Abstract
Lysin motif (LysM) modules are approximately 50 amino acids long and bind to peptidoglycan, chitin and its derivatives. Certain LysM proteins in plant pathogenic and entomopathogenic fungi are shown to scavenge chitin oligosaccharides and thereby dampen host defense reactions. Other LysM proteins can protect the fungal cell wall against hydrolytic enzymes. In this study, we investigated the biological function of LysM proteins in the mycoparasitic fungus Clonostachys rosea. The C. rosea genome contained three genes coding for LysM-containing proteins and gene expression analysis revealed that lysm1 and lysm2 were induced during mycoparasitic interaction with Fusarium graminearum and during colonization of wheat roots. Lysm1 was suppressed in germinating conidia, while lysm2 was induced during growth in chitin or peptidoglycan-containing medium. Deletion of lysm1 and lysm2 resulted in mutants with increased levels of conidiation and conidial germination, but reduced ability to control plant diseases caused by F. graminearum and Botrytis cinerea. The Δlysm2 strain showed a distinct, accelerated mycelial disintegration phenotype accompanied by reduced biomass production and hyphal protection against hydrolytic enzymes including chitinases, suggesting a role of LYSM2 in hyphal protection against chitinases. The Δlysm2 and Δlysm1Δlysm2 strains displayed reduced ability to colonize wheat roots, while only Δlysm1Δlysm2 failed to suppress expression of the wheat defense response genes PR1 and PR4. Based on our data, we propose a role of LYSM1 as a regulator of fungal development and of LYSM2 in cell wall protection against endogenous hydrolytic enzymes, while both are required to suppress plant defense responses. Our findings expand the understanding of the role of LysM proteins in fungal-fungal interactions and biocontrol.
Keywords: antagonism, biocontrol, biological control, Clonostachys rosea, fungal–fungal interactions, LysM effector, LysM protein, mycoparasitism
Introduction
Lysin motif (LysM) domains are approximately 50 amino acids long family 50 carbohydrate-binding modules (CBM50) that bind to N-acetylglucosamine (GlcNAc)-containing glycans such as peptidoglycan, chitin and chitin-like compounds (Buist et al., 2008). LysM-containing proteins can comprise one or multiple LysM modules and show high amino acid sequence variability (Buist et al., 2008; Akcapinar et al., 2015). LysM modules were first discovered in the antimicrobial enzyme lysozyme of a bacteriophage (Garvey et al., 1986), and subsequently identified in proteins from all kingdoms of life including bacteria, fungi, plants and animals (Buist et al., 2008; Kombrink and Thomma, 2013; Akcapinar et al., 2015). In bacteria, LysM modules are found in various proteins including hydrolytic enzymes like peptidoglycan hydrolases, peptidases, chitinases, esterases and nucleotidases, as well as in adhesins required for cell growth and division, autolysis, and virulence (Buist et al., 2008). In plants, LysMs are present in certain cell surface receptor kinases and recognize chitin, peptidoglycan and their derivatives (Antolín-Llovera et al., 2014), which are essential structural components of the fungal and bacterial cell wall, respectively. Also, they are a well-established microbe-associated molecular pattern (MAMP) in fungi and bacteria. LysM modules are also present in plants’ Nod factor receptors and Myc factor receptors that bind Nod factors from symbiotic rhizobia and Myc factors from symbiotic arbuscular mycorrhizal fungi, respectively. Detection of these compounds by transmembrane LysM-containing protein receptors of host plants activate MAMP-triggered defense signaling, against bacterial and fungal pathogens, and symbiotic signaling during interactions between legume plants and nitrogen-fixing bacteria and arbuscular mycorrhiza (Antolín-Llovera et al., 2014).
In fungi, LysM modules can be found either as part of secreted proteins with varying number of LysMs, but without any known catalytic domain (referred to as LysM effectors), or together with known catalytic protein modules (referred to as LysM-containing proteins) (de Jonge and Thomma, 2009; Martinez et al., 2012). Examples of LysM-containing proteins include glycoside hydrolases (chitinases), CyanoVirin-N homology (CVNH) modules, N-acetylmuramoyl-L-alanine amidase modules and polysaccharide deacetylase type 1 modules (de Jonge and Thomma, 2009). Based on evolutionary analyses, the number and consensus pattern of cysteine (Cys) residues in the LysM module divides them into a fungal-specific group and a fungal/bacterial group, respectively (Akcapinar et al., 2015; Cen et al., 2017; Kombrink et al., 2017). The fungal-specific group contains three conserved Cys residues in the LysM module and one Cys residue outside of the module, which putatively forms two disulfide bridges. In contrast, the fungal/bacterial group contains no or only one Cys residue at a different position than those found in fungal-specific LysMs (Akcapinar et al., 2015).
Functional characterization of LysM proteins from fungal plant pathogens revealed that LysM effectors can suppress chitin-triggered immunity in host plants by sequestering the chitin oligosaccharides released from the fungal cell walls, and thereby contribute to fungal virulence (de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Takahara et al., 2016; Cen et al., 2017; Kombrink et al., 2017). Furthermore, LysM effectors Mg3LysM and Vd2LysM (from the septoria tritici blotch fungus, Zymoseptoria tritici, and the soil-borne vascular wilt pathogen, Verticillium dahliae, respectively), are shown to protect fungal hyphae against the hydrolytic activity of host chitinase (Marshall et al., 2011; Cen et al., 2017; Kombrink et al., 2017). In contrast to these findings, deletion of four LysM effector genes that are highly expressed in the blue mold fungus, Penicillium expansum, during the infection of apple, and three core LysM effector genes (Vd4LysM, Vd5LysM and Vd6LysM) conserved in several V. dahliae strains, did not affect their virulence on their host plants (Kombrink et al., 2017; Levin et al., 2017). The biological role of LysM effectors has also been investigated in the insect-pathogenic fungus, Beauveria bassiana. Analysis of gene deletion mutants showed that the LysM effector Blys2 and Blys5 are necessary for the full virulence of B. bassiana against its insect hosts. However, only Blys5 could protect fungal hyphae against chitinase hydrolysis (Cen et al., 2017).
Interestingly, these characterized LysM proteins belong mainly to the fungal/bacterial group and are LysM effectors (de Jonge and Thomma, 2009; Akcapinar et al., 2015). The vast majority of genes encoding LysM effectors and LysM-containing proteins in fungal species with distinct lifestyles, including plant pathogens, saprotrophs and mycotrophs (de Jonge and Thomma, 2009; Gruber et al., 2011; Martinez et al., 2012; Kombrink et al., 2017), remain uncharacterized. Therefore, more functional studies are required to understand (i) whether there is a distinction in function between fungal-specific and fungal/bacterial LysM proteins, and (ii) the role of LysM proteins in filamentous fungi, including those with saprotrophic and mycotrophic life style. Considering the fact that LysM proteins show high diversity in protein length, LysM module number, and amino acid sequence between closely related fungal species and even between allelic proteins (de Jonge and Thomma, 2009; Akcapinar et al., 2015; Cen et al., 2017), it is utmost important to characterize their functions in fungi with diverse life styles in order to gain insights into the range of biological roles of LysM proteins. For example, external application of purified TAL6, which is a fungal-specific LysM effector from the mycoparasitic fungus Trichoderma atroviride, inhibited spore germination of Trichoderma spp. but not of other fungi, suggesting a species-specific role of fungal LysM modules in regulation of fungal growth and development (Seidl-Seiboth et al., 2013). In addition, Tal6 deletion strains showed reduced mycoparasitic ability toward the host fungus (Romero-Contreras et al., 2019). The results from these studies highlight a possible difference in function between fungal-specific and fungal/bacterial LysM effectors (Seidl-Seiboth et al., 2013).
The filamentous fungus Clonostachys rosea (Link: Fr.) Schroers, Samuels, Seifert & W. Gams, comb. nov. (Schroers et al., 1999) has a complex life style as a necrotrophic mycoparasite by killing other fungi and feeding on dead mycelia, as a saprotroph by digesting organic material in soil and as a beneficial plant root colonizer (Sutton et al., 1997; Ravnskov et al., 2006; Karlsson et al., 2015). C. rosea strain IK726 is known to be an effective biological control agent against a broad range of fungal plant pathogens and plant parasitic nematodes (Jensen et al., 2000, 2004, 2016; Cota et al., 2008; Iqbal et al., 2018). The aim of the present study was to investigate the biological role of fungal-specific LysM proteins in C. rosea, with emphasis on their function in mycoparasitic fungal-fungal and beneficial fungal-plant interactions. By generating and analyzing gene deletion and complementation strains, we demonstrate a role of LysM proteins in regulating fungal development and autolysis, in biological control of plant pathogens, in suppression of plant host defense response and in root colonization. Our findings expand the understanding of the role of LysM proteins in biological interactions between filamentous fungi and with plants.
Materials and Methods
Fungal Strains and Culture Conditions
Clonostachys rosea strain IK726 wild type (WT; Knudsen et al., 1995) and mutants derived from it, Botrytis cinerea strain B05.10 (Amselem et al., 2011) and Fusarium graminearum strain 1104-14 (Fusarium collection at the National Veterinary Institute in Norway) were maintained on potato dextrose agar (PDA; Oxoid, Cambridge, United Kingdom) medium at 25°C. SMS medium (Dubey et al., 2012) supplemented with 1% glucose or Czapek-dox (CZ) was used to grow C. rosea for gene expression analysis unless otherwise specified.
Gene Identification and Sequence Analysis
The C. rosea strain IK726 genome version 1 (Karlsson et al., 2015) and version 2 (Broberg et al., 2018), were screened for the presence of genes encoding LysM-containing protein by BLASTP analysis using amino acid sequences of LysM containing proteins from a diverse group of fungi (Supplementary Table S1). Presence of conserved domains were analyzed with Simple modular architecture research tool (SMART) (Bork et al., 2009), InterProScan (Quevillon et al., 2005) and conserved domain search (CDS) (Marchler-Bauer et al., 2011). Presence of Cys residues in specific spacing pattern was analyzed manually. Signal P 4.1 (Petersen et al., 2011) was used to search for signal peptide cleavage sites. Amino acid sequence alignment was performed using ClustalW2 (Larkin et al., 2007) with default settings for multiple sequence alignment. Single nucleotide polymorphisms (SNPs) in lysm1, lysm2 and chiC2 were detected using a procedure described previously (Broberg et al., 2018). A multidimensional scaling (MDS) was performed using R 3.5 on SNPs occurrence in lysm1, lysm2 and chiC2 to visualize the sequence variation across isolates.
Phylogenetic Analysis and Molecular Evolution
LysM module protein sequences were aligned using Muscle (Edgar, 2004) and phylogenetic analysis was performed using maximum likelihood methods implemented in MEGA ver. 6 (Tamura et al., 2013). The WAG (Whelan and Goldman) with G (Gamma distribution) amino acid substitution model (Whelan and Goldman, 2001) was used with pairwise deletion of gaps. Statistical support for branches was assessed by 1000-iteration bootstrap resampling. Recombination breakpoints were identified using the program GARD (Kosakovsky-Pond et al., 2006) implemented in the HyPhy software package (Kosakovsky-Pond et al., 2005), through the Datamonkey webserver (Kosakovsky-Pond et al., 2005). The rate of non-synonymous and synonymous substitutions at each codon, and identification of sites evolving under positive or negative selection, was determined using the random effects maximum likelihood models (REL) using default settings (Kosakovsky-Pond and Frost, 2005a, b). A Bayes factor value ≥ 10 was used as an indication of positive selection at a site.
Gene Expression Analysis
Gene expression analyses of lysm1 and lysm2 were carried out in C. rosea strain IK726 under the following conditions: (1) during dual plate interaction with the fungal preys, B. cinerea or F. graminearum, (2) during interaction with wheat roots, (3) in culture filtrates from F. graminearum, and C. rosea itself, (4) in liquid SMS amended with 1% colloidal chitin or 0.1% peptidoglycan, and (5) in germinated conidia in liquid SMS.
For LYSM1 and LYSM2 gene expression analysis during interactions, C. rosea was confronted with B. cinerea (Cr-Bc) or F. graminearum (Cr-Fg) on agar plates as described previously (Dubey et al., 2014b, 2016). The growing front (7–10 mm) of C. rosea was harvested at mycelial contact (C; early physical contact between the mycelia) and at 24 h post mycelial contact (AC; close physical contact). C. rosea confronted with itself (Cr-Cr) was used as control treatment.
