Abstract
In eukaryotic cells, proteome remodeling is mediated by the ubiquitin-proteasome system, which regulates protein degradation, trafficking, and signaling events in the cell. Interplay between the cellular proteome and ubiquitin is complex and dynamic, and many regulatory features that support this system have only recently come into focus. An unexpected recurring feature in this system is the physical interaction between E3 ubiquitin ligases and deubiquitylases (DUBs). Recent studies have reported on the regulatory significance of DUB-E3 interactions, and it is becoming clear that they play important but complicated roles in the regulation of diverse cellular processes. Here, we summarize the current understanding of interactions between ubiquitin conjugation and deconjugation machineries, and we examine the regulatory logic of these enigmatic complexes.
Keywords: Ubiquitin, deubiquitylase, E3 ubiquitin ligase, polyubiquitin chain editing
The complexity of cellular ubiquitin dynamics
It is widely appreciated that ubiquitin is a multi-functional post-translational modifier of the proteome that regulates many complex cellular processes, including protein quality control, signaling relays, protein trafficking, and cell cycle control [1–6]. Ubiquitin conjugation is mediated by an enzymatic cascade (E1-E2-E3) that culminates in the activity of an E3 ubiquitin ligase, which catalyzes the formation of an isopeptide bond between the C-terminal glycine carboxyl group of ubiquitin to the ε–amino group of a lysine residue on a substrate protein [7]. Conjugation can be reversed by the activity of ubiquitin-specific isopeptidases known as deubiquitylases, or DUBs [8, 9]. The biochemistry of these reactions has been studied for decades, and their roles in regulating diverse cellular processes is well-appreciated [1, 10]. Less well-understood is how conjugation and deconjugation activities are coordinated in the cell to achieve complex regulation of protein homeostasis. As our understanding of the complexity of the ubiquitin code (see Glossary) continues to grow – with increasing appreciation for features of the code such as branching [11, 12] and post-translational modification of polyubiquitin chains [13, 14] – more emphasis must be placed on interrogating how cells manage the complexity of the ubiquitin code and how conjugation and deconjugation activities manage cellular ubiquitin dynamics.
The number of E3 ubiquitin ligases encoded in the human genome (~700) is far greater than the number of DUBs (~100), although new DUBs are still being discovered [15–18]. It is widely appreciated that the balance of ubiquitin conjugation and deconjugation activities is critical for maintaining many complex cellular processes, and it is increasingly evident that many instances of ubiquitin regulation involve the coupling of conjugation and deconjugation machineries. This E3-DUB coupling can facilitate the fine-tuning of a number of ubiquitin polymer variables including chain length and incorporated linkage types, and has potential to regulate in modes ranging from switch-like (binary) to rheostat-like behavior, allowing for sampling of many distinct, probabilistic outcomes. Here, we examine the evidence that DUB-E3 complexes and the coupling of ubiquitin conjugation and deconjugation activities plays an important role in the management of cellular ubiquitin dynamics.
DUBs step to E3s: prevalence of DUB-E3 complexes in eukaryotic cells
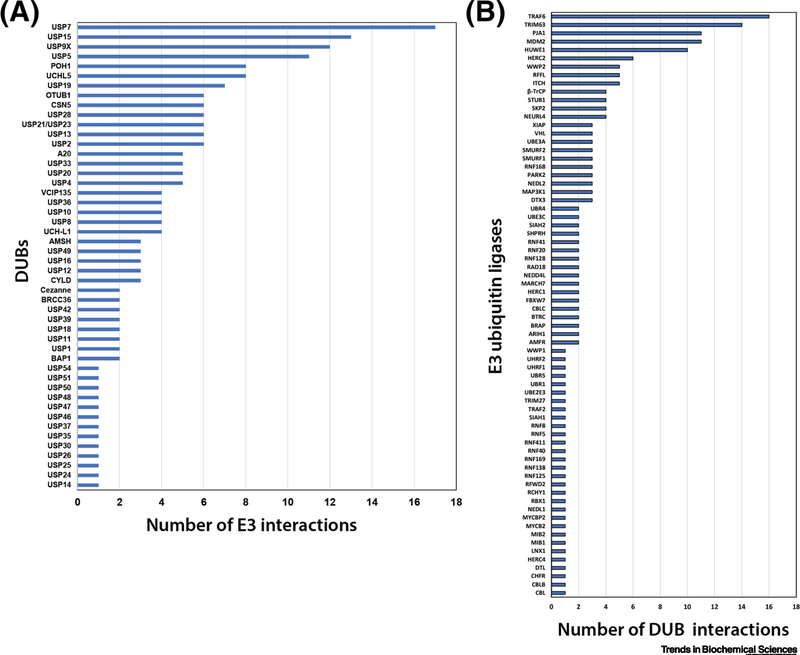
From the current literature, we catalogued 194 physical interactions between human DUBs and E3 ubiquitin ligases reported in the protein interaction repository BioGrid (https://thebiogrid.org/) (Table S1). Of the ~100 DUBs encoded in the human genome, 48 were reported to interact with at least one E3 ubiquitin ligase, suggesting that just under half of all DUBs encoded in the human genome physically associate with E3s (Figure 1 and Table S1). Of these reported interactions, 35 DUBs interact with more than one E3 ubiquitin ligase (Figure 1A). These interactions involve 70 different E3 ubiquitin ligases, accounting for about 10% of all E3s encoded in the human genome [19]. Of these, greater than half of the E3s exhibit interactions with more than one DUB, and several appear to be hubs for DUB interactions (Figure 1B). Of course, this catalog of E3-DUB interactions reported in the literature has caveats including the potential for reporting of false positive interactions, as well as a bias toward reporting of interactions for well-studied or high abundance proteins. Nevertheless, the high prevalence of DUB-E3 interactions in human cells suggests that they play a critical role in cellular ubiquitin dynamics.
Figure 1.
Prevalence of DUB-E3 interactions reported in human cells. A complete catalog of reported human DUB-E3 interactions curated on BioGrid (https://thebiogrid.org/) is provided in Table S1. (A) For each DUB in the DUB-E3 interaction catalog, the number of distinct E3 interactions is plotted. (B) For each E3 in the DUB-E3 interaction catalog, the number of distinct DUB interactions is plotted. A complete listing of these interactions is provided in Table S1.
For comparison, we analyzed the prevalence of reported DUB-E3 interactions in yeast cells. The yeast genome encodes 22 known DUBs and ~60–100 E3 ubiquitin ligases [15, 20]. Of these, we found four yeast DUBs that were reported to interact with a single E3 ubiquitin ligase (Table 1), and one yeast DUB reported to interact with an E3 SUMO ligase [21]. For the remaining 17 DUBs encoded in the yeast genome, there were no reports of interactions with E3s (https://thebiogrid.org/). Furthermore, no yeast DUBs were reported to interact with more than one E3 ubiquitin ligase. The possibility of additional DUB-E3 interactions in yeast cannot be excluded, but given the abundance of high throughput interaction screens performed in yeast, these findings suggest a lower incidence of such interactions in yeast compared to human cells.
