Abstract
Streptomyces is an important treasure trove for natural products discovery. In recent years, many scientists focused on the genetic modification and metabolic regulation of Streptomyces to obtain diverse bioactive compounds with high yields. This review summarized the commonly used regulatory strategies for natural products discovery and overproduction in Streptomyces from three main aspects, including regulator-related strategies, promoter engineering, as well as other strategies employing transposons, signal factors, or feedback regulations. It is expected that the metabolic regulation network of Streptomyces will be elucidated more comprehensively to shed light on natural products research in the future.
Keywords: Gene expression regulation, Natural products, Streptomyces, Biosynthetic gene clusters
1. Introduction
Natural products have served as important raw materials for the pharmaceutical industry for centuries [1]. Semi-synthetic natural products derivatives and synthetic analogs are widely used to treat various clinical diseases, including infectious diseases, cancer, high cholesterol, and transplant rejection [2]. Streptomyces, belonging to the Actinomycetes family, are gram-positive bacterium serving as a rich reservoir for natural products discovery [3,4], such as anthraquinones (e.g., adriamycin), lactones (e.g., rapamycin), and flavonoids (e.g., O-methylated phenylpropanoids) [5]. Since Streptomyces can produce a variety of bioactive natural products, the isolation of fermentation products from Streptomyces have attracted great attentions. However, the life cycle of Streptomyces is complicated, and their morphological differentiation is accompanied by complex physiological changes [6], indicating that the cell metabolism in Streptomyces is under sophisticated regulations.
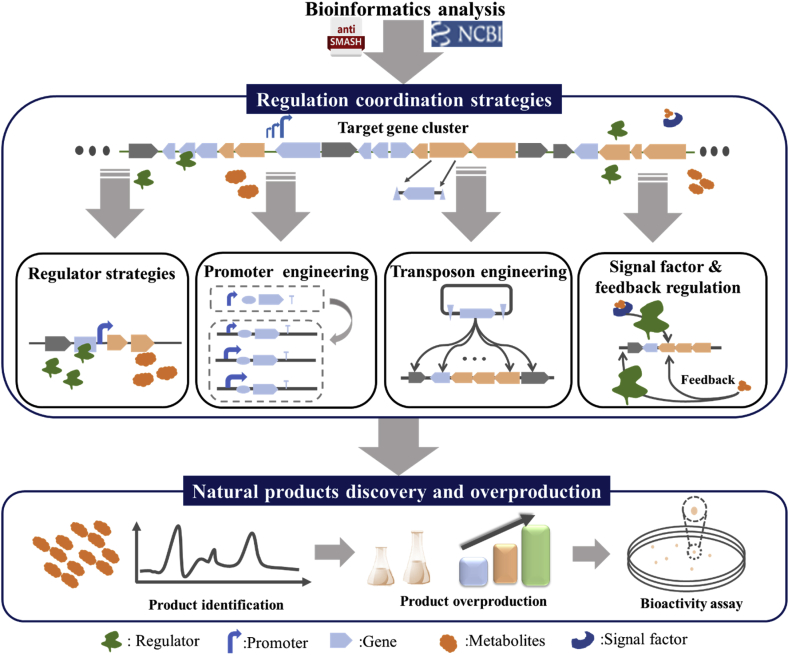
Significant advances in next-generation sequencing technology reveal that Streptomyces is a rich resource for natural products discovery [[7], [8], [9]]. It also discloses that most secondary metabolites gene clusters in Streptomyces are unexpressed or low-expressed under standard laboratory fermentation conditions [2,[10], [11], [12]]. The expression of these gene clusters in Streptomyces is governed by complex and delicate regulatory networks [11,13]. At present, these complex regulatory networks have not been clearly illustrated, which is an obstacle to the excavation of natural products in Streptomyces. After meticulous analysis via bioinformatics tools, researchers have attempted to coordinate the regulatory network by engineering regulatory elements, such as the regulators, promoters, ribosome binding sites, and terminators [14,15]. This review summarized several regulation strategies applied in natural products discovery and overproduction in Streptomyces (Fig. 1). First, strategies employing up-regulation and down-regulation of regulators in Streptomyces are summarized. Then, promoter engineering strategies applied in Streptomyces for natural products discovery were discussed. Finally, we probed into other regulatory engineering methods, such as the utilization of transposons, signal factors, and the feedback regulations. These strategies have efficiently promoted the overproduction of known natural products, as well as the discovery of novel natural products in Streptomyces.
Fig. 1.
Regulation coordination strategies for nature products discovery and overproduction in Streptomyces.
2. Up and down regulation of the regulators
The transcription and translation processes guide the conversion of information from DNA molecules to functional proteins, which is precisely regulated in vivo to ensure the orderliness of cell metabolism. The coordination of important cellular process, such as osmotic pressure related transportation, catabolic process, differentiation, and the expression of natural product gene clusters, depends on complex interactions among various regulatory elements. Regulators are proteins that can directly or indirectly recognize or bind to the cis-acting elements and participate in regulating the transcription activity of target genes. The TetR family regulator is the most widely distributed family in Streptomyces [16]. In addition, there are regulators from other families such as the MarR family, LuxR family, ArgR family and so on. Here, we summarized the regulation strategies based on the regulators.
2.1. Positive regulation strategies
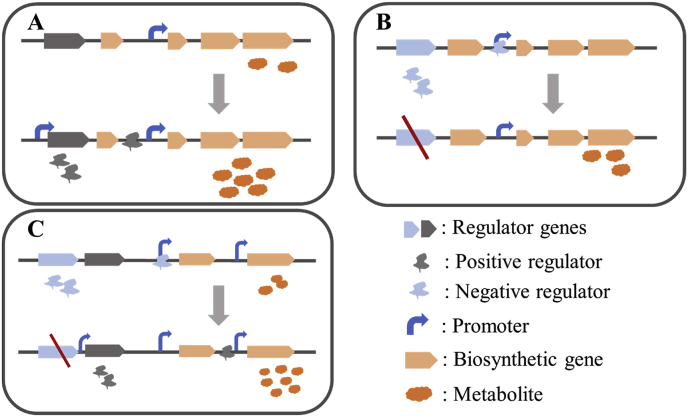
Production of secondary metabolites in Streptomyces is regulated by complex regulatory systems. Among them, the expression of some regulators is positively related to the yield of natural products, which are often called as positive regulators. Overexpression of positive regulators is a commonly used strategy to increase the production of secondary metabolites (Fig. 2A). In 2019, Xu et al. identified a regulator ToyA from the LuxR family, which directly activated the expression of the toyB and toyE operons [17]. They used promoters of different strengths to control the expression of ToyA in S. disatotochromogenes 1628. When promoters SPL57, SPL21, and ermE*p were used, the yield of toyocamycin was 2-fold, 1-fold, and 0.8-fold greater than that of the wild type strain, respectively. In order to understand the biosynthesis of lincomycin, Hou et al. identified a new regulator LmbU in S. lincolnensis NRRL 2936, which promotes the biosynthesis of lincomycin by regulating the transcription of key biosynthetic genes [18]. In another case, after the CtcS regulator knockout, the transcription of several biosynthetic genes was altered, leading to reduced production of tetracycline and chlorotetracycline, which indicates that CtcS is a positive regulator [19]. Chen et al. separately overexpressed genes orf 22 and orf 42 in Streptomyces fungicidicus ATCC 31731, which increased enduracidins titers by about 4-fold and 2.3-fold, respectively [20].
Fig. 2.
Schematic diagram of positive and negative regulations. (A) positive regulation strategy: overexpression of positive regulators to promote the yield of metabolites; (B) negative regulation strategy: inhibition of the negative regulator expression to release the production of metabolites; (C) Combinatorial regulation strategy: coordination of positive and negative regulators to increase the yield of metabolites.
The positive regulators can not only regulate the expression of single gene, but can also coordinate the expression of multiple genes in some cases. A positive regulator HcdR2 belonging to the LuxR family can significantly increase the yield of herbicidin F by enhancing the transcription of several structural genes as well as transporters in the herbicidin biosynthetic gene cluster [21]. Under other circumstances, one regulator can take effects in different strains. For example, Liu et al. introduced the pluripotency regulator gene adpa into Streptomyces ZYJ-6, leading to the highest reported sugar production of 9338 μg/mL [22]. The AdpA regulator was also used in the wild-type S. griseus strain to increase the yield of streptomycin [22], as well as in S. diastatochromogenes 1628 to increase the production of toyocamycin by 120.1% [23].
