Abstract
The protonation state of embedded charged residues in transmembrane proteins (TMPs) can control the onset of protein function. It is understood that interactions between an embedded charged residue and other charged or polar residues in the moiety would influence its pKa, but how the surrounding environment in which the TMP resides affects the pKa of these residues is unclear. Proteorhodopsin (PR), a light-responsive proton pump from marine bacteria, was used as a model to examine externally accessible factors that tune the pKa of its embedded charged residue, specifically its primary proton acceptor D97. The pKa of D97 was compared between PR reconstituted in liposomes with different net headgroup charges and equilibrated in buffer with different ion concentrations. For PR reconstituted in net positively charged compared to net negatively charged liposomes in low-salt buffer solutions, a drop of the apparent pKa from 7.6 to 5.6 was observed, whereas intrinsic pKa modeled with surface pH calculated from Gouy-Chapman predictions found an opposite trend for the pKa change, suggesting that surface pH does not account for the main changes observed in the apparent pKa. This difference in the pKa of D97 observed from PR reconstituted in oppositely charged liposome environments disappeared when the NaCl concentration was increased to 150 mM. We suggest that protein-intrinsic structural properties must play a role in adjusting the local microenvironment around D97 to affect its pKa, as corroborated with observations of changes in protein side-chain and hydration dynamics around the E-F loop of PR. Understanding the effect of externally controllable factors in tuning the pKa of TMP-embedded charged residues is important for bioengineering and biomedical applications relying on TMP systems, in which the onset of functions can be controlled by the protonation state of embedded residues.
Significance
Embedded charged residue plays an important role in gating the functions of natural and engineered proteins. Our work elucidates that the electrostatic environment of liposomes in which proteorhodopsin resides can affect the intrinsic pKa of its embedded D97 residue. The pKa change is accompanied with a structural rearrangement measured by magnetic resonance spectroscopic techniques. Our study adds value to the biophysical understanding of how electrostatic environment can affect the protonation behavior of embedded charged residues through a change of protein-intrinsic structural properties. Changes of both the intrinsic and the apparent pKa of D97 under different electrostatic environments reported here also offer a guideline for optimizing functions of transmembrane proteins that are controlled by a similar mechanism as proteorhodopsin.
Introduction
Protonation or deprotonation of certain charged residues in a transmembrane protein (TMP) controls the onset of many TMP functions, including ion transport (1,2), redox reaction (3), and catalysis (4). For example, an ion transport process of retinal proteins could involve concerted protonation and deprotonation of several charged residues during its reaction cycle to achieve a vectorial transport of ions across lipid bilayers (5, 6, 7). In proteorhodopsin (PR), a light-activated pump transports proton from the intracellular to the extracellular side of a marine bacterial membrane, whereas the key aspartic acid residue D97 controls whether its function can be initiated after photoactivation (8,9). This D97 residue is not located on the surface but buried inside the protein (10). The pKa of such a buried residue inside a TMP can be different from the pKa of the same amino acid in aqueous solution, and the value could vary from protein site to site for the same amino acid (11,12). This pKa generally shifts in the direction that favors the neutral form of the amino acid when these residues are embedded in the hydrophobic interior of the protein because the D96 residue inside bacteriorhodopsin has a high pKa and is mostly protonated (13). Moreover, the pKa of embedded residues was also shown to be affected by the environment in which the TMP resides. For example, the pKa of the embedded D97 residue (pKaD97) of PR can differ by more than one unit by the extent of oligomerization of surfactant-reconstituted PR (14,15), as well as the NaCl concentration in the buffer that contains detergent-constituted PR (16). These results unveiled that the external environment in which TMP resides can affect the pKa of embedded charged residues, but not the underlying mechanism. Identifying the key factors that determine the pKa of an embedded charged residue is important for gaining a biophysical understanding and for tuning the TMP functions in engineering applications with the protein reconstituted in different synthetic host materials (17,18).
The factors that determine the pKa of a protein-embedded charged residue are not well understood, and their study is challenging. Experimental and simulation studies on quantifying the pKa of embedded charged residues have been performed with a water-soluble protein, staphylococcal nuclease (12,19, 20, 21, 22, 23, 24), and a TMP, bacteriorhodopsin (25). By applying continuous constant pH molecular dynamics and virtual mixture of multiple states method (26, 27, 28, 29), these simulation studies successfully reproduced some of the experimental pKa values determined by NMR techniques (30,31), but large discrepancies between simulated and experimental pKa are observed in most cases (28,32, 33, 34). Various studies in the literature suggested water or ion penetration in the vicinity (21,35,36), charge-charge interactions with other ionizable groups (22,25,37), charge-polar interactions (38), and hydrogen bonding of the embedded charged residue to be microenvironmental factors that affect the pKa of embedded charged residues on water-soluble proteins (39). Other studies suggested that the ionization of the embedded charged residue can induce conformational rearrangements of protein structures (40, 41, 42) or that changes in the electrostatic network around the embedded charged residue can induce conformational changes that, in turn, affect the ionization potential of the embedded charged residue (43). Overall, these studies indicated that the microenvironments around the embedded charged residue in proteins can sensitively modulate its pKa, but there is no consensus regarding the mechanisms.
In comparison to water-soluble proteins that are hydrated in bulk water, TMPs are embedded in a lipid bilayer with a densely charged surface formed by lipid headgroups and a low dielectric interior made of hydrophobic lipid hydrocarbon chains (44,45). The protonation state of embedded charged residues in TMPs reconstituted in such an environment could potentially be affected by the charge of lipid headgroups, the orientation of water at the lipid bilayer surface (46), the population of penetrated water inside lipid bilayers, or ion condensation around the surface of lipid bilayers including protons (25). In fact, it has been suggested that differences in the local proton concentration on a charged lipid bilayer surface, which give rise to a spatially varying surface proton concentration compared to the bulk pH, affect the apparent pKa of embedded charged residues in TMPs (47, 48, 49, 50). Motivated by biophysical understanding objectives, we ask in this study whether the deviation of surface proton concentration is the only major factor that affects the protonation behavior of embedded charged residues or whether the change of external factors, such as lipid compositions, buffer and salt type, and/or salt concentration, that affect the local proton concentration can also affect the microenvironment around the embedded charge through structural rearrangement to change its intrinsic pKa.
Specifically, we focus on the question of whether the electrostatic environment of the liposome in which PR resides modulates the microenvironment around the embedded charged residue and, in turn, induces an intrinsic pKa shift. We also need to test whether potential shifts in pKa under different external environments are associated with structural changes of PR, as seen in simulation studies of the water-soluble protein staphylococcal nuclease (41,42), as evidence of whether the observed pKa shift is not only an apparent effect due to surface pH deviation. Here, we focus on the charged aspartic acid D97 inside PR. PR is an excellent model TMP, given its robustness that allows us to explore the effect of a wide range of external environmental conditions on pKaD97 of PR reconstituted in liposomes. Moreover, because D97 is located near the retinal chromophore of PR, the ultraviolet (UV)-visible absorbance of the protein is sensitive to its protonation state (51). The pKaD97 can therefore be determined conveniently by spectrophotometric titration rather than by the powerful, but more challenging, NMR titration method, which requires high resolution of the charged residue of interest (22,37).
We systematically altered the electrostatic environment of PR reconstituted in liposomes by varying the salt type and concentration inside aqueous solution and the net charge of the liposome headgroup composition. Both the apparent pKaD97 and the intrinsic pKaD97, which takes the surface pH difference into account under these different environments, were compared to identify the key factors that affect the pKa of the embedded charged residue D97. We performed continuous-wave electron paramagnetic resonance (cw EPR) and 1H Overhauser dynamic nuclear polarization (ODNP) relaxometry, with a spin label attached to site 174 to reveal possible structural changes in PR associated with the observed pKa shift (52, 53, 54, 55, 56, 57), as site 174 on the E-F loop of PR has been shown to be a sensitive reporter on the light-activated structural movement of PR (58). The insight gained from the embedded charged D97 in PR could be extended to other TMPs activated by the protonation/deprotonation at embedded sites.