Table 1:
Yeast DUB-E3 interactions
| DUB | E3 | Regulatory function of complex | Reference |
|---|---|---|---|
| Otu1 | Ufd2 | Cdc48 simultaneously engages both Ufd2 and Otu1, coupling activities to fine tune substrate ubiquitylation. | [47, 74] |
| Rpn11 | Mms22 | Rpn11 was detected in a proteomics screen for Mms22 interactors. Regulatory significance unknown. | [75] |
| Ubp2 | Rsp5 | Ubp2 interacts with Rsp5 and is reported to antagonize its function and also protects Rsp5 adaptors from degradation. | [33, 34, 62, 76] |
| Ubp15 | Ssm4 | Interaction detected in a high throughput two-hybrid screen. Regulatory significance unknown. | [77] |
E3-DUB Hubs
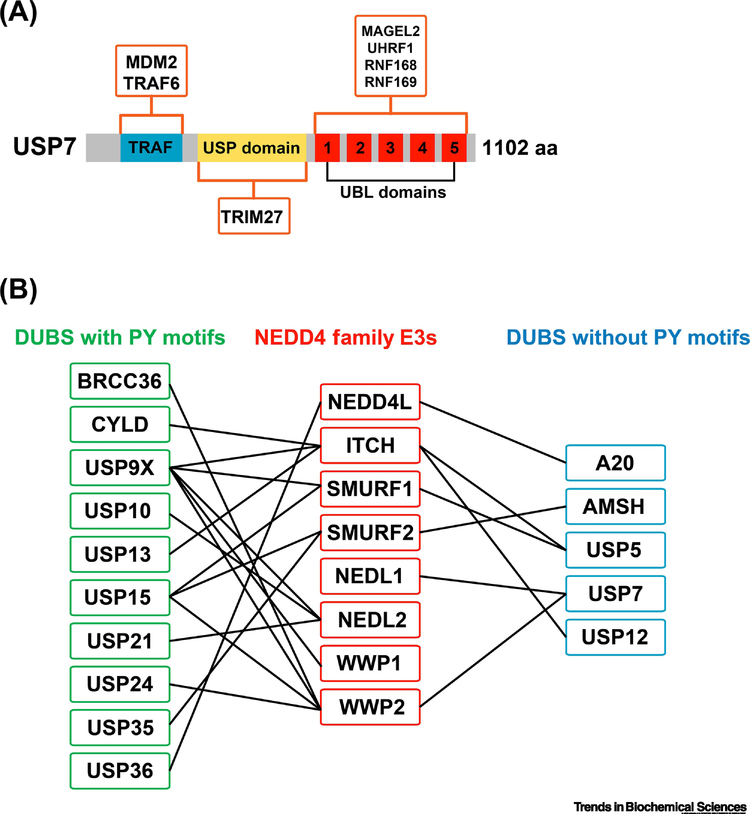
In this catalog of human E3-DUB interactions, several human DUBs, including ubiquitin specific protease 7 (USP7), USP15, and USP9X, were reported to interact with a multitude of E3 ubiquitin ligases. For some of these hubs, the biochemical basis of DUB-E3 interactions has been described. For example, the large number of USP7-E3 interactions appears to be due to a number of factors, including its ability to localize to both cytosol and nucleus [22], as well as the presence of accessory domains including TNF receptor-associated factor (TRAF) and ubiquitin-like (UBL) domains [23–26]. Interestingly, structural determinants of several USP7-E3 interactions have been mapped and characterized [23–26], and it is clear that different domains of USP7 contribute to interactions with multiple E3s (Figure 2A). The interactions between USP7 and several E3 ubiquitin ligases have been recently reviewed [26]. Additionally, USP15 is a well-studied DUB with reported roles in the TGF-β receptor signaling pathway, the NFκB signaling pathway, and the regulation of the canonical WNT pathway [27–29]. USP15 contains an N-terminal domain present in ubiquitin specific proteases (DUSP domain), which facilitates interaction with the COP9 signalosome [30] – a complex that is critical for regulation of many Skp-Cullin-F-box (SCF) E3 ubiquitin ligases. Thus, USP15 may indirectly engage a number of SCF E3 ubiquitin ligases via its interaction with the COP9 signalosome. Indeed, USP15 interactions with the COP9 signalosome are reported to be important for regulation of the NFκB pathway and the canonical WNT pathway [29, 31]. Another DUB that engages multiple E3s is USP9X; USP9X is a very large DUB (the primary sequence is 2,570 amino acids) which harbors a USP catalytic domain, a UBL domain, and four PY (PPXY) motifs [32]. Two C-terminal PY motifs of USP9X have been shown to mediate interactions with the WW domains of NEDD4 family E3 ubiquitin ligases [32], and these PY-WW interactions likely facilitate USP9X interactions with other reported NEDD4 family members (Figure 1A and Table S1, discussed further below).
Figure 2.
E3-DUB hubs in the ubiquitin network. (A) USP7 is the DUB with the most reported E3 interactions. A schematic representation of the USP7 domain structure is depicted, illustrating the location of the TRAF domain (blue), the USP catalytic domain (yellow), and the five C-terminal ubiquitin-like (UBL) domains (red). Orange boxes illustrate interactions where the specific domain of USP7 that binds to the specified E3 has been mapped. (B) Many DUBs are reported to interact with NEDD4 family E3 ubiquitin ligases (boxed in red, middle). Many of the NEDD4-interacting DUBs harbor PY motifs (boxed in green, left), while a few lack PY motifs (boxed in blue, right). The preponderance of NEDD4 family interactions with PY motif-containing DUBs suggests that many occur via WW domain scaffolding, although in most cases experimental evidence on the biochemical basis of interaction is lacking.
This analysis also revealed a number of E3 ubiquitin ligases, including TRAF6, TRIM63, PJA1, MDM2, and HUWE1, are reported to interact with a large number of DUBs (Figure 1B). Some of these proteins are major hubs for many protein-protein interactions (like TRAF6 and MDM2). Indeed, there is substantial overlap between the hub E3s and the hub DUBs. For example, TRAF6, TRIM63, PJA1, MDM2, and HUWE1 each interact with USP7, and all except PJA1 interact with USP15 (Figure 1, Figure 2A, Table S1, and references therein). This is suggestive of significant cross-talk between these major E3-DUB hubs, although it could also result from experimental artifacts due to high expression or non-specific, “sticky” interactions. Ultimately, distinguishing between these possibilities will require rigorous characterization of the biochemical basis and physiological function of these E3-DUB interactions.
One interesting category of E3-DUB interactions clusters around NEDD4 family E3 ubiquitin ligases, a family of HECT E3 ubiquitin ligases that harbor an N-terminal C2 domain and multiple WW scaffolding domains. Of the nine NEDD4 family E3 ubiquitin ligases encoded in the human genome, eight exhibit at least one DUB interaction, and many are reported to interact with multiple DUBs (Figure 2B). One recent analysis of the NEDD4 family member WWP1 detected interactions with DUBs in breast cancer cells [32], and each of these DUBs contained one or more PY motifs capable of engaging the WW domains. Indeed, NEDD4 family members are reported to interact with 15 different DUBs (Figure 1A, Table S1, and references therein), and inspection of primary sequence reveals 10 of these to contain PY motifs (Figure 2B). Furthermore, of the few DUB-E3 complexes reported in S. cerevisiae, one involves the NEDD4 family member Rsp5 and Ubp2 [33, 34], a DUB that contains multiple PY motifs in the primary sequence. Thus, although much work remains to determine if the WW domains of NEDD4 family members generally scaffold interactions with PY-containing DUBs, we predict that these interfaces will be a recurring theme for regulation of NEDD4 family E3s by DUBs across evolution.