For expression analyses of lysm1 and lysm2 in C. rosea during the interaction with wheat roots (Cr-Wr), wheat seeds were surface sterilized with 2% sodium hypochlorite (VWR international, Fontenay-sous-Bois, France) for 10 min under constant shaking condition on a rotary shaker at 100 rpm. The seeds were then washed several times with sterile water to remove traces of chlorine. Surface sterilized seeds were germinated on sterilized moist filter paper placed in 9-cm petri plates (five seeds per plate) in dark at 20°C. Three days old wheat seedlings were inoculated by dipping the roots for 5 min in C. rosea spore suspensions (1e + 07 spore/ml) in sterile condition, transferred back to the filter paper in petri plates and incubated at 20°C in a growth cabinet with 12 h light (CLF plant climates, GmbH, Germany). Roots were harvested 48 h and 96 h post inoculations (hpi).
Gene expression analysis of lysm1 and lysm2 was performed in culture filtrates from F. graminearum, and from C. rosea itself to monitor the response to the exogenous (F. graminearum) and endogenous (C. rosea) secreted factors including chitinases. C. rosea mycelia was cultivated and harvested 2 hpi as described previously (Dubey et al., 2014b, 2016). In brief, an agar plug of C. rosea was pre-cultivated for 5 days in 100 ml flasks containing 20 ml liquid CZ medium on rotary shaker (200 rpm) at 25°C. The mycelia were harvested by filtering through Miracloth. The harvested mycelium was washed with sterile distilled water and then transferred to new flasks containing 20 ml culture filtrates of C. rosea or F. graminearum. Mycelia transferred in fresh CZ medium were used as control treatment. Culture filtrates were obtained by growing C. rosea or F. graminearum in 200 ml liquid CZ medium as described previously (Dubey et al., 2014b).
For gene expression analysis in medium containing chitin or peptidoglycan (Sigma-Aldrich, St. Louis, MO, United States), C. rosea was pre-cultivated for 5 days in SMS medium supplemented with 1% glucose. Mycelia were harvested 3 days post inoculation (dpi), washed with sterilized distilled water and transferred to new flasks containing 20 ml SMS medium where 1% glucose was substituted with 1% colloidal chitin or 0.1% peptidoglycan as a sole carbon source (Dubey et al., 2012). Colloidal chitin was prepared from crab-shell chitin (Sigma-Aldrich, St. Louis, MO, United States) as following the procedure described previously (Seidl et al., 2005). Mycelia transferred in fresh SMS medium with 1% glucose was used as control treatment. Fungal mycelia were collected 2 hpi and washed in distilled water.
For lysm1 and lysm2 gene expression analysis in C. rosea germinated conidia (GC), conidia were harvested in sterile water from the surface of 2 weeks old PDA plates and the concentration was determined under the microscope using a bright line hemocytometer (Sigma-Aldrich, St. Louis, MO, United States). A conidial suspension (1e+07) was inoculated in 50 ml SMS medium to germinate. Samples were harvested 24 hpi by centrifugation. Agar plugs from the growing mycelial front were inoculated in 50 ml SMS medium to grow for 24 h and used as control treatment to compare gene expression in germinating conidia to mycelia. Harvested samples were immediately frozen in liquid nitrogen and stored at −80°C.
Gene expression analysis of PR1 (GenBank accession number: AF384143.1) and PR4 (Wheatwin-1; GenBank accession number: CAA06856.1) gene was performed in wheat roots inoculated with C. rosea WT and lysm deletion strains. The C. rosea – wheat interaction experiment was set up as described above and roots were harvested 120 hpi.
RNA was extracted using the Qiagen RNeasy kit following the manufacturer’s protocol (Qiagen, Hilden, Germany) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, United States). Transcript level was quantified by quantitative reverse transcription PCR (RT-qPCR) using gene specific primer pairs (Supplementary Table S3) as described previously (Kamou et al., 2016). For lysm1 and lysm2 gene expression analysis in C. rosea, relative expression levels for the target gene in relation to actin (GenBank accession number: MT037018; Supplementary Table S3) and β-tubulin gene (GenBank accession number: AF358197.1; Mamarabadi et al., 2008) were calculated from threshold cycle (Ct) values using the 2–ΔΔCt method (Livak and Schmittgen, 2001). For PR1 and PR4 gene expression analysis in wheat roots, gene specific primers described previously (Roberti et al., 2008; Marshall et al., 2011) were used and expression data were normalized to expression of the wheat β-tubulin gene (GenBank accession number: U76744.1). Gene expression analysis was carried out in three or five biological replicates, each based on two technical replicates.
Construction of Deletion Vector, Transformation and Mutant Validation
Three fragment multisite gateway cloning system (Invitrogen, Carlsbad, CA, United States) was used to construct gene deletion vectors. The ∼ 1 kb 5′-flank and 3′-flank regions of lysm1 and lysm2 were amplified from genomic DNA of C. rosea using gene specific primer pairs 568 ups F/568 ups R and 568 ds F/568 ds R; and 6500 ups F/6500 ups R and 6500 ds F/6500ds R, respectively, as indicated in Supplementary Figure S3. Gateway entry clones of the purified 5′-flank and 3′-flank PCR fragments were generated as described by the manufacturer (Invitrogen, Carlsbad, CA, United States). The hygromycin resistance cassette (hygB) generated during our previous studies (Dubey et al., 2012, 2013a) from pCT74 vector, as well as a chlorimuron ethyl resistance cassette (ILV1) generated as a PCR product from the pCB1532 vector (Sweigard et al., 1997), were used. The gateway LR recombination reaction was performed using entry plasmid of respective fragments and destination vector pPm43GW (Karimi et al., 2005) to generate the deletion vectors.
Complementation cassettes for lysm1 and lysm2 were constructed by PCR amplification of the full-length sequence of lysm1 and lysm2 including more than 1 kb upstream and around 500 bp downstream regions from genomic DNA of C. rosea WT using 568 ups F/568 comp R and 6500 ups F/6500 comp R primers, respectively (Supplementary Table S3). The amplified DNA fragments were purified and integrated into destination vector pPm43GW as described above using Gateway cloning technology to generate complementation vectors.
Agrobacterium tumefaciens-mediated transformation (ATMT) was performed based on a previous protocol for C. rosea (Utermark and Karlovsky, 2008). Transformed strains were selected on plates containing either hygromycin for single gene deletion; chlorimuron ethyl for complementation; and both antibiotics, in the case of double gene deletion. Validation of homologous integration of the deletion cassettes in putative transformants were performed using a PCR screening approach with primer combinations targeting the hygB cassette (i.e., for lysm1 or lysm2 deletions) or ILV1 cassette (i.e., for lysm1lysm2 double-deletions) and sequences flanking the deletion cassettes (Supplementary Figure S3), as described previously (Dubey et al., 2013a, b). The PCR positive transformants were tested for mitotic stability, and were purified by two rounds of single spore isolation (Dubey et al., 2012). Transcript level of lysm1 and lysm2 on WT, and respective gene deletion and complementation strains, was determined by RT-PCR using RevertAid premium reverse transcriptase (Fermentas, St. Leon-Rot, Germany) and their respective primer pairs (Supplementary Table S3).
Phenotypic analyses experiments were performed with C. rosea WT, single deletion strain lysm1 (Δlysm1) and lysm2 (Δlysm2) and their respective complemented strains Δlysm1+ and Δlysm2+, and two lysm1lysm2 double deletion strains (Δlysm1Δlysm2A and Δlysm1Δlysm2B). Each experiment included three to five biological replicates (depending on the phenotype), and each experiment was repeated two times with similar results unless otherwise specified.
Phenotypic Analyses
To analyze the growth rate and colony morphology, a three mm agar plug from the growing mycelial front was transferred to solid PDA or CZ and colony diameter was measured after 5 days of growth at 25°C. Colony morphology was observed weekly, photographs were taken by scanning the Petri dishes using Epson Perfection V700 Photo scanner (Epson, Suwa, Japan), and pixel intensity was analyzed with the ImageJ image analysis software (Schneider et al., 2012). Pixel intensity values from the colony morphology of the lysm deletion and complementation strains were normalized to the values of the WT and were used as proxy for mycelial disintegration and biomass measurement.
Conidial yield and germination was determined following the procedure described before (Fatema et al., 2018). For conidiation analysis, conidia were harvested from a 12-day-old plate in 10 ml distilled water, and filtered through Miracloth to remove the mycelial debris. Conidial concentration was determined under the microscope using a bright line hemocytometer (Sigma-Aldrich, St. Louis, MO, United States).
For conidial germination, a conidial suspension (1e+05) was placed on microscope slides with half-strength PDA. The slides were then transferred to nine-cm plates, covered with the lid and incubated at 25°C in dark. After 8 and 24 h of incubation, photographs of the germinating conidia were recorded in a Leica DM5500M Microscope at 20 X magnification equipped with a Leica DFC360FX digital camera (Wetzlar, Germany). The experiment was performed in four biological replicates. For conidia germination, two hundred conidia were counted while 50–100 conidia were used for germ tube measurement for each replicate. The growth rate and conidiation experiment was performed in four biological replicates.
Antagonistic behavior against the phytopathogenic fungi, B. cinerea and F. graminearum, was tested using an in vitro plate confrontation assay on PDA medium as described before (Dubey et al., 2014a, b, 2016). The growth of B. cinerea and F. graminearum was measured daily until their mycelial front touched the C. rosea mycelial front, while the growth of C. rosea strains was measured until the fungus reached another side in the plate. Plates were scanned and the photographs were used to measure the mycelial biomass as described above.
To test the hyphal protection of C. rosea WT, deletion and complemented strains against secreted factors including the hydrolytic enzymes from B. cinerea or F. graminearum, mycelial biomass production of C. rosea WT and deletion strains was measured in culture filtrates from B. cinerea or F. graminearum. The mycelial biomass was measured following procedure described previously (Dubey et al., 2014a, b, 2016). The plate confrontation assay and culture filtrates test was performed in five biological replicates.
Protoplast Formation Assay
Protoplasts formation was done as previously described by Sun et al. (2017), with some modifications. Spores (1e+06) of C. rosea WT and lysm deletion strains were incubated in 250-mL flasks containing 20 mL of potato dextrose broth (PDB) and placed at 26°C for 2 days. The culture was mixed 1:1 with 1.4 M NaCl solution containing 80 mg mL–1 of snailase enzyme mix (Bio Basic Inc., Markham, Ontario Canada) to hydrolyze the mycelia. Chitinase activity of snailase enzyme mix has been shown previously (Boller et al., 1983; Xiao et al., 2007). The cultures were placed in a water bath at 28°C for 3 h. Protoplast release was monitored by removing 200 μL aliquots every 30 min and counting using a hemocytometer (Sigma-Aldrich, St. Louis, MO, United States). The experiment was performed in five replicates.
Enzyme Activity Assays
To measure chitinase activity, conidial suspensions (1e+07) were inoculated into 100 ml flasks containing 50 ml of liquid SMS with 1% glucose and were shaken on rotary shaker (150 rpm) at 25°C. Chitinase activity was measured using (GlcNAc)2 conjugated to 4-methylumbelliferyl (4-MU) as substrates (Sigma-Aldrich, St. Louis, MO, United States) as described previously (Dubey et al., 2012). Fluorescence of released 4-MU was determined by using a luminescence spectrometer model Fluostar Omega (BMG Labtech, Germany) at wave length Ex350/Em455 and gain 1500. The experiment was performed in five biological replicates and each replicate consisted of two technical replicates.
In planta Biocontrol Assay Against B. cinerea and F. graminearum
The biocontrol ability of C. rosea strains against B. cinerea and F. graminearum was evaluated in an Arabidopsis thaliana detached-leaf and fusarium foot rot assay as described previously (Knudsen et al., 1995). Briefly, detached leaves of A. thaliana ecotype Columbia-0 (Col-0) were inoculated with a 5-μl droplet of conidial suspension (1e+06 conidia/ml) derived from C. rosea strains and allowed to dry for 60 min. Then, the same spot was re-inoculated with a 5-μl conidial suspension (1e+06 conidia/ml) of B. cinerea. The diameter of necrotic lesions was measured 60 h post inoculation as described previously (Dubey et al., 2014a, 2016). The experiments were performed in five biological replicates and each replicate consisted of six leaves for each treatment.
For fusarium foot rot assay, surface sterilized wheat seeds were treated with C. rosea conidia (1e+07 conidia/ml) in sterile water, sown in moistened sand, and kept in a growth chamber after pathogen inoculation (Dubey et al., 2014b, 2016). Plants were harvested 3 weeks post inoculation and disease symptoms were scored on 0-4 scale as described before (Knudsen et al., 1995; Dubey et al., 2014b, 2016). The experiment was performed in five biological replicates with 15 plants in each replicate.