Materials and Methods
PR expression and purification
Cysteine-free green-light absorbing PR with a 6x-His tag on its C-terminus were expressed and purified using previously described protocols (58,59). The PR gene was cloned into a pET26b (+) vector (Novagen, Madison, WI) and transformed into a BL21(DE3) Escherichia coli strain for protein expression. The expressed PR was purified by using Ni-NTA resin that specifically binds with the 6x-His tag on PR and eluted by a buffer (50 mM potassium phosphate dibasic buffer, 150 mM potassium chloride, 0.05 wt% n-dodecyl β-D-maltoside (DDM (pH 8.2))) with 300 mM of imidazole. The PR was then buffer exchanged into the same buffer without imidazole and concentrated by Amicon Ultra centrifugal filter units with a 50 kDa MWCO (Millipore, Burlington, MA) for storage or later PR-containing liposome preparation.
Preparation of PR-containing liposomes
The lipid extrusion method was used to prepare large unilamellar vesicles (LUVs). Lipids dissolved in chloroform (Avanti Polar Lipids, Alabaster, AL) were mixed to achieve desired compositions in a glass vial, and the chloroform was then removed through drying under a nitrogen stream and overnight desiccating under vacuum. The dried lipids were resuspended using a HEPES buffer (10 mM HEPES (pH 6.7)) with the desired concentration of additional salt (KCl, NaCl, CaCl2, and MgCl2) as specified in each experimental condition. The mixture was vortexed for at least 10 min under room temperature for complete resuspension, and the lipid solution was then extruded 19 times through a miniextruder installed with a Nuclepore polycarbonate membrane (200 nm pore size; Whatman, Maidstone, UK) to form LUVs.
Purified PR was reconstituted into liposomes to form proteoliposomes using the BioBeads removal method (60). The extruded LUVs were mixed with DDM surfactant solution to achieve a final DDM concentration that is two times the critical micelle concentration (0.0088 w/v %) and shaken for 1 h for lipid-surfactant complex formation. PR was then added to the lipid-surfactant solution with a 1:50 protein/lipid molar ratio and gently shaken for another 1 h. The PR-lipid-DDM solution was then transferred through six vials of microcentrifuge tubes loaded with 140 mg of SM-2 BioBeads (BioRad, Hercules, CA) to remove DDM and drive PR proteoliposome formation.
Optical absorption measurements and analyses
PR pH-dependent optical absorbance change was recorded by a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan), and the data analysis for getting the pKaD97 was done by the processing and fitting algorithm described in previous studies (15). Prepared PR-containing liposomes were diluted by the HEPES buffer with the desired salt concentrations to achieve a total volume of 750 μL and an initial optical density between 0.3 and 0.5 at 520 nm. For all PR samples, optical absorbance spectra between 400 and 750 nm were recorded with a 0.5 nm and under interval at up to 30 different pH values between 4 and 10. Acidic PR, with a protonated D97 residue, has a maximum absorbance at ∼535 nm, which shifts to ∼518 nm upon deprotonation. Instead of directly tracking the maximum absorbance shift under different pH, the pKa was obtained from the pH-dependent change in optical absorbance at 570 nm with a better resolution, fitting by the Henderson-Hasselbalch equation. The details of how the pH-dependent difference absorbance at 570 nm was obtained and converted to the spectral titration curve are included in Fig. S1.
Spin labeling of PR and cw EPR measurement
(1-Oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (MTSL) spin labels (Toronto Research Chemicals, Ontario, Canada) with EPR sensitivity were conjugated to PR through disulfide formation with the selected cysteine mutation site 174 on PR. The cysteine mutation site was introduced to PR using the site-directed mutagenesis technique. After PR with the cysteine mutation was expressed and purified, 1 mM dithiothreitol was added to the PR-containing buffer to ensure the thiol groups were fully reduced before the labeling reaction. The dithiothreitol was then removed by a PD-10 desalting column (GE Healthcare, Chicago, IL) containing Sephadex G-25 resin directly before the labeling reaction. A 10× molar excess of MTSL spin labels was then mixed with PR and shaken for at least 16 h under room temperature for the conjugation reaction to complete. The excess MTSL was removed via size exclusion chromatography, in which PR was run through a HiLoadTM 16/600 SuperdexTM 200 pg column (GE Healthcare) connected to an NGC Medium-Pressure Liquid Chromatography System (BioRad). The prepared spin-labeled PR was then reconstituted into liposomes with the desired condition using the method described above. The x-band (0.35 T) cw EPR spectra from PR-liposome samples were recorded on an EMXPlus spectrometer (Bruker, Billerica, MA), with a microwave power of 20 mW, a modulation amplitude of 1 G, and a total sweep width of 150 G.
ODNP
ODNP measurements on PR reconstituted in liposomes were carried out in the 0.35 T Bruker EMXPlus magnet. A homebuilt NMR probe was added within a TE011 microwave cavity to hold the sample, and the probe was connected to a Bruker Avance 300 NMR spectrometer to detect 1H NMR signal from the sample. PR-liposome samples were loaded in capillary tubes with a PR concentration of 200–500 μM and a volume of 3.5 μL. The 1H NMR signal from water molecules around the spin-labeled site on PR was collected under different powers of microwave irradiation to get the 1H NMR enhancement series. The spin-lattice relaxation time T1 under different microwave irradiation powers was collected by inverse recovery experiments for temperature-correction purpose because of a sample heating issue. The details of the automated ODNP experiment setup are described in Franck et al. (56), and the detailed theory and derivation for getting the reported cross-relaxivity kσ as water dynamic information is provided in the Supporting Materials and Methods.
Results
Determining the pKaD97 of PR using UV-visible spectral titration measurements
The primary proton acceptor D97 is buried in the interior of the PR. Its protonation state affects the maximum absorption wavelength of PR through changes in the environment around the retinal chromophore of PR. This property allows the pKa of the D97 residue (pKaD97) to be measured through UV-visible spectral titration measurements (see details of pKaD97 determination in the Materials and Methods and Fig. S1). Fig. 1 shows exemplary data for pKaD97 determination of WT PR reconstituted in 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) (80/20, mol/mol) liposomes in the HEPES buffer without adding additional salt, along with a curvefit to obtain the corresponding pKaD97 (7.62 ± 0.02). The pKaD97 from wild-type PR (WT PR) was shown to be tunable and varies from 6.2 to 7.6 depending on the sample condition (14,16). Note that the titration curve in Fig. 1 is plotted against the bulk pH in buffers, and the pKaD97 obtained from fitting is the apparent pKa, which does not count in the deviation of surface proton concentration from the bulk pH. To understand the mechanism of how the surrounding environment affects the pKaD97 of PR reconstituted in liposomes, the electrostatic environment of PR-containing liposomes was modulated.
Figure 1.
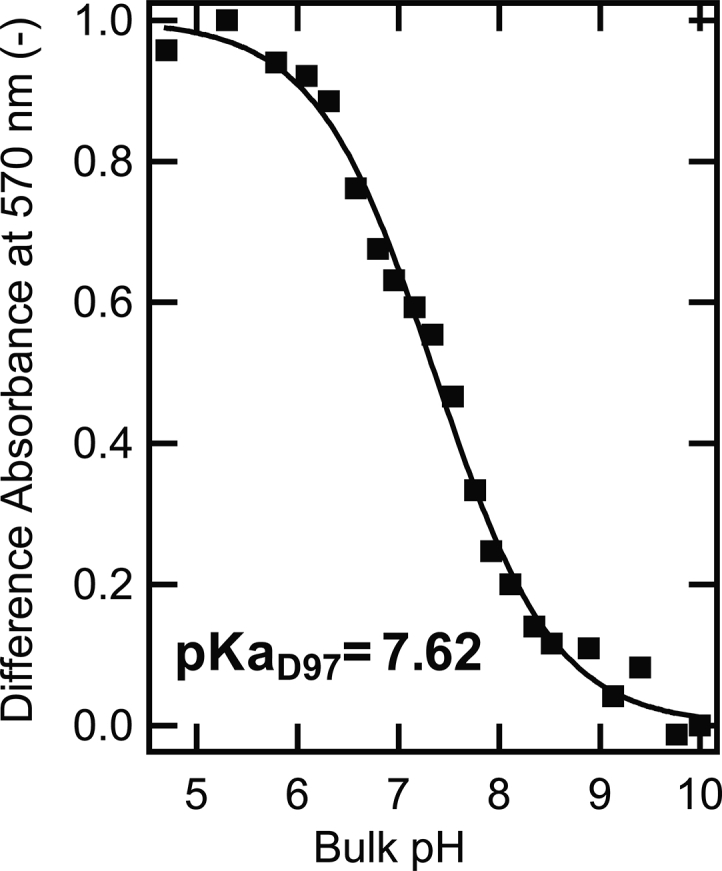
Difference optical absorbance at 570 nm under various bulk pH from WT PR reconstituted in POPC/POPG (80/20, mol/mol) liposomes and in a 10 mM HEPES buffer. The difference absorbance at 570 nm under different pH was normalized to the difference between the maximum and the minimum. The pH-dependent absorbance curve was fitted by the Henderson-Hasselbalch equation to get the apparent pKaD97.