The regulatory logic of DUB-E3 interactions
The prevalence of DUB-E3 interactions suggests they play an important role in tuning the dynamics of cellular ubiquitylation, but the regulatory logic underlying these complexes has not been fully elucidated. There is now strong evidence that coupling the ubiquitin conjugation and deconjugation machineries can contribute to the regulation of shared substrates (Table 2). However, DUB-E3 complexes can also facilitate mutual regulatory circuits (Table 3). These two regulatory modes are not mutually exclusive. Here, we explore the regulatory logic of DUB-E3 interactions using well-characterized examples reported in the literature.
Table 2:
Human DUB-E3 complexes operating on shared substrates
| DUB | E3 | Regulatory function of complex | Reference |
|---|---|---|---|
| DUB-E3 interactions that mediate ubiquitin switches or rheostats on a shared substrate | |||
| UCHL1 | MDM2 | UCHL1 antagonizes MDM2-mediated ubiquitylation of p53 | [78] |
| UCHL1 | TRAF6 | UCHL1 antagonizes TRAF6-mediated ubiquitylation of TRAF3 | [79] |
| USP7 | TRIM27 | USP7 interacts with MAGE-L2-TRIM27 to regulate WASH ubiquitylation and actin polymerization on endosomes | [25] |
| USP9X | WWP1 | USP9X interacts with WWP1 to regulate DVL2 ubiquitylation and WNT signaling | [32] |
| USP10 | RNF168 | USP10 interacts with RNF168 to regulate TOP2α ubiquitylation and chromatin association | [80] |
| USP12 | ITCH | USP12 interacts with ITCH to regulate Notch trafficking and degradation | [81, 82] |
| USP13 | AMFR | USP13 interacts with gp78 to stabilize UBL4A and promote ERAD | [45] |
| USP15 | βTrCP | USP15 interacts with βTrCP to regulate the stability of β-catenin | [29] |
| USP20 | TRAF6 | USP20 interacts with TRAF6 to regulate the ubiquitylation of β-arrestin2 | [83] [58] |
| USP28 | FBXW7 | USP28 interacts with FBXW7 in nucleus to regulate MYC degradation | [84] |
| USP28 | RCHY1 | USP28 interacts with PIRH2 to regulate CHK2 degradation in the DNA damage checkpoint | [85] |
| USP36 | FBXW7 | USP36 interacts with FBXW7 in nucleoli to regulate c-MYC degradation | [86] |
| Polyubiquitin chain editing by DUB-E3 complexes | |||
| A20 | A20 | A20 performs a polyubiquitin chain editing event on RIP, replacing K63-linked polymers with K48-linked polymers, targeting RIP for degradation | [49] |
| CYLD | ITCH | An ITCH-CYLD complex performs a polyubiquitin chain editing event on TAK1, replacing K63-linked polymers with K48-linked polymers, targeting TAK1 for degradation | [53] |
Table 3:
Examples of mutual regulation in the human DUB-E3 interaction network
| DUB | E3 | Regulatory function of complex | References |
|---|---|---|---|
| DUB-E3 interactions that promote the activity of the E3 ubiquitin ligase | |||
| CSN5 | MDM2 | CSN5 deubiquitylates MDM2, thereby regulating p53 stability | [87] |
| OTUB1 | TRAF6 | OTUB1 and OTUB2 interact with TRAF3 and TRAF6 to attenuate innate immune signaling | [57] |
| USP2 | MDM2 | USP2a regulates p53 by deubiquitylating and stabilizing MDM2 | [59] |
| USP4 | HUWE1 | USP4 deubiquitylates HUWE1 resulting in decreased p53 levels | [88] |
| USP4 | TRAF2 | USP4 deubiquitylates TRAF2 to attenuate innate immune signaling | [55] |
| USP4 | TRAF6 | USP4 deubiquitylatesTRAF6 to attenuate innate immune signaling | [55] [56] |
| USP7 | MARCH7 | USP7 deubquitylates and stabilizes MARCH7 in the nucleus | [89] |
| USP7 | MDM2 | USP7 deubiquitylates MDM2 to promote its stability and destabilize p53 | [90, 91] |
| USP7 | RAD18 | USP7 deubiquitylates RAD18 to regulate the DNA damage response | [92] |
| USP7 | RNF168 | USP7 deubiquitylates RNF168 to regulate the DNA damage response | [93] |
| USP7 | RNF169 | USP7 deubiquitylates RNF169 to promote double-strand break repair | [94] |
| USP7 | TRIM27 | USP7 deubiquitylates TRIM27 regulating WASH complex activation | [25] |
| USP7 | UHRF1 | USP7 interaction with UHRF1 promotes its association with chromatin | [95] |
| USP9X | ITCH | USP9X interacts with ITCH and antagonizes autoubiquitylation, promoting its stability | [96–98] |
| USP9X | MARCH7 | USP9X deubquitylates and stabilizes MARCH7 in the cytosol | [89] |
| USP9X | SMURF1 | USP9X interacts with SMURF and antagonizes autoubiquitylation, promoting its stability | [99] |
| USP10 | TRAF6 | USP10 interacts in a complex with TANK and MCPIP1 in order to deubiquitylate TRAF6 and attenuate NFκB activation | [100] |
| USP11 | XIAP | USP11 deubiquitylates XIAP, promoting tumor initiation | [101] |
| USP12 | MDM2 | USP12 negatively regulates p53 stability by deubiquitylating MDM2 | [61] |
| USP15 | SMURF2 | USP15 deubiquitylates SMURF2 to regulate TGFβ receptor signaling | [28, 102] |
| USP19 | XIAP | USP19 regulates the stability of XIAP, enhancing TNFα-induced caspase activation and apoptosis | [103] |
| USP20 | TRAF6 | USP20 negatively regulates NFκB activation by deubiquitylating TRAF6 | [58] |
| USP26 | MDM2 | USP26 deubiquitylates MDM2 resulting in decreased stability of p53 | [60] |
| DUB-E3 interactions that destabilize the DUB by promoting proteasomal degradation | |||
| AMSH | SMURF2 | RNF11 recruits AMSH to Smurf2 for ubiquitination, leading to its degradation by the 26S proteasome | [104] |
| UCHL1 | PARK2 | UCHL1 is ubiquitylated and degraded in a PARK2-dependent manner | [105] |
| USP5 | SMURF1 | USP5 is ubiquitylated and degraded in a USP5-dependent manner | [65] |
| USP20 | VHL | USP20 is ubiquitylated and degraded in a pVHL-dependent manner | [106] |
| USP19 | SIAH1/2 | SIAH1 and SIAH2 promote USP19 ubiquitylation and proteasomal degradation | [107] |
| USP33 | β-TrCP | USP33 is ubiquitylated and degraded in a β-TrCP-dependent manner | [108] |
| USP33 | VHL | USP33 is ubiquitylated and degraded in a pVHL-dependent manner | [106] |
| USP37 | βTrCP | SCFβTrCP ubiquitylates USP37 to promote its degradation and facilitate mitotic entry | [64] |
DUB-E3 complexes operating on a common substrate: switches, rheostats, and quality control
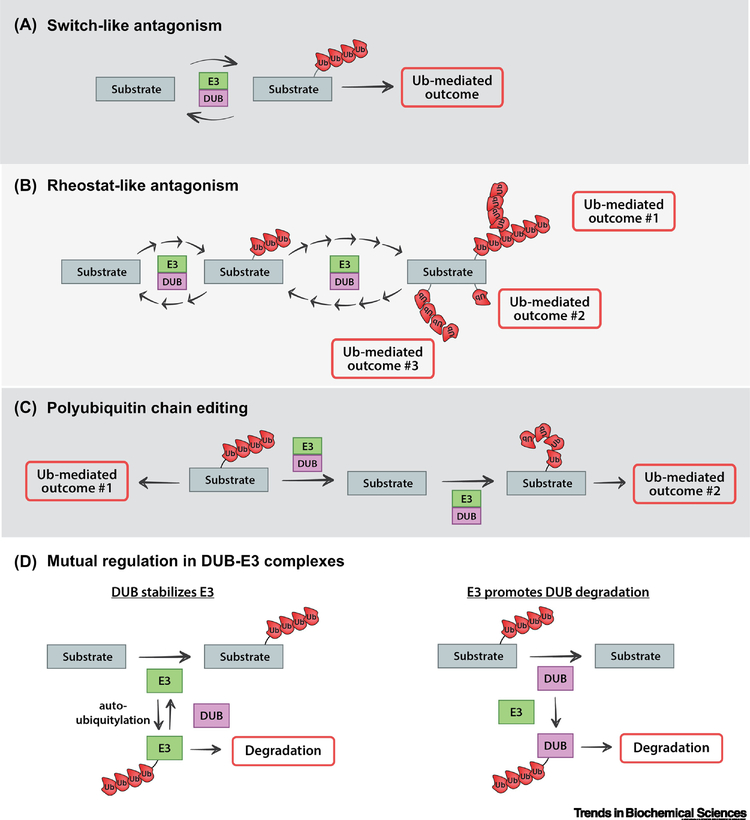
Since DUBs and E3 ubiquitin ligases catalyze opposing reactions, a classical viewpoint predicts that complexing of DUBs and E3s would result in antagonism, with the two activities competing to determine if the substrate achieves a specific ubiquitin-mediated outcome (Figure 3A). There are numerous reports of this type of switch-like regulation resulting from DUB-E3 complexes. One example involves the canonical WNT signaling pathway, which, when activated, signals to stabilize the protein β-catenin, allowing it to accumulate in the nucleus where it interacts with transcriptional regulators and modifies the transcriptional program of the cell [35]. In the absence of WNT ligand, the β-transducin repeat containing protein (βTrCP), an F-Box subunit of an SCF E3 ubiquitin ligase complex, functions to ubiquitylate β-catenin, targeting it for proteasomal degradation and preventing canonical WNT activation [35]. Importantly, the DUB USP15 can interact with βTrCP and thereby antagonize ubiquitylation of β-catenin, limiting its proteasomal degradation to promote canonical WNT activation [29]. Many other examples of such antagonism have been reported in recent years (Table 2 and references therein).