In addition to increasing the production of secondary metabolites, regulators also show considerable potential to activate silent gene clusters. Kang et al. found that the absence of AdpAlin, a pleiotropic transcriptional regulator of S. lincolnensis NRRL 2936, interrupted the biosynthesis of lincomycin [24]. At the same year, Fu et al. confirmed that the CepR regulator served as a transcriptional activator of cephamycin C biosynthesis, which provided a theoretical basis for clavulanic acid production in S. clavuligerus F613-1 [25]. Generally, the activation of the putative operon is achieved by directing regulators to the corresponding promoter regions. For example, StaR is a regulator of the LuxR family, which activates staurosporine biosynthesis by binding to the promoter regions of staO-staC and staG-staN [26]. In another case, a TetR family regulator MilR2 was shown to be involved in the biosynthesis of 5-oxomilbemycin A3/A4 in S. hygroscopicus. Further studies revealed that MilR2 serves as an activator for 5-oxomilbemycin A3/A4 production, and its function is mediated by suppressing the transcription of its upstream hydrolase gene [27]. In another case, researchers overexpressed the PAS regulator from the LuxR family in S. clavuligerus ATCC 27064, which activated the biosynthesis of polyene macrolide antifungal drugs and other antibiotics. Moreover, the production of clavulanic acid, cephalosporin C, and tunicamycin complexes increased by 10, 7, and 5-fold, respectively [28]. Besides, different regulators can coordinate together to activate the expression of cryptic secondary metabolite gene clusters. Bu et al. identified three regulatory factors, Sxim22880, CVNABCSX, and WblASX in Streptomyces FR-008 and S. albus J1074, whose overexpression stimulated the production of new secondary metabolites, revealing the potential of these conserved regulators in activating the recessive secondary metabolite gene cluster in Streptomyces [29].
2.2. Negative regulation strategies
Apart from positive regulators, negative regulators also involve in the biosynthesis of numerous secondary metabolites in microorganisms (Fig. 2B). Negative effects of these regulators were often relieved by suppressing or removing these negative regulators. For instance, Planckaert et al. confirmed that the regulator CebR could inhibit the production of taxtomina. Additionally, they revealed that the CebR deletion mutants grew faster compared to the wild type strain [30]. The ArsR/SmtB family regulator BlmR was an inhibitor for bleomycin production. Chen et al. obtained a 34% increase in bleomycin B2 production by BlmR deletion in S. verticillus [31]. Negative regulators usually hinder product synthesis by binding to the promoters of the key biosynthetic genes. Mao et al. reported for the first time that the transcription regulator DepR2 from the ArsR family could directly interact with the dptEp promoter of the daptomycin gene cluster. DepR2 knockout led to increased daptomycin production by approximately 2.5-fold [32].
Negative regulators may show different regulatory mechanisms. Apart from down-regulating biosynthetic genes, regulators can also be self-regulated. In a previous research, transcriptomics study was combined with Chromatin immune precipitation to explore the in vivo interactions between the regulator AbsA2 and the S. coelicolor genome. They disclosed that AbsA2 performed two different inhibition mechanisms. Besides bounding to several sites in the calcium-dependent antibiotic gene cluster to inhibit the transcription of CdaR, AbsA2 also bound to its own gene to directly inhibits the transcription of itself [33]. Luo et al. introduced a Himar1-based random mutation screening strategy and identified a transcription regulator PhaR. The disruption of PhaR increased the expression of the gene cluster by approximately 2.68-fold, and therefore the daptomycin titer increased approximately 6.14-fold. They found that in addition to directly binding to the promoter of the daptomycin biosynthetic gene cluster, PhaR can also directly bind to its own promoter [34].
Negative regulator can regulate the expression of multiple genes. In 2019, Xu et al. identified a TetR family transcriptional regulator SLCG_2919, which had a negative effect on the biosynthesis of lincomycin in S. lincolnensis LCGL. SLCG_2919 specifically binds to the promoter region of the lincomycin biosynthetic genes (including 25 structural genes, three resistance genes, and one regulatory gene), inhibiting the transcription of these genes [35]. Gou et al. identified a new TetR family transcriptional regulator CalR3, whose disruption resulted in significant increase in calcimycin and cezomycin production, which was 30-fold and 171-fold that of the wild type strain, respectively. Also, they have identified two CalR3 binding sites within the promoter region of calT and calR3 to illustrate the yield change in this research [36]. Zhao et al. introduced a TFD (transcription factor decoy) strategy to successfully activate eight silent BGCs in multiple Streptomyces. In this study, DNA molecules interfering with gene regulations are designed to bind to regulators, thus preventing the latter from binding to their cognate DNA targets. The targeted and high-throughput activation of silent BGCs in Streptomyces demonstrated the potential of TFD strategy for natural product discovery [37].
2.3. Combinatorial regulation strategies
To obtain a high yield of the target product, up-regulation of positive regulators and down regulation of negative regulators are often performed simultaneously (Fig. 2C). The chromomycins gene cluster in S. reseiscleroticus has two representative regulatory factors, the activator SrcmRI and the inhibitor SrcmRII. Overexpression of SrcmRI or disruption of SrcmRII starts the biosynthesis of tryptomycin. Therefore, by deleting SrcmRII and overexpressing SrcmRI, a high-titer tryptomycin production strain was constructed [38]. Similarly, Overexpression of the regulator AcyB2 greatly increased the production of carbomycins, while overexpression of the regulator CbmR hindered the production of carbomycin. Therefore, high-yield of carbomycins was achieved by overexpressing AcyB2 and knocking out CbmR simultaneously [39].
Coordination among regulators with the same effect can also achieve enhancement of natural product production. Simultaneous overexpression of positive regulatory genes ccaR and claR increased the production of clavulanic acid in S. clavuligerus OR by about 43% [40]. In another case, MonH, MonRI, and MonRII co-regulate the expression of the post-PKS genes, which helps to increase monensin production in S. cinnamonensis. A synergistic cascade process was identified, in which MonH up-regulates the transcription of MonRII while MonRII enhances the transcription of MonRI [41]. Bu et al. identified three transcriptional regulatory factors, namely Sxim22880, CVNABCSX, and WblASX, which played positive regulation roles in polycyclic tetramate macrolactams biosynthesis in S. mangrove Xiamen 318. Simultaneous overexpression of these three regulators resulted in a 24.5% increase of polycyclic tetramate macrolactams production [29].
Coordinating the regulators with other functional proteins is also an effective approach to boost product yields. Atratumycin biosynthesis is regulated by a number of factors, including two LuxR-regulated genes, two ABC transporters, and one streptomycin antibiotic-regulated gene (atr32). Yang et al. identified a rare Streptomyces antibiotic regulatory protein Atr32 as a negative regulatory protein. Through rational engineering of these regulatory genes and transporters, the yield of atratumycin was 1.7-fold–2.3-fold greater than that of the wild-type [42].
3. Promoters engineering strategies
Although bioinformatics analysis of the sequenced microbial genome revealed a large number of uncharacterized biosynthetic gene clusters [43,44], it is still difficult to obtain the potential natural products of these gene clusters as most of them are silent under normal laboratory culture conditions. Therefore, tools that can systematically activate silent biosynthetic gene clusters are in urgent need. Promoter engineering is a widely used strategy to activate cryptic biosynthetic gene clusters. Through promoter engineering, native regulatory sequences that are strictly regulated by pathway-specific or pleiotropic regulators are replaced by regulatory sequences with known or controlled characteristics [[45], [46], [47]]. Therefore, promoter engineering owns the potential to serve as a universal tool for natural products discovery and overproduction.
3.1. Promoter structures
Promoter is a DNA sequence located upstream of the 5′ end of a structural gene, allowing the recognition and binding of RNA polymerase. It is a typical cis-acting element that coordinates with transcription factors (trans-acting factors) to regulate the manner, location and level of gene expression. The length of the promoter varies according to the type of the organisms, and the length of a prokaryote promoter is generally 20–200bp.