Root Colonization
Wheat seeds were surface sterilized and germinated as described in the gene expression section. The roots were then inoculated with conidial suspension of C. rosea strains (1e+07 conidia/ml) as described in gene expression section and were co-cultivated for 5 days at room temperature. For each treatment five biological replicates with five seedlings per replicate were used. To quantify the root colonization, the roots were detached from the plant five dpi, washed with water and used for DNA extraction. The concentration of DNA was determined spectrophotometrically using a nanodrop (Thermo Fisher Scientific, Wilmington, DE, United States) and 100 ng was used to quantify the DNA level of C. rosea strains in wheat roots using qPCR (Dubey et al., 2013a). The actin gene was used as target gene for C. rosea, and Hor1 (Nicolaisen et al., 2009) was used as target gene for wheat. Root colonization was expressed as the ratio between actin and Hor1.
To quantify the internal root colonization, water-washed roots were surface sterilized with 2% sodium hypochlorite (VWR international, Fontenay-sous-Bois, France) for 2 min and washed two times with sterilized distilled water to remove the traces of sodium hypochlorite. The optimum concentration of sodium hypochlorite was selected based on successive screening of C. rosea to this compounds (Dubey et al., 2014a). The roots were blot dried, weighed, and homogenized in two ml phosphate buffer pH 7.4 with 0.01% Triton X-100 using a RETSCH MM 400 Mixer Mill (Thermo Fisher Scientific, Wilmington, DE, United States) with 20 HZ for 15 s. Serial dilutions were plated on PDA plates containing Rose Bengal 50 μg/ml, chloramphenicol 25 μg/ml and kanamycin 50 μg/ml (Sigma-Aldrich, St. Louis, MO, United States) to count the colony forming units (cfus).
Statistical Analysis
Analysis of variance (ANOVA) was performed on gene expression and phenotype data using a General Linear Model approach implemented in Statistica version 13 (TIBCO Software Inc., Palo Alto, CA, United States). Pairwise comparisons were made using the Fisher’s or Tukey–Kramer method at the 95% significance level. Student’s t-test was also performed on gene expression data.
Results
Identification and Sequence Analysis of Predicted LysM Proteins
Analysis of the C. rosea strain IK726 genome version 1 (Karlsson et al., 2015) and version 2 (Broberg et al., 2018) using 35 protein sequences with variable number (1 – 7) of LysM modules and one protein sequences with the additional chitin binding module (Supplementary Table S1) from taxonomically different fungal species, identified three genes predicted to encode proteins containing LysM modules. One gene (protein ID: CRV2G00000260) was already identified as chiC2 (GenBank accession number: MT037003), encoding a killer toxin-like chitinase (Tzelepis et al., 2015). The second gene model predicted to encode protein ID CRV2G00003915 was named lysm1 (GenBank accession number: MT037001). Comparisons between the C. rosea ver. 1 and ver. 2 gene models of the third gene revealed a C-terminal abhydrolase module present in ver. 2 but not in ver. 1. RNA-seq read coverage from Karlsson et al. (2015) showed that the ver. 1 gene model (protein ID: BN869_T00006500) was correct and this was named lysm2 (GenBank accession number: MT037002). The characteristics of LYSM1, LYSM2 and CHIC2 are presented in Table 1.
TABLE 1.
Characteristics of the putative LysM proteins in Clonostachys rosea, C. chloroleuca, and C. solani.
| Name/ | Signal peptide/ | Genomic | Coding | No of | Protein | LysM | No. Cys |
| protein ID | Transmembrane§ | sequences | sequences | introns | length | modules§ | residuesλ̄ |
| LYSM1# | Transmembrane (87–109) | 534 bp | 531 bp | 0 | 177 aa | 1 (127–171) | 4 (3) |
| LYSM2 | Signal peptide (1–23) | 1513 bp | 1392 bp | 2 | 464 aa | 2 (235–280; 289–339) | 22 (3; 3) |
| CHIC2 | Signal peptide (1–20) | 2955 bp | 2598 bp | 2 | 866 aa | 2 (309–354; 372–419) | 32 (3; 5) |
| MT037016# | No | 1440 | 1341 | 1 | 447 aa | 1 (121–165) | 7 (1) |
| MT037017# | No | 852 | 828 | 1 | 276 aa | 3 (5–47; 122–160; 229–275) | 11 (3; 3; 3) |
ChtBD1, chitin-binding (CBM18) module; GH18, glycoside hydrolase family 18 module. #Secreted through a non-classical secretion pathway (SecretomeP score ≥ 0.91). §Number in parenthesis denotes the amino acid positions from N-terminus. CHIC2 contains additional modules ChtBD1 and GH18 between the amino acid position 443–449 and 513–862, respectively. λ̄Number in parenthesis denotes the Cys residues present in each LysM module. Gene encoding putative LysM protein with GenBank accession number MT037016, and gene encoding LysM protein GenBank accession number: MT037017 was found only in C. chloroleuca and C. solani, respectively.
Analysis of translated amino acid sequences for the presence of conserved modules using SMART identified a single 45-amino-acid LysM module (PFAM: PFO1476; InterPro: IPR018392) between positions 127-171 and a 23-amino-acid transmembrane region between positions 87-109 for LYSM1. LYSM2 contained two LysM modules (46 and 50 amino acids, respectively) between positions 235-280 and 289-339, and is predicted to contain a signal peptide in the N-terminus (Figure 1). The CHIC2 protein, contained a 20-amino-acid N-terminal signal peptide, two LysM modules between positions 309–354 and 372–419, an additional chitin-binding (CBM18) module, and a catalytic glycoside hydrolase family 18 (GH18) module (Figure 1), conforming to the known structure of subgroup C-II killer toxin-like chitinases (Tzelepis et al., 2015). These results suggest that LYSM2 and CHIC2 are putatively secreted, and LYSM1 is a putative transmembrane protein.
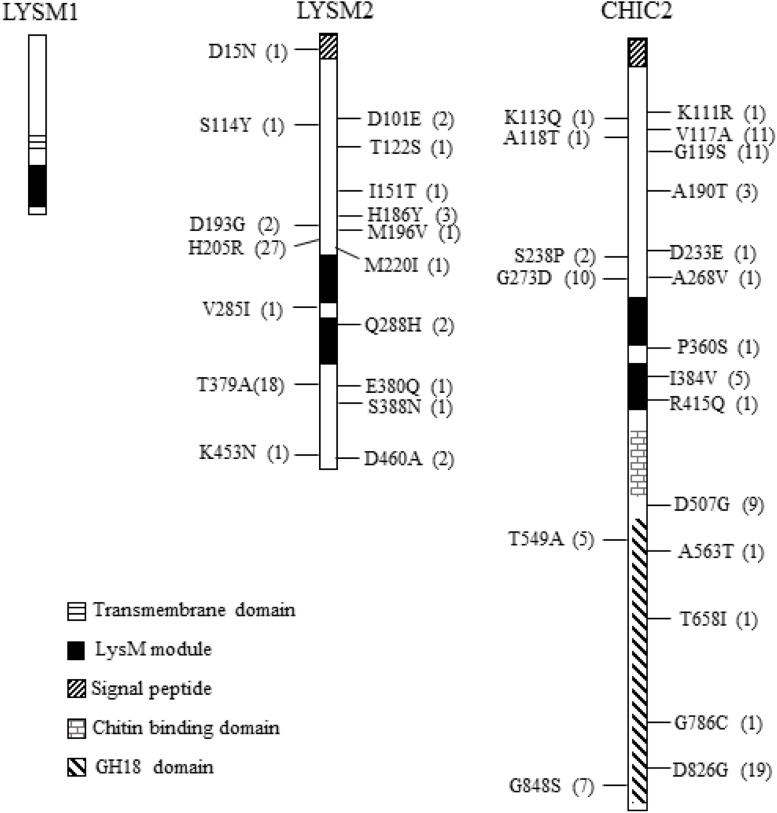
FIGURE 1.
Non-synonymous nucleotide substitution in the lysm1, lysm2, and chiC2 genes in C. rosea strains. LYSM1 has one LysM module between amino acid positions 127–171; LYSM2 and CHIC2 contain two LysM modules between amino acid positions 235–280, 289–339, and 309–354, 372–419, respectively. The single nucleotide polymorphism (SNPs) were compared among worldwide collection of 53 C. rosea strains. The number in parenthesis indicates the number of strains share the same nucleotide polymorphism. Detailed information of all SNPs reported in lysm1, lysm2, and chiC2 can be found in Supplementary Table S2.
The amino acid sequences of LYSM1, LYSM2 and CHIC2 were predicted to contain four, 22 and 32 Cys residues, respectively. From these, three Cys residues were always present in each LysM module and one Cys was always located close, but outside, to the LysM modules suggesting that these LysM modules belonged to the fungal-specific group. Based on our data, LYSM2 is predicted to be LysM effector, and LYSM1 and CHIC2 are LysM-containing proteins (de Jonge and Thomma, 2009). A phylogenetic analysis of the five individual LysM modules from C. rosea, together with LysM modules from other species, was not able to distinguish between internal tandem gene duplication or exon shuffling as the cause of the dual LysM modules present in LYSM2 and CHIC2, due to very low bootstrap support (≤62%, Supplementary Figure S1). In contrast, LysM modules number four, five, six and seven from T. atroviride TAL6 clustered together with 97% bootstrap support, indicating internal gene duplication.
Genome sequence analysis of four different Clonostachys spp. (Broberg et al., unpublished) revealed that C. byssicola, C. rhizophaga, C. chloroleuca, and C. solani each contained three genes orthologous to C. rosea’s lysm1, lysm2, and chiC2. GenBank accession number is provided in Supplementary Table S4. C. chloroleuca and C. solani also contained one additional gene each, predicted to encode a fungal/bacterial (GenBank accession number: MT037016) and fungal-specific (GenBank accession number: MT037017) LysM protein, respectively (Table 1). Sequence alignments of the predicted LysM proteins from five Clonostachys species showed that LYSM1 and CHIC2 were highly conserved between species (98 and 95% amino acid identity, respectively), while LYSM2 showed a lower level (75%) of amino acid identity.
Analysis of LysM Gene Sequence Polymorphisms in Clonostachys rosea
We assessed DNA sequence variation of LysM genes in 53 genome-sequenced strains of C. rosea (Broberg et al., 2018). All these strains contain one copy of lysm1, lysm2 and chic2. Nine synonymous SNPs but no non-synonymous SNPs were identified in lysm1 in 17 strains (Figure 1 and Supplementary Table S2). In lysm2, 14 synonymous and 17 non-synonymous SNPs were identified across three exons involving 38 strains (Figure 1 and Supplementary Table S2). Only four SNPs (three synonymous, one non-synonymous) were located in the two LysM modules (Figure 1 and Supplementary Table S2). In chiC2, 57 synonymous and 20 non-synonymous SNPs were found across two exons of chiC2 in 38 strains, five of which were present in the LysM modules. Two of these SNPs were non-synonymous, I384V and R415Q (Figure 1 and Supplementary Table S2). A MDS analysis revealed a correlation between allele distribution and geographic origin of strains for lysm2 and chic2, but not for lysm1 (Supplementary Figure S2). GARD analysis indicated (P < 0.001) one potential recombination event at position 192 in lysm1 and another potential recombination event at position 531 in chiC2 (Supplementary Figure S2). REL analysis indicated one site evolving under positive selection (Bayes factor = 46) at codon 205 in lysm2 and one site (Bayes factor = 699) at codon 826 in the predicted GH18 module of chiC2 (Figure 1 and Supplementary Figure S2).
Gene Expression Analysis
Gene expression analyses of lysm1 and lysm2 were carried out in C. rosea strain IK726 under conditions relevant for lysm gene expression like fungal-fungal and fungal-plant interactions, in culture media amended with chitin or peptidoglycan as a sole carbon source and during conidia germination.