Effect of concentration of cation in buffer and the net charge of liposomes on apparent pKaD97
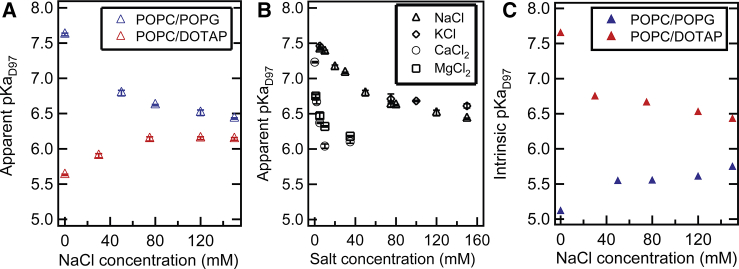
We examined the effect of the external electrostatic environment on the apparent pKaD97 of PR by changing the net charge of liposomes and the concentration of ions in the HEPES buffer. The net charge of liposomes was adjusted by mixing POPC with a zwitterionic headgroup with either POPG with a negatively charged headgroup or 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) with a positively charged headgroup (structures shown in Fig. S2 A). The apparent pKaD97 of PR was measured in the negatively charged POPC/POPG (80/20, mol/mol) liposomes and positively charged POPC/DOTAP (80/20, mol/mol) liposomes in the same HEPES buffer while successively increasing the NaCl concentration from 0 to 150 mM. In the case of negatively charged POPC/POPG (80/20, mol/mol) liposomes, pKaD97 decreased gradually from 7.4 ± 0.02 to 6.4 ± 0.01 when the NaCl concentration in the buffer was increased (Fig. 2 A, blue; spectral titration curves under different NaCl concentrations for the Henderson-Hasselbalch fitting are plotted in Fig. S3 A for reference). In contrary, an opposite incremental trend of pKa shift was observed in the case of positively charged POPC/DOTAP (80/20, mol/mol) liposomes while NaCl concentration was increased from 0 mM (pKaD97 = 5.63 ± 0.01) to 150 mM (pKaD97 = 6.14 ± 0.02) in the HEPES buffer (Fig. 2 A, red; corresponding spectral titration curves are plotted in Fig. S3 B). The sharpest pKaD97 difference, which is an up to ∼2 pH unit change, between the negatively charged POPC/POPG and positively charged POPC/DOTAP liposomes can be observed when there is no additional NaCl in the HEPES buffer. With the presence of more NaCl in the buffer, the difference of measured apparent pKaD97 between PR in these two types of liposomes with opposite net charges became smaller. For the PR-containing liposomes constituted of pure zwitterionic POPC, the measured pKaD97 lay between the ones from positively charged POPC/DOTAP (80/20, mol/mol) liposomes and negatively charged POPC/POPG (80/20, mol/mol) liposomes for both buffers with and without NaCl (Fig. S2 B). These results all follow a systematic trend, in which the apparent pKaD97 shifted toward a lower value when the charge of the liposome surface changed from net negative to net positive.
Figure 2.
(A) Apparent pKaD97 of WT PR reconstituted in negatively charged POPC/POPG (80/20, mol/mol) liposomes (blue triangle) and positively charged (80/20, mol/mol) liposomes (red triangle) in a 10 mM HEPES buffer. The HEPES buffer contained different concentrations of NaCl between 0 and 150 mM. (B) Apparent pKaD97 of WT PR reconstituted in negatively charged POPC/POPG (80/20, mol/mol) liposomes in the HEPES buffer containing different concentrations of NaCl (triangle, same as in A), KCl (diamond), CaCl2 (circle), and MgCl2 (square) is shown. (C) Intrinsic pKaD97 of WT PR reconstituted in negatively charged POPC/POPG (80/20, mol/mol) liposomes (blue triangle) and positively charged (80/20, mol/mol) liposomes (red triangle) in a HEPES buffer containing different concentrations of NaCl is shown.
The effect of adding different salts in the buffer was also tested in the case of PR reconstituted in negatively charged POPC/POPG (80/20, mol/mol) liposomes (Fig. 2 B). Adding KCl (Fig. 2 B, diamond) with a monovalent cation showed a similar trend of pKaD97 change as NaCl. The case of adding salts with divalent cations (MgCl2 and CaCl2) both showed a more abrupt apparent pKaD97 change. The apparent pKaD97 dropped to 6.75 ± 0.03 upon addition of a lower ion concentration (1 mM) of Ca2+ (Fig. 2 B, circle) and declined even further to 6.10 ± 0.02 at the highest Ca2+ concentration (35 mM). This pKa measured under 35 mM Ca2+ is lower than the smallest value (6.43) observed with monovalent cations at much greater NaCl or KCl concentration. A similar trend was observed for Mg2+ (Fig. 2 B, square). Concentrations of divalent cations above 35 mM were not tested because no further change of pKa is expected in that region. Overall, all the salt added can change the apparent pKaD97 of PR reconstituted in negatively charged liposomes. The observed pKa change is steeper for divalent cations (Mg2+ and Ca2+) in comparison with monovalent cations (Na+ and K+).
Effect of concentration of cation in buffer and the net charge of liposomes on intrinsic pKaD97
The electrostatic environment of the PR-embedding liposomes, including the net charge of liposomes and the salt concentration in buffer, dominantly and significantly modulates the apparent pKa of the buried D97 residue. Early bacteriorhodopsin (bR) studies suggested that distribution shift between the protonated and the deprotonated form of its embedded Schiff base counterion residue was due to a deviation of the surface proton concentration from the bulk pH around a charged lipid membrane surface (47,48). This means that the observed pKa change in the spectral titration experiment is only an apparent effect without changing the equilibrium constant. Nevertheless, the electrostatic environment may also cause a structural rearrangement of TMPs to change the microenvironment around the embedded charged residue and, in turn, induce a shift in its intrinsic pKa.
To further prove that the observed pKa shift is not only an apparent effect dictated mainly by surface pH, we used the Gouy-Chapman (GC) model to calculate the surface pH on charged liposomes under the conditions tested in Fig. 2 A (detail derivation included in the Supporting Materials and Methods) (61,62). After calculating the surface pH on charged liposome surfaces and replotting the spectral titration data against the surface pH, the intrinsic pKaD97 obtained from Henderson-Hasselbalch fitting still changed systematically under the electrostatic environment tested (Fig. 2 C, surface potentials predicted from GC included in Table S1; corresponding spectral titration data are plotted in Fig. S4). The greatest deviation in the intrinsic pKaD97 from the apparent pKaD97 was found in buffers without additional NaCl. Furthermore, the intrinsic pKa at 0 mM NaCl is distinctly different for PR in POPC/POPG (80/20, mol/mol) versus POPC/DOTAP (80/20, mol/mol) liposome environments after taking surface proton concentration difference into account (5.11 vs. 7.64). In the presence of more NaCl in the buffer, the intrinsic pKaD97 shifts in the opposite direction to the apparent pKaD97 while the two values converge. This shows us that the electrostatic effect exerted on PR’s pKaD97 by its liposome environment is screened out at high NaCl concentrations. If surface pH were the only factor to affect the measured apparent pKaD97 in Fig. 2 A, then comparable or similar intrinsic pKaD97 would be expected in Fig. 2 C under all conditions tested. In fact, we found an opposite trend for the intrinsic pKaD97 compared to the apparent pKaD97, whereas we also found that the intrinsic pKaD97 clearly changes for PR reconstituted in liposomes with different electrostatic environments. Our results suggest the observed pKaD97 change is not solely an apparent effect caused by the deviation of proton concentrations on charged liposome surfaces compared to the bulk solution but due to actual changes of its acid dissociation constant. It is also noted that all the spectral titration curves, both for the ones plotted against bulk pH (Fig. S3) and surface pH calculated by GC model (Fig. S4), show a similar shape without significant distortion. This can be further supported by comparing the Hill coefficient from the Henderson-Hasselbalch fitting, where a similar Hill coefficient was found between the spectral titration curves plotted against bulk pH or surface pH under the same condition tested (Fig. S5). These results further support our hypothesis that the deviation of surface pH on charged liposome surface is not the only nor the main driving factor affecting the apparent pKaD97 (Fig. 2 A) for PR reconstituted in liposomes with different electrostatic environments.