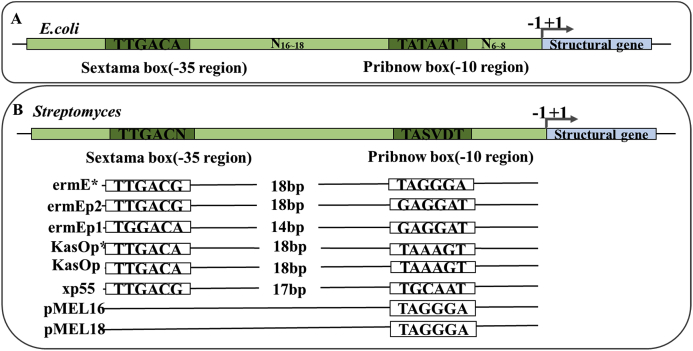
Prokaryotic promoters usually consist of 4 parts (Fig. 3): (1) Transcription initiation sequence: locates in the transcription initiation position and encodes a base complementary to the first nucleotide of a new RNA strand, usually a purine; (2) Pribnow box (−10 region): locates 10 bp upstream of the transcription start site with a conserved consensus sequence TATAAT [48]. It is predicted that the initial TA and the final T plays an important role in RNA polymerase binding in this conserved sequence. A Pribnow-like region, named Hogness region, also exists in eukaryotes [49]; (3) Sextama box (−35 region): a sequence upstream of the −10 region, whose center is about −35bp with a conserved sequence TTGACA [50]. This sequence is the recognition site for RNA polymerase, which largely determines the strength of the promoter; (4) Interval region: the area between the Pribnow box and the Sextama box [50]. When the center position of the two is 16–18 bp, the promoter has a strong transcription function, but if the distance between the two central positions is closer or farther, the initiation of transcription will be weakened.
Fig. 3.
The structure of promoters in prokaryotes. (A) Promoter structure of E. coli, including Pribnow box and Sextama box with conserved sequences, as well as interval region and transcription initiation sequence. (B) Putative Streptomyces promoter structures.
Some Streptomyces promoters are different from the above-mentioned typical prokaryotic promoters. There are several different types of Streptomyces promoters (Fig. 3). (1) The −10 region and −35 region are similar to promoters of E. coli [51]. For example, the erythromycin related promoter ermEp2 [52], and xp55 promoter are similar to the E. coli promoters [53]. (2) The −10 region is similar, while the −35 region is different, some even do not have the −35 region. For example, Manome et al. identified two strong promoter pMEL16 and pMEL18, with similarity to the sequence in −10 region of PI promoter which drives the expression of the thiostrepton-resistance gene, but different in sequences of the upstream region [54]. (3) Another type of Streptomyces promoter does not show any similarity with the known promoter sequences. In one study, 139 promoters were compared and many of them do not display the typical −10 and −35 regions [51].
The nucleotide sequences of the −10 and −35 regions are much less conserved in Gram-positive bacteria [55,56]. Earlier, the consensus sequences of 28 Streptomyces promoters were analyzed, and a consensus sequences TTGAC-(Pu) (where Pu is A or G) for the −35 region and TAG-(Pu)-(Pu)-T for the −10 region were identified [51]. In 2011, researchers constructed a synthetic promoter library. The hexamers TTGACN (where N is A, T, C, or G) and TASVDT (where S is G or C, V is G, A, or C and D is A, T, or G) corresponding to the −35 and −10 consensus sequences were preserved, while a 17 bp spacer region between the −10 and −35 sequences were totally randomized [57,58]. The distance between the −10 region and the −35 region of the Streptomyces promoter varies from 7 bp to 24 bp [59]. In short, the Streptomyces promoters show more diversity compared with the general prokaryotic promoter.
3.2. Methods for promoter identification and characterization
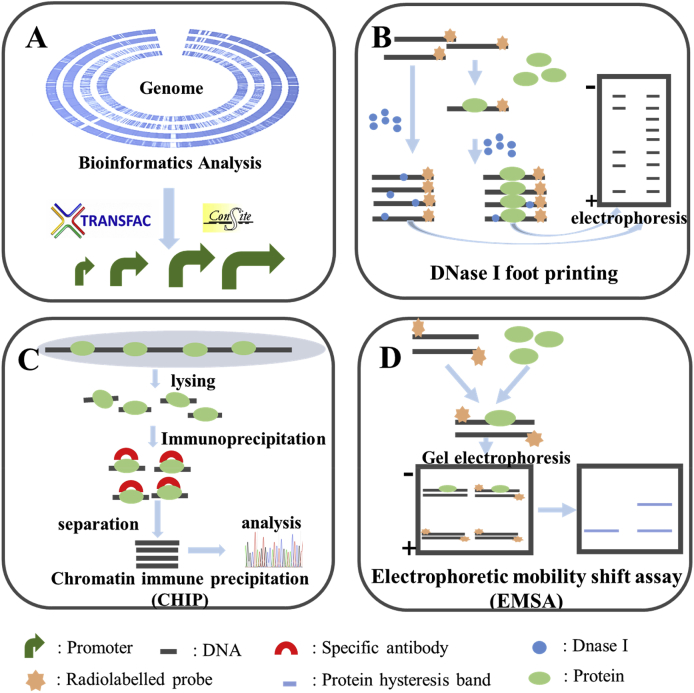
Promoters are important elements involved in gene transcription regulations. Currently, more than 20 databases can be used for prediction as well as analysis of promoter structures and functions. The commonly used databases include BDGP (promoter prediction), BIMAS (Prokaryotic promoter prediction), CONSITE (transcription factor binding site prediction), TRES (transcriptional regulatory factor analysis), TESS (transcription factor binding site prediction), Gene-Regulation (prediction of eukaryotic transcription factor binding sites), TRANSFAC (prediction of transcription factor binding sites) and so on [60]. The commonly used software for promoter identification includes: Core-Promoter Prediction Program, Finding Promoter (NCBI), Neural Network Promoter Prediction, Promoter 2.0, Promoter Scan, and The Markov Chain Promoter Prediction Server [[61], [62], [63]] (Fig. 4A).
Fig. 4.
Methods for promoter identification and characterization. (A) Bioinformatics analysis. (B) DNase I foot printing. DNA will not be degraded by DNase I once binding with a protein. This can be used to predict the binding sites of transcription factors and promoters. (C) Chromatin immune precipitation. DNA fragment bounds to the target protein will be specifically enriched, which can be used to analyze the promoter sequences. (D) Electrophoretic mobility shift assay. Protein-DNA complexes migrate slower than DNA alone. This can be used to identify promoter binding proteins.
Besides bioinformatics prediction, other experimental methods for promoter identification and characterization are also available, including the classic DNase I foot printing method and the Chromatin immune precipitation (ChIP). DNase I foot printing can be used to accurately predict the binding sites of transcription factors and promoters (Fig. 4B) [64]. The basic principle is that DNA will not be degraded by DNase I while binding with a protein. At present, ChIP [65] is the only method to study the interactions between DNA and proteins in vivo (Fig. 4C). Its basic principle is to fix the protein-DNA complex in a living cell and randomly cut it into small chromatin fragments within a certain length range. DNA fragments bound to the target protein will be specifically enriched after precipitated via immunological methods. Finally, DNA sequences of promoters can be obtained through purification and sequencing of the target fragments.
Other methods for promoter function prediction and verification are also based on the principle of protein-DNA interactions, such as the electrophoretic mobility shift assay (EMSA). Electrophoretic mobility shift assay (EMSA) [66] is a typical method to study the interaction between promoters and proteins (Fig. 4D). The basic principle is that the electrophoretic migration speed of the protein-DNA/RNA complex is slower than DNAs/RNAs without protein binding. This method can detect the interaction of nucleic acids (DNA or RNA) with binding proteins, therefore it has been widely used to identify promoter functions via known proteins [67,68].
It is of practical significance in optimization of the above-mentioned methods. For example, combining with gene chip and high-throughput sequencing, ChIP has been developed to ChIP-on chip and ChIP-seq [69] technology. Lewis et al. used ChIP-on chip technology to uncover the binding of absA2 to the promoter regions of redZ and actII-orfIV, which affects the production of calcium-dependent antibiotic [33]. Pepe et al. applied ChIP-sequencing in combination with RNA-sequencing to study the binding sites of HspR and identified the special conservation sequence of the promoter that HspR bound [70]. Through the combination of EMSA and DNase I foot printing assays, the PhoP binding sequence consisting of 11 nucleotide direct repeat units was identified [68]. In another research, FkbR1 was found to bind to the intergenic region of fkbR1-fkbE via EMSA and ChIP-qPCR assays together, providing a foundation for subsequent engineering of the biosynthesis of ascomycin [71]. The advances in the development of effective DNA-protein interaction technologies shed lights on the identification and characterization of promoters, laying a foundation for future promoter engineering.