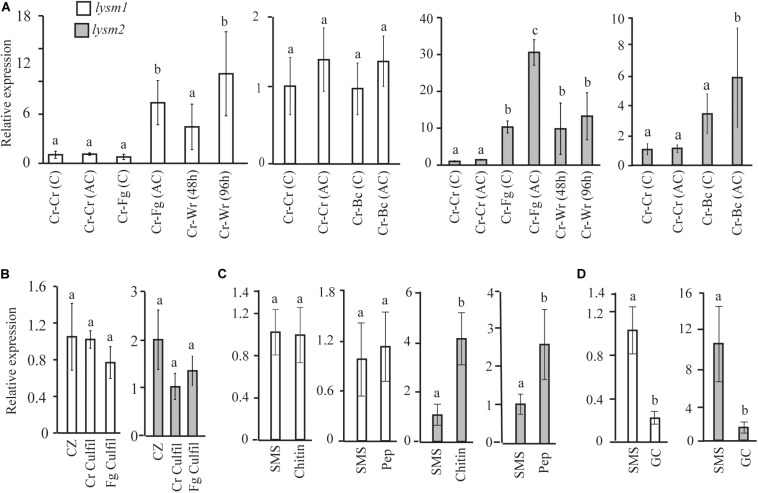
lysm1 and lysm2 gene expression was investigated in C. rosea during dual plate interaction with the fungal prey B. cinerea or F. graminearum at two time points: at the mycelial contact stage and at 24 h after mycelial contact. Gene expression data showed no significant difference in lysm1 expression in C. rosea at neither contact nor after contact stages during interaction with B. cinerea, compared with a C. rosea self-interaction control (Cr-Cr); however, a significant 7.4-fold (P ≤ 0.012) increase in lysm1 expression was recorded during interaction with F. graminearum at 24 h after mycelial contact, compared with Cr-Cr (Figure 2A). Expression of lysm2 was 5.8- fold higher (P ≤ 0.023) during interaction with B. cinerea after the mycelial contact stage, and with F. graminearum at both contact (10.3-fold) and after contact (30.8-fold) stages, compared with Cr-Cr (Figure 2A).
FIGURE 2.
Expression analysis of lysm1 and lysm2 in C. rosea. (A) During in vitro interaction with B. cinerea (Cr-Bc) or F. graminearum (Cr-Fg) at two time points: at contact (C) and at 24 h after contact (AC) stage, and wheat root (Cr-Wr) at 48 and 96 hpi. C. rosea confronted with itself (Cr-Cr) was used as control. (B) Gene expression in self (Cr Culfil) or F. graminearum (Fg Culfil) culture filtrates, mycelia grown in liquid Czapek-dox (CZ) was used as control. (C) Gene expression analysis in C. rosea mycelia grown in liquid SMS medium amended with 1% colloidal chitin or 0.1% peptidoglycan (Pep) as a sole carbon source. SMS medium amended with 1% glucose was used as control. (D) Gene expression analysis during conidial germination. Liquid SMS with 1% glucose was used for the conidial germination. C. rosea mycelia grown in SMS with 1% glucose was used as control. Relative expression level based on RT-qPCR was calculated as the ratio between the target lysm gene and β-tubulin gene using 2–ΔΔCt method (Livak and Schmittgen, 2001), and compared with the respective control. Error bar represent standard deviation based on three or five biological replicates. Statistically significant differences (P ≤ 0.05) in gene expression between treatments were determined using Fisher’s exact test or T-test and are indicated by different letters.
For lysm1 and lysm2 gene expression analyses in C. rosea during interaction with wheat roots, samples were harvested at 48 and 96 hpi. Expression of lysm1 was significantly higher (10.4-fold; P ≤ 0.012) only during colonization of wheat roots 96 hpi, while expression of lysm2 was 9.8-and 13.2-fold higher (P ≤ 0.023) during colonization of wheat roots at both time points, compared with Cr-Cr (Figure 2A).
lysm1 and lysm2 expression analysis was performed in culture filtrates from F. graminearum and from C. rosea itself to investigate the effect of secreted factors including the chitinases on expression of LYSM genes. The culture filtrate from C. rosea itself was used to investigate the possible role of LYSM proteins in protection against self-produced chitinases, and also to investigate common or species specific response in lysm1 and lysm2 expression. No differences in lysm1 or lysm2 expression were found in C. rosea mycelia grown in self-culture filtrates or F. graminearum culture filtrates, compared with the control treatment (Figure 2B).
Expression of lysm1 and lysm2 was further assessed in C. rosea grown in liquid SMS medium amended with 1% colloidal chitin or 0.1% peptidoglycan (Sigma-Aldrich, St. Louis, MO, United States) and compared to SMS medium with 1% glucose. Expression of lysm2, but not lysm1, was significantly higher (P ≤ 0.009) when C. rosea was grown in SMS medium containing colloidal chitin (4.0-fold) or peptidoglycan (2.6-fold) as a sole carbon source, compared with SMS medium with glucose (Figure 2C).
Gene expression of lysm1 and lysm2 in germinated conidia in SMS with 1% glucose was compared with the C. rosea mycelia grown under same conditions. Expression of lysm1 and lysm2 was 4.5-fold and 7.1-fold low (P ≤ 0.019), respectively, during conidial germination compared with expression in C. rosea mycelia (Figure 2D). Expression of lysm1 and lysm2 was observed in all tested conditions.
Generation of Gene Deletion and Complementation Strains
To characterize the biological role of LYSM 1 and LYSM2 in C. rosea, single lysm1 and lysm2, and double Δlysm1Δlysm2 deletion strains were generated through ATMT. Single lysm1 and lysm2 deletion mutants were generated by replacing lysm1 and lysm2 with the hygromycin resistance gene cassette hygB (Supplementary Figure S3A). A double Δlysm1Δlysm2 deletion strain was generated by replacing lysm2 with the sulfonylurea resistance gene selection cassette (ILV1) in a Δlysm1 strain. Successful gene replacement in hygromycin-/sulfonylurea-resistant transformants were confirmed by PCR using primers located within the hygB/ILV1 cassette, together with primers located upstream or downstream of the construct (Supplementary Figure S3A) as described in our previous works (Dubey et al., 2013a, b). The expected size of PCR fragments were amplified in Δlysm1, Δlysm2 and Δlysm1Δlysm2 strains, while no amplification was observed in the WT (Supplementary Figures S3B–D). Furthermore, RT-PCR experiments using primers specific to the lysm1 and lysm2 sequence demonstrated the complete loss of lysm1and lysm2 transcript in each individual and double deletion mutant, while expression of lysm1 and lysm2 were detected in WT (Supplementary Figures S3B,C).
Single Δlysm1 and Δlysm2 strains were complemented with lysm1 and lysm2 through ATMT, respectively. Successful integration of the ILV1 selection cassette in mitotically stable mutants was confirmed by PCR amplification of ILV1. RT-PCR from randomly selected ILV1 positive Δlysm1 and Δlysm2 strains using lysm1- and lysm2-specific primer pairs, demonstrated restored lysm1 or lysm2 transcription in lysm1 and lysm2 complemented (Δlysm1+; Δlysm2+) strains, respectively, while no transcripts were detected in the parental deletion strains (Supplementary Figures S3E,F).
Phenotypic Analyses
Deletion of lysm1 or lysm2 Affects Conidial Germination and Mycelial Integrity
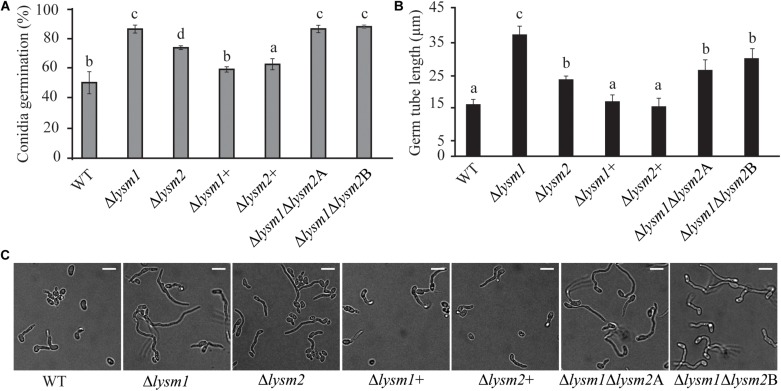
Deletion of lysm1, lysm2, or lysm1lysm2 did not affect the mycelial growth rate and biomass production on PDA or in PDB medium 5 dpi, respectively. Conidial germination and germ tube length were determined after eight h and 24 h of incubation on PDA. After eight h of incubation, lysm deletion strains had significantly (P ≤ 0.007) higher conidial germination (1.5-fold – 1.7-fold) and germ tube length (1.5-fold – 2.3-fold) compared with the WT (Figures 3A–C). However, these differences were no longer detected (P ≥ 0.24) after prolonged incubation for 24 h (Supplementary Figure S4). The conidial germination of the Δlysm1 strain was significantly (P < 0.001) higher compared with the Δlysm2 strain (Figure 3A), while conidial germ tube length of the Δlysm1 strain was significantly (P < 0.001) higher compared with both the Δlysm2 and the Δlysm1Δlysm2 strains (Figures 3B,C). Complementation strains Δlysm1+ and Δlysm2+ showed partial restoration of normal conidial germination levels, and complete restoration of germ tube length (Figures 3A–C).
FIGURE 3.
Analysis of conidial germination and germ tube length 8 h post inoculation on half-strength PDA medium. (A,C) Frequency of germinating conidia was determined by counting the number of germinating and non-germinating conidia. (B,C) Germ tube length was measured with the ImageJ image analysis software (Schneider et al., 2012). Scale bar: 10 μm. Error bars represent standard deviation based on four biological replicates. Different letters indicate statistically significant differences (P ≤ 0.05) within the experiments based on Fisher’s exact test.
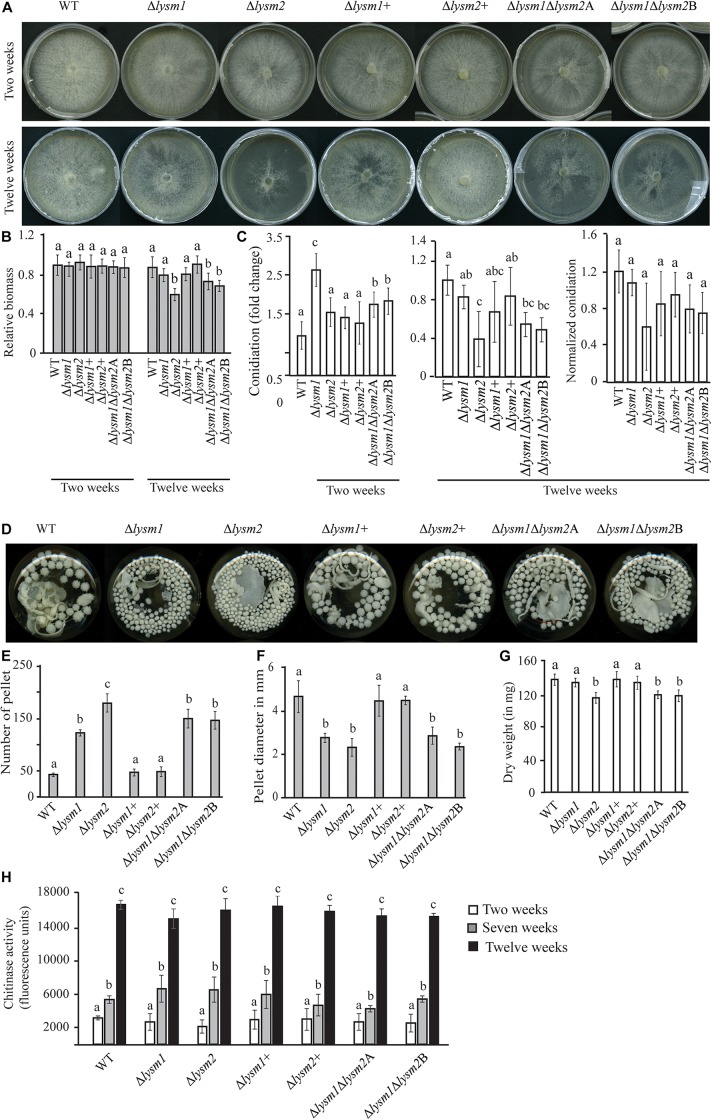
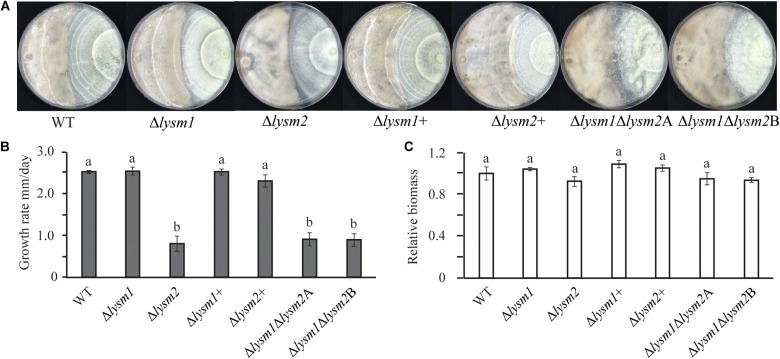
Mycelial colony morphology on PDA was recorded weekly during a 13-week period. An accelerated mycelial disintegration/lysis phenotype leading to significant (P ≤ 0.034) reduction in mycelial biomass was observed 6 weeks post inoculation (wpi) and onward in the Δlysm2 and Δlysm1Δlysm2 deletion strains, compared with the WT strain (Supplementary Figure S5). Twelve wpi, the Δlysm2 and Δlysm1Δlysm2 strains displayed severe (P ≤ 0.005) disintegration/lysis of the colony and reduction in the biomass compared to that of the WT (Figures 4A,B and Supplementary Figure S5). The experiment was repeated three times with the same result.