We then went on to test other possible mechanisms of how the external electrostatic environment in which PR resides may affect the intrinsic pKa of the embedded D97 residue. Because the protonation behavior of embedded residues can be determined by their local microenvironment or the conformation of proteins, one of the most plausible mechanisms is direct long-range or mediated short-range electrostatic static interactions between charged lipid headgroups and the embedded charged residues, here specifically D97 (17,25). If changes in the external factors that change the embedded residue pKa, under conditions that do not alter the charge state of D97, are accompanied by structural rearrangements of the protein, this would suggest that the electrostatic effect of the external factors are coupled to the protein-internal electrostatic network. Hence, we next investigate whether a structural rearrangement of PR can be observed under different electrostatic environments by using EPR techniques with spin labels on PR, and measurements were performed at a sufficiently high pH, at which the majority of D97 remains deprotonated.
Revealing the structure of PR E-F loop under different electrostatic environment using cw EPR
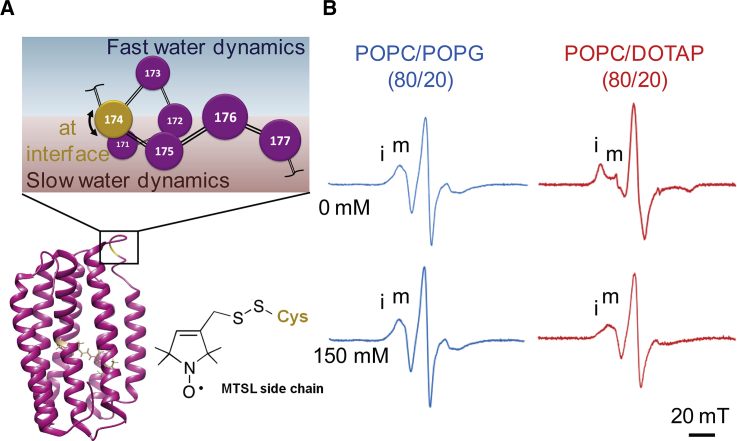
cw EPR is a common technique for obtaining protein local structural information. EPR sensitive spin labels with an unpaired electron (e.g., MTSL) are covalently linked to the selected cysteine mutation sites. Protein structural changes around the labeled site can be reflected in the cw EPR lineshape change if the spin label experiences altered dynamics in the subnanosecond to tens of nanoseconds regime (52,63). For MTSL-labeled PR, site 174 on the E-F loop has been reported to be an interfacial site that showed cw EPR lineshape change due to tertiary interaction after PR experienced a light-activated structural movement (Fig. 3 A; (58)). Specifically, an increase of the immobile component “i” and a reduction of the mobile component “m” on the cw EPR spectrum was observed upon photoactivating PR (58). Here, we applied site-directed mutagenesis and spin labeling on the same site 174 (Fig. 3 A) to detect possible structural rearrangement associated with the observed intrinsic pKaD97 change for PR under different electrostatic environments.
Figure 3.
(A) PR structure and a schematic diagram of the α-helical structure of its E-F loop. The spin-labeled 174 was shown to be located at the interface between the region with fast water dynamics (blue shading) and the region with slow water dynamics (red shading). The chemical structure of the MTSL spin label is also shown along the side. (B) cw EPR spectra of PR spin labeled at E-F loop site 174 reconstituted in negatively charged POPC/POPG (80/20, mol/mol) liposomes (blue spectra) or positively charged POPC/DOTAP (80/20, mol/mol) liposomes (red spectra) in 10 mM HEPES buffers with 0 mM NaCl or with 150 mM NaCl are shown. All the samples were equilibrated at pH 8.5.
X-band cw EPR spectra of PR spin labeled at site 174 were measured and compared between two differently charged liposomes (negatively charged POPC/POPG versus positively charged POPC/DOTAP) at two different NaCl concentration (0 mM vs. 150 mM) in HEPES buffer and at pH 8.5. For all cw EPR spectra shown here, the locations of the restricted immobile (“i”) component and the faster-moving mobile (“m”) component are highlighted at the low-field region of the spectra (Fig. 3 B). The spectral amplitude of these two components conveys the extent of tertiary contacts that the nitroxide label on site 174 experienced. PR reconstituted in negatively charged POPC/POPG liposomes in a HEPES buffer with 0 mM NaCl that showed the lowest intrinsic pKaD97 (5.11 ± 0.02) is the sample that revealed a dominance of the faster-moving mobile (“m”) component in the cw EPR spectrum (Fig. 3 B, top left). In contrast, the sample under the condition with the highest intrinsic pKaD97 (7.64 ± 0.01, positively charged POPC/DOTAP, 0 mM NaCl) revealed a prominent immobile component (“i”) and a concurrent reduction of the mobile component (“m”) (Fig. 3 B, top right). PR reconstituted in the same positively charged liposome but in the presence of 150 mM NaCl in the HEPES buffer revealed a recovery of the mobile (“m”) component (Fig. 3 B, bottom right), although still less mobile compared to the condition that yielded the lowest intrinsic pKaD97 for PR. No significant cw EPR lineshape difference was observed when varying the NaCl concentrations (0 mM vs. 150 mM) for the PR sample reconstituted in negatively charged POPC/POPG liposomes (Fig. 3 B, blue spectra). We also measured and compared the pKaD97 of the PR with MTSL spin labels on site 174 to WT PR (data not shown). No significant difference was observed between the spin-labeled variant and the WT PR. Taken together, these results show a trend of a cw EPR lineshape change at site 174 that coincides with the observed intrinsic pKaD97 shift, with a higher fraction of the immobile component when the PR has a higher intrinsic pKaD97. These observations suggest that the same changes in the electrostatic environment that modulate the pKaD97 of the buried D97 residue also cause structural modulation in PR’s peripheral E-F loop. We note that all PR-liposome samples studied by cw EPR were equilibrated at pH 8.5 so that at least 88% or more of the buried D97 is deprotonated (even for the highest apparent pKaD97 = 7.62). Thus, the changes seen in cw EPR lineshapes are not a consequence of structural changes that follow the protonation of D97. It is possible that the electrostatic environment couple to D97 is mediated by ordered surface water and is reflected in the E-F loop spin-label dynamics. Hence, next we examine the surface water diffusion dynamics around site 174.
Measuring the hydration dynamics around site 174 on PR E-F loop under different electrostatic environments using ODNP relaxometries
ODNP relaxometry measures the surface water dynamics within 1 nm of the spin-labeled surface and is sensitive to changes in the hydrogen bond network of water surrounding the spin label but also to subtle protein structural changes that reflect themselves in changes in the dynamics of the coupled hydration water to the protein surface (64). The local hydration dynamics <10 Å around the MTSL-labeled site 174 on PR were measured and compared under the four conditions used in the previous section. In ODNP, local water dynamic information is extracted from the electron-1H spin cross-relaxivity kσ that is sensitive to the dynamic of diffusing, loosely bounded (approximately tens of picoseconds timescale) water obtained from 1H NMR signal enhancements of water molecules around the attached spin label on PR under various microwave irradiation powers (53,54,56). Details of the theory and the analysis are included in the Supporting Materials and Methods (57). These fitted kσ can reveal changes in the hydration dynamics associated with subtle structural movements around site 174 (Fig. 3 A) with greater sensitivity compared to cw EPR lineshape analysis (58).
