Abstract
Gastric cancer (GC) is one of the leading causes of cancer-related death worldwide. The role of the microorganisms in gastric tumorigenesis attracts much attention in recent years. These microorganisms include bacteria, virus, and fungi. Among them, Helicobacter pylori (H. pylori) infection is by far the most important risk factor for GC development, with special reference to the early-onset cases. H. pylori targets multiple cellular components by utilizing various virulence factors to modulate the host proliferation, apoptosis, migration, and inflammatory response. Epstein–Barr virus (EBV) serves as another major risk factor in gastric carcinogenesis. The virus protein, EBER noncoding RNA, and EBV miRNAs contribute to the tumorigenesis by modulating host genome methylation and gene expression. In this review, we summarized the related reports about the colonized microorganism in the stomach and discussed their specific roles in gastric tumorigenesis. Meanwhile, we highlighted the therapeutic significance of eradicating the microorganisms in GC treatment.
Subject terms: Gastric cancer, Cancer immunotherapy
Introduction
Gastric cancer (GC) is the second leading cause of cancer-related death in the world [1]. GC mainly occurs in Asia, Latin America, and Central and Eastern Europe, however, it is no longer a common disease in North America and part of Western Europe [2]. GC can be separated into two types according to the locus, gastric adenocarcinomas and gastro-esophageal-junction adenocarcinomas [3]. Gastric adenocarcinoma can also be subdivided histologically into intestinal and diffuse types by Lauren’s classification. In 2014, The Cancer Genome Atlas (TCGA) research network has described a comprehensive molecular evaluation on 295 primary gastric adenocarcinomas. They proposed a molecular classification dividing GC into four subtypes: positive for Epstein–Barr virus (EBV) (9%), microsatellite unstable tumors (22%), genomically stable tumors (20%), and chromosomally unstable tumors (50%) [4]. In 2015, the Asian Cancer Research Group (ACRG) proposed another molecular classification associated with clinical outcome and defined GC as four distinct molecular subtypes: microsatellite instability (MSI), microsatellite stable with epithelial-to-mesenchymal transition features (MSS/EMT), MSS/TP53 mutant (MSS/TP53+), and MSS/TP53 wild type (MSS/TP53–) [5]. Identification of these subtypes sheds new lights on the prognosis and clinical treatment [6].
More than 15% of the tumor cases were attributed to infectious pathogens. The proportion was even higher in less developed countries or regions (22.9%) [7]. The infectious pathogens include viruses, bacteria, and parasites. Among the pathogens, Helicobacter pylori (H. pylori), human papillomavirus, hepatitis B virus (HBV), and hepatitis C virus together attributed to 2 million new cancer cases worldwide in 2012. They induced the tumorigenicity of the stomach, liver, and cervix. Of note, HBV and H. pylori have most vicious contributions to the tumor burden in China [8]. H. pylori and EBV are the most well-known pathogens in gastric carcinogenesis. H. pylori is an important risk factor found in 65–80% of primary GCs, while EBV leads to 10% of the GC cases. Besides, it has been reported other microorganisms are also associated with gastric malignancies.
Accompanied with the development of strategies for manipulating infectious agents, opportunities are emerging to prevent and cure the infection-related cancers. Here, we comprehensively reviewed the role of microbiome in promoting gastric carcinogenesis.
Bacteriome in gastric carcinogenesis
Because of the acid production, stomach was thought as a sterile organ previously. However, in recent years, culture independent methods have been developed to facilitate the identification of various bacteria species in human stomach. It is believed that apart from the predominant bacteria H. pylori, multiple kinds of bacteria were coexisting in human stomach, although little is known about their associations with GC progression.
Infection of H. pylori
H. pylori infection is the most popular chronical bacterial infection worldwide. More than 50% of the world population are infected with H. pylori, however, over 80% of infections are asymptomatic [9]. The transmission of H. pylori is implicated with fecal/oral, oral/oral, or gastric/oral pathways [10]. Part of the infections develop coexisting gastritis for several years, and the persistent infection might develop into gastric atrophy followed by intestinal metaplasia, dysplasia, and eventually adenocarcinoma [6]. World Health Organization designates H. pylori as a class I carcinogen because of its chronic infection as the strongest risk factor for gastric adenocarcinoma. It was estimated that 90% of all noncardia GCs are associated with H. pylori [11]. A study with 1526 Japanese population found the increasing risk of GC development in patients infected with H. pylori compared with the uninfected ones [12]. The eradication of H. pylori significantly decreased the occurrence of GC, suggesting that H. pylori might influence early stages in gastric carcinogenesis [13].
Molecular pathogenesis of H. pylori-related GC
Environmental factors have long been considered to play dispensable roles in GC. High salt intake was found significantly associated with GC especially in the context of H. pylori infection and atrophic gastritis [14]. It was also believed that the risk of GC increased in the subjects with both smoking habit and H. pylori infection [15]. It has been puzzling about H. pylori infection, although half of the population infected with H. pylori worldwide, only a minority of colonized individuals (1–2%) develops tumors. The low morbidity indicates the impact of different strains in tumor initiation and development.
Different strains of H. pylori play diverse roles in driving GC. H. pylori can be subdivided into bacterial oncoprotein cytotoxin-associated gene A (CagA) positive and CagA negative strains. In a meta-analysis, patients infected with CagA positive strains demonstrate a higher risk of GC [16], which was consistent with previous reports that individuals with CagA antibodies have a higher risk of tumor [17–20]. Transgenic mice bearing CagA appears gastric neoplasms development, confirming that CagA is a bacteric oncoprotein [21]. However, the mechanism seems particularly complex. H. pylori injects CagA into the host gastric epithelial cells with the activation of integrin [22]. Moreover, CagA undergoes tyrosine phosphorylation by Src family kinases or Abl kinase and subsequently activates multiple signaling pathways. For instance, phosphorylated CagA interacts with activated SHP2. CagA–SHP2 potentiates the magnitude of Erk-MAP kinase signaling in both Ras-dependent and Ras-independent manners [23]. CagA–SHP2 also dephosphorylates focal adhesion kinase (FAK) and mediates cell–extracellular matrix interaction. Both signaling lead to a cellular morphological change, which is called hummingbird phenotype, thus to increase the cell migration abilities [24]. In addition, nonphosphorylated CagA impairs intracellular signaling networks. The nonphosphorylated intracellular CagA interacts with E-cadherin to disrupt the E-cadherin–β-catenin complex. It thus induces nuclear β-catenin accumulation, allowing transcription of the target genes associated with carcinogenesis. Meanwhile, CagA was reported to directly activate β-catenin by interacting with MET and activating PI3K–AKT signaling [25, 26]. CagA activates the signal transducer and activator of transcription 3 (STAT3) pathway. The activated STAT3 pathway is driven by the host immune response and is associated with H. pylori-induced gastritis and cancer progression, independent of CagA phosphorylation [27–29]. In a recent study, CagA also binds to 25 known factors in the host cells to hijack various signaling pathways related to inflammation, proliferation, genetic instability, cell polarity, and apoptosis [30]. Apart from CagA, the Cag secretion system also delivers H. pylori peptidoglycan into the host cells through outer membrane vesicles. The peptidoglycan subsequently activates PI3K–AKT and regulates cell migration, proliferation, and apoptosis [31].
Apart from Cag, vacuolating toxin A (VacA) is another major virulence determinant of H. pylori. H. pylori gene vacA encodes the secreted protein VacA. VacA has been reported to link to multiple cellular processes, such as vacuolation, membrane-channel formation, apoptosis, proinflammatory response, and malignancy [32]. Although all of the H. pylori strains contain vacA, there is variation in the vacA structure. Among them, s1m1i1d1 type strains are strongly associated with gastric adenocarcinoma. Nakayama et al. reported that VacA activates β-catenin through PI3K-dependent manner [33].
Approximately, 4% of the H. pylori genome encodes integral outer membrane proteins (OMPs) [34, 35], which are subdivided as 5 families [36]. Some of them functioned as adherence factors, such as sialic acid-binding adhesin, blood-group-antigen-binding adhesin, adherence-associated lipoprotein A and B, outer inflammatory protein A (OipA), and Helicobacter OMP Q. Most of them are linked with poor clinical outcomes. OipA was identified as a proinflammatory response inducing protein and knockout of this gene can reduce interleukin (IL)-8 production [37]. In patient samples, it was confirmed that OipA was significantly associated with gastric inflammation and IL-8 levels [38]. Basically, OipA is involved in the attachment of H. pylori to gastric epithelial cells, which is important for the initiation and development of GC. In addition, inactivation of OipA decreases the incidence of carcinoma by attenuating β-catenin nuclear translocation [39].
The aberrant host genetic changes are also crucial for the interaction of H. pylori and gastric epithelium cells. Polymorphisms in IL-1β and its endogenous receptor antagonist affect gastric mucosal IL-1β production in response to infection of H. pylori and are associated with GC occurrence [40–42]. In addition, the combination of HLA class II and IL-10–592A/C polymorphisms affect the susceptibility to GC development in H. pylori-infected Japanese individuals [43].
The causal relationship between inflammation and cancer has been well recognized. An individual infected with H. pylori has a bigger chance to develop chronic inflammation. H. pylori utilizes virulence factors CagA, VacA, and peptidoglycan to upregulate proinflammatory cytokines such as IL-1, IL-6, IL-8, TNF-α, and NF-κB, to activate NF-κB signaling cascade in gastric epithelial cells and circulating immune cells [44]. The production of cytokine triggers activation and migration of leukocytes, and regulation cascade of cytokines, chemokine, and adhesions. Granulocyte-macrophage colony-stimulating factor, a growth factor facilitating white cell differentiation, was found in H. pylori-infected antral biopsies and human gastric epithelial cells [45]. Besides, inflammation modulators cyclooxygenase-2, which convers arachidonic acid to prostaglandins to induce inflammatory reactions, was significantly higher in H. pylori-infected gastric epithelia cells [46]. Apart from the cytokine release, lipopolysaccharide (LPS), VacA, and H. pylori neutrophil activating protein contribute to induce reactive oxygen species (ROS) or reactive nitrogen species (RNS) in gastric epithelial cells and inflammatory cells. The generation of intracellular ROS and RNS are found relating to the pathogenesis of H. pylori-associated GC. In addition, H. pylori-induced chronic inflammation leads to aberrant DNA methylation, which is the major cause of H. pylori-associated GC. On the other hand, when H. pylori-induced inflammation was suppressed by cyclosporine A in animal model, induction of aberrant DNA methylation was also suppressed [47, 48]. Methylation on tumor-suppressor genes can inactivate the gene expression and promotes cancer development. For example, promoter methylation in E-cadherin, an epithelial marker, has been detected in H. pylori-infected stomach [49]. Regarding to these studies, H. pylori-induced chronic inflammation is essential for both initiation and the development of GC.