3.3. Promoter engineering in natural products discovery and overproduction
Promoter engineering is an effective regulatory strategy to increase the yield of natural products or activate the expression of silent gene clusters (Table 1). The commonly used “plug-and-play” method in synthetic biology often employs different promoters, which have been widely applied to activate silent biosynthetic gene clusters and increase the production of microbial secondary metabolites [72].
Table 1.
Applications of promoter engineering.
| Strain | effect | promoter | Reference |
|---|---|---|---|
| S.avermitilis | Lycopene yield was increased to 82 mg/g dry cell weight | Sp44 | [79] |
| S. hygroscopicus XM201 | Geldanamycin yield was increased by 88% | 5063p | [80] |
| S. lividans TK24 | Ptac*RBS3 activity is 17.6-fold than that of Ptac and 3.6-fold than that of PkasO*R15 | Ptac*RBS3 | [81] |
| S. albus J1074 | Production of the blue pigment indigoidine and activation of the biosynthesis of 6-epi-alteramides A/B | ermE*p | [82] |
| S. lividans TK24 | Gene expression control at the posttranslational level in Actinobacteria | An RBS selector in vivo | [83] |
| S. lividans | The production of transglutaminases reached 5.73 U/mL | TGase promoter | [84] |
| S. avermitilis MA-4680 | Chitobiase activity was 24-fold higher | xylA | [85] |
| S. fradiae CGMCC 4.576 | Neomycin production was increased by 36% | PkasO* | [86] |
| S. natalensis | Activation of pimaricin biosynthesis | pimM | [87] |
| S. coelicolorA3(2) | Actinorhodin production tripled | SPL-20 | [88] |
| S. rimosus M527 | β-glucuronidase activity increased by 2.2-fold | SPL-21 | [89] |
| S. lividans | Enzymatic activity of Phospholipase D reached 69.12 U/mL | Ptip | [90] |
Directly replacement of native promoter with a well-characterized one is one of the most popular promoter engineering strategies. Xu et al. replaced the promoter of toyF gene in S. diastatochromo 1628 with the SPL-21 promoter and the yield of toyocamycin was 2-fold greater than that of the wild-type, reaching 489.7 mg/L [73]. Promoter engineering can also be combined with regulator related strategies to achieve increased yield. Wang et al. identified the promoter thlM4p based on microarray analysis, and found that it was 7-fold more active than the commonly used promoter ermE*p. The application of the thlM4p promoter to drive the expression of the regulator gene scnRII leading to 30% higher production of natamycin [74]. At the same year, they identified another strong promoter, groESp, through proteomic analysis of the natamycin-producing strain, Chattanogenesis L10. Under the control of groESp, the yield of natamycin was approximately 20% higher [75]. This study also revealed that proteomics is an effective method for promoter identification, which can be widely applied to other Streptomyces species.
In addition to increasing secondary metabolites production, promoter engineering can also be used to activate silent gene clusters. Zhou et al. transferred the constitutive promoter ermE*p to marine-derived S. chattanoogensis L10, thereby specifically activating the production of chattamycin B, which showed significant antitumor and antibacterial activities [76]. Horbal et al. described a cluster reconstruction method that replaced the native promoter with a randomly generated constitutive synthetic promoter. Depending on this method, they optimized the titer of botulinycin and characterized new derivatives that had not been described previously [77]. Saha et al. used promoter engineering and heterologous expression to activate silent gene clusters in marine Streptomyces SCSIO 02999, they successfully identified six new antitumor polycyclic tetraamino macrolactam antibiotics pactamide A–F [78].
Since promoter engineering is an efficient strategy for natural products discovery and overproduction, the development of these methods grows fast. In 2017, researchers combined the CRISPR-Cas9 technology with promoter engineering by using CRISPR technology to efficiently and accurately introduce foreign promoters into the Streptomyces genome to activate silent biosynthetic gene clusters [91]. Ji et al. constructed a library of Streptomyces regulatory sequences and used the blue pigment indigo gene cluster to rapidly screen the library. In subsequent applications, they inserted four regulatory sequences to the silent actinomycin gene cluster and successfully activated it in S. albus J1074 [92]. Other than constitutive promoters, inducible promoters can also be applied in dynamic regulation of gene expression. Li et al. identified several natural inducible promoters and applied them to increase the production of actinomycin and oxytetracycline in S. coelicolor M145 by 1.3-fold and 9.1-fold, respectively [93]. In summary, the continuous development of synthetic biology technologies will further increase the applications of promoter engineering.
4. Other gene expression regulation strategies
4.1. Engineering the transposon related elements
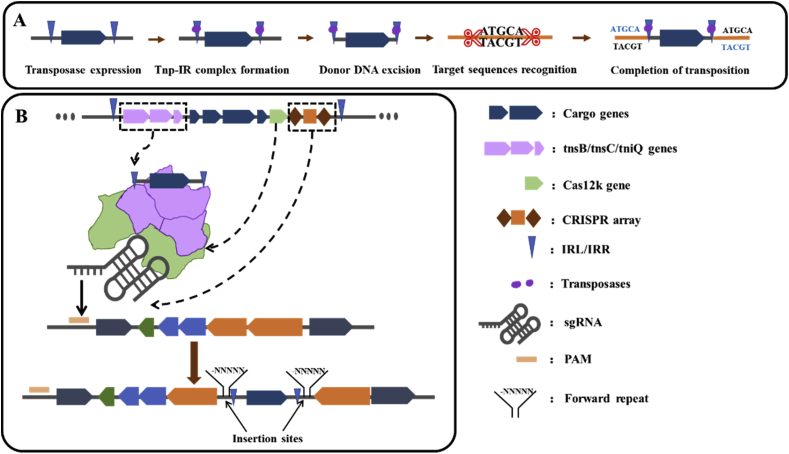
Transposable Elements (TEs), also known as mobile genes or jumping genes, are DNA sequences that can be copied and displaced on the chromosomal DNA [94]. There are three main types of transposons: Insertion Sequence (IS), Composite Transposon (Tn), and Transposable Phage [95]. TEs have cis-regulatory elements that can be replicated throughout the genome, therefore the occurrence of the transposition process can lead to the spread of such regulatory elements throughout the genome, which could increase the potential for simultaneous regulation of different genes and promote the spatiotemporal innovation of genes [96]. The general transposition processes are as follows [97,98] (Fig. 5A): (i) Transposase expression: TEs are regulated by their corresponding transposases, whose expression marks the beginning of the transposition process. (ii) Formation of a transposase-inverted repeat (transposase-IR) complex: The transposase recognizes and binds the inverted repeats at opposite ends of the transposon and combines to form a transposase-IR complex. (iii) Donor DNA excision: the transposase-IR complex has the ability to recognize and excis the donor DNA, making the transposable gene fall off. (iv) Target sequences recognition: the target is identified and attacked by transposable gene, generating staggered cuts at both ends of the attacked sites. (v) Completion of transposition: polymerase and ligase are recruited to fill the DNA gap and form an approximately 4–9 bp repeat sequence, completing the “gene jumping” process [95].
Fig. 5.
Transposon transposition process and the RNA-guided DNA transposition. (A) Five steps of transposon “gene jumping”. (B) The process of RNA-guided DNA transposition. The ShCAST complex consisting of Cas12 k, TnsB, TnsC, and TniQ mediates the insertion of DNA 60–66 bp downstream of the PAM. The IRL and IRR sequences of the transposon and other cargo genes were inserted into the DNA with a 5 bp forward repeat.