FIGURE 4.
Phenotypic characterizations of C. rosea WT, deletion and complementation strains. (A,B) Deletion of lysm2 affects hyphal protection. After 2 weeks of inoculation no visible difference in mycelial disintegration was found between the WT and the deletion strains. However, after 12 weeks of inoculation, mycelial disintegration was distinct in the Δlysm2 and Δlysm1Δlysm2 deletion strains. (C) Conidiation of WT, lysm deletion and complementation strains on PDA medium after 2 and 12 weeks of inoculation. The number of conidia from each treatment was normalized to their respective mycelial biomass. (D–F) Deletion of lysm1 or lysm2 affects the mycelial pellet formation in liquid media. (G) Biomass production in C. rosea WT and deletion strains 12 weeks post inoculation. Biomass production was determined measuring mycelial dry weight. (H) Chitinase activity analysis in culture filtrates from C. rosea WT and the deletion strains 2 weeks, 7 weeks and 12 wpi in liquid SMS medium with 1% glucose. Error bars represent standard deviation based on five biological replicates. Different letters indicate statistically significant differences based on Tukey HSD method at the 95% significance level.
Conidial production of C. rosea WT and lysm deletion strains was determined two and 12 wpi by counting the conidia harvested in equal amounts of water. Two wpi, 1.6-fold – 2.6-fold higher (P ≤ 0.001) conidial numbers were recorded in all lysm deletion strains compared with the WT (Figure 4C), indicating that deletion of lysm accelerated conidial production and hence increased conidial yield in the lysm deletion strains 2 wpi. However, the differences in conidial yield between the WT and the Δlysm1 strain were diminished 12 wpi, and were significantly (P ≤ 0.001) lower in the Δlysm2 and the Δlysm1Δlysm2 strains compared with the WT (Figure 4C). In order to confirm whether reduced number of conidia in the Δlysm2 and Δlysm1Δlysm2 strains was related with their mycelial disintegration, the number of conidia produced in the WT and the deletion strains was normalized with their respective mycelial biomass at 12 wpi. After normalization, no significant (P ≥ 0.368) difference in number of conidia was found between the WT and the deletion strains (Figure 4C).
A similar experiment was performed using submerged liquid cultures, where conidia from the WT and lysm mutant strains were inoculated in SMS medium with 1% glucose as a carbon source and incubated at a rotary shaker (150–200 rpm) for 12 weeks. Already after one wpi, a clear difference in size of the mycelial pellets was observed between the WT and the deletion strains. The mycelial pellets of Δlysm1, Δlysm2, and Δlysm1Δlysm2 were 39% – 50% smaller in size, but more numerous, compared with the WT and the complemented strains (Figures 4D–F). In addition, a visible hyphal disintegration was observed in the cultures of the WT and the deletion strains over time, which became more pronounced in cultures of the Δlysm2 and the Δlysm1Δlysm2 strains compared with the WT. In addition, mycelial biomass of the Δlysm2 and the Δlysm1Δlysm2 strains was 16–20% reduced (P ≤ 0.001) compared with the WT 12 wpi (Figure 4G).
In order to test whether biomass reduction were associated with the hydrolytic activity of secreted enzymes, chitinase activity was measured in the culture filtrates from the WT and the deletion strains 2, 7, and 12 wpi. Chitinase activity increased significantly (P ≤ 0.020) over time for all strains (Figure 4H). Chitinase activity was also compared between the WT and the deletion strains. No significant differences in chitinase activity were recorded in the culture filtrates from the WT and the deletion strains two, seven and 12 wpi (Figure 4H).
LYAM1 and LYSM2 Contribute to Hyphal Protection Against Chitinases
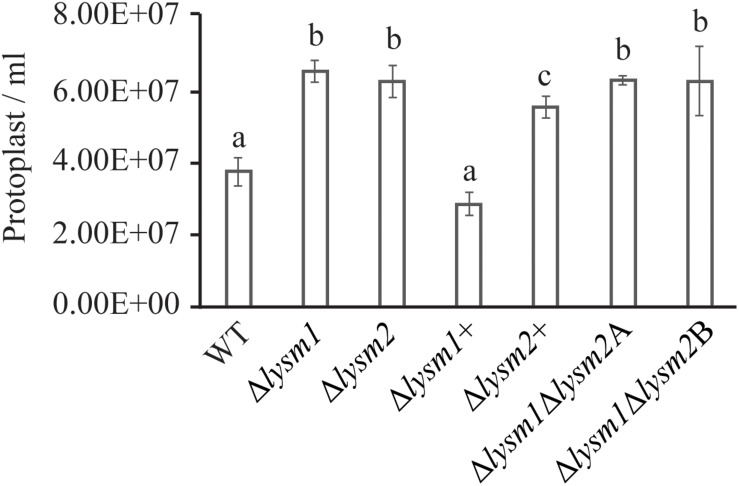
Protoplast release has been used to measure the cell wall stability and hyphal protection against the hydrolytic activity of chitinases (Cen et al., 2017; Romero-Contreras et al., 2019). We evaluated the role of LYSM1 and LYSM2 in C. rosea hyphal protection by measuring the protoplast release after treating the mycelia with lytic enzyme mix including the chitinases. The result showed higher number (P ≤ 0.002) of protoplast were released from the mycelia of Δlysm1, Δlysm2 and Δlysm1Δlysm2 strains compared to the WT strains (Figure 5). No significant difference was found in protoplast release between either of single LYSM deletion strains and double deletion strains. Complementation strains Δlysm1+ and Δlysm2+ showed complete and partial restoration of protoplast release, respectively. The result suggest that LYSM1 and LYSM2 contribute to hyphal protection against exogenous chitinases.
FIGURE 5.
Cell wall protection assay of C. rosea strains against snailase enzyme mix. Protoplast release from C. rosea and deletion strains was counted after 60 min of incubation. The experiment was performed in five replicates. Different letters indicate statistically significant differences based on Fisher’s exact test at the 95% significance level.
Deletion of lysm2 Reduces Antagonistic Ability Toward Botrytis cinerea
Antagonism of C. rosea WT and deletion strains was measured using a dual-culture-plate confrontation assay and a culture-filtrate assay. During dual-culture interactions, C. rosea initially reduces the mycelial growth of the fungal prey, and then, after mycelial contact, starts to overgrow and conidiate on the mycelia of the prey fungus (i.e., F. graminearum and B. cinerea (Dubey et al., 2014a, b, 2016). No differences in growth rate or mycelial biomass of F. graminearum or B. cinerea were recorded during in vitro dual-plate confrontation with either single or double lysm deletion strains, compared with the WT (Supplementary Figure S6). Similarly, no differences in growth rate or mycelial biomass between C. rosea WT and lysm deletion strains were measured during the same conditions (Supplementary Figure S6). However, on average, a 3.0-fold reduced (P ≤ 0.001) growth rate on B. cinerea mycelia (overgrowth rate) was observed in the Δlysm2 and the Δlysm1Δlysm2 strains compared to the growth rate of the WT (Figures 6A,B). A similar experiment against F. graminearum showed no difference in antagonism between the WT and the single or double lysm deletion strains (Supplementary Figure S6). In order to study whether the reduced ability to overgrow B. cinerea was related with a reduction in mycelial biomass of the Δlysm2 and the Δlysm1Δlysm2 strains, mycelial biomass of C. rosea WT and lysm deletion strains was quantified. No significant differences in mycelial biomass was recorded between the WT and lysm deletion strains (Figure 6C).
FIGURE 6.
Mycoparasitism analyses of C. rosea (right side in the plate) strains against B. cinerea (left side in the plate). (A) The Δlysm2 and Δlysm1Δlysm2 strains showed reduced ability to grow on B. cinerea. (B) Growth rate (overgrow) of C. rosea WT and deletion strains on B. cinerea. The growth of C. rosea on B. cinerea was measured from the point of mycelial contact. (C) Mycelial biomass of C. rosea WT and deletion strains 3 weeks post inoculation. Mycelial biomass was quantified by measuring the pixel intensity of the photographs using ImageJ image analysis software (Schneider et al., 2012). The experiment was performed in four biological replicates and photographs of representative plates were taken 3 weeks post inoculation.
In order to investigate a role of LYSM protein in tolerance against secreted chitinases, mycelial biomass of C. rosea WT and lysm deletion strains was measured in the culture filtrates from the prey fungus B. cinerea or F. graminearum. Similar to the result from the dual-culture experiment, no significant differences in mycelial biomass was found between the WT and the lysm deletion strains when grown in culture filtrates from either B. cinerea or F. graminearum (Supplementary Figure S6).
LYSM1 and LYSM2 Contributes to Biocontrol of Botrytis cinerea and Fusarium graminearum
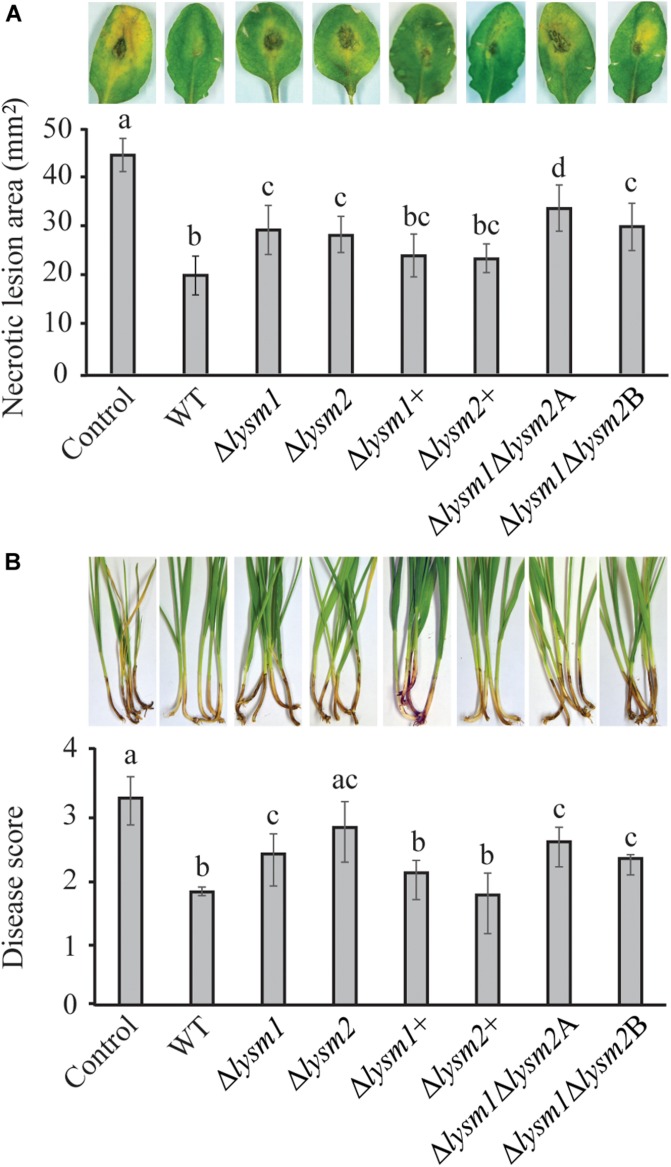
The biocontrol performance of C. rosea WT and lysm deletion strains against the necrotrophic pathogen B. cinerea was determined on detached Arabidopsis thaliana leaves. Significant (P ≤ 0.022) increases of 28.6% – 40% in necrotic lesion area were measured on leaves pre-inoculated with conidia from the Δlysm1, Δlysm2 and the Δlysm1Δlysm2 strains, compared to the necrotic lesion area on leaves pre-inoculated with C. rosea WT spores (Figure 7A). However, complementation strains Δlysm1+ and Δlysm2+ showed partial restoration of the WT phenotype.
FIGURE 7.
In vivo bioassay to test the biocontrol ability of C. rosea strains against B. cinerea and F. graminearum. (A) Measurement of B. cinerea necrotic lesions on detached leaves of A. thaliana plants. The leaves were inoculated with C. rosea strains 60 min before application of B. cinerea and allowed to interact for 60 h. Only pathogen inoculated leaves were used as control. Necrotic lesion area was measured under the microscope using DeltaPix camera and software. The exprement was performed in five biological replicates with six leaves in each replicate in each treatment. (B) Foot rot disease on wheat using sand seedling test. Seedlings were harvested 3 weeks post inoculation and disease symptoms were scored on 0–4 scale. The experiment was performed in five biological replicates with 15 plants in each replicate. Different letters indicate statistically significant differences (P ≤ 0.05) within experiments based on the Fisher’s exact test.