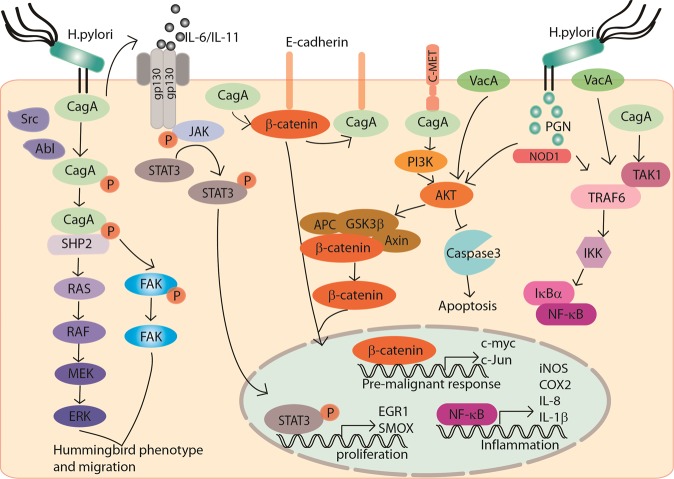
The potential molecular network of H. pylori and oncogenic signaling pathways in gastric carcinogenesis are summarized in Fig. 1.
Fig. 1. Molecular pathogenesis of H. pylori in gastric carcinogenesis.
The MEK–ERK and FAK signaling pathways are activated by phosphorylated CagA to mediate hummingbird phenotype of the epithelial cells and promote cell migration. The β-catenin is activated by nonphosphorylated intracellular CagA by disruption of the E-cadherin–β-catenin complexes or PI3K–AKT signaling. CagA activates JAK-STAT3 pathway by releasing IL-6/IL11 and activating gp130. Nuclear translocation of STAT3 initiates gene expression for cell proliferation. H. pylori peptidoglycan and VacA potentiate PI3K–AKT signaling to promote epithelial cell migration, increase proliferation, and reduce apoptosis. CagA, VacA, and peptidoglycan coordinate to activate NF-κB signaling cascade thus to transcriptionally upregulate proinflammatory cytokines such as IL-1 and IL-8, and promote inflammation.
Host immunity in H. pylori-related GC
The host immune system is the formidable barrier to prevent H. pylori infection. The immune system includes innate immune response and adaptive immune response. Innate immune response is the first-line defense. Epithelial cells, dendritic cells, monocytes, macrophages, and neutrophils could play important roles in defending H. pylori infection. Pathogen-associated molecular patterns of H. pylori, such as, peptidoglycan, LPS, lipoproteins, and flagellins are recognized by pattern recognition receptors (PRRs). Toll-like receptors, C-type lectin receptors, NOD-like receptors, and RIG-like receptors are members of the PRR family. The engagement of PRR then triggers the activation of multiple signaling cascades that culminate in NF-κB activation and immune effectors production. Such an immune response could induce a chronic inflammation, which has been shown closely associated with molecular pathogenesis of H. pylori-related GC.
However, in adaptive immune response, H. pylori can be recognized and presented by antigen-presenting cells (APCs), such as dendritic cell [50], neutrophil, macrophage, and epithelial cells [51]. The APCs produce cytokines to stimulate naive CD4+ T cells and induce antigen-specific responses in Th1 cells [52, 53] and Th17 cells [54–56]. The Th1 cells and Th17 cells are critical for the control of H. pylori infection, however they are also associated with increased gastritis as well as GC [54, 57–60]. At the same times, the T regulatory (Treg) cell response is also observed, which drives immune tolerance and suppresses Th1- and Th17-mediated immunity against H. pylori infection [61, 62]. It has been reported that B cells and antibodies are not required for clearing the H. pylori, rather, they might be detrimental to elimination of the bacteria [63].
Diagnosis and treatment of H. pylori
H. pylori should be tested in patients with dyspepsia if the local H. pylori prevalence exceeds 10%. The testing can be performed by noninvasive and invasive methods. The noninvasive methods include the urea breath tests and fecal antigen test. Serologic test and invasive testing strategies require upper endoscopy, biopsy urease (campylobacter-like organism) test, histologic assessment, and culture [64].
The eradication of H. pylori dramatically decreases the presence of premalignant lesions and reduce the GC risk in infected individuals. Anti-H. pylori therapy is an effective means for GC prevention and there are various proposed treatment regimens for H. pylori eradication [65]. Traditional treatment regimens include standard triple therapy (PPI, amoxicillin, and clarithromycin), bismuth quadruple PBMT therapy (PPI, bismuth, metronidazole, and tetracycline), or a treatment including PPI, clarithromycin, and metronidazole. However, with increasing clarithromycin resistance, another regimen concomitant nonbismuth therapy PAMC (PPI, amoxicillin, metronidazole, and clarithromycin) was proposed. The first-line treatment was recommended with a 14-day course of either concomitant PAMC therapy or bismuth quadruple PBMT therapy, according to the 2016 Toronto Consensus guidelines [66]. The 2016 Maastricht V/Florence Consensus Report recommends first-line treatment with a 14-day course of bismuth quadruple PBMT therapy or concomitant PAMC therapy in high clarithromycin resistance areas (>15% resistance). A standard triple therapy or bismuth quadruple PBMT therapy in low clarithromycin resistance (<15% resistance) areas is also proposed by this report [67].
Other bacteria in GC
In 2006, Bik et al. used a small subunit 16S rDNA clone library approach identified 128 phylotypes belonging to five phyla (Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, and Fusobacteria) in 23 human gastric biopsies [68]. Lately, 133 phylotypes were identified by Li et al. and 59 families were detected by Delgado et al. [69, 70], which were quite similar from both phyla and genera level. It reflects the significance of the bacterial homeostasis in stomach.
Loss of bacterial homeostasis might be a reason in driving GC progression. Coker et al. reported that microbial composition was changed, and bacterial interactions were different across stages of gastric carcinogenesis, indicating the presence of microbial dysbiosis in gastric carcinogenesis. They also found potential roles of some microbial such as Peptostreptococcus stomatis, Dialister pneumosintes, Slackia exigua, Parvimonas micra, and Streptococcus anginosus in GC progression [71]. It was also reported that a consistent increase of lactic acid bacteria promotes GC by a number of mechanisms such as supply of exogenous lactate, production of ROS, and N-nitroso compounds, as well as anti-H. pylori properties [72].
Notably, H. pylori and other bacteria might affect each other in the stomach, but the causality has not yet been clearly explained. As currently known, bacteria colonies in the stomach could affect the outcome of H. pylori infection and the progression of GC. On the other side, H. pylori infection may influence the density of bacteria. In animal model, long-term H. pylori infection affects the bacterial composition of the gastric microbiota. Maldonado-Contreras et al. reported a higher abundance of Proteobacteria, Spirochetes, and Acidobacteria, and a decreased abundance of Actinobacteria, Bacteroidetes, and Firmicutes in H. pylori-positive patients compared with H. pylori-negative subjects [73]. A microbial diversity analysis showed that compared with negative subjects, both of the species and Shannon index were increased in subjects with past or current H. pylori-infected subjects, indicating the alterations of fecal microbiota, especially Bacteroidetes, Firmicutes, and Proteobacteria, may be involved in the process of H. pylori-related gastric lesion progression [74]. However, some reports indicated that chronic H. pylori infection does not alter the microbiota of stomach [68, 71, 75, 76], suggesting the relationship between H. pylori infection and the gastric microbiota dysbiosis is still controversial [77, 78].
EBV in gastric carcinogenesis
The mammalian virome is constituted of viruses that infect host cells, virus-derived elements in human chromosomes, and viruses that infect the broad array of other types of organisms [79]. It was reported that EBV, CMV, and HHV6 can be detected in gastric tumors [80]. Among them, EBV is the most prominent one.
The structure of EBV
More than 90% of adults have been infected by EBV [81], and it is asymptomatic in the majority of carriers. However, some of the infections can cause infectious mononucleosis. EBV is classified as a group I carcinogen by the International Agency for Research on Cancer, since the latently infection estimated to be responsible for 200,000 cancers cases worldwide [82], such as Burkitt lymphoma, hemophagocytic lymphohistiocytosis, Hodgkin’s lymphoma, GC, and nasopharyngeal carcinoma (NPC). Until now, approved vaccines for EBV have not been available. However, a vaccine targeting the EBV glycoprotein gp350 has been developed to reduce the incidence of infectious mononucleosis and the efficacy has been proved [83].
EBV belongs to Herpesviridae containing an ~172 kb liner form dsDNA genome. The expression products cover 80 proteins and 46 functional small-untranslated RNAs. EBV prefers to infect B cell and epithelial cells. After entry, like all kind of herpesviruses, EBV has two distinct life cycles: lytic replication and latency. However, upon EBV de novo infection, it takes latency infection firstly. During latency, viral genomes exist as extrachromosomal episomes in the nucleus and only express some latent proteins (EBV-determined nuclear antigen 1 (EBNA1), 2, 3A, 3B, 3C, and EBNA-LP; latent membrane protein 1 (LMP1) and LMP2), noncoding RNA (EBER1 and EBER2), and viral miRNAs (BHRF1-miRNA and BART-miRNA) (Fig. 2a). EBV latency is categorized by three latency types (latency I–III), which have different latency protein expression patterns depending on the type of cell infected. Several different kinds of latency were shown schematically (Fig. 2b). The lytic infection is triggered by several factors from the latent state. Then, nearly 80 proteins are encoded to facilitate the viral particle formation and release into the extracellular space.
Fig. 2. Different forms of EBV latency.
a Schematic illustration of the EBV genome and latent genes. b Latent gene expression spectrum in different forms of latency. EBVaGC belongs to latency I but LMP2A can be detected in approximately half of cases.