In recent years, the association between the Tn7-like transposon and the CRISPR/Cas system has been reported successively [99,100], proving that the evolution of the CRISPR/Cas system was closely related with the mobile elements [101]. Tn7 transposon that exists in E. coli is regulated by five transposases, TnsA-E [[102], [103], [104]]. Different from the Tn7 transposon, Tn7-like transposon lacks TnsE and TnsD, but possesses a homolog of TnsE named TniQ [100]. Klompe et al. discovered that the TniQ-Cascade complex exists in the mini-IF CRISPR/Cas system of Vibrio cholerae, where TniQ bound to the Cas6 subunit. This study represented the first example of a type I crRNA-guided effector complex that directly interacts with non-Cas proteins [105]. They subsequently structurally explained that the CRISPR system is mainly responsible for the recognition of target DNA, with the transposase TniQ for the transposition insertion [106]. However, how the TniQ-Cascade complex achieves “transposition” after identifying the target sequence is still inconclusive. Later, Zhang et al. characterized the CRISPR-related transposase of the cyanobacteria Scytonema hofmanni (ShCAST), which consists of a Tn7-like transposase subunit and a V–K CRISPR effector (Cas12k). Transposase genes tnsB, tnsC, and tniQ are on one side while cas12k and CRISPR arrays are on the other side (Fig. 5B). In this system, the Cas protein and the transposase were fused together. After Cas12k binding to the PAM region, the transposon inserts a fragment 60–66 bp downstream of the PAM region (Fig. 5B). ShCAST integrated DNA into unique locations in the E. coli genome with a frequency of up to 80% [107], establishing a new paradigm for precise DNA insertion.
Due to the randomness and autonomy of transposition process, increasing the transposition efficiency and frequency leads to a wide range of mutations in the genome. The transposons widely used in Streptomyces are IS204 and Tn5, which are derived from Nocardia asteroides and E. coli, respectively. Zhang et al. developed an efficient transposable elements delivery vehicle derived from IS204. A large mutation library has been established to screen important regulatory genes for natural product biosynthetic pathways. Twenty-five S. coelicolor mutants were obtained, one of those revealed an unknown gene in the undecylprodigiosyn (red) biosynthesis pathway, rrdA. This method achieves efficient gene delivery and becomes an attractive method for identifying remote positive and negative regulatory genes for natural products biosynthetic gene clusters [97].
Genome-wide mutagenesis using transposons can identify new genes and pathways that affect antibiotic production. Scientists have modified the Tn5 transposon in S. coelicolor. They confirmed that the transposable insertion was randomly happened in the Streptomyces genome [[108], [109], [110]]. Xu et al. mutated 5 positions of the Tn5 transposase (E54K, M56A, P242A, E345K, and L372P) to construct a highly-active mini-Tn5 transposase system [110]. A library of 50,000 independent mutants based on the highly-active Tn5 transposase system was constructed and 551 genes altering the production of actinorhodin were identified, more than half of which were new effectors [109]. In summary, highly active and efficient transposons can be applied to genome-wide mutagenesis study in Streptomyces, with the potential to identify important genes and pathways that may affect the metabolic regulation network of Streptomyces from an overall perspective.
4.2. Regulation strategies using signal factors
In Streptomyces, morphological development and secondary metabolism are simultaneously affected by multiple nutritional factors and also controlled by extracellular signal molecules. It is reported that most Actinomycetes may use γ-butyrolactone (GBL) to control the production of antibiotics, which is known as the “streptomycin hormone” [111]. The most well characterized signal factor of GBL is “Factor A”, discovered in the early 1960s [112], which was identified to be associated with the production of streptomycin in S. griseus in 1967 [113]. ArpA is the receptor for Factor A, which recognizes and binds a 22 bp palindrome of DNA. ArpA usually binds with the promoter adpA to form an ArpA-DNA complex, thus inhibits the transcription of adpA and decreases the production of streptomycin. When Factor A is added, Factor A will bind to ArpA, releasing the transcription of adpA [111].
Recio et al. showed that limited endogenous PI [2,3-diamino-2,3-bis (hydroxymethyl) −1,4-butanediol] factors restricted the biosynthesis of pimaricin in wild-type strains. They restored the biosynthesis of pimaricin in mutant S. natalensis strain by supplementing Factor A or PI, which stimulated pimaricin production by 33% more [114]. In another case, two regulatory genes (jadR2 and jadR3) encoding homologues of the γ-butyrolactone receptor were identified. Zou et al. purified a JadR3 interacting molecule SVB1, which has the same structure as γ-butyrolactone SCB3 in S. coelicolor [115]. The authors stated that the addition of SVB1 or extraction from S. coelicolor to the mutant strain could restore jadomycin production. This study indicated that the binding of JadR3 and SVB1 plays an important role in controlling jadomycin biosynthesis; on the other hand, it provided new insights into the γ-butyrolactone/receptor system. Zhang et al. found that GBL can be used as an interspecies signal in Streptomyces. They expressed the GBL biosynthetic gene deriving from S.coelicolor M145 in S. albidoflavus J1074 to synthesize Streptomyces coelicolor butanolides (SCBs) [116]. This showed that GBL has great potential in natural products biosynthesis regulations.
4.3. Regulation strategies involving feedback regulations
In natural products biosynthesis, some intermediates may cause feedback regulation to coordinate the expression of biosynthetic genes. For example, the polyketide gene cluster aur1 is responsible for the production of auricin in S. aureofaciens CCM3239 [117]. Auricin and its intermediates could bind to the pathway-specific activator Aur1P, thus inhibited the expression of aur1. This process is obviously related to the acidity of the fermentation environment, which can be intervened by stabilizing the acidic conditions [118]. SsaA is a key activator for sansanmycin biosynthesis. SsaA strictly controls the production of sansanmycins via sensing the accumulation of the final products [119]. The regulator AtrA in S. globisporus serves as a transcriptional activator for actinorhodin biosynthesis in S. coelicolor [120]. The pleiotropic regulator AtrA could coordinate the production of lidamycin, which is inhibited by a biosynthetic intermediate. The activity of AtrA is also regulated by actinorhodin concentrations [121].
In addition to the aforementioned products feedback regulation, some biosynthetic processes require the participation of multiple inhibitors and activators, forming a complex regulatory network. For example, pristinamycin production in S. pristinaespiralis Pr11 is tightly co-regulated by a γ-butyrolactone receptor gene (spbR), two TetR repressor genes (papR3 and papR5), three Streptomyces antibiotic regulatory genes (papR1, papR2, and papR4), and a response regulator gene (papR6) [122]. In the absence of γ-butyrolactone, the auto-regulator SbpR inhibits the expression of all SARPs (Streptomyces Antibiotic Regulatory Protein) in this pathway, including the pristinamycin biosynthetic pathway. In the presence of a critical concentration of γ-butyrolactone, the inhibition is alleviated. PapR1 and PapR2, as the main activators of the pristinamycin biosynthetic gene cluster, activate the transcription of the pristinamycin structural genes. Repressor PapR3 inhibits the transcription of papR4 and papR5, while PapR5 suppresses the transcription of papR1 and papR4 [122]. Deletion of papR5 in combination with overexpression of papR4 and papR6 increased pristinamycin-II titers approximately 1.5-fold than that of the parental strain. At the same time, pristinamycin-II titer increased more than 5-fold by adding macroreticular resin to lessen end-product feedback inhibition and toxic effects [123]. Similarly, the resistance of S. aureus to oxytetracycline is controlled by genes encoding ribosome protection protein (OtrA) and efflux proteins (OtrB and OtrC), which in turn affects the production of oxytetracycline. Yin et al. adopted a three-protein co-expression strategy to improve the synergistic effect of drug resistance and increased the production of oxytetracycline by approximately 2-fold [124].
5. Summary and perspectives
With the rapid development in bioinformatics studies, more and more biosynthetic gene clusters (BGCs) and their related regulatory elements are predicted. Accordingly, to obtain diverse natural products, scientists have developed multiple strategies to coordinate the complex metabolic network in Streptomyces. This review mainly summarized the regulation strategies for Streptomyces natural products biosynthesis from three aspects: regulatory factors, promoters, and others (transposons, signal factors, and feedback regulations). The applications of these strategies should also improve the efficiency of genetic manipulation in Streptomyces. With the development of the CRISPR technology, there will be predictable improvements in this area.
Generally, synergistic effects among several regulatory elements exist to regulate certain metabolic pathways [14]. Therefore, understanding the relationship between different regulatory networks may open up the opportunities for new strategies. Some regulators involved in processes such as bacterial growth and spore production may also indirectly affect the regulation of secondary metabolites biosynthetic pathways. In addition, some competing pathways may lead to the loss of carbon sources, thereby reducing the production of target products. Based on current situations, a better understanding of the regulation in Streptomyces is in urgent need to help us to develop comprehensive regulation strategies. In short, rational application of regulatory strategies will help the discovery and overproduction of valuable natural products, and is expected to open a new century for natural product research.