The biocontrol ability of C. rosea strains was further assessed against F. graminearum foot rot disease on wheat plants using a growth chamber sand seedling test. Our result showed 21% – 43% increase (P ≤ 0.031) of disease severity in wheat seedlings previously seed coated with conidia from the Δlysm1, Δlysm2 and the Δlysm1Δlysm2 strains, compared with seedlings from seeds coated with WT C. rosea (Figure 7B). In all cases, complementation of lysm deletions strains restored the biocontrol phenotypes to WT levels. With the exception for biocontrol of fusarium foot rot by the Δlysm2 strain, all other lysm deletion strains did not completely abolish the biocontrol effect as disease symptoms were significantly (P ≤ 0.006) milder compared with the symptoms from control treatments (pathogen inoculation without any C. rosea strain) (Figures 7A,B).
With the exception for biocontrol of B. cinerea by the Δlysm1Δlysm2A strain, no significant differences in biocontrol ability were recorded between any of the lysm deletion strains against the tested pathogens.
LYSM1 and LYSM2 Are Jointly Required for Suppression of Defense Gene Expression in Wheat
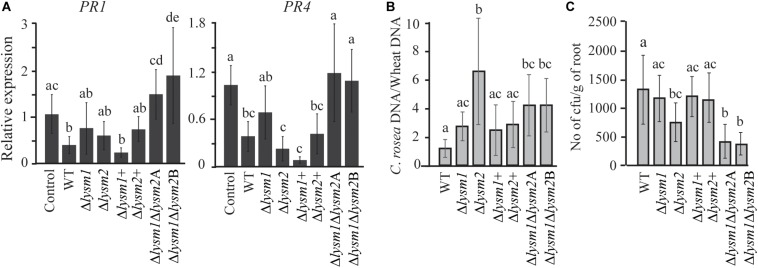
To investigate whether LYSM1 and LYSM2 interfere with plant defense responses, expression of wheat defense response genes PR1 and PR4 (Marshall et al., 2011) was measured with RT-qPCR in wheat roots inoculated with C. rosea WT, lysm deletion and complementation strains. Our data showed a significant 2.63-fold and 2.73-fold (P ≤ 0.032) suppression in expression of PR1 and PR4 genes, respectively, in wheat roots colonized by C. rosea WT, compared with the non-inoculated control (Figure 7A). However, the double deletion strains Δlysm1Δlysm2 failed to suppress expression of both genes in wheat roots to C. rosea WT levels, in contrast with the single Δlysm1 and Δlysm2 mutants (Figure 8A).
FIGURE 8.
Expression analysis of defense related genes and root colonization assay in wheat. (A) Gene expression analysis of defense response related genes in wheat roots colonized by C. rosea strains. Total RNA isolated from wheat roots 5 days post inoculation of WT and Δlysm strains and was used for RT-qPCR. Expression level of PR1 and PR4 genes was normalized to expression of the wheat β-tubulin (Marshall et al., 2011). Relative expression level based on RT-qPCR was calculated using 2–ΔΔCt method (Livak and Schmittgen, 2001), and compared with the water inoculated control. (B) Determination of C. rosea root colonization in wheat roots, 5 days post inoculation, by quantifying DNA level using RT-qPCR. C. rosea colonization is expressed as the ratio between C. rosea DNA and wheat DNA. For DNA quantification actin and Hor1 were used as target gene for C. rosea and wheat, respectively. (C) Internal root colonization by C. rosea strains. Surface sterilized roots were homogenized in phosphate buffer and serial dilutions were plated on Rose Bengal selection plates under sterile condition at 25°C. Colony forming units (cfus) were counted 3 days post plating. Statistically significant differences (P ≤ 0.05) in gene expression between treatments were determined using Fisher’s exact test and are indicated by different letters. Error bars represent standard deviation based on five biological replicates.
Furthermore, in order to analyze the contribution of LYSM1 and LYSM2 in root colonization, the biomass of C. rosea strains on wheat roots was measured 5 dpi by quantifying the ratio between C. rosea DNA and wheat DNA with qPCR. Significantly higher C. rosea/wheat DNA ratios were detected on wheat roots inoculated with the Δlysm2 strain (5.5-fold; P < 0.001) or the Δlysm1Δlysm2 strains (4.2-fold; P ≤ 0.026), compared with roots inoculated with the WT (Figure 7B), suggesting increased biomass of the Δlysm2 and Δlysm1Δlysm2 strains, compared with the WT. No significant differences in C. rosea/wheat DNA ratio were found between WT and Δlysm1, Δlysm1+ or Δlysm2+ inoculated wheat roots (Figure 8B).
Colony forming units (cfus) of surface sterilized roots were determined to measure the internal root colonization levels of C. rosea WT and deletion strains. A significant reduction (P < 0.001) of 47 or 70% in cfus from the roots inoculated with the Δlysm2 or the Δlysm1Δlysm2 strains were found, compared to the roots inoculated with the WT (Figure 8C). No significant differences in cfus were found between the WT and the Δlysm1, Δlysm1+ or Δlysm2+ inoculated wheat roots (Figure 8C).
Expression Analysis of lysm1, lysm2, and chiC2 in the WT and the lysm Deletion Strains
In order to study if deletion of lysm1 results in regulatory feed-back loops that influence expression levels of lysm2 and vice versa, gene expression analysis was performed using RNA extracted from mycelia of WT, Δlysm1 and Δlysm2 strains. RT-qPCR gene expression analysis revealed no significant (P = 0.26) difference in lysm1 expression between mycelia of the WT and the Δlysm2 strain (Supplementary Figure S7A), and similarly, no significant (P = 0.053) difference in lysm2 expression was observed between the WT and the Δlysm1 strain (Supplementary Figure S7B). Expression levels of chiC2 were also analyzed in C. rosea WT, and deletion strains including the complementation and the double deletion strains. Similar to lysm1 and lysm2, no significant difference (P ≥ 0.245) in expression of chiC2 was found between the WT and the deletion strains (Supplementary Figure S7C).
Discussion
Filamentous fungi generally contain multiple genes coding for LysM proteins (de Jonge and Thomma, 2009; Cen et al., 2017), especially in plant pathogens such as F. graminearum (13 genes), F. oxysporum (16 genes), F. verticillioides (17 genes) and in insect pathogens such as B. bassiana (12 genes) and Metarhizium robertsii (13 genes) (de Jonge and Thomma, 2009; Cen et al., 2017). Even certain mycoparasitic and plant beneficial fungi, such as T. atroviride and T. virens, contain high numbers of LysM-encoding genes (12 and 18 genes, respectively). Furthermore, the diversity in LysM gene copy number can vary considerably even between closely related fungal species (de Jonge and Thomma, 2009; Akcapinar et al., 2015; Cen et al., 2017; Kombrink et al., 2017). For instance, genomes of plant root colonizing and mycoparasitic Trichoderma spp. possess an increased number of genes encoding secreted LysM proteins compared with primarily saprophytic Trichoderma spp. (Kombrink and Thomma, 2013; Mendoza-Mendoza et al., 2018). Similarly, the generalist insect pathogen M. robertsii contains 13 LysM genes, whereas only four genes are present in the specialist species M. acridum (Cen et al., 2017). In addition to gene copy number variation, previous studies also report high sequence variation (sometimes driven by positive selection) and modular structure in LysM proteins (Akcapinar et al., 2015; Cen et al., 2017; Kombrink et al., 2017). Taken together, this indicates that ecological niche adaptation drives the evolution of the fungal LysM gene family through a birth-and-death process followed by sequence diversification (Wapinski et al., 2007; Marshall et al., 2011; Cen et al., 2017).
The relatively low number of three LysM genes in the mycoparasitic and plant beneficial fungus, C. rosea, therefore suggests important ecological differences between C. rosea and related hypocrealean plant and insect pathogens. Especially, the difference between C. rosea and Trichoderma spp. is striking, given the apparent similarity in their ecology as necrotrophic mycoparasites and plant beneficial symbionts, and suggests that the role of LysM proteins in C. rosea may differ to those in Trichoderma spp. One important aspect of the LysM gene family in C. rosea is the predicted membrane localization of LYSM1, which is rare in fungi and may suggest a function as a receptor.
Analysis of sequence polymorphism showed that lysm1 is highly conserved, both between C. rosea strains and between Clonostachys species, indicating a conserved function. In contrast, the higher levels of sequence polymorphisms and the presence of codons predicted to evolve under positive selection suggests different evolutionary trajectories of lysm2 and chiC2, compared with lysm1. However, these results should be interpreted with care as the presence of recombination may interfere with the analysis of molecular evolution. Our results are in line with previous data from the leaf pathogen Z. tritici where codons in MgLysM1 and MgLysM3 were shown to evolve under positive selection (Marshall et al., 2011). Different degrees of sequence polymorphisms were also reported in core LysM effectors in the soil borne pathogen, V. dahlia, while no evidence of positive or negative selection was found (Kombrink et al., 2017).
What can then be the function of LYSM1 and LYSM2? The expansion of the LysM gene family in plant, insect pathogenic and mycoparasitic fungi is hypothesized to be associated with their important role in dampening the host defense response by chitin sequestration, consequently avoiding recognition during the interactions with their hosts (de Jonge and Thomma, 2009; Kombrink and Thomma, 2013; Romero-Contreras et al., 2019). This function has been proven for several LysM effectors, for instance Ecp6 from the tomato leaf mold pathogen, Cladosporium fulvum (de Jonge et al., 2010), Mg3LysM from the septoria leaf blotch fungus, Z. tritici (Marshall et al., 2011), Slp1 from the rice blast pathogen, Magnaporthe oryzae (Mentlak et al., 2012), ChELP1 and ChELP2 from the Brassicaceae anthracnose fungus, Colletotrichum higginsianum (Takahara et al., 2016), Vd2LysM from the soil borne vascular wilt fungal pathogen, V. dahliae (Kombrink et al., 2017), Blys2 and Blys5 in the entomopathogen, B. bassiana (Cen et al., 2017). Other LysM effectors are reported to protect the fungal cell wall against chitinases produced by the host, for example in the case of Mg1LysM and Mg3LysM from Z. tritici (Marshall et al., 2011), and Blys5 from B. bassiana (Cen et al., 2017). Furthermore, LysM effector Tal6 from the mycoparasitic fungus T. atroviride has been implicated in mycoparasitic interaction and modulation of host defense response (Romero-Contreras et al., 2019).
The repressed expression of C. rosea lysm1 and lysm2 in germinating conidia, compared with filamentous growth, fits well with a possible function as an inhibitor of spore germination. This hypothesis is further supported by the fact that lysm1 deletion strains displayed increased conidiation, conidial germination and germ tube length, not only compared with the WT but also compared with the Δlysm2 strain. This result is in line of previous findings from T. atroviride, where external application of a purified LysM protein TAL6 displayed a strong inhibitory effect on spore germination in T. atroviride and T. reesei, but not in more distantly related fungi, providing evidence for a role of LysM proteins in regulating developmental processes in fungi (Seidl-Seiboth et al., 2013). Based on the lack of binding of TAL6 to unmodified chito-oligosaccharides or fungal cell wall material, Seidl-Seiboth and co-authors hypothesized that the germination inhibitor function of TAL6 may be exerted through binding of modified chito-oligosaccharides that acts as autoregulatory signaling molecules (Seidl-Seiboth et al., 2013). In the sporulating bacterium, Bacillus subtillis, the two LysM proteins safA and SpoVID localize to the cortex-coat interface of developing spores and interacts with each other and with coat components to induce formation of a protein complex that regulates spore formation (Costa et al., 2006). Although we, after several attempts, were not able to overexpress C. rosea LYSM1 and LYSM2 and test its chitin-binding capacity, we cannot exclude the possibility that LYSM proteins exerts its activity through direct binding of C. rosea cell wall chitin (Romero-Contreras et al., 2019). Irrespectively of the underlying mechanism, the observed alterations in mycelial pellet size in Δlysm1 strain suggest a disturbance in cell wall structure as it is known that fungal pellet formation is the result of spore aggregation that in turn depends on spore cell wall components and their interactions (Zhang and Zhang, 2016). Our results confirm the previous consideration that LysM proteins may act in physiological processes by affecting cell wall modification (Kombrink and Thomma, 2013).