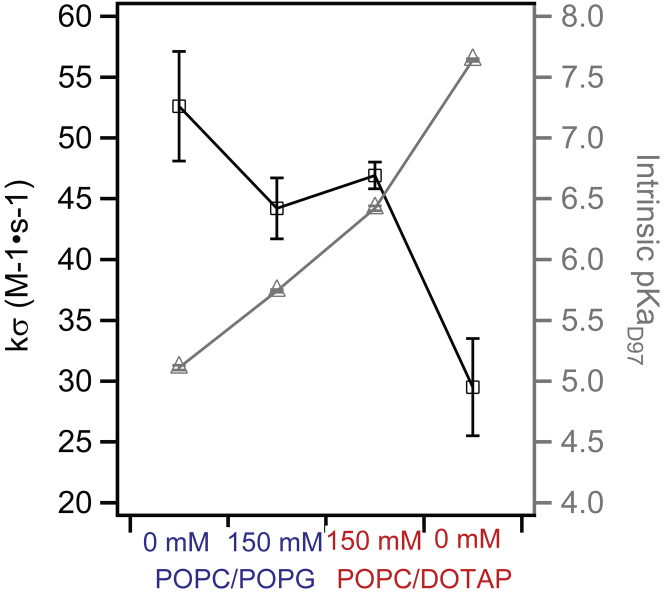
The cross-relaxivity kσ around site 174 of PR reconstituted in two different liposome environments (negatively charged POPC/POPG versus positively charged POPC/DOTAP) in two different HEPES buffers (0 mM NaCl vs. 150 mM) were measured and compared (Fig. 4). Interestingly, we found consistent trends for changes in kσ with that in intrinsic pKaD97. Overall, the condition (positively charged POPC/DOTAP, 0 mM NaCl) with the highest intrinsic pKaD97 yielded the lowest kσ among the conditions tested, which agrees with a more immobile cw EPR spectrum (Fig. 3 B). For PR reconstituted in positively charged POPC/DOTAP liposomes, 150 mM NaCl in the HEPES buffer led to a higher kσ (46.9 ± 1.1 s−1) compared to with 0 mM NaCl (kσ = 29.5 ± 4.0 s−1), accompanying the decrease in intrinsic pKaD97 from 7.64 ± 0.01 to 6.42 ± 0.02 with 150 mM NaCl. For PR reconstituted in negatively charged POPC/POPG liposomes, the presence of 150 mM NaCl in the HEPES buffer led to a lower kσ (44.2 ± 2.5 s−1) at site 174 in comparison to the condition without additional NaCl salt (kσ = 52.6 ± 4.5 s−1). This reduction in kσ accompanies an increase in intrinsic pKaD97 from 5.11 ± 0.02 to 5.74 ± 0.02. Here, the ODNP measurements revealed a change in the hydration environment of site 174 that was not reflected in the cw EPR lineshape between these two conditions.
Figure 4.
Cross-relaxivities kσ that reflect the hydration dynamics (approximately tens of picoseconds timescale) around MTSL-labeled site 174 measured by ODNP on PR reconstituted in negatively charged POPC/POPG (80/20, mol/mol) liposomes or positively charged POPC/DOTAP (80/20, mol/mol) liposomes, in HEPES buffers with 0 mM NaCl or with 150 mM NaCl. The intrinsic pKaD97 under each condition is also plotted along the side as a reference (triangle). To see this figure in color, go online.
Control measurements were done on a model single α-helical WALP23 peptide, with an MTSL spin label conjugated to the cysteine residue right above its α-helix at the lipid-water interface to mimic the location of site 174 on PR in lipid bilayers (the sequence of the C-WALP23 peptide and the location of the labeled site are described in Fig. S7 A). When 150 mM NaCl was added to the solution, the hydration dynamics measured on MTSL-labeled C-WALP in POPC/POPG (80/20, mol/mol) liposomes increased (Fig. S7 B), in contrast to observing a decreased hydration dynamic at site 174 for PR reconstituted in the same negatively charged liposomes (Fig. 4). This observation shows that the change in hydration dynamics observed at site 174 is not a reflection of changes in hydration dynamics on the lipid bilayer surface because of nonspecific screening of electrostatic effects. We note that high intrinsic pKaD97 implies an environment that stabilizes a neutral form of the embedded D97 residue in PR, which is accompanied by the stiffening of site 174 on the E-F loop and the slowing down of the hydration layer near this site. Whatever the biophysical and structural basis of this observation, the external electrostatic modulation that alters the intrinsic pKaD97 of PR is accompanied by changes in its structure, independent of the protonation or deprotonation of D97.
Discussion
Significant and systematic changes in both the apparent and the intrinsic pKaD97 were found for PR reconstituted in liposomes while we varied the net charge of phospholipid headgroups in the liposomes, as well as the ion type and their concentration in the buffer solution (Fig. 2). It is noteworthy that the apparent pKaD97 of PR embedded in 0.05 wt% DDM detergent micelles, unlike in liposomes, only showed a small dependence on the NaCl concentration in buffers (Fig. S8 A). This difference may be due to differences in the surface potential between PR reconstituted in liposomes and detergent micelles (62). In the liposome environment, PR, together with charged and zwitterionic lipids, creates a well-defined planar surface that could strengthen the electrostatic modulation induced by the presence of salt in buffer, whereas PR in detergent micelle creates a relatively weaker surface potential that may lead to a smaller dependence of the apparent pKaD97 on the NaCl concentration in buffers. Sharaabi et al. reported similar dependencies of pKaD97 on the NaCl concentration in buffers, but with a larger effect size for PR reconstituted in DDM detergent micelles (Fig S8 B; (16)). This difference in the magnitude can have diverse origins. PR oligomer distribution has been shown to significantly affect the pKaD97 of PR reconstituted in detergent micelles (14,15), but not in liposomes (unpublished data), so that different degree of oligomerization observed in different detergent concentration could concurrently affect the measured pKaD97 (15).
Because the addition of cations in buffer can change both the extent of hydration and the electrostatic environments of PR-containing liposomes (65,66), some might attribute the observed pKaD97 shift to be a result of the hydration change in liposomes. To clarify this, we examined several modulators that adjust only the extent of hydration of the PR-containing liposomes, one at a time. Overall, the applied modulators that are known to change the extent of hydration of PR-containing liposomes by changing water pore formation probability did not induce a significant apparent pKaD97 shift (Table S2). These observations suggest that even though the addition of cations in buffer can also change the hydration of lipid bilayers, it may not exert dominant effects on the pKa of the embedded D97 inside PR or that the experimental conditions and measurements are not sensitive to the relevant changes. In addition, other possible factors, including the oligomer distribution of PR in liposomes or the compositions of liposomes without changing their net charge, that could be modulators of the pKaD97 of PR were also examined, but no significant change in pKaD97 was observed (data not shown). These together suggest the electrostatic environment around PR reconstituted in liposomes should be the major factor determining its pKaD97.
Other retinal protein studies also revealed a transition in the protonation-deprotonation equilibrium of its embedded proton acceptor that depended on the ion concentration in the buffer. These studies inferred that the transition is induced either by ion-specific binding to the membrane protein or by a deviation of the surface pH on the membrane surface. When adding 150 mM of NaCl into the buffer, we found an opposite trend for the pKaD97 shift between PR reconstituted in positively charged liposomes and negatively charged liposomes (Fig. 2). This observation excludes ion-specific bindings as the key mechanism because pKa shift in the same direction should occur with specific binding, regardless of the liposome’s net surface charge. Furthermore, a much higher cation concentration was required to induce a measurable pKa shift in the PR studied here compared to bR. With bR, a complete transition of pKa was observed when 10 μM of Ca2+ was added to the purple membrane sample containing a bR concentration of 1.6 μM, which corresponds to only a sixfold molar excess of ions (67,68). In this study, the same transition reaches saturation with Ca2+ concentration in the 10 mM range, corresponding to a three orders of magnitude higher ion concentration compared to the PR concentration of 10 μM (Fig. 2 B). On the other hand, although a steeper apparent pKaD97 change was found for divalent cations compared to monovalent cations, this could be due to stronger association between the divalent cations and the negatively charged POPG lipid headgroups compared to monovalent cations (69). This stronger association may effectively screen the negative charge on liposome surfaces at lower ion concentration and so unify surface and bulk proton concentrations. This result suggests the deviation of surface from bulk pH due to interactions between the proton and charged lipid surface to be one of the most plausible factors that affects the apparent pKa of embedded charge residue.
Interestingly, the intrinsic pKaD97 modeled with surface pH calculated for charged liposome surfaces using GC models showed an opposite trend from the apparent pKaD97 because the net charge of liposomes and the concentration of NaCl in buffers were altered. Furthermore, the intrinsic pKaD97 was found to be still significantly modulated under different external electrostatic environments. These observations further prove that surface pH on charged liposome surfaces cannot be the main factor to give rise to changes in the apparent pKaD97 (Fig. 2, A and C). Even though the GC model might not quantitatively describe the surface potential of liposome systems with mixed lipids and in solution with different electrolyte concentrations, the difference between the experimentally determined surface potential ψ0 for POPC/POPG (80/20, mol/mol) liposomes in a buffer with 50 mM electrolyte (ψ0 = −102 mV) reported in the literature (62) and the one predicted by the GC model (ψ0 = −72 mV) under similar conditions are still comparable, suggesting that the GC-derived surface potential and pH represent physical trends. In the future, a more careful study, probably directly measuring the surface pH on PR-contained liposomes, will be required to obtain a more precise surface potential and intrinsic pKaD97.