CRediT authorship contribution statement
Qun Zhou: Writing - original draft. Shuqing Ning: Writing - original draft. Yunzi Luo: Writing - review & editing, Project administration.
Declaration of competing interest
None.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFA0903300), the Natural Science Foundation of Tianjin Province (19JCYBJC24200), and the National Natural Science Foundation of China (81502966).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Ran Liu Z.D., Liu Tiangang. Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab Eng. 2018;50:74–84. doi: 10.1016/j.ymben.2018.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13(8):509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Q., Wang L., Luo Y. Recent advances in natural products exploitation in Streptomyces via synthetic biology. Eng Life Sci. 2019;19(6):452–462. doi: 10.1002/elsc.201800137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Y. Systematic identification of a panel of strong constitutive promoters from Streptomyces albus. ACS Synth Biol. 2015;4(9):1001–1010. doi: 10.1021/acssynbio.5b00016. [DOI] [PubMed] [Google Scholar]
- 5.Barka E.A. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genilloud O. Actinomycetes: still a source of novel antibiotics. Nat Prod Rep. 2017;34(10):1203–1232. doi: 10.1039/c7np00026j. [DOI] [PubMed] [Google Scholar]
- 7.Weixin Tao A.Y., Deng Zixin, Sun Yuhui. CRISPR/Cas9-Based editing of Streptomyces for discovery, characterization, and production of natural products. Front Microbiol. 2018;9(1660):1–8. doi: 10.3389/fmicb.2018.01660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aigle B. Genome mining of Streptomyces ambofaciens. J Ind Microbiol Biotechnol. 2014;41(2):251–263. doi: 10.1007/s10295-013-1379-y. [DOI] [PubMed] [Google Scholar]
- 9.Nee W.A.a.A. Genome mining for the search and discovery of bioactive compounds: the Streptomyces paradigm. FEMS Microbiol Lett. 2018;365(24):1–20. doi: 10.1093/femsle/fny240/5108151. [DOI] [PubMed] [Google Scholar]
- 10.Saibin Zhu Y.D.a.Y.H. The application of ribosome engineering to natural product discovery and yield improvement. Antibiotics (Berlin) 2019;8(3):133–149. doi: 10.3390/antibiotics8030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas C., McLean B.W., Hutchings Matthew I., Devin Rebecca. Dissolution of the disparate: Co-ordinate regulation in antibiotic biosynthesis. Antibiotics (Berlin) 2019;8(2):83–100. doi: 10.3390/antibiotics8020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhijie Yang J.H., Wei Xin, Ju Jianhua, Ma Junying. Exploration and genome mining of natural products from marine Streptomyces. Appl Microbiol Biotechnol. 2019;104(1):67–76. doi: 10.1007/s00253-019-10227-0. [DOI] [PubMed] [Google Scholar]
- 13.Weber S.Y.L.T. Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol Adv. 2019;37(6):1–15. doi: 10.1016/j.biotechadv.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Lyu H.N. Harnessing diverse transcriptional regulators for natural product discovery in fungi. Nat Prod Rep. 2019;37(1):6–16. doi: 10.1039/c8np00027a. [DOI] [PubMed] [Google Scholar]
- 15.Baltz R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol. 2016;43(2–3):343–370. doi: 10.1007/s10295-015-1682-x. [DOI] [PubMed] [Google Scholar]
- 16.Cuthbertson L., Nodwell J.R. The TetR family of regulators. Microbiol Mol Biol Rev : MMBR (Microbiol Mol Biol Rev) 2013;77(3):440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J. ToyA, a positive pathway-specific regulator for toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Appl Microbiol Biotechnol. 2019;103(17):7071–7084. doi: 10.1007/s00253-019-09959-w. [DOI] [PubMed] [Google Scholar]
- 18.Hou B. The novel transcriptional regulator LmbU promotes lincomycin biosynthesis through regulating expression of its target genes in Streptomyces lincolnensis. J Bacteriol. 2018;200(2) doi: 10.1128/jb.00447-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong L. CtcS, a MarR family regulator, regulates chlortetracycline biosynthesis. BMC Microbiol. 2019;19(1):268–279. doi: 10.1186/s12866-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y.W. Characterization of three regulatory genes involved in enduracidin biosynthesis and improvement of enduracidin production in Streptomyces fungicidicus. J Appl Microbiol. 2019;127(6):1698–1705. doi: 10.1111/jam.14417. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y. Exploring novel herbicidin analogues by transcriptional regulator overexpression and MS/MS molecular networking. Microb Cell Factories. 2019;18(1):175. doi: 10.1186/s12934-019-1225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X. Dynamic changes of metabolomics and expression of candicidin biosynthesis gene cluster caused by the presence of a pleiotropic regulator AdpA in Streptomyces ZYJ-6. Bioproc Biosyst Eng. 2019;42(8):1353–1365. doi: 10.1007/s00449-019-02135-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang J. AdpAsd, a positive regulator for morphological development and toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Curr Microbiol. 2018;75(10):1345–1351. doi: 10.1007/s00284-018-1529-6. [DOI] [PubMed] [Google Scholar]
- 24.Kang Y. AdpAlin, a pleiotropic transcriptional regulator, is involved in the cascade regulation of lincomycin biosynthesis in Streptomyces lincolnensis. Front Microbiol. 2019;10:2428. doi: 10.3389/fmicb.2019.02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu J. The two-component system CepRS regulates the cephamycin C biosynthesis in Streptomyces clavuligerus F613-1. Amb Express. 2019;9(1):118. doi: 10.1186/s13568-019-0844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan H. Important role of a LAL regulator StaR in the staurosporine biosynthesis and high-production of Streptomyces fradiae CGMCC 4.576. Sci China Life Sci. 2019:1638–1654. doi: 10.1007/s11427-019-1597-6. [DOI] [PubMed] [Google Scholar]
- 27.Wei K. MilR2, a novel TetR family regulator involved in 5-oxomilbemycin A3/A4 biosynthesis in Streptomyces hygroscopicus. Appl Microbiol Biotechnol. 2018;102(20):8841–8853. doi: 10.1007/s00253-018-9280-2. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Burgo Y. Activation of secondary metabolite gene clusters in Streptomyces clavuligerus by the PimM regulator of Streptomyces natalensis. Front Microbiol. 2019;10:580. doi: 10.3389/fmicb.2019.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bu X.L. Three transcriptional regulators positively regulate the biosynthesis of polycyclic tetramate macrolactams in Streptomyces xiamenensis 318. Appl Microbiol Biotechnol. 2019:701–711. doi: 10.1007/s00253-019-10269-4. [DOI] [PubMed] [Google Scholar]
- 30.Planckaert S. Proteomic response to thaxtomin phytotoxin elicitor cellobiose and to deletion of cellulose utilization regulator CebR in Streptomyces scabies. J Proteome Res. 2018;17(11):3837–3852. doi: 10.1021/acs.jproteome.8b00528. [DOI] [PubMed] [Google Scholar]
- 31.Chen H. Negative regulation of bleomycins biosynthesis by ArsR/SmtB family repressor BlmR in Streptomyces verticillus. Appl Microbiol Biotechnol. 2019;103(16):6629–6644. doi: 10.1007/s00253-019-09923-8. [DOI] [PubMed] [Google Scholar]
- 32.Mao X.M., Luo S., Li Y.Q. Negative regulation of daptomycin production by DepR2, an ArsR-family transcriptional factor. J Ind Microbiol Biotechnol. 2017;44(12):1653–1658. doi: 10.1007/s10295-017-1983-3. [DOI] [PubMed] [Google Scholar]
- 33.Lewis R.A. Genome-wide analysis of the role of the antibiotic biosynthesis regulator AbsA2 in Streptomyces coelicolor A3(2) PloS One. 2019;14(4):673–696. doi: 10.1371/journal.pone.0200673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo S. Transposon-based identification of a negative regulator for the antibiotic hyper-production in Streptomyces. Appl Microbiol Biotechnol. 2018;102(15):6581–6592. doi: 10.1007/s00253-018-9103-5. [DOI] [PubMed] [Google Scholar]
- 35.Xu Y. TetR-type regulator SLCG_2919 is a negative regulator of lincomycin biosynthesis in Streptomyces lincolnensis. Appl Environ Microbiol. 2019;85(1):274–288. doi: 10.1128/aem.02091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gou L. A novel TetR family transcriptional regulator, CalR3, negatively controls calcimycin biosynthesis in Streptomyces chartreusis NRRL 3882. Front Microbiol. 2017;8:2371. doi: 10.3389/fmicb.2017.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang B. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat Chem Biol. 2019;15(2):111–114. doi: 10.1038/s41589-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L. Manipulation of two regulatory genes for efficient production of chromomycins in Streptomyces reseiscleroticus. J Biol Eng. 2018;12:9. doi: 10.1186/s13036-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong J. Identification of two regulatory genes involved in carbomycin biosynthesis in Streptomyces thermotolerans. Arch Microbiol. 2017;199(7):1023–1033. doi: 10.1007/s00203-017-1376-z. [DOI] [PubMed] [Google Scholar]