Establishment of EBV infection in stomach epithelial cells
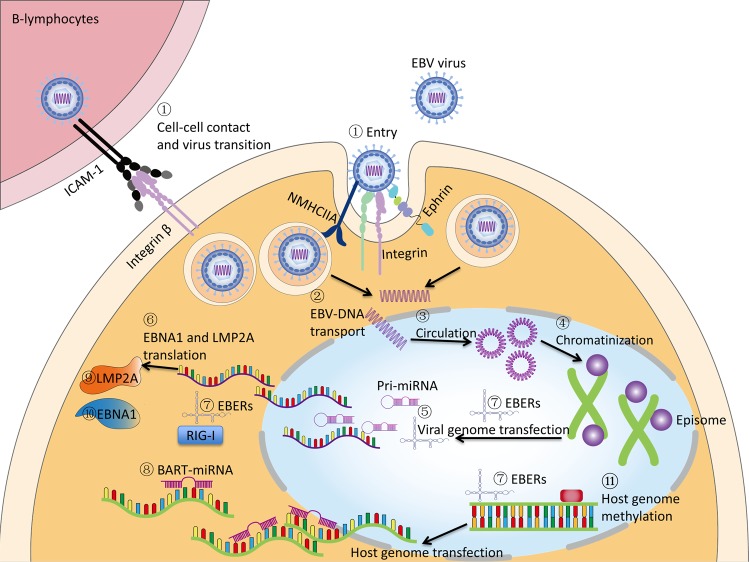
The first puzzle about EBV-associated gastric carcinoma (EBVaGC) is how EBV infects gastric epithelial cells, as the EBV infection often occurs in B lymphocytes and the oral epithelium. It is possible that the EBV-contained saliva is ingested and EBV infects the epithelial cells directly. Another explanation is that EBV is reactivated somehow in B lymphocytes in stomach and released to infect epithelial cells [84]. Ephrin receptor A2 as well as integrins and nonmuscle myosin heavy chain IIA (NMHCIIA) serve as cofactors and play an important role in EBV epithelial cell entry [85–88]. Coculturing of epithelial cells with EBV-positive lymphocyte cells showed about 800 fold higher efficiency of infection than cell-free infection, suggesting the possibility of direct cell-to-cell mediated virus infection [89]. It was proposed that EBV-infected lymphocytes contacts with epithelial cells via integrin β1/β2, and then promotes cell-to-cell contact by translocating intracellular adhesion molecule-1 to the cell surface. At last, the viral particle is transmitted by clathrin-mediated endocytosis pathway [90]. After endocytosis, the EBV-DNA is transported to nucleus, where the naked linear DNA genomes are assembled into a functional circular mini-chromosome. After circulation, viral genome chromatinization can effectively protect it from DNA damage and offer tight regulation of gene expression [91]. The CpG motifs of viral genome are widely methylated and by this way, latent infection is successfully established. The infection and latency processes were summarized in Fig. 3.
Fig. 3. EBV life cycle in stomach epithelial cells and the oncogenic properties in gastric carcinoma.
① Dissociative EBV from saliva or B cell enters stomach epithelial cells with the help of host receptor such as integrins, ephrin receptor A2, and NMHCIIA. Interaction of B cell and epithelial cell also facilitates the entry of EBV. ② Naked EBV-DNA is transported to nucleus, and then goes through ③ circulation, ④ chromatination, and CpG methylation. The latent infection is established followed by viral genome ⑤ transcription and ⑥ translation. The transcription products include ⑦ EBERs and ⑧ BART-miRNAs. The translational products are ⑨ LMP2A and ⑩ EBNA1. The oncogenic factors corporately promote gastric tumorigenesis. EBV can also induce ⑪ globally genomic methylation of the host cells.
It is well known that EBV LMP1 and nuclear antigen 2 (EBNA2) play major roles in EBV-induced oncogenesis. However both of them were rarely detected in gastric adenocarcinoma cells [92–94]. Instead, EBNA1 expression was confirmed [93, 95]. It was reported that transcription of EBNA was initiated from EBNA promoters, Qp but not Cp or Wp, which may result in the absent expression of EBNA2 [96]. In addition, BZLF1 is expressed in a proportion of the tumors, suggesting the switch from latent to lytic infection [92].
The histopathological features of EBVaGC
In 1990, Burke et al. firstly detected EBV in lymphoepithelial carcinoma of the stomach, which was similar to undifferentiated nasopharyngeal lymphoepithelioma [97]. However, Shibata subsequently found that EBV is involved not only in the rare gastric lymphoepithelioma-like cancers, but also in gastric adenocarcinomas. They demonstrated that the EBV genomes were specifically present within the gastric carcinoma cells and adjacent dysplastic epithelium but were absent in surrounding normal cells [98, 99]. The result was confirmed by polymerase chain reaction (PCR) and in situ hybridization (ISH) in variety of studies [92, 94, 100–103]. Since the EBV-positive tumor cells were from a single clonal proliferation [93, 102, 104], and EBV was not generally detected in normal stromal cells, metaplasia, gastric mucosa, and lymphocytes [93, 99, 103, 104], EBV infection was believed to occur in the dysplastic phase and related to gastric carcinogenesis. In gastric carcinoma with lymphoid stroma, all cases are EBV-positive tumors. However, in gastric adenocarcinomas, only a small fraction of the cases shows EBV positive. It is believed that EBV plays distinct roles in etiology of these two types of GC [92, 98].
EBVaGC-associated mortality was estimated to be 70,000 worldwide each year [105]. Epidemiological studies show that male EBV-positive GC patients were twice than female [99, 106] and type 1 strain is most prevalent one in gastric carcinoma [107, 108]. EBVaGC has distinctive clinical characteristics compared with EBV-negative cases. EBVaGC often appears in the upper part of the stomach and has a diffuse-type histology with lymphoid infiltration [109]. By analyzing individual-level data on 4599 GC patients from 13 studies, it was demonstrated that EBV positivity is a powerful prognostic indicator of GC. In addition, the report also indicated that patients with EBV-positive GC had a better survival than EBV-negative ones [110], because of the high degree of homogeneity in EBVaGCs compared with EBV-negative cases. Furthermore, most of the altered genes in EBVaGCs are immune response related genes leading to more immune cells to migrate into the microenvironment, compared with EBV-negative GC. The recruitment of immune cells contributes to the better clinical outcome for EBVaGC cases [111]. Besides, CD204-positive M2-type tumor-associated macrophages, which were associated with the aggressive behavior of tumors, exhibit low density in EBVaGCs, partly explaining the favorable outcomes [112].
Recently, comprehensive molecular characterization of GC presents several distinct molecular features and epigenetic alterations of EBVaGC, including lack of TP53 mutations, frequent PI3K mutations, and a high degree of CpG methylation in the tumor cell genome [113].
The molecular pathogenesis of EBVaGC
To date, the mechanism of EBVaGC has not yet been comprehensively deciphered. In general, virologic aspects and genetic abnormalities of host cells co-potentiate the tumor development. As for virologic aspects, since EBV-positive GC is in latency type I, only EBERs, EBNA1, and miR-BARTs are highly expressed, while LMP2A could be detected in some cases [96, 114]. Meanwhile, genetic abnormalities of host cells caused by EBV infection, such as aberrant DNA methylation, attract more and more attention these years. The methylation of CpG DNA of the host genome is also caused by the establishment of EBV latent infection and the expression of the EBV latent genes.
Promoting roles of virologic genes in GC pathogenesis
EBERs are viral nonpolyadenylated RNA, which is abundantly expressed in latently EBV-infected cells. Because of their abundance, EBERs serve as the most reliable and sensitive target by ISH to detect EBV infection in tissues. It plays a role in cell proliferation, apoptosis, and antiviral innate immunity. However, only a few studies investigated the roles of EBERs in EBV-mediated oncogenesis. EBER1 upregulates the expression of insulin growth factor 1, which promotes proliferation of EBVaGC cells [115]. Another work showed that EBERs induce chemoresistance and enhance cellular migration in coordination with IL-6-STAT3 signaling pathway [116]. EBERs as well as BARF0, EBNA1, and LMP2A contribute to the downregulation of miR-200 family, resulting in E-cadherin expression reduction, which is a crucial step in the carcinogenesis of EBVaGC [117].
EBNA1 is an essential molecule for EBV latency infection. It binds to viral oriP sequence in a sequence dependent manner and tethers EBV episomes onto host cell chromosomes, which is essential for episomal maintenance. EBNA1 also functions as a transactivator of the viral genes. In EBVaGC, EBNA1 enhances tumorigenicity in mouse model [118]. It was also reported to cause loss of promyelocitic leukemia (PML) nuclear bodies (NBs), resulting in impaired responses to DNA damage and promotion of cell survival [119]. In addition, EBNA1 induces ROS accumulation mediated by miR-34a and NOX2 to regulate the tumor cell viability [120].
LMP2A was detected in half of the EBVaGC cases [121]. Fukayama et al. found that LMP2A activates the NF-κB-survivin pathway to rescue EBV-infected epithelial cells from serum deprivation, which may play a role in the progression of EBV-infected GC [122]. By using a recombinant adenoviral expression vector, Liu et al. found that LMP2A plays an important role in pathogenesis of EBVaGC through regulating cyclin E expression and S phase cell ratio [123]. Besides, LMP2A mediates Notch signaling to elevate mitochondrial fission and promote cellular migration [124]. In addition, LMP2A could also downregulate HLA to evade the immune response of the malignant cells [125]. It can activate PI3K/Akt pathway to mediate the transformation process and inhibit TGFβ1-induced apoptosis, which provides a clonal selective advantage for EBV-infected cells during tumor development [126, 127]. LMP2A upregulates miR-155–5p though NF-κB pathway and this will lead to the inhibition of Smad2 and p-Smad2 [128]. Apart from the direct modulating effects on tumorigenesis, LMP2A also promotes malignancy by inducing epigenetic modifications of the host genome [129].