Taking all observations together, we hypothesize that long-range interactions or mediated short-range interactions between charged lipid headgroups and the embedded charged residue inside protein must be at play, in addition to the effects of surface potential and pH, for altering its pKa. We gleaned protein structural information by cw EPR lineshape analysis and ODNP relaxometry measurements using PR spin labeled at site 174. Both methods revealed that the addition in the intrinsic pKaD97 is accompanied by changes in site 174 toward a more immobile environment, both in terms of the dynamics of the spin label according to cw EPR and the surrounding hydration water according to ODNP (Figs. 3 and 4). In other words, the observed change in intrinsic pKaD97 may be associated with structural rearrangement of PR induced by changes in the electrostatic environment.
We propose several possible mechanisms to explain the connection between the modulation of pKaD97 and the structural rearrangement of PR observed from the E-F loop. The first possibility could be a change of local hydrogen bond interactions between the embedded charged residue of interest and surrounding residues, as its importance on tuning the pKa of embedded residue has been revealed by previous retinal protein studies (17,25,39). In the case of PR, the conserved H75-D97 interaction was proposed to stabilize the protonated D97 and hence increase the pKaD97 (39). Mutation of H75 to different amino acids that cannot form hydrogen bonds with D97 brought down the pKaD97. The observed pKaD97 shift in this study may be due to a perturbation of the H75-D97 contact after PR was reconstituted in different electrostatic environments. A similar interaction compared to the H75-D97 contact in PR was also found in rhodopsin. The ionic lock between E134 and R135 has been suggested to stabilize the inactive MII state of rhodopsin, whereas reconstituting rhodopsin in either pure positively charged liposomes or triblock copolymer membranes with high cationic charge density can break down this ionic lock to drive a transition of rhodopsin to its active state (17). In the said rhodopsin study, the charge neutralization between positively charged DOTAP lipid headgroups and the deprotonated E134 was proposed to drive this transition. Their MD simulation results showed a direct contact between site E134 and the DOTAP lipid headgroups, made possible by the shallow location of E134 near the interface between the lipid bilayer and the aqueous phase. However, the D97 residue in PR is embedded inside the protein interior rather than exposed near the lipid bilayer-aqueous interface. Hence, even if the external electrostatic environment caused a modulation in the H75-D97 interaction, it must be mediated by long-range electrostatic interactions involving the protein. In future studies, the modulation in the H75-D97 distance must be directly examined as a function of changing external electrostatic environments that modulated pKaD97 by solid-state NMR spectroscopy but at a sufficiently high pH to keep D97 deprotonated in the different environments.
The second possibility is that the observed structural rearrangements can affect the relative positions between D97 and other surrounding polar or charged residues, the aforementioned H75 being one candidate, and hence its pKaD97. Theoretical studies have suggested that the charge density of an active site inside an enzyme can be affected by the positions of critical residues via electrostatic preorganization (43). In studies of bR, the orientation of positively charged R82 in simulation was found to affect the protonation state of other embedded charged residues on this protein, including its Schiff base primary proton acceptor D85 (25). In the case of PR, a weaker coupling was found between the analogous R94 and the primary proton acceptor D97 because mutating this positively charged arginine into other neutral amino acids did not affect its pKaD97 (70). However, remote residues beyond the proximal H75 have, in fact, been implicated in the modulation of pKaD97, namely site 178 on the E-F loop, whose mutations have been found to significantly affect the pKaD97 (51). NMR studies revealed that the A178R mutation modulated the structure of PR, affecting the chemical shifts of up to 31 residues from site 178, including the D97 residue (71). These results demonstrate the importance of the E-F loop on the structure and function of PR, including the microenvironment around D97 that determines its pKa. Our observation of E-F loop movement registered around 174 is thus consistent with the past studies identifying long-range interaction between site 178 and the embedded D97 residue. However, high-resolution structure studies under different electrostatic environments are needed to answer these questions conclusively.
We discuss one more possible mechanism. The water molecules hydrating the surface of PR may also be reoriented by the external electrostatic environment and so affect the protonation state of the embedded D97. In the studies of bR, a hydrogen bond network between structural water molecules inside the protein and two aspartic acids has been suggested to affect the protonation or the deprotonation of its D85 residue, which reveals the importance of protein-bound water’s orientation in determining the protonation state of embedded charged residues (72). Although such structural water has not been resolved from existing PR structures (10,73), a basis for this speculation is given by the direct observation of changes in surface water dynamics under conditions in this study where pKaD97 is altered (shown in Fig. 4). Clearly, a more careful study is required to better understand the effect of changes in the internal protein structure and hydration layer landscape after PR is reconstituted in different electrostatic environments.
Conclusion
Our study found that both the apparent and intrinsic pKa of an embedded charged residue D97 inside a transmembrane protein PR can be tuned by changing the ion concentration in the buffer or the net charge of liposomes in which PR is reconstituted. Although the deviation of local pH on charged liposome surfaces from bulk pH could be one of the factors to affect the measured apparent pKa, the intrinsic pKa modeled with the GC equation was still strongly modulated under different external electrostatic environment, likely through long-range interactions between charged lipid headgroups and the embedded charged residue. These long-range interactions are likely mediated by protein structural changes and/or ordering of water, as hinted by changes in structural and hydration dynamics properties at site 174 of the E-F loop of PR according to cw EPR lineshape and ODNP measurements. This work does not interrogate the underlying mechanism and structural basis of the intrinsic pKa modulation but underscores the importance of the environments, including the valency and the concentration of cations in buffer or the net charge of surrounding biomimetic environment, in modulating the activity and function of liposome-reconstituted TMPs, especially for those with functions modulated by the protonation state of embedded charges. This insight is critical for the study of many different TMPs in the context of biomedical and bioengineering research.
Author Contributions
C.-T.H. and S.H. designed the experiments. S.H. supervised the study. C.-T.H., J.S., T.C., and C.P. performed experiments, and C.-T.H. carried out data analysis. C.-T.H. and S.H. wrote and revised the manuscript.
Acknowledgments
S.H. and C.-T.H. gratefully acknowledge the support from the National Science Foundation through Molecular and Cellular Biology grants NSF-MCB 1617025. This work was sponsored by the U.S. Army Research Office and accomplished under cooperative agreement W911NF-19-2-0026 for the Institute for Collaborative Biotechnologies. Support for the ODNP studies was promoted through the Deutsche Forschungsgemeinschaft as part of the “Exzellenzstrategie des Bundes und der Länder” EXC 2033, project number 390677874, RESOLV.
Editor: Michael Brown.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.02.027.
Supporting Citations
References (74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86) appear in the Supporting Material.