- 40.Cho H.S. Improved production of clavulanic acid by reverse engineering and overexpression of the regulatory genes in an industrial Streptomyces clavuligerus strain. J Ind Microbiol Biotechnol. 2019;46(8):1205–1215. doi: 10.1007/s10295-019-02196-0. [DOI] [PubMed] [Google Scholar]
- 41.Tang Z.K. Characterization of three pathway-specific regulators for high production of monensin in Streptomyces cinnamonensis. Appl Microbiol Biotechnol. 2017;101(15):6083–6097. doi: 10.1007/s00253-017-8353-y. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z. Characterization of the noncanonical regulatory and transporter genes in atratumycin biosynthesis and production in a heterologous host. Mar Drugs. 2019;17(10):576–587. doi: 10.3390/md17100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cimermancic P. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doroghazi J.R. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10(11):963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang H.S., Charlop-Powers Z., Brady S.F. Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth Biol. 2016;5(9):1002–1010. doi: 10.1021/acssynbio.6b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat Commun. 2013;4:2894. doi: 10.1038/ncomms3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montiel D. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc Natl Acad Sci U S A. 2015;112(29):8953–8958. doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lifton R.P. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harbor Symp Quant Biol. 1978;42(2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- 50.Lanzer M., Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci U S A. 1988;85(23):8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strohl W.R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20(5):961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bibb M.J., Janssen G.R., Ward J.M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1–3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 53.Buttner M.J. RNA polymerase heterogeneity in Streptomyces coelicolor A3(2) Mol Microbiol. 1989;3(11):1653–1659. doi: 10.1111/j.1365-2958.1989.tb00151.x. [DOI] [PubMed] [Google Scholar]
- 54.Manome T., Hoshino E. Cloning of DNA fragments containing Streptomyces promoter activity. J Antibiot (Tokyo) 1987;40(10):1440–1447. doi: 10.7164/antibiotics.40.1440. [DOI] [PubMed] [Google Scholar]
- 55.Patek M. Corynebacterium glutamicum promoters: a practical approach. Microb Biotechnol. 2013;6(2):103–117. doi: 10.1111/1751-7915.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patek M., Nesvera J. Sigma factors and promoters in Corynebacterium glutamicum. J Biotechnol. 2011;154(2–3):101–113. doi: 10.1016/j.jbiotec.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 57.Myronovskyi M., Luzhetskyy A. Native and engineered promoters in natural product discovery. Nat Prod Rep. 2016;33(8):1006–1019. doi: 10.1039/c6np00002a. [DOI] [PubMed] [Google Scholar]
- 58.Seghezzi N. The construction of a library of synthetic promoters revealed some specific features of strong Streptomyces promoters. Appl Microbiol Biotechnol. 2011;90(2):615–623. doi: 10.1007/s00253-010-3018-0. [DOI] [PubMed] [Google Scholar]
- 59.Feitelson J.S. An improved plasmid for the isolation and analysis of Streptomyces promoters. Gene. 1988;66(1):159–162. doi: 10.1016/0378-1119(88)90233-8. [DOI] [PubMed] [Google Scholar]
- 60.Kolchanov N.A. Transcription regulatory regions database (TRRD): its status in 2002. Nucleic Acids Res. 2002;30(1):312–317. doi: 10.1093/nar/30.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horton P.B., Kanehisa M. An assessment of neural network and statistical approaches for prediction of E. coli promoter sites. Nucleic Acids Res. 1992;20(16):4331–4338. doi: 10.1093/nar/20.16.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zwir I., Harari O., Groisman E.A. Gene promoter scan methodology for identifying and classifying coregulated promoters. Methods Enzymol. 2007;422:361–385. doi: 10.1016/s0076-6879(06)22018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vandenbon A. Markov chain-based promoter structure modeling for tissue-specific expression pattern prediction. DNA Res. 2008;15(1):3–11. doi: 10.1093/dnares/dsm034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galas D.J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solomon M.J., Larsen P.L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 66.Hellman L.M., Fried M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Y., Ye B.C. GlnR and PhoP regulate beta-glucosidases involved in cellulose digestion in response to nitrogen and phosphate availability. Microbiology. 2018;164(5):779–789. doi: 10.1099/mic.0.000654. [DOI] [PubMed] [Google Scholar]
- 68.Martin J.F., Ramos A., Liras P. Regulation of geldanamycin biosynthesis by cluster-situated transcription factors and the master regulator PhoP. Antibiotics (Berlin) 2019;8(3):353–369. doi: 10.3390/antibiotics8030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson D.S. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 70.Pepe S. The Helicobacter pylori heat-shock repressor HspR: definition of its direct regulon and characterization of the cooperative DNA-binding mechanism on its own promoter. Front Microbiol. 2018;9:1887–1903. doi: 10.3389/fmicb.2018.01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song K. Engineering of the LysR family transcriptional regulator FkbR1 and its target gene to improve ascomycin production. Appl Microbiol Biotechnol. 2017;101(11):4581–4592. doi: 10.1007/s00253-017-8242-4. [DOI] [PubMed] [Google Scholar]
- 72.Xu N., Wei L., Liu J. Recent advances in the applications of promoter engineering for the optimization of metabolite biosynthesis. World J Microbiol Biotechnol. 2019;35(2):33–43. doi: 10.1007/s11274-019-2606-0. [DOI] [PubMed] [Google Scholar]
- 73.Xu X. Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J Microbiol Biotechnol. 2017;33(2):30–38. doi: 10.1007/s11274-016-2194-1. [DOI] [PubMed] [Google Scholar]
- 74.Wang K. Transcriptome-based identification of a strong promoter for hyper-production of natamycin in Streptomyces. Curr Microbiol. 2019;76(1):95–99. doi: 10.1007/s00284-018-1589-7. [DOI] [PubMed] [Google Scholar]
- 75.Wang K. Identification of a secondary metabolism-responsive promoter by proteomics for over-production of natamycin in Streptomyces. Arch Microbiol. 2019;201(10):1459–1464. doi: 10.1007/s00203-019-01710-3. [DOI] [PubMed] [Google Scholar]
- 76.Zhou Z. Genome mining-directed activation of a silent angucycline biosynthetic gene cluster in Streptomyces chattanoogensis. Chembiochem. 2015;16(3):496–502. doi: 10.1002/cbic.201402577. [DOI] [PubMed] [Google Scholar]
- 77.Horbal L. Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab Eng. 2018;49:299–315. doi: 10.1016/j.ymben.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 78.Saha S. Activation and characterization of a cryptic gene cluster reveals a cyclization cascade for polycyclic tetramate macrolactams. Chem Sci. 2017;8(2):1607–1612. doi: 10.1039/c6sc03875a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai C. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc Natl Acad Sci U S A. 2015;112(39):12181–12186. doi: 10.1073/pnas.1511027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang X. Improved PKS gene expression with strong endogenous promoter resulted in geldanamycin yield increase. Biotechnol J. 2017;12(11) doi: 10.1002/biot.201700321. [DOI] [PubMed] [Google Scholar]
