jackson P, Frank M, and Rock CO (2013) Phosphatidylglycerol homeostasis in glycerol-phosphate auxotrophs of Staphylococcus aureus. BMC Microbiol. 13, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautio M, Eerola E, Vaisanen-Tunkelrott ML, Molitoris D, Lawson P, Collins MD, and Jousimies-Somer H (2003) Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a new genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and description of Alistipes finegoldii sp. nov., from human sources. Syst. Appl. Microbiol 26, 182–188. [DOI] [PubMed] [Google Scholar]
- Rizza V, Tucker AN, and White DC (1970) Lipids of Bacteroides melaninogenicus. J. Bacteriol 101, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson RM, Yao J, Gajewski S, Kumar G, Martin EW, Rock CO, and White SW (2017) A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer. Nat. Struct. Mol. Biol 24, 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TSB, Raes J, and Bork P (2018) The human gut microbiome: from association to modulation. Cell 172, 1198–1215. [DOI] [PubMed] [Google Scholar]
- Schonfeld P, and Wojtczak L (2016) Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J. Lipid Res. 57, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, and Geiger O (2016) Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev 40, 133–159. [DOI] [PubMed] [Google Scholar]
- Vance DE, Goldberg I, Mitsuhashi O, Bloch K, Omura S, and Nomura S (1972) Inhibition of fatty acid synthetases by the antibiotic cerulenin. Biochem. Biophys. Res. Commun 48, 649–656. [DOI] [PubMed] [Google Scholar]
- Walker A, Pfitzner B, Harir M, Schaubeck M, Calasan J, Heinzmann SS, Turaev D, Rattei T, Endesfelder D, Castell WZ, Haller D, Schmid M, Hartmann A, and Schmitt-Kopplin P (2017) Sulfonolipids as novel metabolite markers of Alistipes and Odoribacter affected by high-fat diets. Sci. Rep 7, 11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters WA, Xu Z, and Knight R (2014) Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RH (1984) Biosynthesis of the sulfonolipid 2-amino-3-hydroxy-15-methylhexadecane-1-sulfonic acid in the gliding bacterium Cytophaga johnsonae. J. Bacteriol 159, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Bruhn DF, Frank MW, Lee RE, and Rock CO (2016) Activation of exogenous fatty acids to acyl-acyl carrier protein cannot bypass FabI inhibition in Neisseria. J. Biol. Chem 291, 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Cherian PT, Frank MW, and Rock CO (2015a) Chlamydia trachomatis relies on autonomous phospholipid synthesis for membrane biogenesis. J. Biol. Chem 290, 18874–18888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Dodson VJ, Frank MW, and Rock CO (2015b) Chlamydia trachomatis scavenges host fatty acids for phospholipid synthesis via an acyl-acyl carrier protein synthetase. J. Biol. Chem 290, 22163–22173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Maxwell JB, and Rock CO (2013) Resistance to AFN-1252 arises from missense mutations in Staphylococcus aureus enoyl-acyl carrier protein reductase (FabI). J. Biol. Chem 288, 36261–36271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, and Rock CO (2013) Phosphatidic acid synthesis in bacteria. Biochim. Biophys. Acta 1831, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, and Rock CO (2015) How bacterial pathogens eat host lipids: Implications for the development of fatty acid synthesis therapeutics. J. Biol. Chem 290, 5940–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]