Supporting Material
References
- 1.Luecke H., Richter H.-T., Lanyi J.K. Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 2.Yoshikawa S., Shinzawa-Itoh K., Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 3.Churg A.K., Warshel A. Control of the redox potential of cytochrome c and microscopic dielectric effects in proteins. Biochemistry. 1986;25:1675–1681. doi: 10.1021/bi00355a035. [DOI] [PubMed] [Google Scholar]
- 4.Warshel A., Sharma P.K., Parson W.W. Modeling electrostatic effects in proteins. Biochim. Biophys. Acta. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Brown L.S., Kamikubo H., Lanyi J.K. A local electrostatic change is the cause of the large-scale protein conformation shift in bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1997;94:5040–5044. doi: 10.1073/pnas.94.10.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancaster C.R.D., Michel H. The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB. Structure. 1997;5:1339–1359. doi: 10.1016/s0969-2126(97)00285-2. [DOI] [PubMed] [Google Scholar]
- 7.Lanyi J.K. Bacteriorhodopsin. Annu. Rev. Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 8.Váró G., Brown L.S., Lanyi J.K. Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 2003;84:1202–1207. doi: 10.1016/S0006-3495(03)74934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakatos M., Lanyi J.K., Váró G. The photochemical reaction cycle of proteorhodopsin at low pH. Biophys. J. 2003;84:3252–3256. doi: 10.1016/S0006-3495(03)70049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reckel S., Gottstein D., Dötsch V. Solution NMR structure of proteorhodopsin. Angew. Chem. Int.Engl. 2011;50:11942–11946. doi: 10.1002/anie.201105648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimsley G.R., Scholtz J.M., Pace C.N. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isom D.G., Castañeda C.A., García-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. USA. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerwert K., Hess B., Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1989;86:4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain S., Kinnebrew M., Han S. Functional consequences of the oligomeric assembly of proteorhodopsin. J. Mol. Biol. 2015;427:1278–1290. doi: 10.1016/j.jmb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Idso M.N., Baxter N.R., Han S. Proteorhodopsin function is primarily mediated by oligomerization in different micellar surfactant solutions. J. Phys. Chem. B. 2019;123:4180–4192. doi: 10.1021/acs.jpcb.9b00922. [DOI] [PubMed] [Google Scholar]
- 16.Sharaabi Y., Brumfeld V., Sheves M. Binding of anions to proteorhodopsin affects the Asp97 pK(a) Biochemistry. 2010;49:4457–4465. doi: 10.1021/bi901746b. [DOI] [PubMed] [Google Scholar]
- 17.Chawla U., Jiang Y., Liang H. A usual G-protein-coupled receptor in unusual membranes. Angew. Chem. Int.Engl. 2016;55:588–592. doi: 10.1002/anie.201508648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahnke J.P., Idso M.N., Chmelka B.F. Functionally active membrane proteins incorporated in mesostructured silica films. J. Am. Chem. Soc. 2018;140:3892–3906. doi: 10.1021/jacs.7b06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stites W.E., Gittis A.G., Shortle D. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J. Mol. Biol. 1991;221:7–14. doi: 10.1016/0022-2836(91)80195-z. [DOI] [PubMed] [Google Scholar]
- 20.García-Moreno B., Dwyer J.J., Stites W.E. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys. Chem. 1997;64:211–224. doi: 10.1016/s0301-4622(96)02238-7. [DOI] [PubMed] [Google Scholar]
- 21.Dwyer J.J., Gittis A.G., García-Moreno E B. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys. J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harms M.J., Schlessman J.L., García-Moreno B. A buried lysine that titrates with a normal pKa: role of conformational flexibility at the protein-water interface as a determinant of pKa values. Protein Sci. 2008;17:833–845. doi: 10.1110/ps.073397708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takayama Y., Castañeda C.A., Iwahara J. Direct evidence for deprotonation of a lysine side chain buried in the hydrophobic core of a protein. J. Am. Chem. Soc. 2008;130:6714–6715. doi: 10.1021/ja801731g. [DOI] [PubMed] [Google Scholar]
- 24.Goh G.B., García-Moreno E B., Brooks C.L., III The high dielectric constant of staphylococcal nuclease is encoded in its structural architecture. J. Am. Chem. Soc. 2011;133:20072–20075. doi: 10.1021/ja2084866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J. Mol. Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 26.Wallace J.A., Wang Y., Shen J.K. Toward accurate prediction of pKa values for internal protein residues: the importance of conformational relaxation and desolvation energy. Proteins. 2011;79:3364–3373. doi: 10.1002/prot.23080. [DOI] [PubMed] [Google Scholar]
- 27.Wu X., Brooks B.R. A virtual mixture approach to the study of multistate equilibrium: application to constant ph simulation in explicit water. PLoS Comput. Biol. 2015;11:e1004480. doi: 10.1371/journal.pcbi.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X., Lee J., Brooks B.R. Origin of pKa shifts of internal lysine residues in SNase studied via equal-molar VMMS simulations in explicit water. J. Phys. Chem. B. 2017;121:3318–3330. doi: 10.1021/acs.jpcb.6b08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunner M.R., Baker N.A. Continuum electrostatics approaches to calculating pKas and Ems in proteins. Methods Enzymol. 2016;578:1–20. doi: 10.1016/bs.mie.2016.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markley J.L. Observation of histidine residues in proteins by nuclear magnetic resonance spectroscopy. Acc. Chem. Res. 1975;8:70–80. [Google Scholar]
- 31.Forsyth W.R., Antosiewicz J.M., Robertson A.D. Empirical relationships between protein structure and carboxyl pKa values in proteins. Proteins. 2002;48:388–403. doi: 10.1002/prot.10174. [DOI] [PubMed] [Google Scholar]
- 32.Alexov E., Mehler E.L., Word J.M. Progress in the prediction of pKa values in proteins. Proteins. 2011;79:3260–3275. doi: 10.1002/prot.23189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorobjev Y.N., Scheraga H.A., Vila J.A. Coupled molecular dynamics and continuum electrostatic method to compute the ionization pKa’s of proteins as a function of pH. Test on a large set of proteins. J. Biomol. Struct. Dyn. 2018;36:561–574. doi: 10.1080/07391102.2017.1288169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cvitkovic J.P., Pauplis C.D., Kaminski G.A. PKA17-A coarse-grain grid-based methodology and web-based software for predicting protein pK a shifts. J. Comput. Chem. 2019;40:1718–1726. doi: 10.1002/jcc.25826. [DOI] [PubMed] [Google Scholar]
- 35.Kougentakis C.M., Grasso E.M., García-Moreno E B. Anomalous properties of Lys residues buried in the hydrophobic interior of a protein revealed with 15N-detect NMR spectroscopy. J. Phys. Chem. Lett. 2018;9:383–387. doi: 10.1021/acs.jpclett.7b02668. [DOI] [PubMed] [Google Scholar]
- 36.Wu X., Brooks B.R. Hydronium ions accompanying buried acidic residues lead to high apparent dielectric constants in the interior of proteins. J. Phys. Chem. B. 2018;122:6215–6223. doi: 10.1021/acs.jpcb.8b04584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harms M.J., Castañeda C.A., García-Moreno E B. The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors. J. Mol. Biol. 2009;389:34–47. doi: 10.1016/j.jmb.2009.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Mao J., Gunner M.R. Are acidic and basic groups in buried proteins predicted to be ionized? J. Mol. Biol. 2005;348:1283–1298. doi: 10.1016/j.jmb.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 39.Hempelmann F., Hölper S., Glaubitz C. His75-Asp97 cluster in green proteorhodopsin. J. Am. Chem. Soc. 2011;133:4645–4654. doi: 10.1021/ja111116a. [DOI] [PubMed] [Google Scholar]
- 40.Alexov E.G., Gunner M.R. Incorporating protein conformational flexibility into the calculation of pH-dependent protein properties. Biophys. J. 1997;72:2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Damjanović A., Brooks B.R., García-Moreno B. Conformational relaxation and water penetration coupled to ionization of internal groups in proteins. J. Phys. Chem. A. 2011;115:4042–4053. doi: 10.1021/jp110373f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J., Swails J., Roitberg A.E. A coupled ionization-conformational equilibrium is required to understand the properties of ionizable residues in the hydrophobic interior of staphylococcal nuclease. J. Am. Chem. Soc. 2018;140:1639–1648. doi: 10.1021/jacs.7b08569. [DOI] [PubMed] [Google Scholar]
- 43.Morgenstern A., Jaszai M., Alexandrova A.N. Quantified electrostatic preorganization in enzymes using the geometry of the electron charge density. Chem. Sci. (Camb.) 2017;8:5010–5018. doi: 10.1039/c7sc01301a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee A.G. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 45.Peetla C., Stine A., Labhasetwar V. Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery. Mol. Pharm. 2009;6:1264–1276. doi: 10.1021/mp9000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y., Okur H.I., Roke S. Zwitterionic and charged lipids form remarkably different structures on nanoscale oil droplets in aqueous solution. Langmuir. 2018;34:1042–1050. doi: 10.1021/acs.langmuir.7b02896. [DOI] [PubMed] [Google Scholar]
