Abstract
Background
Studies on the impact of the skin microbiota on human health have been gaining more attention. Bacteria are associated with various diseases, although certain strains of bacteria, which are known as probiotics, are considered beneficial. Mixtures of several bacteria (bacterial cocktail) isolated from targeted organs have shown promising modulatory activities for use in skin therapeutics. The objectives of this study were to determine and identify the microbial communities on the skin that can potentially be used as probiotics, as determined by bacterial isolation and cultivation, followed by next-generation sequencing (NGS).
Results
Samples were collected by swabbing on forehead and cheek skin. Genomic DNA from bacterial swab samples were directly extracted to be further processed into NGS. Cultivation of skin bacteria was carried out in subsequent medium. Thus, around twenty bacterial isolates with different characteristics were selected and identified by both culture-based method and 16sRNA sequencing. We found that Actinobacteria and Firmicutes are the most abundant phylum present on the skin as presented by NGS data, which constitute to 67% and 28.59% of the whole bacterial population, consecutively. However, Staphylococcus hominis, Staphylococcus warneri, and Micrococcus luteus (AN MK968325.1; AN MK968315.1; and MK968318.1 respectively) were able to be obtained in the samples of cultivable, and could be potentially developed as probiotics in skin microbiome therapeutic as well as for postbiotic formulation.
Conclusion
Skin microbiome is considered to provide several probiotics for skin therapeutic. However, some opportunistic pathogens were discovered in this study population. Thus, the promising formula of bacterial cocktail for skin microbiome therapeutic must be thoroughly elucidated to avoid unwanted species. Our study is the first human skin microbiome profile of Indonesia resulted from a Next Generation Sequencing as an effort to show a representative of tropical country profile.
Keywords: Bioinformatics, Microbiology, Genetics, Micrococcus luteus, Microbiome therapeutics, Probiotics, Postbiotics, Skin, Staphylococcus hominis, Staphylococcus warneri
Bioinformatics; Microbiology; Genetics; Micrococcus luteus; Microbiome therapeutics; Probiotics; Postbiotics; Skin; Staphylococcus hominis; Staphylococcus warneri.
1. Introduction
The human skin is home to large and diverse populations of bacteria, fungi and viruses that collectively compose the skin microbiota [1]. The skin provides diverse microenvironments that vary in acidity, temperature, moisture and sebum content [2]. Cutaneous invaginations and appendages, including the sweat glands (eccrine and apocrine), sebaceous glands and hair follicles, are occupied by their own unique residents [3]. For example, Propionibacterium sp. are the dominant microorganisms in sebaceous areas, whereas Staphylococcus and Corynebacterium are the most abundant species colonising moist areas [1]. The dry sites are reportedly home to a mixed population of microorganisms dominated by Gram-negative bacteria [4].
Recent advances in the function of the skin microbiome have revealed a strong symbiotic relationship between the microbiota of the skin and host. The dysbiosis of the microbiome and other factors influence the surface microbiota and can affect homeostasis and keratinocyte function. The symbiotic relationship between the skin microbiota and host can be used as a guide for microbial exploration for the development of active pharmaceuticals for use as cosmetics and skin care [5].
Conventionally, skin microbial communities are explored with the use of cultivation methods and continued by examinations of physiological and biochemical characteristics, which are relatively time-consuming. Thus, to circumvent the bias imposed by cell culture and to capture the diversity of the microbiome as a whole, investigators began applying sequencing methods using sequence variations in conserved taxonomic markers as molecular fingerprints to identify members of microbial communities [1].
As fecal transplant of gut microbiome has been proven with a successful story to treat certain disease that is approved by FDA [6, 7], skin microbiome may become an interesting therapeutic option in diseases affecting the skin such as psoriasis or acne vulgaris [8]. Interestingly, the modulation of the skin microbiome composition was reported successful which has been carried out by adding mixtures of different skin microbiome components [8], or also known as bacterial cocktail.
The idea was to develop novel therapeutic approach which focuses on the act of molecules that are secreted, modulated, or degraded by the microbiome obtained from skin of healthy people to treat unhealthy ones in a same background population of people.