- 81.Zhao M. Engineering diverse eubacteria promoters for robust Gene expression in Streptomyces lividans. J Biotechnol. 2019;289:93–102. doi: 10.1016/j.jbiotec.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 82.Olano C. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb Biotechnol. 2014;7(3):242–256. doi: 10.1111/1751-7915.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Horbal L., Siegl T., Luzhetskyy A. A set of synthetic versatile genetic control elements for the efficient expression of genes in Actinobacteria. Sci Rep. 2018;8(1):491. doi: 10.1038/s41598-017-18846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu S. Improving the active expression of transglutaminase in Streptomyces lividans by promoter engineering and codon optimization. BMC Biotechnol. 2016;16(1):75–84. doi: 10.1186/s12896-016-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noguchi Y. Development of a strictly regulated xylose-induced expression system in Streptomyces. Microb Cell Factories. 2018;17(1):151–160. doi: 10.1186/s12934-018-0991-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zheng J. Enhancement of neomycin production by engineering the entire biosynthetic gene cluster and feeding key precursors in Streptomyces fradiae CGMCC 4.576. Appl Microbiol Biotechnol. 2019;103(5):2263–2275. doi: 10.1007/s00253-018-09597-8. [DOI] [PubMed] [Google Scholar]
- 87.Barreales E.G. Promoter engineering reveals the importance of heptameric direct repeats for DNA binding by Streptomyces antibiotic regulatory protein-large ATP-binding regulator of the LuxR family (SARP-LAL) regulators in Streptomyces natalensis. Appl Environ Microbiol. 2018;84(10) doi: 10.1128/AEM.00246-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sohoni S.V. Synthetic promoter library for modulation of actinorhodin production in Streptomyces coelicolor A3(2) PloS One. 2014;9(6) doi: 10.1371/journal.pone.0099701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song Z.Q. Development and optimization of an intergeneric conjugation system and analysis of promoter activity in Streptomyces rimosus M527. J Zhejiang Univ - Sci B. 2019;20(11):891–900. doi: 10.1631/jzus.B1900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tao X. Comparison of the expression of phospholipase D from Streptomyces halstedii in different hosts and its over-expression in Streptomyces lividans. FEMS Microbiol Lett. 2019;366(5) doi: 10.1093/femsle/fnz051. [DOI] [PubMed] [Google Scholar]
- 91.Zhang M.M. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol. 2017;13:607–609. doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ji C.H., Kim J.P., Kang H.S. Library of synthetic Streptomyces regulatory sequences for use in promoter engineering of natural product biosynthetic gene clusters. ACS Synth Biol. 2018;7(8):1946–1955. doi: 10.1021/acssynbio.8b00175. [DOI] [PubMed] [Google Scholar]
- 93.Li S. An autoregulated fine-tuning strategy for titer improvement of secondary metabolites using native promoters in Streptomyces. ACS Synth Biol. 2018;7(2):522–530. doi: 10.1021/acssynbio.7b00318. [DOI] [PubMed] [Google Scholar]
- 94.Green B. Insertion site preference of mu, Tn5, and Tn7 transposons. Mobile DNA. 2012;3(3):1–6. doi: 10.1186/1759-8753-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodriguez-Valera F., Martin-Cuadrado A.B., Lopez-Perez M. Flexible genomic islands as drivers of genome evolution. Curr Opin Microbiol. 2016;31:154–160. doi: 10.1016/j.mib.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 96.Rebollo R., Romanish M.T., Mager D.L. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 97.Zhang X. Efficient transposition of IS204-derived plasmids in Streptomyces coelicolor. J Microbiol Methods. 2012;88(1):67–72. doi: 10.1016/j.mimet.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 98.Ason B., Reznikoff W.S. Mutational analysis of the base flipping event found in Tn5 transposition. J Biol Chem. 2002;277(13):11284–11291. doi: 10.1074/jbc.M111119200. [DOI] [PubMed] [Google Scholar]
- 99.Faure G. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol. 2019;17(8):513–525. doi: 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McDonald N.D. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genom. 2019;20(1):105. doi: 10.1186/s12864-019-5439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peters J.E. Tn7. Microbiol Spectr. 2014;2(5) doi: 10.1128/microbiolspec.MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- 103.Shi Q. Conformational toggling controls target site choice for the heteromeric transposase element Tn7. Nucleic Acids Res. 2015;43(22):10734–10745. doi: 10.1093/nar/gkv913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sarnovsky R.J., May E.W., Craig N.L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15(22):6348–6361. doi: 10.1002/j.1460-2075.1996.tb01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klompe S.E. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571(7764):219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 106.Halpin-Healy T.S. Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Nature. 2020;577(7789):271–274. doi: 10.1038/s41586-019-1849-0. [DOI] [PubMed] [Google Scholar]
- 107.Strecker J. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Petzke L., Luzhetskyy A. In vivo Tn5-based transposon mutagenesis of Streptomycetes. Appl Microbiol Biotechnol. 2009;83(5):979–986. doi: 10.1007/s00253-009-2047-z. [DOI] [PubMed] [Google Scholar]
- 109.Xu Z. Genome-wide mutagenesis links multiple metabolic pathways with actinorhodin production in Streptomyces coelicolor. Appl Environ Microbiol. 2019;85(7) doi: 10.1128/AEM.03005-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu Z. Large-Scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl Environ Microbiol. 2017;83(6) doi: 10.1128/AEM.02889-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xia H. The regulatory cascades of antibiotic production in Streptomyces. World J Microbiol Biotechnol. 2020;36(1):13. doi: 10.1007/s11274-019-2789-4. [DOI] [PubMed] [Google Scholar]
- 112.Kitani S. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc Natl Acad Sci U S A. 2011;108(39):16410–16415. doi: 10.1073/pnas.1113908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thao N.B. Discovering potential Streptomyces hormone producers by using disruptants of essential biosynthetic genes as indicator strains. J Antibiot (Tokyo) 2017;70(10):1004–1008. doi: 10.1038/ja.2017.85. [DOI] [PubMed] [Google Scholar]
- 114.Recio E. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J Biol Chem. 2004;279(40):41586–41593. doi: 10.1074/jbc.M402340200. [DOI] [PubMed] [Google Scholar]
- 115.Zou Z. A gamma-butyrolactone-sensing activator/repressor, JadR3, controls a regulatory mini-network for jadomycin biosynthesis. Mol Microbiol. 2014;94(3):490–505. doi: 10.1111/mmi.12752. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y. Activation of paulomycinproduction by exogenous gamma-butyrolactone signaling molecules in Streptomyces albidoflavus J1074. Appl Microbiol Biotechnol. 2020;104(4):1695–1705. doi: 10.1007/s00253-019-10329-9. [DOI] [PubMed] [Google Scholar]
- 117.Novakova R. The role of the TetR-family transcriptional regulator Aur1R in negative regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology. 2010;156(Pt 8):2374–2383. doi: 10.1099/mic.0.037895-0. [DOI] [PubMed] [Google Scholar]
- 118.Kutas P. Strict control of auricin production in Streptomyces aureofaciens CCM 3239 involves a feedback mechanism. Appl Microbiol Biotechnol. 2013;97(6):2413–2421. doi: 10.1007/s00253-012-4505-2. [DOI] [PubMed] [Google Scholar]
- 119.Li Q. SsaA, a member of a novel class of transcriptional regulators, controls sansanmycin production in Streptomyces sp. strain SS through a feedback mechanism. J Bacteriol. 2013;195(10):2232–2243. doi: 10.1128/JB.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uguru G.C. Transcriptional activation of the pathway-specific regulator of the actinorhodin biosynthetic genes in Streptomyces coelicolor. Mol Microbiol. 2005;58(1):131–150. doi: 10.1111/j.1365-2958.2005.04817.x. [DOI] [PubMed] [Google Scholar]
- 121.Li X. Binding of a biosynthetic intermediate to AtrA modulates the production of lidamycin by Streptomyces globisporus. Mol Microbiol. 2015;96(6):1257–1271. doi: 10.1111/mmi.13004. [DOI] [PubMed] [Google Scholar]
- 122.Mast Y. A complex signaling cascade governs pristinamycin biosynthesis in Streptomyces pristinaespiralis. Appl Environ Microbiol. 2015;81(19):6621–6636. doi: 10.1128/aem.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li L. A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metab Eng. 2015;29:12–25. doi: 10.1016/j.ymben.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 124.Yin S. Improvement of oxytetracycline production mediated via cooperation of resistance genes in Streptomyces rimosus. Sci China Life Sci. 2017;60(9):992–999. doi: 10.1007/s11427-017-9121-4. [DOI] [PubMed] [Google Scholar]