The higher expression of lysm2 during growth on chitin-containing media hints on a possible function of LYSM2 in protecting C. rosea hyphae against endogenous chitinases secreted during external chitin degradation. This hypothesis gain further support by the fact that lysm2 is located adjacent to the killer toxin-like chitinase gene chiC1 in the C. rosea genome (Karlsson et al., 2015; Broberg et al., 2018), and that the transcriptional analyses of lysm2 in this study and chiC1 in a previous study (Tzelepis et al., 2015) showed that both genes are co-induced in medium containing chitin as the sole carbon source. There are several examples of chitinase genes located physically close to co-regulated LysM protein-encoding genes (Gruber et al., 2011; Martinez et al., 2012), indicating an inter-related function of chitinases and LysM proteins (Gruber and Seidl-Seiboth, 2012).
The most distinctive phenotype in the lysm2 gene deletions strain is the gradual increase in hyphal disintegration over time. Mycelial fragmentation and lysis due to secreted chitinases and other fungal cell wall-degrading enzymes has been demonstrated in other filamentous fungi like Aspergillus nidulans, A. fumigatus and T. atroviride (Carsolio et al., 1999; Yamazaki et al., 2007; Shin et al., 2009). However, the disintegration/lysis phenotype in the Δlysm2 strain is not related with increased production of chitinolytic enzymes, as the chitinase activity in Δlysm2 liquid cultures is similar to that of the WT. Similarly, this phenotype is not related to the susceptibility of the deletion strains to chitinases, as Δlysm1 strain that showed similar reduced hyphal protection against chitinases, only a weak disintegration phenotype can be observed. Instead, the most plausible explanation is that LYSM2 acts by protecting the C. rosea cell wall against its own chitinases.
Due to the role of LysM effectors in biotic interactions in plant pathogenic, entomopathogenic and mycoparasitic fungi, we hypothesized that LYSM1 and LYSM2 are involved in mycoparasitic interactions and plant root colonization by C. rosea. The significantly higher expression of lysm1 and lysm2 during interaction with F. graminearum, B. cinerea, and during the colonization of wheat roots, supports a role of the corresponding proteins in biotic interactions in C. rosea. We note that lysm1 is expressed higher later during the interactions compared with lysm2, perhaps reflecting the putative plasma membrane-localization of LYSM1 that requires close interaction for contact with external inducers like chitin derivatives.
Our in vivo bioassay experiment with Δlysm1 and Δlysm2 strains showed that LYSM1 and LYSM2 are both required for biocontrol of gray mold on A. thaliana caused by B. cinerea and of fusarium foot rot disease on wheat caused by F. graminearum. For LYSM1, the proposed function as a regulator of fungal development and conidial germination does not contradict the reduced biocontrol phenotype. In both assays, C. rosea was applied as conidia and any factor that interferes with the normal spore germination program may also influence the highly complex process of biological control. For LYSM2, it is not surprising if the proposed function in protecting the C. rosea cell wall against hydrolytic enzymes also influence biocontrol. Mycoparasitism and biocontrol is a highly complex process that requires signaling between the two combatants, secretion of extracellular enzymes and secondary metabolites, and protection against the counterattack from the fungal prey (Karlsson et al., 2018). Therefore, it is not difficult to envision that a compromised cell wall in the Δlysm1 and Δlysm2 strains, exemplified by the disintegration and pellet size alteration phenotypes, will have a negative effect on biocontrol in C. rosea. From a more mechanistic aspect, secreted hydrolytic enzymes including chitinases are also produced by the prey fungus during competitive fungal-fungal interactions (Langner and Göhre, 2016). However, the lack of regulation of lysm1 and lysm2 gene expression in culture filtrates from F. graminearum, together with the lack of reduced growth of lysm deletion strains in culture filtrates from B. cinerea and F. graminearum, suggests that the reduced biocontrol ability is not primarily associated with secreted factors from the fungal prey. For LYSM2, the failure to overgrow B. cinerea in dual plate assays, but not F. graminearum, shows a certain level of specificity in LYSM2 function. This may be due to protection against fungal cell wall-degrading enzymes from B. cinerea or C. rosea itself that are specifically induced during their interaction, but not during growth in single cultures. Similar results showing host-specificity during mycoparasitism were reported previously in a study involving another mycoparasitic fungus, T. virens, in which deletion of the MAP kinase genes TmkA or TmkB resulted in a mutant with reduced ability to overgrow Sclerotium rolfsii, but not Rhizoctonia solani and Pythium ultimum (Kumar et al., 2010). Furthermore, nox1-overexpressed mutants of T. harzianum showed higher mycoparasitic activity toward P. ultimum, but not against B. cinerea (Montero-Barrientos et al., 2011).
Based on the expression analysis of the defense response marker genes PR1 and PR4 in wheat roots, we show that the C. rosea WT strain is able to suppress the pattern-triggered immunity (PTI) response in wheat. This is in line with previous findings where a transient suppression in peroxidase (higher expression during plant defense) activity was reported in C. rosea-treated wheat coleoptiles and roots (Roberti et al., 2008). Similarly, biocontrol Trichoderma spp. transiently suppresses PTI in A. thaliana by down-regulating genes associated with salicylic acid and jasmonic acid-related genes, and genes coding for plant cytochrome P450 monooxygenases that facilitate synthesis and metabolism of antimicrobial compounds (Morán-Diez et al., 2012; Brotman et al., 2013). Suppression of the host PTI response to establish root colonization has been shown in several plant beneficial microbes (Zamioudis and Pieterse, 2012). Deletion of lysm1 or lysm2 does not influence this ability; however, the failure of the Δlysm1Δlysm2 double deletion strains to suppress expression of PR1 and PR4 to the same extent as the WT strain provides an indication that both LYSM proteins jointly influence this ability. The increased ratio between C. rosea and wheat DNA during root surface colonization and reduced ability of internal root colonization of the Δlysm2 strain, shows that LYSM2 also influences the interaction of C. rosea with plants; although, the exact mechanism will require extensive microscopic investigations of the root colonization process (Saraiva et al., 2015). The role of the LYSM2 protein in internal root colonization is in line with several examples from plant pathogenic fungi where deletion of LysM effector genes reduces their ability to infect the host plant (de Jonge and Thomma, 2009; de Jonge et al., 2010; Marshall et al., 2011; Mentlak et al., 2012; Kombrink and Thomma, 2013; Takahara et al., 2016). The similarity between LYSM2 and previously reported LysM effectors from plant pathogenic fungi in terms of interaction with plants, is intriguing considering the fact that LysM effectors from plant pathogenic fungi belong to the fungal/bacterial group, while C. rosea LYSM2 has a different modular structure than classical LysM effectors and belong to the fungal-specific LysM group.
In summary, we here characterize two LysM proteins in the mycoparasitic fungus C. rosea and show that both proteins influence conidial germination and the stability of the hyphae. Both proteins also influence the ability of C. rosea to control plant pathogenic fungi. Although the phenotypes of LYSM1 and LYSM2 are partially overlapping, differences in gene expression, predicted modular structure and the severity of the phenotypes suggest different functions; LYSM1 primarily as a regulator of fungal development and LYSM2 primarily in protecting the cell wall against hydrolytic enzymes and interactions with plant.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
MD, DJ, and MK conceived and designed the experiments. MD and HV performed the experiments. MD, MB, and MK analyzed the data. DJ and MK contributed to the reagents, materials, and analysis tools. MD and MK wrote the manuscript. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was financially supported by the Department of Forest Mycology and Plant Pathology, Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) (Grant Numbers 220-2014-289, 942-2015-1128, and 2018-01420), and the Centre for Biological Control (CBC) at the Swedish University of Agricultural Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00679/full#supplementary-material
References
- Akcapinar G. B., Kappel L., Sezerman O. U., Seidl-Seiboth V. (2015). Molecular diversity of LysM carbohydrate binding motifs in fungi. Curr. Genet. 61 103–113. 10.1007/s00294-014-0471-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J., Cuomo C. A., van Kan J. A. L., Viaud M., Benito E. P., Couloux A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín-Llovera M., Petutsching E. K., Ried M. K., Lipka V., Nürnberger T., Robatzek S., et al. (2014). Knowing your friends and foes - plant receptor-like kinases as initiators of symbiosis or defence. New Phytol. 204 791–802. 10.1111/nph.13117 [DOI] [PubMed] [Google Scholar]
- Boller T., Gehri A., Vögeli U. (1983). Chitinase in bean leaves: induction by ethylene, purification, properties, and possible function. Planta 157 22–31. 10.1007/BF00394536 [DOI] [PubMed] [Google Scholar]
- Bork P., Letunic I., Doerks T. (2009). SMART 6: recent updates and new developments. Nucleic Acids Res. 37 D229–D232. 10.1093/nar/gkn808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg M., Dubey M., Sun M., Ihrmark K., Schroers H., Li S., et al. (2018). Out in the cold: identification of genomic regions associated with cold tolerance in the biocontrol fungus Clonostachys rosea through genome-wide association mapping. Front. Microbiol. 22:2844. 10.3389/fmicb.2018.02844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotman Y., Landau U., Cuadros-Inostroza Á, Takayuki T., Fernie A. R., Chet I., et al. (2013). Trichoderma-plant root colonization: escaping early plant defense responses and activation of the antioxidant machinery for saline stress tolerance. PLoS Pathog. 9:e1003221. 10.1371/journal.ppat.1003221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist G., Steen A., Kok J., Kuipers O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido) glycans. Mol. Microbiol. 68 838–847. 10.1111/j.1365-2958.2008.06211.x [DOI] [PubMed] [Google Scholar]
- Carsolio C., Benhamou N., Haran S., Cortés C., Gutiérrez A., Chet I., et al. (1999). Role of the Trichoderma harzianum endochitinase gene, ech42, in mycoparasitism. Appl. Environ. Microbiol. 65 929–935. 10.1128/aem.65.3.929-935.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen K., Li B., Lu Y., Zhang S., Wang C. (2017). Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 13:e1006604. 10.1371/journal.ppat.1006604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T., Isidro A. L., Moran C. P., Jr., Henriques A. O. (2006). Interaction between coat morphogenetic proteins SafA and SpoVID. J. Bacteriol. 188 7731–7741. 10.1128/jb.00761-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota V. L., Maffia L. A., Mizubuti E. S. G., Macedo P. E. F., Antunes R. F. (2008). Biological control of strawberry gray mold by Clonostachys rosea under field conditions. Biol. Control 46 515–522. 10.1016/j.biocontrol.2008.04.023 [DOI] [Google Scholar]
- de Jonge R., Thomma B. P. (2009). Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17 151–157. 10.1016/j.tim.2009.01.002 [DOI] [PubMed] [Google Scholar]
- de Jonge R., van Esse H. P., Kombrink A., Shinya T., Desaki Y., Bours R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329 953–955. 10.1126/science.1190859 [DOI] [PubMed] [Google Scholar]
- Dubey M. K., Jensen D. F., Karlsson M. (2014a). Hydrophobins are required for conidial hydrophobicity and plant root colonization in the fungal biocontrol agent Clonostachys rosea. BMC Microbiol. 14:18. 10.1186/1471-2180-14-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey M. K., Jensen D. F., Karlsson M. (2014b). An ATP-binding cassette pleiotropic drug transporter protein is required for xenobiotic tolerance and antagonism in the fungal biocontrol agent Clonostachys rosea. Mol. Plant Microbe Interact. 27 725–732. 10.1094/MPMI-12-13-0365-R [DOI] [PubMed] [Google Scholar]
- Dubey M., Jensen D. F., Karlsson M. (2016). The ABC transporter ABCG29 is involved in H2O2 tolerance and biocontrol traits in the fungus Clonostachys rosea. Mol. Genet. Genomics 291 677–686. 10.1007/s00438-015-1139-y [DOI] [PubMed] [Google Scholar]
- Dubey M. K., Broberg A., Jensen D. F., Karlsson M. (2013a). Role of the methylcitrate cycle in growth, antagonism and induction of systemic defence responses in the fungal biocontrol agent Trichoderma atroviride. Microbiology 159 2492–2500. 10.1099/mic.0.070466-0 [DOI] [PubMed] [Google Scholar]