Recent studies imply that miR-BARTs contribute to EBV-associated epithelial carcinogenesis. The miR-BARTs are abundantly expressed in EBV-infected GCs cell line, but not in EBV-transformed lymphocytes [4, 130]. By using EBV-infected AGS cell line (AGS-EBV), the expression of miR-BARTs was quite rich but the expression of the viral protein was limited [131, 132]. EBV miRNAs contribute to the initiation and development of EBVaGC by targeting multiple host proteins to mediate cell proliferation, transformation, senescence, apoptosis, and immune response. A comprehensive profiling of EBV miRNAs in EBVaGC was constructed by Tsai et al. and they found the deletion of miR-BART9 could increase E-cadherin expression and decrease proliferative and invasive ability [133]. BART3–3p plays an important role in inhibiting the senescence of GC cells by targeting TP53 [134]. As for apoptosis, it was reported that BART5–3p directly targets TP53, leading to acceleration of the cell cycle progress and inhibition of cell apoptosis [135]. Besides, EBV encoded miR-BART5 could target p53 upregulated modulator of apoptosis (PUMA), which is a proapoptotic protein belonging to the Bcl-2 family, to counteract apoptosis and promote cellular survival [136]. In addition to PUMA, it was reported that miR-BART9, 11, and 12 strongly downregulate Bim, which is also a member of Bcl-2 family [137]. By comprehensively profiling the expression of EBV miRNAs in EBVaGC tissues, EBV-miR-BART4–5p was found to play a role in gastric carcinogenesis through apoptosis regulation by suppressing the proapoptotic protein Bid (the BH3-interacting domain death agonist) [138]. MiR-BART20–5p contributes to tumorigenesis of EBVaGC by directly interacting with 3′UTR of BAD [139]. Different from proteins, EBV-microRNAs could escape immune recognition as well as inhibit the immune response by directly suppressing the function of some antiviral host factors to facilitate the establishment of latent EBV infection. For example, EBV miRNA BART16 have been reported to suppress type I IFN signaling [140]. The oncogenic proteins and miR-BARTs in EBVaGC were summarized in Table 1.
Table 1.
EBV genes, functional roles and their targets in gastric tumorigenesis.
| Gene name | Functional roles | Refs |
|---|---|---|
| EBER | Induces insulin growth factor 1 expression and promote cell proliferation | [115] |
| Induces chemoresistance and promotes cell migration | [116] | |
| Downregulates mature miR-200 family thus to reduce E-cadherin expression | [117] | |
| EBNA1 | Causes the loss of PML NBs and impairs responses to DNA damage | [119] |
| Induces ROS accumulation to regulate cell viability | [120] | |
| LMP2A | Activates NF-κB-survivin pathway to rescue EBV-infected epithelial cells from serum deprivation | [122] |
| Regulates cyclin E expression and S phase cell ratio | [123] | |
| Elevates mitochondrial fission and promotes cellular migration through Notch pathway | [124] | |
| Downregulates HLA to evade immune response | [125] | |
| Activates phosphatidylinositol 3-kinase/Akt pathways to mediate transformation and inhibits transforming growth factor-beta 1-induced apoptosis | [126, 127] | |
| Promotes cell malignant by inducing epigenetic changes of host genome | [129] | |
| Upregulates miR-155–5p, and targets Smad2 and p-Smad2 to regulate TGF-β pathway | [128] | |
| miR-BARTs | miR-BART9 decreases E-cadherin expression and upregulates proliferation | [133] |
| miR-BART3–3p inhibits the senescence of gastric cancer cells by targeting TP53 | [134] | |
| miR-BART5–3p targets the tumor-suppressor gene TP53, leading to acceleration of the cell cycle progress and inhibition of cell apoptosis | [135] | |
| miR-BART5 targets PUMA, counteracts apoptosis and promotes cellular survival | [136] | |
| miR-BART9, 11, and 12 downregulate Bim expression | [137] | |
| miR-BART4–5p suppresses the proapoptotic protein Bid to regulate apoptosis | [138] | |
| miR-BART20–5p interacts with 3’UTR of BAD to contribute to tumorigenesis | [139] | |
| miR-BART16 suppresses type I IFN signaling | [140] |
Genetic and epigenetic abnormalities of host cells in EBVaGC
Multiple abnormalities of the EBVaGC cells have been identified. Among them, high frequency and nonrandom DNA methylation attract most attentions [141, 142]. However, the mechanisms are not fully elucidated yet. LMP2A was confirmed to mediate this process. LMP2A induces the STAT3 phosphorylation followed by DNMT1 transcriptionally activation and PTEN promoter methylation, indicating LMP2A plays an essential role in the development and maintenance of EBV-associated cancer [143]. Besides, a resistance factor against DNA methylation namely TET2 was suppressed to contribute to DNA methylation acquisition during EBV infection [144]. Variety of tumor-suppressor genes have been identified to be methylated during EBV infection, such as p16, p14, APC, SSTR1, FHIT, CRBP1, WWOX, DLC-1, AQP3, REC8, TP73, BLU, FSD1, BCL7A, MARK1, SCRN1, and NKX3.1 [129, 145–151]. The developed high-throughput sequencing makes it possible to reveal the EBV-induced DNA hypermethylation comprehensively. Using methyl-DNA immunoprecipitation microarray assays, Zhao et al. found 886 genes involved in cancer-related pathways were aberrantly promoter-hypermethylated in EBV-positive AGS cells [152]. They also employed whole-genome, transcriptome, and epigenome sequence analyses of EBV-infected or noninfected AGS cells together with primary samples to comprehensively reveal that EBV infection alters host gene expression through methylation and affects five prominent networks [153]. Apart from the methylation of host cells, EBV could promote vasculogenic mimicry formation, a new tumor vascular paradigm independent of endothelial cells, in NPC and GC cells through the PI3K/AKT/mTOR/HIF-1α axis [154].
EBV infects ~95% of people, however only part of the population develops tumors, indicating that molecular abnormalities of host cells are also equally important in the EBV-associated tumorigenesis. As for EBVaGC, high-frequency mutations of PIK3CA, ARID1A, and BCOR have been identified. Interestingly, TP53 mutation, which counts the most frequent mutation type in cancers, is extremely rare [113]. The amplification of JAK2, PD-L1, and PD-L2 were also revealed as prominent molecular features [155, 156].
Host immunity in EBV-positive GC
By using gene expression profile analysis, it was found that the prominent changes in EBVaGCs are immune response genes, which might allow EBVaGC to recruit reactive immune cells [111]. In fact, EBVaGC is characterized with the high density of CD8+ T cells and low density of CD204+ macrophages [112, 157, 158]. The robust present of infiltrating immune cells and specific microenvironments partially contribute to antitumor immunity [159].
However, the tumor cells in EBVaGC evade the immune response through multiple strategies. It was reported that indoleamine 2,3-dioxygenase (IDO1), a potent immune-inhibitory molecule, was upregulated in EBVaGC to resistance tumor immune response [130, 160]. In addition, Tregs were recruited by CCL22 produced by EBVaGC cells to counteract the antitumor response of CD8+ T cells [161]. EBVaGC also exhibits higher levels of programmed death ligand 1 (PD-L1) expression in carcinoma cells and the infiltrated immune cells [162, 163]. As tumor cells employ PD-L1 to evade antitumor immunity through interaction with programmed cell death protein 1 on the surface of T cells, the high expression of PD-L1 in EBVaGC is thought to contribute to the tumor progression [164].
The diagnosis and treatment of EBVaGC
By measuring immune-related proteins in plasma of patients with EBV-positive tumors and EBV-negative tumors, Camargo et al. found some chemokines and PD-L1 in plasma that could be used for the diagnosis of EBV status [165]. The plasma EBV-DNA load in EBVaGC patients decreases when the patients show response to the treatment, while load increases when the disease progresses, suggesting that plasma EBV-DNA serves as an ideal marker in predicting recurrence and chemotherapy response [166].
EBVaGC, MSI-high GC, intestinal type GC as a surrogate for chromosomal instability, diffuse type as a surrogate for genomically stable was classified as four different subtypes of GC proposed by TCGA [113]. The molecular subtypes of GC are also correlated with the immune subtype [167, 168], suggesting the TCGA classification could be further employed in future immunotherapy trials. The ACRG classification also revealed four molecular subtypes with clinical outcome. MSI subtype has the best prognosis and lowest recurrence rate followed by MSS/TP53+ and MSS/TP53−, while the MSS/EMT subtype demonstrates the worst prognosis and highest recurrence rate among the four subtypes. In ACRG classification, EBVaGCs are more frequently found in the MSS/TP53+ group than in the other groups, indicating a modest survival and recurrence [5].
Patients with EBV-positive tumors showed high responses to pembrolizumab treatment in a phase II trial of metastatic GC [169]. The satisfied response might rely on that EBVaGC expresses high levels of PD-L1 [165, 170] and exhibits more tumor infiltrating lymphocytes (TILs) [163, 167, 171, 172]. The amount of TILs has been reported to be associated with improved overall survival in GC patients [173]. In a research of advanced GC patients treated with nivolumab, only 25% of patients (1/4) demonstrated good response, and this might be because not all EBV-positive tumors show high PD-L1 expression [174]. Evaluating both EBV status and PD-L1 expression is necessary for predicting clinical benefit of anti-PD-L1 therapy [175]. To some extent, the result indicates that EBV is a potential biomarker for selecting patients with better response to PD-L1 treatment [176]. In addition to PD-L1, Kim et al. combined PI3K/mTOR dual inhibitor CMG002, together with the autophagy inhibitor CQ, to provide enhanced therapeutic efficacy against EBVaGC [177].
Fungus in gastric carcinogenesis
Fungus is a kind of eukaryotic microorganism, which is widely distributed worldwide. It was identified that more than 400 species of fungus associated with human beings. These years, the incidence of invasive fungal infections has experienced a dramatic increase globally.
Fungus is detectable in the digestive tract of about 70% of healthy adults in an analysis by using culture dependent methods. Most of them belong to Candida genus, and the number of fungus in the human stomach is 0–102 CFU/mL [178, 179]. Another research using PCR amplification of bacterial 16S ribosomal RNA genes and fungal internal transcribed spacers identified two fungal genera, Candida and Phialemonium, in gastric fluid from 25 clinically patients [180].