Figure 3.
The regulatory logic of coupling ubiquitin conjugation and deconjugation activities. DUB-E3 complexes can operate on a shared substrate, resulting regulation of substrate protein fate that can be switch-like or rheostat-like. (A) Switch-like behavior results from direct antagonism in a DUB-E3 complex and can lead to ubiquitin-driven outcomes that are binary in nature. In such cases, E3 activity must overcome the counter-acting DUB activity to achieve a ubiquitin-mediated outcome. (B) Rheostat-like behavior can also result from antagonism in a DUB-E3 complex when fine-tuning of E3 and DUB activities results the substrate transiting through different states with varying degrees of ubiquitylation, leading to graded responses and/or multi-variate outcomes. (C) DUB-E3 complexes can mediate polyubiquitin chain editing. This occurs when the DUB-E3 complex operates sequentially on a polyubiquitylated substrate, with the DUB activity removing the extant polyubiquitin chain and the E3 activity adding a polyubiquitin chain of a different linkage type. This type of chain remodeling can have the effect of altering the fate of the substrate protein. (D) DUB-E3 complexes can also lead to mutual regulation of DUB and E3 stability. Specifically, DUBs can protect an interacting E3 from the potential degradation that can result from autoubiquitylation (left) while an E3 can potentially ubiquitylate an interacting DUB and thereby alter its activity or promote its degradation (right).
While complexing DUBs with E3s has the potential to result in direct antagonism that generates a switch-like behavior, it also has the potential to facilitate other forms of regulation. For example, the coordinated regulation of E3 and DUB activities on a shared substrate can behave more like a rheostat to provide graded regulatory outcomes within the dynamic environment of the cell (Figure 3B). For example, the WNT signal transducing protein disheveled (DVL2) is subject to complex regulation by multiple E3 ubiquitin ligases and DUBs [32, 36–38], and it was recently reported that an interaction between USP9X and WWP1 regulates DVL2 participation in canonical WNT and non-canonical WNT signaling pathways [32]. Biochemically, these two coupled activities finely tune the extent of ubiquitylation on DVL2, which regulates not only proteasomal degradation but also DVL2 interactions with canonical WNT and non-canonical WNT factors [32]. A similar type of tuning was reported to occur on the Wiscott-Aldrich Syndrome protein family homolog (WASH) complex, which nucleates actin polymerization on endosomes and regulates endosomal protein recycling [25]. In this case, the MAGE-L2-TRIM27 E3 ubiquitin ligase complex, which contains the DUB USP7, regulates K63-linked ubiquitylation of the WASH complex. Importantly, the coupling of DUB and E3 activities in this complex contributes to a ubiquitin rheostat on the WASH complex, which is critical for fine tuning of actin polymerization on endosomal membranes [25]. Thus, ubiquitin rheostats play important roles in complex cellular processes subject to fine regulation – like localized actin polymerization or pathway specification in cell signaling.
Coupling of DUBs and E3s into protein complexes that function as switches or rheostats also plays an important role in protein quality control (QC). A looming question central to cellular protein QC relates to the ability of E3 ubiquitin ligases to successfully triage potential substrates and discriminate between folded and misfolded proteins, a non-trivial task considering the large number of potential misfolded proteins that a QC E3 may encounter. DUB-E3 interactions may facilitate such triage by allowing QC E3s to sample large numbers of potentially misfolded substrates with associated DUB activities serving to limit the commitment to degradative outcomes. Importantly, recent work has revealed that protein quality control at the endoplasmic reticulum (ER) involving ER-associated degradation (ERAD) requires not only the activity of E3 ubiquitin ligases but also the activity of associated DUBs [39–43]. Using in vitro reconstitution biochemistry and kinetic modeling to characterize how the HIV-encoded virulence factor Vpu promotes the specific degradation of host CD4 by ERAD, one study revealed that the presence of DUB activities converts subtle differences in substrate-E3 engagement into larger differences in poly-ubiquitination of the substrate [44]. Although this study does not specifically account for or model direct interactions between DUBs and E3s, it underscores the potential of DUB-E3 interactions to aid in the ability of quality control pathways to discriminate between folded and misfolded substrates [44]. Another example of the importance of DUB-E3 interactions in protein quality control involves an E3 ubiquitin ligase called autocrine motility factor receptor (AMFR) (also called gp78), which ubiquitylates ERAD pathway substrates, as well as ERAD pathway components, including UBL4A [45], a part of the BAG6 complex that helps chaperone retrotranslocated ERAD substrates to the proteasome for degradation [46]. Importantly, USP13 associates with AMFR to prevent polyubiquitylation of UBL4A, protecting the BAG6 complex from proteasomal degradation and maintaining proper ERAD pathway function [45]. Thus, the association of USP13 with AMFR prevents collateral damage in the ERAD pathway by promoting the distinction between bona fide ERAD substrates and protein complexes that ERAD pathway E3s must engage to ensure proper substrate shuttling for degradation. In yeast, DUB-E3 complexes appear to play a similar role, as evidenced by the interaction of Ufd3 (a ubiquitin ligase) and Otu1 (a DUB) on Cdc48 complexes that regulate proteasomal degradation of substrates [47]. These findings suggest that an important and evolutionarily conserved function of DUB-E3 coupling may relate to protein quality control and the discrimination between folded and misfolded substrates.