In this study, determination of skin microbiota population from healthy Indonesian male and female was first conducted by employing Next Generation Sequencing (NGS) of total DNA obtained from direct swab sample targetting 16s rDNA. Further, in order to obtain cultivable bacteria, the same swab samples were cultured to obtain single colonies; single colonies species determination were carried out by 16s rDNA Sanger sequencing.
2. Materials and methods
2.1. Participants
Skin bacterial samples were obtained from the healthy skin of eight young male and female volunteers (age range, 17–25 years) from Depok, West Java, Indonesia, who provided written informed consent. All participants were free from dermatologic diseases (i.e. atopic dermatitis and psoriasis) and were not using antimicrobials. The study participants were asked not to wash their faces for 5 h prior to sampling and not to use cosmetics or skin care products on the day of sampling.
2.2. Collection of skin swab samples
Sampling was carried out in a sterile room by swabbing on forehead and cheek skin of eight healthy young participants aged 17–25 years (four males and four females) with two sterile cotton swabs pre-moistened by recovery diluent (0.9% NaCl and 0.1% Tween-20). All participants were free from dermatologic diseases (i.e. atopic dermatitis, psoriasis) and were not on antimicrobial treatment. They were asked not to wash their face 5 h prior to sampling and not to wear cosmetics and skin care products on the sampling day. The study was approved by the Ethics Committee of the Faculty of Medicine, Universitas Indonesia No. 0049/UN2.F1/ETIK/2019, Protocol Number 19-01-0069 (Supplementary file 2). A written and signed informed consent was obtained from each participant prior to sample collection.
The cotton buds of the swabs were cut aseptically, put into 1 mL of recovery diluent in 2-mL microcentrifuge tubes and then homogenised by vortexing [63]. For Next Generation Sequencing (NGS) analysis, sampling was carried out four times on different days for each subject between February and April 2019 to maximise the recovery of uncultured skin bacteria.
2.3. Bacterial isolation and identification of sample from swab
Liquid samples were serially diluted to 10−1 and 10−2 with recovery diluent. Each diluted sample was then spread on two blood agar plates with the use of glass beads. One plate was cultured at 37 °C for 2 days to culture aerobic bacteria, whereas the other plate was cultured in an anaerobic jar at 37 °C for 2 days to culture anaerobic bacteria. After 48 h of culture, the number of colonies on each medium was counted using a plate counter. Colonies with relatively high abundances were selected for visual identification by colony morphologies and microscopically by Gram staining. The selected colonies were subcultured in 7 mL of tryptic soy broth at 37 °C for 48 h under aerobic and anaerobic conditions. Afterwards, 600 μL of liquid culture was transferred into a sterile 2-mL microcentrifuge tube, and 600 μL of 85% sterile glycerol was added to prepare a frozen stock that was stored at −80 °C.
2.4. DNA extraction
Liquid cultures of bacterial isolates in tryptic soy broth were centrifuged at 14000 × g for 2 min. Genomic DNA (gDNA) was extracted from the bacterial pellet using the Presto™ Mini gDNA Bacteria kit (Geneaid Biotech, Ltd., New Taipei City, Taiwan) in accordance with the manufacturer's protocol. For NGS, liquid samples in recovery diluent were collected into one microcentrifuge tube and centrifuged at 14000 × g for 2 min, and gDNA was directly extracted from the bacterial pellet.
2.5. Skin microbiome analysis by NGS of 16S rRNA
The concentration and purity of the gDNA were visualised on 1% agarose gels. According to the concentration, the gDNA was diluted to 1 ng/μL with sterile water. The V3–V4 regions of 16S rRNA (16S) were amplified using specific primers (i.e. 16S V4: 515F-806R). Polymerase chain reaction (PCR) analysis was conducted using Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA). The PCR products were mixed with the same volume of 1× loading buffer (containing SYB green fluorescent cyanine dye) and then separated by electrophoresis on 2% agarose gels. Samples with bright bands of 400–450 bp were chosen for further analysis. gDNA libraries were generated using the Ion Plus Fragment Library Kit (Thermo Fisher Scientific, Waltham, MA, USA) and quantified by Qubit fluorometric quantitation and quantitative PCR. The samples were sequenced using the Ion S5 XL System (Thermo Fisher Scientific).