Generally, host immune system could tolerate fungus colonization and defend its invasion. However, the infection will occur when the balance is disturbed by systemic immunosuppressive such as the acquired immune deficiency syndrome, leukemia and HSCT, solid organ transplantation and immunosuppressant therapy, anti-microbial and steroid treatments, total parenteral nutrition, iatrogenic catheters and mechanical ventilation, malignant tumors, chemoradiotherapy, and diabetes mellitus [181, 182]. Besides, GI mucosal lesions and surgical procedures can also lead to GI fungal infection [181]. In a gastro-esophageal candidiasis detection by histological examination of biopsies from 465 patients, it was thought that the candidiasis is usually secondary to mucosal damage [183]. Candidiasis was detected in 54.2% of the gastric ulcer cases and 10.3% of the chronic gastritis cases. As for GC, the candidiasis was present in 20% of patients [179, 183].
Although the infection of fungal microorganisms in GC is only in rare cases, it is necessary to eliminate opportunistic infection of Candida to reduce the significant morbidity and mortality.
Future directions
Although EBV-related and H. polyri-related GCs are classified into different categories, it should be reminded that the stomach is an organ with multiple microorganism coexistence, which means that disease is promoted by multiple microorganisms. In fact, apart from direct promoting gastric carcinogenesis, H. pylori potentiates the transformation of the gastric mucosa into a hypochlorhidric environment, which further allow other microbes to colonize. In addition, coinfection with EBV and H. pylori in pediatric patients are associated with more severe inflammation than those with H. pylori infection alone [184]. Although the underlying mechanism has been partially suggested, such as host SHP1 phosphatase, antagonist of CagA, is downregulated by EBV-induced promoter hypermethylation [185], the synergistic oncogenic effects of two or more infectious agents remain to be further explored in the future studies. In recent years, the researches about the microbiota in gastrointestinal attract more and more attentions. However, the studies on the virome and fungus in stomach cancer are still in infancy. As enormous viruses and fungi do exist in human body including our gastrointestinal tract, it is imperative to understand the relationship between virome/fungi infection and stomach health.
Acknowledgements
We acknowledge the technical support from Core Utilities of Cancer Genomics and Pathobiology of Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong.
Funding
The current manuscript is supported by Research Grants Council of the Hong Kong Special Administrative Region, China [Project nos.: CUHK 14100019, 14118518 (for GRF projects)], and CUHK Direct Grant for Research (2018.002) from The Chinese University of Hong Kong.
Author contributions
WK and KFT provided direction and instruction in preparing this manuscript. YZ and JZ reviewed the literatures and drafted this manuscript. ASLC and JY reviewed the manuscript and made significant revisions on the drafts.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanan Zhao, Jinglin Zhang
Contributor Information
Ka Fai To, Email: kfto@cuhk.edu.hk.
Wei Kang, Email: weikang@cuhk.edu.hk.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, et al. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–44. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Colquhoun A, Arnold M, Ferlay J, Goodman KJ, Forman D, Soerjomataram I. Global patterns of cardia and non-cardia gastric cancer incidence in 2012. Gut. 2015;64:1881–8. doi: 10.1136/gutjnl-2014-308915. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. [DOI] [PMC free article] [PubMed]
- 5.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 6.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 7.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–15. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 8.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–e616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 9.Blaser MJ. Who are we? Indigenous microbes and the ecology of human diseases. EMBO Rep. 2006;7:956–60. doi: 10.1038/sj.embor.7400812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry S, de la Luz Sanchez M, Yang S, Haggerty TD, Hurst P, Perez-Perez G, et al. Gastroenteritis and transmission of Helicobacter pylori infection in households. Emerg Infect Dis. 2006;12:1701–8. doi: 10.3201/eid1211.060086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoicov C, Saffari R, Cai X, Hasyagar C, Houghton J. Molecular biology of gastric cancer: Helicobacter infection and gastric adenocarcinoma: bacterial and host factors responsible for altered growth signaling. Gene. 2004;341:1–17. doi: 10.1016/j.gene.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784–9. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 13.Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, et al. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291:187–94. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 14.Shikata K, Kiyohara Y, Kubo M, Yonemoto K, Ninomiya T, Shirota T, et al. A prospective study of dietary salt intake and gastric cancer incidence in a defined Japanese population: the Hisayama study. Int J Cancer. 2006;119:196–201. doi: 10.1002/ijc.21822. [DOI] [PubMed] [Google Scholar]
- 15.Shikata K, Doi Y, Yonemoto K, Arima H, Ninomiya T, Kubo M, et al. Population-based prospective study of the combined influence of cigarette smoking and Helicobacter pylori infection on gastric cancer incidence: the Hisayama Study. Am J Epidemiol. 2008;168:1409–15. doi: 10.1093/aje/kwn276. [DOI] [PubMed] [Google Scholar]
- 16.Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003;125:1636–44. doi: 10.1053/j.gastro.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Enroth H, Kraaz W, Engstrand L, Nyren O, Rohan T. Helicobacter pylori strain types and risk of gastric cancer: a case-control study. Cancer Epidemiol, Biomark Prev. 2000;9:981–5. [PubMed] [Google Scholar]
- 18.Rudi J, Kolb C, Maiwald M, Zuna I, von Herbay A, Galle PR, et al. Serum antibodies against Helicobacter pylori proteins VacA and CagA are associated with increased risk for gastric adenocarcinoma. Dig Dis Sci. 1997;42:1652–9. doi: 10.1023/a:1018849112533. [DOI] [PubMed] [Google Scholar]
- 19.Yamaoka Y, Kodama T, Kashima K, Graham DY. Antibody against Helicobacter pylori CagA and VacA and the risk for gastric cancer. J Clin Pathol. 1999;52:215–8. doi: 10.1136/jcp.52.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003–8. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–6. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 23.Higashi H, Nakaya A, Tsutsumi R, Yokoyama K, Fujii Y, Ishikawa S, et al. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J Biol Chem. 2004;279:17205–16. doi: 10.1074/jbc.M309964200. [DOI] [PubMed] [Google Scholar]
- 24.Tsutsumi R, Takahashi A, Azuma T, Higashi H, Hatakeyama M. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol Cell Biol. 2006;26:261–76. doi: 10.1128/MCB.26.1.261-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata-Kamiya N, Kurashima Y, Teishikata Y, Yamahashi Y, Saito Y, Higashi H, et al. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26:4617–26. doi: 10.1038/sj.onc.1210251. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Jackson CB, Judd LM, Menheniott TR, Kronborg I, Dow C, Yeomans ND, et al. Augmented gp130-mediated cytokine signalling accompanies human gastric cancer progression. J Pathol. 2007;213:140–51. doi: 10.1002/path.2218. [DOI] [PubMed] [Google Scholar]
- 28.Bronte-Tinkew DM, Terebiznik M, Franco A, Ang M, Ahn D, Mimuro H, et al. Helicobacter pylori cytotoxin-associated gene A activates the signal transducer and activator of transcription 3 pathway in vitro and in vivo. Cancer Res. 2009;69:632–9. doi: 10.1158/0008-5472.CAN-08-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menheniott TR, Judd LM, Giraud AS. STAT3: a critical component in the response to Helicobacter pylori infection. Cell Microbiol. 2015;17:1570–82. doi: 10.1111/cmi.12518. [DOI] [PubMed] [Google Scholar]
- 30.Knorr J, Ricci V, Hatakeyama M, Backert S. Classification of Helicobacter pylori virulence factors: is CagA a toxin or not? Trends Microbiol. 2019;27:731–8. [DOI] [PubMed]
- 31.Nagy TA, Frey MR, Yan F, Israel DA, Polk DB, Peek RM., Jr Helicobacter pylori regulates cellular migration and apoptosis by activation of phosphatidylinositol 3-kinase signaling. J Infect Dis. 2009;199:641–51. doi: 10.1086/596660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629–41. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama M, Hisatsune J, Yamasaki E, Isomoto H, Kurazono H, Hatakeyama M, et al. Helicobacter pylori VacA-induced inhibition of GSK3 through the PI3K/Akt signaling pathway. J Biol Chem. 2009;284:1612–9. doi: 10.1074/jbc.M806981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 35.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 36.Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68:4155–68. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci USA. 2000;97:7533–8. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–24. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 39.Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68:379–87. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 41.El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193–201. doi: 10.1016/s0016-5085(03)00157-4. [DOI] [PubMed] [Google Scholar]
- 42.Hwang IR, Kodama T, Kikuchi S, Sakai K, Peterson LE, Graham DY, et al. Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1beta production in Helicobacter pylori infection. Gastroenterology. 2002;123:1793–803. doi: 10.1053/gast.2002.37043. [DOI] [PubMed] [Google Scholar]
- 43.Ando T, Ishikawa T, Kato H, Yoshida N, Naito Y, Kokura S, et al. Synergistic effect of HLA class II loci and cytokine gene polymorphisms on the risk of gastric cancer in Japanese patients with Helicobacter pylori infection. Int J Cancer. 2009;125:2595–602. doi: 10.1002/ijc.24666. [DOI] [PubMed] [Google Scholar]