DUBs and E3s working together to edit polyubiquitin chains
One interesting and poorly-understood example of a regulatory function for DUB-E3 complexes involves ubiquitin chain editing. In ubiquitin chain editing, a DUB removes polyubiquitin of a specific linkage-type on a substrate and an E3 subsequently adds a different polyubiquitin chain, resulting in a net change of linkage type and an altered fate for the substrate (Figure 3C and Table 2). One example of ubiquitin chain editing involves a key attenuator of NFκB signaling called A20, a multi-functional protein that contains both a DUB domain and an E3 ubiquitin ligase domain, and its substrate receptor interacting protein (RIP), a signal transducing protein that associates with the tumor necrosis factor (TNF) receptor 1 (TNFR1) signaling complex [48, 49].The TNF receptor signaling pathway is an upstream activator of NκFB signaling, and the active TNFR1 signaling complex is stabilized by K63-linked polyubiquitylation of RIP [48, 50]. To attenuate NFκB signaling, the DUB domain of A20 removes K63-linked polyubiquitin chains from RIP and the E3 ligase domain of A20 subsequently conjugates K48-linked polyubiquitin chains to RIP, targeting it for proteasomal degradation and thus inhibiting NFĸB signaling [49].
Another example of ubiquitin chain editing within the NFκB signaling pathway involves the kinase TAK1, another signal transducing protein that can be activated by TNFR signaling to promote NκFB activation. Upon TNF activation, TAK1 undergoes K63-linked polyubiquitylation via the activity of TRAF2/TRAF6, and this K63-polyubiquitination is required for NκFB activation [51, 52]. Interaction between the NEDD4 family E3 ubiquitin ligase ITCH and the DUB CYLD provides a key attenuation step for this level of the inflammatory response, with CYLD removing K63-linked polyubiquitin chains from active TAK1 and ITCH catalyzing the formation of K48-linked polyubiquitin chains onto TAK1, targeting it for degradation by the proteasome [53]. Only a few examples of such polyubiquitin chain editing complexes have been reported in the literature – perhaps owing to the difficulty associated with their discovery and biochemical characterization. However, given the emerging complexity of polyubiquitylation – particularly with chain branching and layered post-translational modifications – we predict that chain editing on substrates will emerge as a key feature of DUB-E3 complexes.
Mutual Regulatory Circuits in DUB-E3 complexes
There are many examples of E3 ubiquitin ligases performing autoubiquitylation as a form of self-regulation to achieve negative feedback [54]. Therefore, it is not surprising that one function of DUB-E3 complexes appears to be to promote E3 stability or function by antagonizing autoubiquitylation (Figure 3D and Table 3). For example, several DUBs, including USP4 [55, 56], OTUB1 [57], and USP20 [58], are reported to interact with and deubiquitylate autoubiquitylated TRAF6, an important attenuation step in the NFκB pathway. Additionally, several DUBs, including USP2 [59], USP26 [60], and USP12 [61], operate on autoubiquitylated MDM2 to promote its stability and effectively antagonize p53. Further, the yeast DUB Ubp2 has long been known to interact with and antagonize the activity of Rsp5 by reversing ubiquitylation of its substrates [33, 34], but more recent studies have implicated Ubp2 in the stabilization of adaptors required for Rsp5 activity [62]. In these instances, rather than antagonizing E3 activity, DUB interactions can positively regulate E3 activity by antagonizing autoubiquitylation and promoting E3 stability. Thus, without empirical evidence, it is difficult to predict if a DUB-E3 interaction will stabilize or de-stabilize a given E3 substrate.
There are also examples of E3 ubiquitin ligases interacting with DUBs to promote their ubiquitylation, thereby regulating DUB stability or activity (Figure 3D and Table 3). The DUB USP37 has been implicated as a key cell cycle regulator, controlling both the G1/S transition (by operating on cyclin A) [63] and mitotic entry [64]. This regulation is achieved, in part, by association with the E3 ubiquitin ligases APCCDH1 and SCFβTrCP which drives the ubiquitylation and degradation of USP37 [64], although how USP37 degradation regulates mitotic entry remains to be elucidated. USP5 is a DUB known to play a key role in promoting the production of TNF-α during the inflammatory response, and its interaction with the NEDD4 family E3 ubiquitin ligase SMURF1 promotes its ubiquitylation and proteasomal degradation and thus attenuates TNF-α production [65]. From these examples, it is clear that the regulation occurring in DUB-E3 complexes can go both ways, depending on the context.
DUB-E3 complex logic: the system is baroque
The regulatory logic of DUB-E3 complexes becomes even more complex when considering that the scenarios outlined above, including operation on a shared substrate to achieve switches, rheostats, or ubiquitin chain editing, and mutual regulatory circuits, are not mutually exclusive. Indeed, many characterized DUB-E3 complexes exhibit properties that correspond to different modes of regulation. For example, in addition to functioning to finely tune ubiquitylation of the WASH complex, USP7 also helps to stabilize the MAGE-L2-TRIM27 E3 ubiquitin ligase complex by antagonizing TRIM27 autoubiquitylation [66]. Because of this, we predict that many DUB-E3 complexes will exhibit multi-faceted regulatory capabilities incorporating many of the elements discussed above. This complexity is compounded even further when considering that multiple E3s or multiple DUBs may be associating with each other or shared substrates. For example, the DUB USP2a interacts with and deubiquitylates MDM2 in order to promote MDM2 stability and p53 degradation [59]. This differs from USP7, which can deubiquitylate both MDM2 and p53 in a context-dependent manner and can thus function to promote or antagonize p53 stability [26]. Thus, in some cases, multiple DUBs can interact with the same substrate to elicit different outcomes.
The functional significance of a DUB-E3 complex can also shift in a context-dependent manner. A clear example that highlights this phenomenon involves USP7, a DUB known to interact with and regulate multiple E3 ubiquitin ligases (Figure 1A, Figure 2A, and Table S1) [26]. One well-characterized regulatory interactor of USP7 is the E3 ubiquitin ligase MDM2. Under normal conditions, MDM2 functions to regulate levels of the transcription factor p53 [67]; in these conditions, USP7 interacts in a complex with MDM2 and p53 to reverse autoubiquitylation events on MDM2 and ultimately drive p53 degradation. However, under stress conditions, USP7 preferentially deubiquitylates and stabilizes p53 in order to activate an apoptotic pathway [26]. This switch in USP7 function results from altered associations with regulatory proteins under stress conditions [68]. Similarly, USP7 complexed with the E3 UHRF1 and its substrate DNA methyltransferase-1 (DNMT1) can function to deubiquitylate and stabilize UHRF1 or DNMT1 in a manner that is cell-cycle dependent [69]. Based on these examples, it will be important to determine if other functions of USP7 (like regulation of WASH-mediated actin polymerization [25]) also involve context-dependent toggling between operating on an E3 (e.g., MAGE-L2-TRIM27) and on a shared substrate (e.g., the WASH complex). These examples illustrate that the regulatory outputs of DUB-E3 complexes can be multi-faceted, context-dependent, and regulated in response to the cell cycle or conditions of stress.