2.6. 16S rDNA PCR and sequencing of cultivable bacterial isolates
Two universal primers (27F: 5′-AGA GTT TGA TCM TGG CTC AG-3′ and 534R: 5′-ATT ACC GCG GCT GCT GG-3′) [47] were used to amplify the bacterial 16S rRNA gene-coding region. Amplification reactions were performed in a total volume of 50 μL containing 6 μL of extracted gDNA as the template, 6.5 μL of ddH2O, 25 μL of 2× KOD Fx Neo Buffer, 10 μL of 2 mM deoxyribonucleotide triphosphate, 0.75 μL of 20 mM primers 27F and 534R and 1 μL of KOD Fx Neo polymerase (Toyobo Co., Ltd., Osaka, Japan). PCR amplification was performed using a standard thermocycler (Biometra GmbH, Göttingen, Germany) under the following parameters: 95 °C for 5 min, followed by 30 cycles at 95 °C for 30 s, 63 °C for 30 s, 68 °C for 40 s and a final extension at 68 °C for 6 min. The DNA sequences of the PCR products were analysed using the Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/BLAST) for species identification.
2.7. Nucleotide sequence accession numbers
The 0.4–0.5-kb sequences of the 16S rDNA of 13 isolates determined in this study were deposited in the GenBank database under the accession numbers listed in Supplementary file 1.
2.8. Limitations
We limited our study to 20 isolates obtained from 8 subjects for cultivable method and 6 subjects for NGS profiling, thus statistical bias was possible.
3. Results
3.1. Microbiome profile
Skin microbiota samples from both female and male volunteers were analysed using the NGS technique. A total of 129744 reads in the 16S rRNA gene library were grouped into operational taxonomic units (OTUs) based on the genetic distances on a neighbour-joining tree (Figure 1). A GraphlAn figure portraying the taxonomy tree of each group separated by their phyla was generated (Figure 2). The top 10 genera with the greatest relative abundances included Corynebacterium, Brevibacterium, Brachybacterium, Kocuria, Micrococcus, Propionibacterium, Truepera, Staphylococcus, Streptococcus and Paracoccus (Figure 3). From the top 10 genera, 21 bacterial species with the highest relative abundance were obtained. We categorized the bacteria as commensal, opportunistic pathogen, or potential probiotic according to the literature study as described in Table 1.
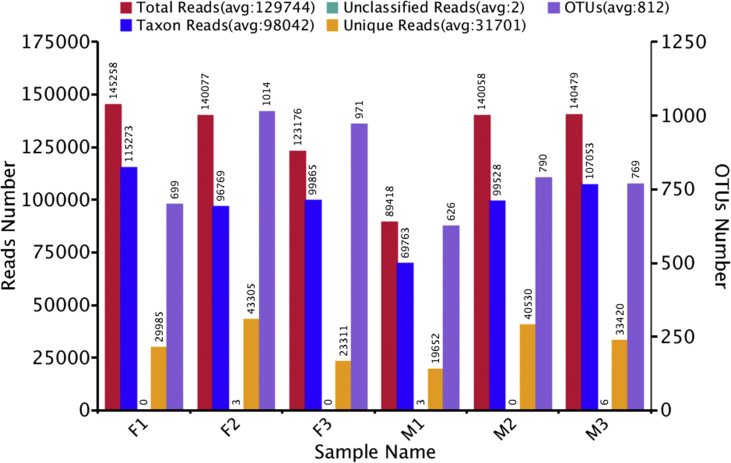
Figure 1.
Reads and OTUs number of each samples. Female samples (F1, F2, F3) showed higher reads and OTUs number compared to male samples (M1, M2, M3).