- 44.Lamb A, Chen LF. Role of the Helicobacter pylori-induced inflammatory response in the development of gastric cancer. J Cell Biochem. 2013;114:491–7. doi: 10.1002/jcb.24389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beales IL, Calam J. Helicobacter pylori stimulates granulocyte-macrophage colony-stimulating factor (GM-CSF) production from cultured antral biopsies and a human gastric epithelial cell line. Eur J Gastroenterol Hepatol. 1997;9:451–5. doi: 10.1097/00042737-199705000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Cho SO, Lim JW, Kim KH, Kim H. Involvement of Ras and AP-1 in Helicobacter pylori-induced expression of COX-2 and iNOS in gastric epithelial AGS cells. Dig Dis Sci. 2010;55:988–96. doi: 10.1007/s10620-009-0828-y. [DOI] [PubMed] [Google Scholar]
- 47.Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–40. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 48.Yamashita S, Nanjo S, Rehnberg E, Iida N, Takeshima H, Ando T, et al. Distinct DNA methylation targets by aging and chronic inflammation: a pilot study using gastric mucosa infected with Helicobacter pylori. Clin Epigenetics. 2019;11:191. doi: 10.1186/s13148-019-0789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, et al. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216–21. doi: 10.1158/1078-0432.CCR-05-2442. [DOI] [PubMed] [Google Scholar]
- 50.Bimczok D, Clements RH, Waites KB, Novak L, Eckhoff DE, Mannon PJ, et al. Human primary gastric dendritic cells induce a Th1 response to H. pylori. Mucosal Immunol. 2010;3:260–9. doi: 10.1038/mi.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye G, Barrera C, Fan X, Gourley WK, Crowe SE, Ernst PB, et al. Expression of B7-1 and B7-2 costimulatory molecules by human gastric epithelial cells: potential role in CD4+ T cell activation during Helicobacter pylori infection. J Clin Investig. 1997;99:1628–36. doi: 10.1172/JCI119325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Elios MM, Manghetti M, De Carli M, Costa F, Baldari CT, Burroni D, et al. T helper 1 effector cells specific for Helicobacter pylori in the gastric antrum of patients with peptic ulcer disease. J Immunol. 1997;158:962–7. [PubMed] [Google Scholar]
- 53.Hafsi N, Voland P, Schwendy S, Rad R, Reindl W, Gerhard M, et al. Human dendritic cells respond to Helicobacter pylori, promoting NK cell and Th1-effector responses in vitro. J Immunol. 2004;173:1249–57. doi: 10.4049/jimmunol.173.2.1249. [DOI] [PubMed] [Google Scholar]
- 54.DeLyria ES, Redline RW, Blanchard TG. Vaccination of mice against H. pylori induces a strong Th-17 response and immunity that is neutrophil dependent. Gastroenterology. 2009;136:247–56. doi: 10.1053/j.gastro.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfe RR, Jahoor F. Recovery of labeled CO2 during the infusion of C-1- vs C-2-labeled acetate: implications for tracer studies of substrate oxidation. Am J Clin Nutr. 1990;51:248–52. doi: 10.1093/ajcn/51.2.248. [DOI] [PubMed] [Google Scholar]
- 56.Zhuang Y, Shi Y, Liu XF, Zhang JY, Liu T, Fan X, et al. Helicobacter pylori-infected macrophages induce Th17 cell differentiation. Immunobiology. 2011;216:200–7. doi: 10.1016/j.imbio.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Eaton KA, Mefford M, Thevenot T. The role of T cell subsets and cytokines in the pathogenesis of Helicobacter pylori gastritis in mice. J Immunol. 2001;166:7456–61. doi: 10.4049/jimmunol.166.12.7456. [DOI] [PubMed] [Google Scholar]
- 58.Eaton KA, Mefford ME. Cure of Helicobacter pylori infection and resolution of gastritis by adoptive transfer of splenocytes in mice. Infect Immun. 2001;69:1025–31. doi: 10.1128/IAI.69.2.1025-1031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akhiani AA, Pappo J, Kabok Z, Schon K, Gao W, Franzen LE, et al. Protection against Helicobacter pylori infection following immunization is IL-12-dependent and mediated by Th1 cells. J Immunol. 2002;169:6977–84. doi: 10.4049/jimmunol.169.12.6977. [DOI] [PubMed] [Google Scholar]
- 60.Stoicov C, Fan X, Liu JH, Bowen G, Whary M, Kurt-Jones E, et al. T-bet knockout prevents Helicobacter felis-induced gastric cancer. J Immunol. 2009;183:642–9. doi: 10.4049/jimmunol.0900511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lundgren A, Suri-Payer E, Enarsson K, Svennerholm AM, Lundin BS. Helicobacter pylori-specific CD4+ CD25 high regulatory T cells suppress memory T-cell responses to H. pylori in infected individuals. Infect Immun. 2003;71:1755–62. doi: 10.1128/IAI.71.4.1755-1762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson K, Kenefeck R, Pidgeon EL, Shakib S, Patel S, Polson RJ, et al. Helicobacter pylori-induced peptic ulcer disease is associated with inadequate regulatory T cell responses. Gut. 2008;57:1375–85. doi: 10.1136/gut.2007.137539. [DOI] [PubMed] [Google Scholar]
- 63.Akhiani AA, Schon K, Franzen LE, Pappo J, Lycke N. Helicobacter pylori-specific antibodies impair the development of gastritis, facilitate bacterial colonization, and counteract resistance against infection. J Immunol. 2004;172:5024–33. doi: 10.4049/jimmunol.172.8.5024. [DOI] [PubMed] [Google Scholar]
- 64.Kamboj AK, Cotter TG, Oxentenko AS. Helicobacter pylori: the past, present, and future in management. Mayo Clin Proc. 2017;92:599–604. doi: 10.1016/j.mayocp.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 65.Wroblewski LE, Peek RM, Jr., Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–39. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fallone CA, Chiba N, van Zanten SV, Fischbach L, Gisbert JP, Hunt RH, et al. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology. 2016;151:51–69 e14. doi: 10.1053/j.gastro.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 67.Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6–30. doi: 10.1136/gutjnl-2016-312288. [DOI] [PubMed] [Google Scholar]
- 68.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–7. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS ONE. 2009;4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delgado S, Cabrera-Rubio R, Mira A, Suarez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65:763–72. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 71.Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024–32. doi: 10.1136/gutjnl-2017-314281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinasco K, Mitchell HM, Kaakoush NO, Castano-Rodriguez N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochimica et Biophysica Acta Rev Cancer. 2019;1872:188309. doi: 10.1016/j.bbcan.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–9. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao JJ, Zhang Y, Gerhard M, Mejias-Luque R, Zhang L, Vieth M, et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol. 2018;8:202. doi: 10.3389/fcimb.2018.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan MP, Kaparakis M, Galic M, Pedersen J, Pearse M, Wijburg OL, et al. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl Environ Microbiol. 2007;73:1010–3. doi: 10.1128/AEM.01675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khosravi Y, Dieye Y, Poh BH, Ng CG, Loke MF, Goh KL, et al. Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. Sci World J. 2014;2014:610421. doi: 10.1155/2014/610421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nardone G, Compare D. The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases. U Eur Gastroenterol J. 2015;3:255–60. doi: 10.1177/2050640614566846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engstrand L, Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol. 2013;27:39–45. doi: 10.1016/j.bpg.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 79.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–50. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cantalupo PG, Katz JP, Pipas JM. Viral sequences in human cancer. Virology. 2018;513:208–16. doi: 10.1016/j.virol.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lieberman PM. Virology. Epstein-Barr virus turns 50. Science. 2014;343:1323–5. doi: 10.1126/science.1252786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–44. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 83.Sokal EM, Hoppenbrouwers K, Vandermeulen C, Moutschen M, Leonard P, Moreels A, et al. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J Infect Dis. 2007;196:1749–53. doi: 10.1086/523813. [DOI] [PubMed] [Google Scholar]
- 84.Hutt-Fletcher LM. The long and complicated relationship between Epstein-Barr virus and epithelial cells. J Virol. 2017; 91:e01677-16. [DOI] [PMC free article] [PubMed]
- 85.Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX, et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol. 2018;3:1–8. doi: 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 86.Chesnokova LS, Hutt-Fletcher LM. Fusion of Epstein-Barr virus with epithelial cells can be triggered by alphavbeta5 in addition to alphavbeta6 and alphavbeta8, and integrin binding triggers a conformational change in glycoproteins gHgL. J Virol. 2011;85:13214–23. doi: 10.1128/JVI.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc Natl Acad Sci USA. 2009;106:20464–9. doi: 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiong D, Du Y, Wang HB, Zhao B, Zhang H, Li Y, et al. Nonmuscle myosin heavy chain IIA mediates Epstein-Barr virus infection of nasopharyngeal epithelial cells. Proc Natl Acad Sci USA. 2015;112:11036–41. doi: 10.1073/pnas.1513359112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371–8. doi: 10.1128/jvi.72.5.4371-4378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nanbo A, Kachi K, Yoshiyama H, Ohba Y. Epstein-Barr virus exploits host endocytic machinery for cell-to-cell viral transmission rather than a virological synapse. J Gen Virol. 2016;97:2989–3006. doi: 10.1099/jgv.0.000605. [DOI] [PubMed] [Google Scholar]
- 91.Lieberman PM. Keeping it quiet: chromatin control of gammaherpesvirus latency. Nat Rev Microbiol. 2013;11:863–75. doi: 10.1038/nrmicro3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rowlands DC, Ito M, Mangham DC, Reynolds G, Herbst H, Hallissey MT, et al. Epstein-Barr virus and carcinomas: rare association of the virus with gastric adenocarcinomas. Br J Cancer. 1993;68:1014–9. doi: 10.1038/bjc.1993.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA. 1994;91:9131–5. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ott G, Kirchner T, Muller-Hermelink HK. Monoclonal Epstein-Barr virus genomes but lack of EBV-related protein expression in different types of gastric carcinoma. Histopathology. 1994;25:323–9. doi: 10.1111/j.1365-2559.1994.tb01350.x. [DOI] [PubMed] [Google Scholar]