Concluding Remarks
All of the components of the ubiquitin system contribute to a highly dynamic ubiquitin code with layers of complexity that are still not fully understood. This complexity is ultimately managed by coordination of the conjugation and deconjugation machinery, which appears to be achieved, at least in part, by interactions between E3 ubiquitin ligases and DUBs. From the examples discussed in this review, it is evident that DUB-E3 complexes play important roles in many complex biological processes, including the regulation of cell signaling and protein quality control. While it is clear that DUB-E3 interactions have the potential to provide many layers of regulation within the ubiquitin network, there are many known DUB-E3 interactions that have not been characterized functionally or biochemically. Thus, given the potential for artifacts leading to false-positive interactions, many of the reported DUB-E3 interactions (Table S1) (especially those reported in high-throughput studies) will require rigorous biochemical validation alongside substantial evidence that such interactions have regulatory significance. Considering that many different DUBs and E3s are currently being investigated as potential therapeutic targets for various diseases, further investigation of DUB-E3 complexes and their regulatory function in cells will have important implications for targeting DUBs and E3s in drug development (see Outstanding Questions).
OUTSTANDING QUESTIONS.
How are DUB-E3 complexes leveraged in quality control pathways? How does the balance of activities in DUB-E3 complexes facilitate discrimination between folded and misfolded substrates in ubiquitylation pathways tasked with protein quality maintenance in the cell?
To what extent does DUB-E3 coupling contribute to the complexity of polyubiquitin chains? Do DUB-E3 interactions facilitate (or restrict) the formation of branched polymers? Do post-translational modifications of ubiquitin (i.e., phosphorylation, acetylation) contribute to the tuning and balance of coupled DUB-E3 activities?
What is the structural basis for DUB-E3 coupling throughout the ubiquitin system? What biochemical scaffolds are used to facilitate these interactions on a broad scale? What is the stoichiometry of these interactions in a cellular context?
In consideration of DUBs and E3s as potential drug targets – how do DUB-E3 interactions affect the outcomes associated with inhibiting a specific DUB or E3? For example, does pharmacological inhibition of a DUB (counter-intuitively) trigger stabilization of protein substrates - based on autoubiquitylation and degradation of an interacting E3?
Why is the coupling of antagonistic activities so prevalent in the ubiquitin system compared to other regulatory networks (e.g., phosphorylation/dephosphorylation)? What regulatory features are enabled by antagonistic coupling? What other networks operate with broad coupling of antagonistic activities?
To our knowledge, the coupling of antagonistic activities is not a prevalent feature of other post-translational regulatory networks. For example, protein kinases and phosphatases do not commonly interact in complexes; this may be related to the inherently binary and digital nature of phosphorylation (compared to the more analog nature of ubiquitin modification). One interesting example of coupled antagonistic activities occurs in phosphatidylinositol phosphorylation where the kinase that generates PI(3,5)P2 on the vacuole/lysosome associates with the PI(3,5)P2 phosphatase FIG4 [70, 71]. Interestingly, this complex is conserved from yeast to man and the presence of the phosphatase is required for kinase activity, as well as conversion of PI(3,5)P2 to PI(3)P [70, 72, 73]. While the regulatory significance of this complex is still being investigated, we propose that such coupling of antagonistic activities will occur in other regulatory networks. Thus, the regulatory logic of DUB-E3 complexes has the potential to inform other regulatory networks that also rely on coupling of antagonistic activities.
Supplementary Material
HIGHLIGHTS.
Physical interactions between the ubiquitin conjugation (E3 ubiquitin ligases) and deconjugation (deubiquitylases; DUBs) machineries are prevalent in eukaryotic cells and contribute to the regulation of complex signaling networks and protein quality control.
DUB-E3 interactions can result in various regulatory outcomes, including ubiquitin switches and rheostats, polyubiquitin chain editing on a shared substrate, or mutual regulation of ubiquitylation and stability.
The regulatory logic of DUB-E3 interactions is complex, and how such an interaction contributes to the outcome of a substrate is difficult to predict.
ACKNOWLEDGEMENTS
We are very grateful to T. Graham for thoughtful comments and critical reading of this manuscript. We are also grateful to E. MacGurn for help with graphic design. J.A.M. is grateful for support from the NIH (R00GM101077, R21AG053562 and R01GM118491) and support from Vanderbilt University.
GLOSSARY
- Canonical WNT signaling
a signaling pathway that senses extracellular WNT ligands and transduces signals to regulate the stability of β-catenin. When stabilized, β-catenin translocates into the nucleus to regulate transcription
- COP9 Signalosome
a multiprotein complex with isopeptidase activity that catalyzes the de-conjugation of NEDD8 (a UBL) from the cullin subunit of SCF E3 ubiquitin ligases. Such NEDD8 conjugation/deconjugation cycles are critical regulators of SCF function in cells
- Endosomes
membrane-bound organelles that often serve as intermediate sorting compartments for trafficking of proteins between the Golgi complex, the plasma membrane, and the lysosome
- Endoplasmic reticulum associated degradation (ERAD)
a ubiquitin-dependent quality control pathway that targets misfolded endoplasmic reticulum proteins for degradation by the proteasome
- NEDD4 family E3 ubiquitin ligases
An evolutionarily conserved family of E3 ubiquitin ligases containing a C-terminal HECT catalytic domain, an N-terminal C2 domain that binds to lipids, and multiple internal WW domains that function in protein interaction scaffolding
- NFκB signaling pathway
NFκB is a transcription factor that activates various immune responses, including the inflammatory response. In the absence of pathway activation, an inhibitory factor called IκB binds and sequesters NFκB in the cytosol. Many different inputs can activate the ubiquitin-dependent degradation of IκB, resulting in translocation of NFκB into the nucleus and transcriptional activation of its targets
- Non-canonical WNT signaling
pathways that sense extracellular WNT ligands and transduce signals into the cytosol independent of the regulation of β-catenin stabilization
- Rheostat
in contrast to “on/off” switches, molecular rheostats are mechanisms for graded regulation of protein function. Molecular rheostats allow for fine tuning of cellular responses, in contrast to the binary nature of an on/off switch
- SCF E3 ubiquitin ligases
Skp-Cullin-F-box complexes are a large family of E3 ubiquitin ligases that share a common modular assembly based on the scaffolding properties of cullin family proteins. SCF complexes bring E2-ubiquitin complexes into close proximity with substrates, thus catalyzing the formation of an isopeptide bond between ubiquitin and a substrate
- SUMO
small ubiquitin-like modifier is a UBL that can be covalently conjugated to substrates to regulate protein function
- TGF-β receptor signaling pathway
transforming growth factor beta is a cytokine that bind and activate surface TGF-β receptors, which are receptor protein kinases that activate a downstream signaling cascade which can regulate various cellular processes including proliferation and differentiation
- TNF receptor signaling pathway
Tumor Necrosis Factor is a cytokine sensed by cell surface receptors that signal to regulate a variety of immune responses
- Ubiquitin code
Ubiquitin conjugation can result in polymers of varying lengths containing linkages at any one of seven internal lysine residues, or the N-terminus. Polymers can be homogenous, contained mixed linkages, or can be branched with multiple linkages arising from a single ubiquitin moiety. Furthermore, ubiquitin itself is subject to post-translational modifications such as phosphorylation or acetylation. All of this leads to a complex code, that ultimately modifies protein function and stability according to the interactions a polymer facilitates with ubiquitin binding domains
- UBL
a protein or domain that contains a ubiquitin-like fold. Some UBL proteins (e.g., SUMO) can undergo conjugation and deconjugation biochemistry, while other UBLs contribute to non-covalent regulation of protein function