- 47.Szundi I., Stoeckenius W. Effect of lipid surface charges on the purple-to-blue transition of bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1987;84:3681–3684. doi: 10.1073/pnas.84.11.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szundi I., Stoeckenius W. Purple-to-blue transition of bacteriorhodopsin in a neutral lipid environment. Biophys. J. 1988;54:227–232. doi: 10.1016/S0006-3495(88)82951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szundi I., Stoeckenius W. Surface pH controls purple-to-blue transition of bacteriorhodopsin. A theoretical model of purple membrane surface. Biophys. J. 1989;56:369–383. doi: 10.1016/S0006-3495(89)82683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voinov M.A., Smirnov A.I. Ionizable nitroxides for studying local electrostatic properties of lipid bilayers and protein systems by EPR. Methods Enzymol. 2015;564:191–217. doi: 10.1016/bs.mie.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada K., Kawanabe A., Kandori H. Importance of alanine at position 178 in proteorhodopsin for absorption of prevalent ambient light in the marine environment. Biochemistry. 2010;49:2416–2423. doi: 10.1021/bi9020204. [DOI] [PubMed] [Google Scholar]
- 52.Columbus L., Hubbell W.L. A new spin on protein dynamics. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 53.Armstrong B.D., Han S. A new model for Overhauser enhanced nuclear magnetic resonance using nitroxide radicals. J. Chem. Phys. 2007;127:104508. doi: 10.1063/1.2770465. [DOI] [PubMed] [Google Scholar]
- 54.Armstrong B.D., Han S. Overhauser dynamic nuclear polarization to study local water dynamics. J. Am. Chem. Soc. 2009;131:4641–4647. doi: 10.1021/ja809259q. [DOI] [PubMed] [Google Scholar]
- 55.McHaourab H.S., Steed P.R., Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franck J.M., Pavlova A., Han S. Quantitative cw Overhauser effect dynamic nuclear polarization for the analysis of local water dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2013;74:33–56. doi: 10.1016/j.pnmrs.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biller J.R., Barnes R., Han S. Perspective of Overhauser dynamic nuclear polarization for the study of soft materials. Curr. Opin. Colloid Interface Sci. 2018;33:72–85. [Google Scholar]
- 58.Hussain S., Franck J.M., Han S. Transmembrane protein activation refined by site-specific hydration dynamics. Angew. Chem. Int.Engl. 2013;52:1953–1958. doi: 10.1002/anie.201206147. [DOI] [PubMed] [Google Scholar]
- 59.Stone K.M., Voska J., Han S. Structural insight into proteorhodopsin oligomers. Biophys. J. 2013;104:472–481. doi: 10.1016/j.bpj.2012.11.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tunuguntla R., Bangar M., Noy A. Lipid bilayer composition can influence the orientation of proteorhodopsin in artificial membranes. Biophys. J. 2013;105:1388–1396. doi: 10.1016/j.bpj.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y., Botelho A.V., Brown M.F. Electrostatic properties of membrane lipids coupled to metarhodopsin II formation in visual transduction. J. Am. Chem. Soc. 2002;124:7690–7701. doi: 10.1021/ja0200488. [DOI] [PubMed] [Google Scholar]
- 62.Voinov M.A., Rivera-Rivera I., Smirnov A.I. Surface electrostatics of lipid bilayers by EPR of a pH-sensitive spin-labeled lipid. Biophys. J. 2013;104:106–116. doi: 10.1016/j.bpj.2012.11.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hubbell W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 64.Barnes R., Sun S., Han S. Spatially heterogeneous surface water diffusivity around structured protein surfaces at equilibrium. J. Am. Chem. Soc. 2017;139:17890–17901. doi: 10.1021/jacs.7b08606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Váró G., Brown L.S., Lanyi J.K. Binding of calcium ions to bacteriorhodopsin. Biophys. J. 1999;76:3219–3226. doi: 10.1016/S0006-3495(99)77473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J., Kang T.H., Han S. Ion specific effects: decoupling ion-ion and ion-water interactions. Phys. Chem. Chem. Phys. 2015;17:8306–8322. doi: 10.1039/c4cp05992a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang C.-H., Chen J.-G., Ebrey T. Cation binding by bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1985;82:396–400. doi: 10.1073/pnas.82.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eliash T., Weiner L., Sheves M. Specific binding sites for cations in bacteriorhodopsin. Biophys. J. 2001;81:1155–1162. doi: 10.1016/S0006-3495(01)75772-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cevc G. Membrane electrostatics. Biochim. Biophys. Acta. 1990;1031:311–382. doi: 10.1016/0304-4157(90)90015-5. [DOI] [PubMed] [Google Scholar]
- 70.Partha R., Krebs R., Braiman M.S. Weakened coupling of conserved arginine to the proteorhodopsin chromophore and its counterion implies structural differences from bacteriorhodopsin. Biochim. Biophys. Acta. 2005;1708:6–12. doi: 10.1016/j.bbabio.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 71.Mehler M., Scholz F., Glaubitz C. The EF loop in green proteorhodopsin affects conformation and photocycle dynamics. Biophys. J. 2013;105:385–397. doi: 10.1016/j.bpj.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gerwert K., Freier E., Wolf S. The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta. 2014;1837:606–613. doi: 10.1016/j.bbabio.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Ran T., Ozorowski G., Luecke H. Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1965–1980. doi: 10.1107/S0907444913017575. [DOI] [PubMed] [Google Scholar]
- 74.Petrache H.I., Tristram-Nagle S., Nagle J.F. Fluid phase structure of EPC and DMPC bilayers. Chem. Phys. Lipids. 1998;95:83–94. doi: 10.1016/s0009-3084(98)00068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jansen M., Blume A. A comparative study of diffusive and osmotic water permeation across bilayers composed of phospholipids with different head groups and fatty acyl chains. Biophys. J. 1995;68:997–1008. doi: 10.1016/S0006-3495(95)80275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen R.D., Che K., Robinson B.H. A ruler for determining the position of proteins in membranes. J. Am. Chem. Soc. 2005;127:6430–6442. doi: 10.1021/ja042782s. [DOI] [PubMed] [Google Scholar]
- 77.Holt A., Killian J.A. Orientation and dynamics of transmembrane peptides: the power of simple models. Eur. Biophys. J. 2010;39:609–621. doi: 10.1007/s00249-009-0567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beranova L., Cwiklik L., Jungwirth P. Oxidation changes physical properties of phospholipid bilayers: fluorescence spectroscopy and molecular simulations. Langmuir. 2010;26:6140–6144. doi: 10.1021/la100657a. [DOI] [PubMed] [Google Scholar]
- 79.Lis M., Wizert A., Cwiklik L. The effect of lipid oxidation on the water permeability of phospholipids bilayers. Phys. Chem. Chem. Phys. 2011;13:17555–17563. doi: 10.1039/c1cp21009b. [DOI] [PubMed] [Google Scholar]
- 80.Cheng C.-Y., Varkey J., Han S. Hydration dynamics as an intrinsic ruler for refining protein structure at lipid membrane interfaces. Proc. Natl. Acad. Sci. USA. 2013;110:16838–16843. doi: 10.1073/pnas.1307678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lueders P., Jäger H., Yulikov M. Distance measurements on orthogonally spin-labeled membrane spanning WALP23 polypeptides. J. Phys. Chem. B. 2013;117:2061–2068. doi: 10.1021/jp311287t. [DOI] [PubMed] [Google Scholar]
- 82.Först G., Cwiklik L., Hof M. Interactions of beta-blockers with model lipid membranes: molecular view of the interaction of acebutolol, oxprenolol, and propranolol with phosphatidylcholine vesicles by time-dependent fluorescence shift and molecular dynamics simulations. Eur. J. Pharm. Biopharm. 2014;87:559–569. doi: 10.1016/j.ejpb.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 83.Pavlova A., Cheng C.-Y., Han S. Protein structural and surface water rearrangement constitute major events in the earliest aggregation stages of tau. Proc. Natl. Acad. Sci. USA. 2016;113:E127–E136. doi: 10.1073/pnas.1504415113. Published online December 28, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Segawa T.F., Doppelbauer M., Jeschke G. Water accessibility in a membrane-inserting peptide comparing Overhauser DNP and pulse EPR methods. J. Chem. Phys. 2016;144:194201. doi: 10.1063/1.4948988. [DOI] [PubMed] [Google Scholar]
- 85.Dreier L.B., Nagata Y., Bonn M. Saturation of charge-induced water alignment at model membrane surfaces. Sci. Adv. 2018;4:eaap7415. doi: 10.1126/sciadv.aap7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lueders P., Jäger H., Yulikov M. Multiple pathway relaxation enhancement in the system composed of three paramagnetic species: nitroxide radical–Ln3+–O2. J. Phys. Chem. Lett. 2012;3:1336–1340. doi: 10.1021/jz300316q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.