- 95.Murray PG, Niedobitek G, Kremmer E, Grasser F, Reynolds GM, Cruchley A, et al. In situ detection of the Epstein-Barr virus-encoded nuclear antigen 1 in oral hairy leukoplakia and virus-associated carcinomas. J Pathol. 1996;178:44–47. doi: 10.1002/(SICI)1096-9896(199601)178:1<44::AID-PATH471>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 96.Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, et al. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer. 1996;74:625–31. doi: 10.1038/bjc.1996.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol. 1990;3:377–80. [PubMed] [Google Scholar]
- 98.Tokunaga M, Land CE, Uemura Y, Tokudome T, Tanaka S, Sato E. Epstein-Barr virus in gastric carcinoma. Am J Pathol. 1993;143:1250–4. [PMC free article] [PubMed] [Google Scholar]
- 99.Shibata D, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol. 1992;140:769–74. [PMC free article] [PubMed] [Google Scholar]
- 100.Leoncini L, Vindigni C, Megha T, Funto I, Pacenti L, Musaro M, et al. Epstein-Barr virus and gastric cancer: data and unanswered questions. Int J Cancer. 1993;53:898–901. doi: 10.1002/ijc.2910530605. [DOI] [PubMed] [Google Scholar]
- 101.Shibata D, Hawes D, Stemmermann GN, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma among Japanese Americans in Hawaii. Cancer Epidemiol, Biomark Prev. 1993;2:213–7. [PubMed] [Google Scholar]
- 102.Harn HJ, Chang JY, Wang MW, Ho LI, Lee HS, Chiang JH, et al. Epstein-Barr virus-associated gastric adenocarcinoma in Taiwan. Hum Pathol. 1995;26:267–71. doi: 10.1016/0046-8177(95)90056-x. [DOI] [PubMed] [Google Scholar]
- 103.Yuen ST, Chung LP, Leung SY, Luk IS, Chan SY, Ho J. In situ detection of Epstein-Barr virus in gastric and colorectal adenocarcinomas. Am J Surg Pathol. 1994;18:1158–63. doi: 10.1097/00000478-199411000-00010. [DOI] [PubMed] [Google Scholar]
- 104.Gulley ML, Pulitzer DR, Eagan PA, Schneider BG. Epstein-Barr virus infection is an early event in gastric carcinogenesis and is independent of bcl-2 expression and p53 accumulation. Hum Pathol. 1996;27:20–27. doi: 10.1016/s0046-8177(96)90133-1. [DOI] [PubMed] [Google Scholar]
- 105.Khan G, Hashim MJ. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990-2010. Infect Agents Cancer. 2014;9:38. doi: 10.1186/1750-9378-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murphy G, Pfeiffer R, Camargo MC, Rabkin CS. Meta-analysis shows that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009;137:824–33. doi: 10.1053/j.gastro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schuster V, Ott G, Seidenspinner S, Kreth HW. Common Epstein-Barr virus (EBV) type-1 variant strains in both malignant and benign EBV-associated disorders. Blood. 1996;87:1579–85. [PubMed] [Google Scholar]
- 108.Sidagis J, Ueno K, Tokunaga M, Ohyama M, Eizuru Y. Molecular epidemiology of Epstein-Barr virus (EBV) in EBV-related malignancies. Int J Cancer. 1997;72:72–76. doi: 10.1002/(sici)1097-0215(19970703)72:1<72::aid-ijc11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 109.Yanai H, Nishikawa J, Mizugaki Y, Shimizu N, Takada K, Matsusaki K, et al. Endoscopic and pathologic features of Epstein-Barr virus-associated gastric carcinoma. Gastrointest Endosc. 1997;45:236–42. doi: 10.1016/s0016-5107(97)70265-7. [DOI] [PubMed] [Google Scholar]
- 110.Camargo MC, Kim WH, Chiaravalli AM, Kim KM, Corvalan AH, Matsuo K, et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: an international pooled analysis. Gut. 2014;63:236–43. doi: 10.1136/gutjnl-2013-304531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim SY, Park C, Kim HJ, Park J, Hwang J, Kim JI, et al. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology. 2015;148:137–47 e139. doi: 10.1053/j.gastro.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 112.Ichimura T, Abe H, Morikawa T, Yamashita H, Ishikawa S, Ushiku T, et al. Low density of CD204-positive M2-type tumor-associated macrophages in Epstein-Barr virus-associated gastric cancer: a clinicopathologic study with digital image analysis. Hum Pathol. 2016;56:74–80. doi: 10.1016/j.humpath.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 113.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luo B, Wang Y, Wang XF, Liang H, Yan LP, Huang BH, et al. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J Gastroenterol. 2005;11:629–33. doi: 10.3748/wjg.v11.i5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iwakiri D, Eizuru Y, Tokunaga M, Takada K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 2003;63:7062–7. [PubMed] [Google Scholar]
- 116.Banerjee AS, Pal AD, Banerjee S. Epstein-Barr virus-encoded small non-coding RNAs induce cancer cell chemoresistance and migration. Virology. 2013;443:294–305. doi: 10.1016/j.virol.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 117.Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, et al. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70:4719–27. doi: 10.1158/0008-5472.CAN-09-4620. [DOI] [PubMed] [Google Scholar]
- 118.Cheng TC, Hsieh SS, Hsu WL, Chen YF, Ho HH, Sheu LF. Expression of Epstein-Barr nuclear antigen 1 in gastric carcinoma cells is associated with enhanced tumorigenicity and reduced cisplatin sensitivity. Int J Oncol. 2010;36:151–60. [PubMed] [Google Scholar]
- 119.Sivachandran N, Dawson CW, Young LS, Liu FF, Middeldorp J, Frappier L. Contributions of the Epstein-Barr virus EBNA1 protein to gastric carcinoma. J Virol. 2012;86:60–68. doi: 10.1128/JVI.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim SM, Hur DY, Hong SW, Kim JH. EBV-encoded EBNA1 regulates cell viability by modulating miR34a-NOX2-ROS signaling in gastric cancer cells. Biochem Biophys Res Commun. 2017;494:550–5. doi: 10.1016/j.bbrc.2017.10.095. [DOI] [PubMed] [Google Scholar]
- 121.Minamitani T, Yasui T, Ma Y, Zhou H, Okuzaki D, Tsai CY, et al. Evasion of affinity-based selection in germinal centers by Epstein-Barr virus LMP2A. Proc Natl Acad Sci USA. 2015;112:11612–7. doi: 10.1073/pnas.1514484112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hino R, Uozaki H, Inoue Y, Shintani Y, Ushiku T, Sakatani T, et al. Survival advantage of EBV-associated gastric carcinoma: survivin up-regulation by viral latent membrane protein 2A. Cancer Res. 2008;68:1427–35. doi: 10.1158/0008-5472.CAN-07-3027. [DOI] [PubMed] [Google Scholar]
- 123.Liu X, Gao Y, Luo B, Zhao Y. Construction and antiapoptosis activities of recombinant adenoviral expression vector carrying EBV latent membrane protein 2A. Gastroenterol Res Pract. 2011;2011:182832. doi: 10.1155/2011/182832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pal AD, Basak NP, Banerjee AS, Banerjee S. Epstein-Barr virus latent membrane protein-2A alters mitochondrial dynamics promoting cellular migration mediated by Notch signaling pathway. Carcinogenesis. 2014;35:1592–601. doi: 10.1093/carcin/bgu069. [DOI] [PubMed] [Google Scholar]
- 125.Deb Pal A, Banerjee S. Epstein-Barr virus latent membrane protein 2A mediated activation of sonic Hedgehog pathway induces HLA class Ia downregulation in gastric cancer cells. Virology. 2015;484:22–32. doi: 10.1016/j.virol.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 126.Fukuda M, Longnecker R. Latent membrane protein 2A inhibits transforming growth factor-beta 1-induced apoptosis through the phosphatidylinositol 3-kinase/Akt pathway. J Virol. 2004;78:1697–705. doi: 10.1128/JVI.78.4.1697-1705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fukuda M, Longnecker R. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt Pathway. J Virol. 2007;81:9299–306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J Biol Chem. 2010;285:41328–36. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang J, Liu W, Zhang X, Zhang Y, Xiao H, Luo B. LMP2A induces DNA methylation and expression repression of AQP3 in EBV-associated gastric carcinoma. Virology. 2019;534:87–95. doi: 10.1016/j.virol.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 130.Strong MJ, Xu G, Coco J, Baribault C, Vinay DS, Lacey MR, et al. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog. 2013;9:e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marquitz AR, Mathur A, Shair KH, Raab-Traub N. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc Natl Acad Sci USA. 2012;109:9593–8. doi: 10.1073/pnas.1202910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J Virol. 2014;88:1389–93. doi: 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tsai CY, Liu YY, Liu KH, Hsu JT, Chen TC, Chiu CT, et al. Comprehensive profiling of virus microRNAs of Epstein-Barr virus-associated gastric carcinoma: highlighting the interactions of ebv-Bart9 and host tumor cells. J Gastroenterol Hepatol. 2017;32:82–91. doi: 10.1111/jgh.13432. [DOI] [PubMed] [Google Scholar]
- 134.Wang J, Zheng X, Qin Z, Wei L, Lu Y, Peng Q, et al. Epstein-Barr virus miR-BART3-3p promotes tumorigenesis by regulating the senescence pathway in gastric cancer. J Biol Chem. 2019;294:4854–66. doi: 10.1074/jbc.RA118.006853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y, et al. Epstein-Barr virus microRNA miR-BART5-3p inhibits p53 expression. J Virol. 2018;92:e01022-18. [DOI] [PMC free article] [PubMed]
- 136.Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–60. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shinozaki-Ushiku A, Kunita A, Isogai M, Hibiya T, Ushiku T, Takada K, et al. Profiling of virus-encoded microRNAs in Epstein-Barr virus-associated gastric carcinoma and their roles in gastric carcinogenesis. J Virol. 2015;89:5581–91. doi: 10.1128/JVI.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim H, Choi H, Lee SK. Epstein-Barr virus miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett. 2015;356:733–42. doi: 10.1016/j.canlet.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 140.Hooykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D, et al. EBV microRNA BART16 suppresses type I IFN signaling. J Immunol. 2017;198:4062–73. doi: 10.4049/jimmunol.1501605. [DOI] [PubMed] [Google Scholar]
- 141.Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, et al. Epstein-Barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787–94. doi: 10.1016/S0002-9440(10)64901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–97. doi: 10.1158/0008-5472.CAN-11-1349. [DOI] [PubMed] [Google Scholar]
- 143.Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–74. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- 144.Namba-Fukuyo H, Funata S, Matsusaka K, Fukuyo M, Rahmutulla B, Mano Y, et al. TET2 functions as a resistance factor against DNA methylation acquisition during Epstein-Barr virus infection. Oncotarget. 2016;7:81512–26. doi: 10.18632/oncotarget.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Geddert H, zur Hausen A, Gabbert HE, Sarbia M. EBV-infection in cardiac and non-cardiac gastric adenocarcinomas is associated with promoter methylation of p16, p14 and APC, but not hMLH1. Cell Oncol. 2011;34:209–14. doi: 10.1007/s13402-011-0028-6. [DOI] [PubMed] [Google Scholar]