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Reyes-Turcu FE et al. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78, 363–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petroski MD and Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 (1), 9–20. [DOI] [PubMed] [Google Scholar]
- 3.MacGurn JA et al. (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem 81, 231–59. [DOI] [PubMed] [Google Scholar]
- 4.Skaar JR et al. (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14 (6), 369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston GM and Brodsky JL (2017) The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem J 474 (4), 445–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacGurn JA (2014) Garbage on, garbage off: new insights into plasma membrane protein quality control. Curr Opin Cell Biol 29C, 92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deol KK et al. (2019) Enzymatic Logic of Ubiquitin Chain Assembly. Front Physiol 10, 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mevissen TET and Komander D (2017) Mechanisms of Deubiquitinase Specificity and Regulation. Annu Rev Biochem 86, 159–192. [DOI] [PubMed] [Google Scholar]
- 9.Komander D et al. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10 (8), 550–63. [DOI] [PubMed] [Google Scholar]
- 10.Sahtoe DD and Sixma TK (2015) Layers of DUB regulation. Trends Biochem Sci 40 (8), 456–67. [DOI] [PubMed] [Google Scholar]
- 11.Swatek KN et al. (2019) Insights into ubiquitin chain architecture using Ub-clipping. Nature 572 (7770), 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer HJ and Rape M (2014) Enhanced protein degradation by branched ubiquitin chains. Cell 157 (4), 910–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L and Luo ZQ (2019) Post-translational regulation of ubiquitin signaling. J Cell Biol 218 (6), 1776–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swatek KN and Komander D (2016) Ubiquitin modifications. Cell Res 26 (4), 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdul Rehman SA et al. (2016) MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell 63 (1), 146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haahr P et al. (2018) ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol Cell 70 (1), 165–174.e6. [DOI] [PubMed] [Google Scholar]
- 17.Hermanns T et al. (2018) A family of unconventional deubiquitinases with modular chain specificity determinants. Nat Commun 9 (1), 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwasna D et al. (2018) Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol Cell 70 (1), 150–164.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshaies RJ and Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78, 399–434. [DOI] [PubMed] [Google Scholar]
- 20.Finley D et al. (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192 (2), 319–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srikumar T et al. (2013) A global S. cerevisiae small ubiquitin-related modifier (SUMO) system interactome. Mol Syst Biol 9, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawat R et al. (2019) Nuclear deubiquitination in the spotlight: the multifaceted nature of USP7 biology in disease. Curr Opin Cell Biol 58, 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng Y et al. (2006) Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol 13 (3), 285–91. [DOI] [PubMed] [Google Scholar]
- 24.Hu M et al. (2006) Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol 4 (2), e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao YH et al. (2015) USP7 Acts as a Molecular Rheostat to Promote WASH-Dependent Endosomal Protein Recycling and Is Mutated in a Human Neurodevelopmental Disorder. Mol Cell 59 (6), 956–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim RQ and Sixma TK (2017) Regulation of USP7: A High Incidence of E3 Complexes. J Mol Biol 429 (22), 3395–3408. [DOI] [PubMed] [Google Scholar]
- 27.Chou CK et al. (2017) The Regulations of Deubiquitinase USP15 and Its Pathophysiological Mechanisms in Diseases. Int J Mol Sci 18 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichhorn PJ et al. (2012) USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat Med 18 (3), 429–35. [DOI] [PubMed] [Google Scholar]
- 29.Greenblatt MB et al. (2016) MEKK2 mediates an alternative β-catenin pathway that promotes bone formation. Proc Natl Acad Sci U S A 113 (9), E1226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hetfeld BK et al. (2005) The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol 15 (13), 1217–21. [DOI] [PubMed] [Google Scholar]
- 31.Schweitzer K et al. (2007) CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J 26 (6), 1532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen CP et al. (2019) USP9X Deubiquitylates DVL2 to Regulate WNT Pathway Specification. Cell Rep 28 (4), 1074–1089.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kee Y et al. (2005) The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J 24 (13), 2414–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kee Y et al. (2006) The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem 281 (48), 36724–31. [DOI] [PubMed] [Google Scholar]
- 35.Marikawa Y and Elinson RP (1998) beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech Dev 77 (1), 75–80. [DOI] [PubMed] [Google Scholar]
- 36.Angers S et al. (2006) The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wntbeta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol 8 (4), 348–57. [DOI] [PubMed] [Google Scholar]
- 37.Tauriello DV et al. (2010) Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell 37 (5), 607–19. [DOI] [PubMed] [Google Scholar]
- 38.Ding Y et al. (2013) HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J Biol Chem 288 (12), 8289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst R et al. (2009) The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell 36 (1), 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q et al. (2006) Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol 174 (7), 963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong X and Pittman RN (2006) Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet 15 (16), 2409–20. [DOI] [PubMed] [Google Scholar]
- 42.Blount JR et al. (2012) Ubiquitin-specific protease 25 functions in Endoplasmic Reticulumassociated degradation. PLoS One 7 (5), e36542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson BG et al. (2019) Cycles of autoubiquitination and deubiquitination regulate the ERAD ubiquitin ligase Hrd1. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang ZR et al. (2013) Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell 154 (3), 609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y et al. (2014) USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation. Elife 3, e01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q et al. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell 42 (6), 758–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rumpf S and Jentsch S (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell 21 (2), 261–9. [DOI] [PubMed] [Google Scholar]
- 48.Wertz I and Dixit V (2014) A20--a bipartite ubiquitin editing enzyme with immunoregulatory potential. Adv Exp Med Biol 809, 1–12. [DOI] [PubMed] [Google Scholar]
- 49.Wertz IE et al. (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430 (7000), 694–9. [DOI] [PubMed] [Google Scholar]
- 50.Harhaj EW and Dixit VM (2012) Regulation of NF-κB by deubiquitinases. Immunol Rev 246 (1), 107–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan Y et al. (2010) Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J Biol Chem 285 (8), 5347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Y et al. (2011) TAK1 Lys-158 but not Lys-209 is required for IL-1β-induced Lys63-linked TAK1 polyubiquitination and IKK/NF-κB activation. Cell Signal 23 (4), 660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmed N et al. (2011) The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol 12 (12), 1176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Bie P and Ciechanover A (2011) Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ 18 (9), 1393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao N et al. (2012) Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFα-induced cancer cell migration. Biochem J 441 (3), 979–86. [DOI] [PubMed] [Google Scholar]