- Dubey M. K., Broberg A., Sooriyaarachchi S., Ubhayasekera W., Jensen D. F., Karlsson M. (2013b). The glyoxylate cycle is involved in pleotropic phenotypes, antagonism and induction of plant defence responses in the fungal biocontrol agent Trichoderma atroviride. Fungal Genet. Biol. 58-59 33–41. 10.1016/j.fgb.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Dubey M. K., Ubhayasekera W., Sandgren M., Jensen D. F., Karlsson M. (2012). Disruption of the Eng18B ENGase gene in the fungal biocontrol agent Trichoderma atroviride affects growth, conidiation and antagonistic ability. PLoS One 7:e36152. 10.1371/journal.pone.0036152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatema U., Broberg A., Jensen D. F., Karlsson M., Dubey M. (2018). The Polyketide synthase genes are involved in antagonism and biocontrol in the fungal biocontrol agent Clonostachys rosea. Sci. Rep. 8:15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey K. J., Saedi M. S., Ito J. (1986). Nucleotide sequence of Bacillus phage phi 29 genes 14 and 15: homology of gene 15 with other phage lysozymes. Nucleic Acids Res. 14 10001–10008. 10.1093/nar/14.24.10001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S., Seidl-Seiboth V. (2012). Self versus non-self: fungal cell wall degradation in Trichoderma. Microbiology 158 26–34. 10.1099/mic.0.052613-0 [DOI] [PubMed] [Google Scholar]
- Gruber S., Vaaje-Kolstad G., Matarese F., Lopez-Mondejar R., Kubicek C. P., Seidl-Seiboth V. (2011). Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology 21 122–133. 10.1093/glycob/cwq142 [DOI] [PubMed] [Google Scholar]
- Iqbal M., Dubey M., McEwan K., Menzel U., Franko M. A., Viketoft M., et al. (2018). Evaluation of Clonostachys rosea for control of plant-parasitic nematodes in soil and in roots of carrot and wheat. Phytopathology 108 52–59. 10.1094/PHYTO-03-17-0091-R [DOI] [PubMed] [Google Scholar]
- Jensen B., Knudsen I. M. B., Jensen D. F. (2000). Biological seed treatment of cereals with fresh and long-term stored formulations of Clonostachys rosea: biocontrol efficacy against Fusarium culmorum. Eur. J. Plant Pathol. 106 233–242. [Google Scholar]
- Jensen B., Knudsen I. M. B., Madsen M., Jensen D. F. (2004). Biopriming of infected carrot seed with an antagonist, Clonostachys rosea, selected for control of seed borne Alternaria spp. Phytopathology 94 551–560. 10.1094/PHYTO.2004.94.6.551 [DOI] [PubMed] [Google Scholar]
- Jensen B., Lübeck P. S., Jørgensen H. J. L. (2016). Clonostachys rosea reduces spot blotch in barley by inhibiting prepenetration growth and sporulation of Bipolaris sorokiniana without inducing resistance. Pest Manag. Sci. 72 2231–2239. 10.1002/ps.4260 [DOI] [PubMed] [Google Scholar]
- Kamou N. N., Dubey M., Tzelepis G., Menexes G., Papadakis E. N., Karlsson M., et al. (2016). Investigating the compatibility of the biocontrol agent Clonostachys rosea IK726 with prodigiosin-producing Serratia rubidaea S55 and phenazine-producing Pseudomonas chlororaphis ToZa7. Arch. Microbiol. 198 369–377. 10.1007/s00203-016-1198-4 [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10 103–105. 10.1016/j.tplants.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Karlsson M., Atanasova L., Jensen D. F., Zeilinger S. (2018). “Necrotrophic mycoparasites and their genomes. Microbiol Spectrum,” in The Fungal Kingdom, eds Heitman J., Howlett B., Crous P., Stukenbrock E., James T., Gow N. (Washington, DC: ASM Press; ), 1005–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M., Durling M. B., Choi J., Kosawang C., Lackner G., Tzelepis G. D., et al. (2015). Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol. Evol. 7 465–480. 10.1093/gbe/evu292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen I. M. B., Hockenhull J., Jensen D. F. (1995). Biocontrol of seedling diseases of barley and wheat caused by Fusarium culmorum and Bipolaris sorokiniana: effects of selected fungal antagonists on growth and yield components. Plant Pathol. 44 467–477. 10.1111/j.1365-3059.1995.tb01669.x [DOI] [Google Scholar]
- Kombrink A., Rovenich H., Shi-Kunne X., Rojas-Padilla E., van den Berg G. C., Domazakis E., et al. (2017). Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol. Plant Pathol. 18 596–608. 10.1111/mpp.12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink A., Thomma B. P. H. J. (2013). LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 9:e1003769. 10.1371/journal.ppat.1003769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky-Pond S., Frost S. (2005a). Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21 2531–2533. 10.1093/bioinformatics/bti320 [DOI] [PubMed] [Google Scholar]
- Kosakovsky-Pond S., Frost S. (2005b). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22 1208–1222. 10.1093/molbev/msi105 [DOI] [PubMed] [Google Scholar]
- Kosakovsky-Pond S., Frost S., Muse S. V. (2005). HyPhy: hypothesis testing using phylogenies. Bioinformatics 21 676–679. 10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- Kosakovsky-Pond S., Posada D., Gravenor M., Woelk C., Frost S. (2006). Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23 1891–1901. 10.1093/molbev/msl051 [DOI] [PubMed] [Google Scholar]
- Kumar A., Scher K., Mukherjee M., Pardovitz-Kedmi E., Sible G. V., Singh U. S., et al. (2010). Overlapping and distinct functions of two Trichoderma virens MAP kinases in cell-wall integrity, antagonistic properties and repression of conidiation. Biochem. Biophys. Res. Commun. 398 765–770. 10.1016/j.bbrc.2010.07.020 [DOI] [PubMed] [Google Scholar]
- Langner T., Göhre V. (2016). Fungal chitinases: function regulation and potential roles in plant/pathogen interactions. Curr. Genet. 62 243–254. 10.1007/s00294-015-0530-x [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Levin E., Ballester A. R., Raphael G., Feigenberg O., Liu Y., Norelli J., et al. (2017). Identification and characterization of LysM effectors in Penicillium expansum. PLoS One 12:e0186023. 10.1371/journal.pone.0186023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39 D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R., Kombrink A., Motteram J., Loza-Reyes E., Lucas J., Hammond-Kosack K. E., et al. (2011). Analysis of two in planta expressed LysM effector homologues from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156 756–769. 10.1104/pp.111.176347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D. A., Oliver B. G., Graser Y., Goldberg J. M., Li W., Martinez-Rossi N. M., et al. (2012). Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio 3:e00259-12. 10.1128/mBio.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamarabadi M., Jensen B., Jensen D. F., Lübeck M. (2008). Real-time RT-PCR expression analysis of chitinase and endoglucanase genes in the three-way interaction between the biocontrol strain Clonostachys rosea IK726, Botrytis cinerea and strawberry. Fems. Microbiol. Lett. 285, 101–110. 10.1111/j.1574-6968.2008.01228.x [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Zaid R., Lawry R., Hermosa R., Monte E., Horwitz B. A., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32 62–85. 10.1016/j.fbr.2017.12.001 [DOI] [Google Scholar]
- Mentlak T. A., Kombrink A., Shinya T., Ryder L. S., Otomo I., Saitoh H., et al. (2012). Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24 322–335. 10.1105/tpc.111.092957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Barrientos M., Hermosa R., Cardoza R. E., Gutiérrez S., Monte E. (2011). Functional analysis of the Trichoderma harzianum nox1 gene, encoding an NADPH oxidase, relates production of reactive oxygen species to specific biocontrol activity against Pythium ultimum. Appl. Environ. Microbiol. 77 3009–3016. 10.1128/AEM.02486-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán-Diez E., Rubio B., Dominguez S., Hermosa R., Monte E., Nicolas C. (2012). Transcriptomic response of Arabidopsis thaliana after 24 h incubation with the biocontrol fungus Trichoderma harzianum. J. Plant Physiol. 169 614–620. 10.1016/j.jplph.2011.12.016 [DOI] [PubMed] [Google Scholar]
- Nicolaisen M., Suproniene S., Nielsen L. K., Lazzaro I., Spliid N. H., Justesen A. F. (2009). Real-time PCR for quantification of eleven individual Fusarium species in cereals. J. Microbiol. Methods 76 234–240. 10.1016/j.mimet.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33 W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskov S., Jensen B., Knudsen I. M. B., Bodker L., Jensen D. F., Karlinski L., et al. (2006). Soil inoculation with the biocontrol agent Clonostachys rosea and the mycorrhizal fungus Glomus intraradices results in mutual inhibition, plant growth promotion and alteration of soil microbial communities. Soil Biol. Biochem. 38 3453–3462. 10.1016/j.soilbio.2006.06.003 [DOI] [Google Scholar]
- Roberti R., Veronesi A. R., Cesari A., Cascone A., Berardino I., Bertini L., et al. (2008). Induction of PR proteins and resistance by the biocontrol agent Clonostachys rosea in wheat plants infected with Fusarium culmorum. Plant Sci. 175 339–347. 10.1016/j.plantsci.2008.05.003 [DOI] [Google Scholar]
- Romero-Contreras Y. J., Ramírez-Valdespino C. A., Guzmán-Guzmán P., Macías-Segoviano J. I., Villagómez-Castro J. C., Olmedo-Monfil V. (2019). Tal6 from Trichoderma atroviride is a LysM effector involved in Mycoparasitism and plant association. Front. Microbiol. 10:2231. 10.3389/fmicb.2019.02231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva R. M., Czymmek K. J., Borges ÁV., Caires N. P., Maffia L. A. (2015). Confocal microscopy study to understand Clonostachys rosea and Botrytis cinerea interactions in tomato plants. Biocontrol Sci. Technol. 25 56–71. [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroers H. J., Samuels G. J., Seifert K. A., Gams W. (1999). Classification of the mycoparasite Gliocladium roseum in Clonostachys as C. rosea, its relationship to Bionectria ochroleuca, and notes on other Gliocladium-like fungi. Mycologia 91 365–385. 10.1080/00275514.1999.12061028 [DOI] [Google Scholar]
- Seidl V., Huemer B., Seiboth B., Kubicek C. P. (2005). A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. FEBS J. 272 5923–5939. 10.1111/j.1742-4658.2005.04994.x [DOI] [PubMed] [Google Scholar]
- Seidl-Seiboth V., Zach S., Frischmann A., Spadiut O., Dietsch C., Herwig C., et al. (2013). Spore germination of Trichoderma atroviride is inhibited by its LysM protein TAL6. FEBS J. 280 1226–1236. 10.1111/febs.12113 [DOI] [PubMed] [Google Scholar]
- Shin K. S., Kwon N. J., Kim Y. H., Park H. S., Kwon G. S., Yu J. H. (2009). Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot. Cell 8 738–746. 10.1128/EC.00368-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. B., Sun M. H., Zhou M., Li S. D. (2017). Transformation of the endochitinase gene Chi67-1 in Clonostachys rosea 67-1 increases its biocontrol activity against Sclerotinia sclerotiorum. AMB Express 7:1. 10.1186/s13568-016-0313-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton J. C., Li D.-W., Peng G., Yu H., Zhang P., Valdebenito-Sanhueza R. M. (1997). Gliocladium roseum – a versatile adversary of Botrytis cinerea in crops. Plant Dis. 81 316–328. [DOI] [PubMed] [Google Scholar]
- Sweigard J. A., Chumley F., Carroll A. N., Farrall L., Valent B. (1997). A series of vectors for fungal transformation. Fungal Genet. Newsl. 44 52–53. 10.4148/1941-4765.1287 [DOI] [Google Scholar]
- Takahara H., Hacquard S., Kombrink A., Hughes B., Halder V., Robin G. P., et al. (2016). Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol. 211 1323–1337. 10.1111/nph.13994 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis G., Dubey M., Jensen D. F., Karlsson M. (2015). Identifying glycoside hydrolase family 18 genes in the mycoparasitic fungal species Clonostachys rosea. Microbiology 161 1407–1419. 10.1099/mic.0.000096 [DOI] [PubMed] [Google Scholar]
- Utermark J., Karlovsky P. (2008). Genetic transformation of filamentous fungi by Agrobacterium tumefaciens. Nat. Protoc. 10.1038/nprot.2008.83 [DOI] [Google Scholar]
- Wapinski I., Pfeffer A., Friedman N., Regev A. (2007). Natural history and evolutionary principles of gene duplication in fungi. Nature 449 54–61. 10.1038/nature06107 [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18 691–699. 10.1093/oxfordjournals.molbev.a003851 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Li X., Yang X., Luo M., Hou L., Guo S., et al. (2007). Cloning and characterization of a balsam pear class I chitinase gene (Mcchit1) and its ectopic expression enhances fungal resistance in transgenic plants. Biosci. Biotechnol. Biochem. 71 1211–1219. 10.1271/bbb.60658 [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Yamazaki D., Takaya N., Takagi M., Ohta A., Horiuchi H. (2007). A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr. Genet. 51 89–98. 10.1007/s00294-006-0109-7 [DOI] [PubMed] [Google Scholar]
- Zamioudis C., Pieterse C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25 139–150. 10.1094/MPMI-06-11-0179 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang J. (2016). The filamentous fungal pellet and forces driving its formation. Crit. Rev. Biotechnol. 36 1066–1077. 10.3109/07388551.2015.1084262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.