- 146.Zhao J, Liang Q, Cheung KF, Kang W, Dong Y, Lung RW, et al. Somatostatin receptor 1, a novel EBV-associated CpG hypermethylated gene, contributes to the pathogenesis of EBV-associated gastric cancer. Br J Cancer. 2013;108:2557–64. doi: 10.1038/bjc.2013.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.He D, Zhang YW, Zhang NN, Zhou L, Chen JN, Jiang Y, et al. Aberrant gene promoter methylation of p16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr virus-associated gastric carcinomas. Med Oncol. 2015;32:92. doi: 10.1007/s12032-015-0525-y. [DOI] [PubMed] [Google Scholar]
- 148.Yu J, Liang Q, Wang J, Wang K, Gao J, Zhang J, et al. REC8 functions as a tumor suppressor and is epigenetically downregulated in gastric cancer, especially in EBV-positive subtype. Oncogene. 2017;36:182–93. doi: 10.1038/onc.2016.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Okada T, Nakamura M, Nishikawa J, Sakai K, Zhang Y, Saito M, et al. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas. Cancer Sci. 2013;104:1309–14. doi: 10.1111/cas.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ushiku T, Chong JM, Uozaki H, Hino R, Chang MS, Sudo M, et al. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2007;120:60–66. doi: 10.1002/ijc.22275. [DOI] [PubMed] [Google Scholar]
- 151.Farrell PJ. Epstein-Barr virus and cancer. Annu Rev Pathol. 2019;14:29–53. doi: 10.1146/annurev-pathmechdis-012418-013023. [DOI] [PubMed] [Google Scholar]
- 152.Zhao J, Liang Q, Cheung KF, Kang W, Lung RW, Tong JH, et al. Genome-wide identification of Epstein-Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer. 2013;119:304–12. doi: 10.1002/cncr.27724. [DOI] [PubMed] [Google Scholar]
- 153.Liang Q, Yao X, Tang S, Zhang J, Yau TO, Li X, et al. Integrative identification of Epstein-Barr virus-associated mutations and epigenetic alterations in gastric cancer. Gastroenterology. 2014;147:1350–62 e1354. doi: 10.1053/j.gastro.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 154.Xiang T, Lin YX, Ma W, Zhang HJ, Chen KM, He GP, et al. Vasculogenic mimicry formation in EBV-associated epithelial malignancies. Nat Commun. 2018;9:5009. doi: 10.1038/s41467-018-07308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dong M, Wang HY, Zhao XX, Chen JN, Zhang YW, Huang Y, et al. Expression and prognostic roles of PIK3CA, JAK2, PD-L1, and PD-L2 in Epstein-Barr virus-associated gastric carcinoma. Hum Pathol. 2016;53:25–34. doi: 10.1016/j.humpath.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 156.Saito R, Abe H, Kunita A, Yamashita H, Seto Y, Fukayama M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1+ immune cells in Epstein-Barr virus-associated gastric cancer: the prognostic implications. Mod Pathol. 2017;30:427–39. doi: 10.1038/modpathol.2016.202. [DOI] [PubMed] [Google Scholar]
- 157.van Beek J, zur Hausen A, Snel SN, Berkhof J, Kranenbarg EK, van de Velde CJ, et al. Morphological evidence of an activated cytotoxic T-cell infiltrate in EBV-positive gastric carcinoma preventing lymph node metastases. Am J Surg Pathol. 2006;30:59–65. doi: 10.1097/01.pas.0000176428.06629.1e. [DOI] [PubMed] [Google Scholar]
- 158.Saiki Y, Ohtani H, Naito Y, Miyazawa M, Nagura H. Immunophenotypic characterization of Epstein-Barr virus-associated gastric carcinoma: massive infiltration by proliferating CD8+ T-lymphocytes. Lab Investig. 1996;75:67–76. [PubMed] [Google Scholar]
- 159.Song HJ, Srivastava A, Lee J, Kim YS, Kim KM, Ki Kang W, et al. Host inflammatory response predicts survival of patients with Epstein-Barr virus-associated gastric carcinoma. Gastroenterology. 2010;139:84–92 e82. doi: 10.1053/j.gastro.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 160.Lu S, Wang LJ, Lombardo K, Kwak Y, Kim WH, Resnick MB. Expression of indoleamine 2, 3-dioxygenase 1 (IDO1) and tryptophanyl-tRNA synthetase (WARS) in gastric cancer molecular subtypes. Appl Immunohistochem Mol Morphol. 2019. 10.1097/PAI.0000000000000761. [DOI] [PMC free article] [PubMed]
- 161.Zhang NN, Chen JN, Xiao L, Tang F, Zhang ZG, Zhang YW, et al. Accumulation mechanisms of CD4(+)CD25(+)FOXP3(+) regulatory T cells in EBV-associated gastric carcinoma. Sci Rep. 2015;5:18057. doi: 10.1038/srep18057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Derks S, Liao X, Chiaravalli AM, Xu X, Camargo MC, Solcia E, et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget. 2016;7:32925–32. doi: 10.18632/oncotarget.9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Saito R, Abe H, Kunita A, Yamashita H, Seto Y, Fukayama M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1(+) immune cells in Epstein-Barr virus-associated gastric cancer: the prognostic implications. Mod Pathol. 2017;30:427–39. doi: 10.1038/modpathol.2016.202. [DOI] [PubMed] [Google Scholar]
- 164.Cho J, Kang MS, Kim KM. Epstein-Barr virus-associated gastric carcinoma and specific features of the accompanying immune response. J Gastric Cancer. 2016;16:1–7. doi: 10.5230/jgc.2016.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Camargo MC, Sivins A, Isajevs S, Folkmanis V, Rudzite D, Gulley ML, et al. Associations of Epstein-Barr virus-positive gastric adenocarcinoma with circulating mediators of inflammation and immune response. Cancers. 2018;10:E284. [DOI] [PMC free article] [PubMed]
- 166.Qiu MZ, He CY, Lu SX, Guan WL, Wang F, Wang XJ, et al. Prospective observation: clinical utility of plasma Epstein-Barr virus DNA load in EBV-associated gastric carcinoma patients. Int J Cancer. 2019;146:272–80. [DOI] [PubMed]
- 167.Kim TS, da Silva E, Coit DG, Tang LH. Intratumoral immune response to gastric cancer varies by molecular and histologic subtype. Am J Surg Pathol. 2019;43:851–60. doi: 10.1097/PAS.0000000000001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim JY, Kim WG, Kwon CH, Park DY. Differences in immune contextures among different molecular subtypes of gastric cancer and their prognostic impact. Gastric Cancer. 2019;22:1164–75. [DOI] [PubMed]
- 169.Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449–58. doi: 10.1038/s41591-018-0101-z. [DOI] [PubMed] [Google Scholar]
- 170.Sasaki S, Nishikawa J, Sakai K, Iizasa H, Yoshiyama H, Yanagihara M, et al. EBV-associated gastric cancer evades T-cell immunity by PD-1/PD-L1 interactions. Gastric Cancer. 2019;22:486–96. doi: 10.1007/s10120-018-0880-4. [DOI] [PubMed] [Google Scholar]
- 171.Panda A, Mehnert JM, Hirshfield KM, Riedlinger G, Damare S, Saunders T, et al. Immune activation and benefit from avelumab in EBV-positive gastric cancer. J Natl Cancer Inst. 2018;110:316–20. doi: 10.1093/jnci/djx213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xing X, Guo J, Ding G, Li B, Dong B, Feng Q, et al. Analysis of PD1, PDL1, PDL2 expression and T cells infiltration in 1014 gastric cancer patients. Oncoimmunology. 2018;7:e1356144. doi: 10.1080/2162402X.2017.1356144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yu PC, Long D, Liao CC, Zhang S. Association between density of tumor-infiltrating lymphocytes and prognoses of patients with gastric cancer. Medicine. 2018;97:e11387. doi: 10.1097/MD.0000000000011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kawazoe A, Shitara K, Kuboki Y, Bando H, Kojima T, Yoshino T, et al. Clinicopathological features of 22C3 PD-L1 expression with mismatch repair, Epstein-Barr virus status, and cancer genome alterations in metastatic gastric cancer. Gastric Cancer. 2019;22:69–76. doi: 10.1007/s10120-018-0843-9. [DOI] [PubMed] [Google Scholar]
- 175.Mishima S, Kawazoe A, Nakamura Y, Sasaki A, Kotani D, Kuboki Y, et al. Clinicopathological and molecular features of responders to nivolumab for patients with advanced gastric cancer. J Immunother Cancer. 2019;7:24. doi: 10.1186/s40425-019-0514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Koemans WJ, Chalabi M, van Sandick JW, van Dieren JM, Kodach LL. Beyond the PD-L1 horizon: In search for a good biomarker to predict success of immunotherapy in gastric and esophageal adenocarcinoma. Cancer Lett. 2019;442:279–86. doi: 10.1016/j.canlet.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 177.Kim MY, Kruger AJ, Jeong JY, Kim J, Shin PK, Kim SY, et al. Combination therapy with a PI3K/mTOR dual inhibitor and chloroquine enhances synergistic apoptotic cell death in Epstein-Barr virus-infected gastric cancer cells. Mol Cells. 2019;42:448–59. doi: 10.14348/molcells.2019.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schulze J, Sonnenborn U. Yeasts in the gut: from commensals to infectious agents. Dtsch Arzteblatt Int. 2009;106:837–42. doi: 10.3238/arztebl.2009.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zwolinska-Wcislo M, Budak A, Bogdal J, Trojanowska D, Stachura J. Fungal colonization of gastric mucosa and its clinical relevance. Med Sci Monit. 2001;7:982–8. [PubMed] [Google Scholar]
- 180.von Rosenvinge EC, Song Y, White JR, Maddox C, Blanchard T, Fricke WF. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 2013;7:1354–66. doi: 10.1038/ismej.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH. Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther. 2014;39:751–66. doi: 10.1111/apt.12665. [DOI] [PubMed] [Google Scholar]
- 182.Lamps LW, Lai KK, Milner DA., Jr Fungal infections of the gastrointestinal tract in the immunocompromised host: an update. Adv Anat Pathol. 2014;21:217–27. doi: 10.1097/PAP.0000000000000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Scott BB, Jenkins D. Gastro-oesophageal candidiasis. Gut. 1982;23:137–9. doi: 10.1136/gut.23.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cardenas-Mondragon MG, Carreon-Talavera R, Camorlinga-Ponce M, Gomez-Delgado A, Torres J, Fuentes-Panana EM. Epstein Barr virus and Helicobacter pylori co-infection are positively associated with severe gastritis in pediatric patients. PLoS ONE. 2013;8:e62850. doi: 10.1371/journal.pone.0062850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Saju P, Murata-Kamiya N, Hayashi T, Senda Y, Nagase L, Noda S, et al. Host SHP1 phosphatase antagonizes Helicobacter pylori CagA and can be downregulated by Epstein-Barr virus. Nat Microbiol. 2016;1:16026. doi: 10.1038/nmicrobiol.2016.26. [DOI] [PubMed] [Google Scholar]