- 56.Zhou F et al. (2012) Ubiquitin-specific protease 4 mitigates Toll-like/interleukin-1 receptor signaling and regulates innate immune activation. J Biol Chem 287 (14), 11002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S et al. (2010) Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem 285 (7), 4291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasunaga J et al. (2011) Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-kappaB signaling. J Virol 85 (13), 6212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevenson LF et al. (2007) The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J 26 (4), 976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lahav-Baratz S et al. (2017) The testis-specific USP26 is a deubiquitinating enzyme of the ubiquitin ligase Mdm2. Biochem Biophys Res Commun 482 (1), 106–111. [DOI] [PubMed] [Google Scholar]
- 61.McClurg UL et al. (2018) Molecular mechanism of the TP53-MDM2-AR-AKT signalling network regulation by USP12. Oncogene 37 (34), 4679–4691. [DOI] [PubMed] [Google Scholar]
- 62.Ho HC et al. (2017) Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs. Mol Biol Cell 28 (9), 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang X et al. (2011) Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell 42 (4), 511–23. [DOI] [PubMed] [Google Scholar]
- 64.Burrows AC et al. (2012) Skp1-Cul1-F-box ubiquitin ligase (SCF(βTrCP))-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. J Biol Chem 287 (46), 39021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qian G et al. (2016) Smurf1 represses TNF-α production through ubiquitination and destabilization of USP5. Biochem Biophys Res Commun 474 (3), 491–496. [DOI] [PubMed] [Google Scholar]
- 66.Hao YH et al. (2013) Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152 (5), 1051–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prives C (1998) Signaling to p53: breaking the MDM2-p53 circuit. Cell 95 (1), 5–8. [DOI] [PubMed] [Google Scholar]
- 68.Tavana O and Gu W (2017) Modulation of the p53/MDM2 interplay by HAUSP inhibitors. J Mol Cell Biol 9 (1), 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Felle M et al. (2011) The USP7/Dnmt1 complex stimulates the DNA methylation activity of Dnmt1 and regulates the stability of UHRF1. Nucleic Acids Res 39 (19), 8355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Botelho RJ et al. (2008) Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell 19 (10), 4273–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin N et al. (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J 27 (24), 3221–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duex JE et al. (2006) Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 5 (4), 723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duex JE et al. (2006) The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol 172 (5), 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu H et al. (2008) High-quality binary protein interaction map of the yeast interactome network. Science 322 (5898), 104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buser R et al. (2016) The Replisome-Coupled E3 Ubiquitin Ligase Rtt101Mms22 Counteracts Mrc1 Function to Tolerate Genotoxic Stress. PLoS Genet 12 (2), e1005843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lam MH et al. (2009) Interaction of the deubiquitinating enzyme Ubp2 and the e3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS One 4 (1), e4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hesselberth JR et al. (2006) Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol 7 (4), R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li L et al. (2010) The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res 16 (11), 2949–58. [DOI] [PubMed] [Google Scholar]
- 79.Karim R et al. (2013) Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PLoS Pathog 9 (5), e1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guturi KKN et al. (2016) RNF168 and USP10 regulate topoisomerase IIα function via opposing effects on its ubiquitylation. Nat Commun 7, 12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jahan AS et al. (2016) Usp12 stabilizes the T-cell receptor complex at the cell surface during signaling. Proc Natl Acad Sci U S A 113 (6), E705–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moretti J et al. (2012) The ubiquitin-specific protease 12 (USP12) is a negative regulator of notch signaling acting on notch receptor trafficking toward degradation. J Biol Chem 287 (35), 29429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jean-Charles PY et al. (2016) Ubiquitin-specific Protease 20 Regulates the Reciprocal Functions of β-Arrestin2 in Toll-like Receptor 4-promoted Nuclear Factor κB (NFκB) Activation. J Biol Chem 291 (14), 7450–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Popov N et al. (2007) The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol 9 (7), 765–74. [DOI] [PubMed] [Google Scholar]
- 85.Bohgaki M et al. (2013) The E3 ligase PIRH2 polyubiquitylates CHK2 and regulates its turnover. Cell Death Differ 20 (6), 812–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun XX et al. (2015) The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci U S A 112 (12), 3734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang XC et al. (2008) Roles for CSN5 in control of p53/MDM2 activities. J Cell Biochem 103 (4), 1219–30. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X et al. (2011) USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J 30 (11), 2177–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nathan JA et al. (2008) The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9 (7), 1130–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meulmeester E et al. (2005) Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 18 (5), 565–76. [DOI] [PubMed] [Google Scholar]
- 91.Li M et al. (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13 (6), 879–86. [DOI] [PubMed] [Google Scholar]
- 92.Zlatanou A et al. (2016) USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene 35 (8), 965–76. [DOI] [PubMed] [Google Scholar]
- 93.Zhu Q et al. (2015) USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168. Cell Cycle 14 (9), 1413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.An L et al. (2017) Dual-utility NLS drives RNF169-dependent DNA damage responses. Proc Natl Acad Sci U S A 114 (14), E2872–E2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang ZM et al. (2015) An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell Rep 12 (9), 1400–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mouchantaf R et al. (2006) The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem 281 (50), 38738–47. [DOI] [PubMed] [Google Scholar]
- 97.O’Connor HF et al. (2015) Ubiquitin-Activated Interaction Traps (UBAITs) identify E3 ligase binding partners. EMBO Rep 16 (12), 1699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Azakir BA and Angers A (2009) Reciprocal regulation of the ubiquitin ligase Itch and the epidermal growth factor receptor signaling. Cell Signal 21 (8), 1326–36. [DOI] [PubMed] [Google Scholar]
- 99.Xie Y et al. (2013) Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem 288 (5), 2976–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang W et al. (2015) TRAF Family Member-associated NF-κB Activator (TANK) Inhibits Genotoxic Nuclear Factor κB Activation by Facilitating Deubiquitinase USP10-dependent Deubiquitination of TRAF6 Ligase. J Biol Chem 290 (21), 13372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Z et al. (2017) Regulation of XIAP Turnover Reveals a Role for USP11 in Promotion of Tumorigenesis. EBioMedicine 15, 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iyengar PV et al. (2015) USP15 regulates SMURF2 kinetics through C-lobe mediated deubiquitination. Sci Rep 5, 14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mei Y et al. (2011) The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J Biol Chem 286 (41), 35380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H and Seth A (2004) An RNF11: Smurf2 complex mediates ubiquitination of the AMSH protein. Oncogene 23 (10), 1801–8. [DOI] [PubMed] [Google Scholar]
- 105.McKeon JE et al. (2015) Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell Mol Life Sci 72 (9), 1811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Z et al. (2002) Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun 294 (3), 700–9. [DOI] [PubMed] [Google Scholar]
- 107.Velasco K et al. (2013) An N-terminal SIAH-interacting motif regulates the stability of the ubiquitin specific protease (USP)-19. Biochem Biophys Res Commun 433 (4), 390–5. [DOI] [PubMed] [Google Scholar]
- 108.Cheng Q et al. (2017) Deubiquitinase USP33 is negatively regulated by β-TrCP through ubiquitin-dependent proteolysis. Exp Cell Res 356 (1), 1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.