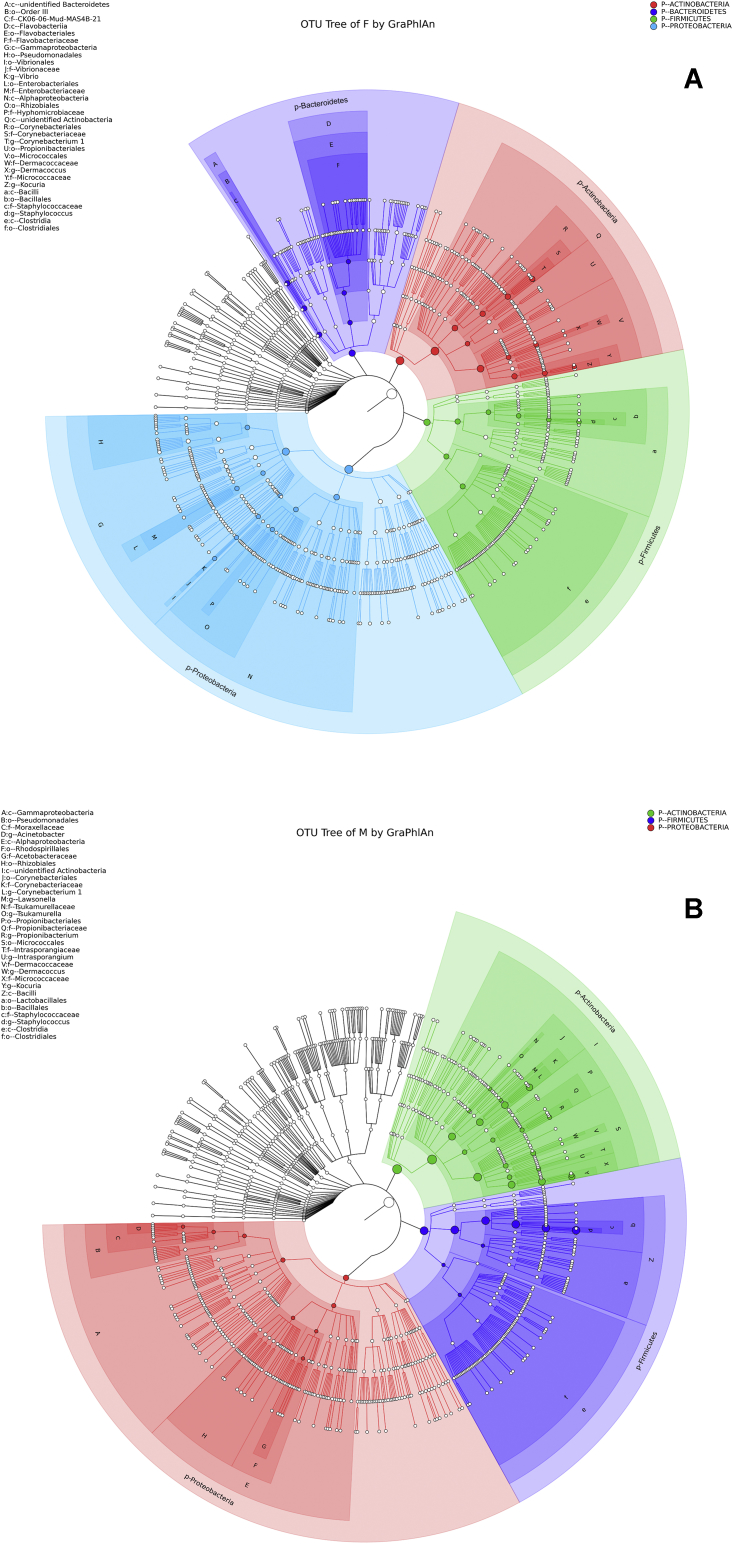
Figure 2.
OTU annotation tree construct of female sample group (A) and male sample group (B). The size of circles signifies the abundance of species. Solid circles stand for the top 40 species in high abundance. In female sample group, the most abundant species were from phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria marked by red, indigo, blue, and green colors respectively. In male sample group the most abundant species were from phyla Actinobacteria, Firmicutes, and Proteobacteria, marked by green, indigo, and red colors respectively.
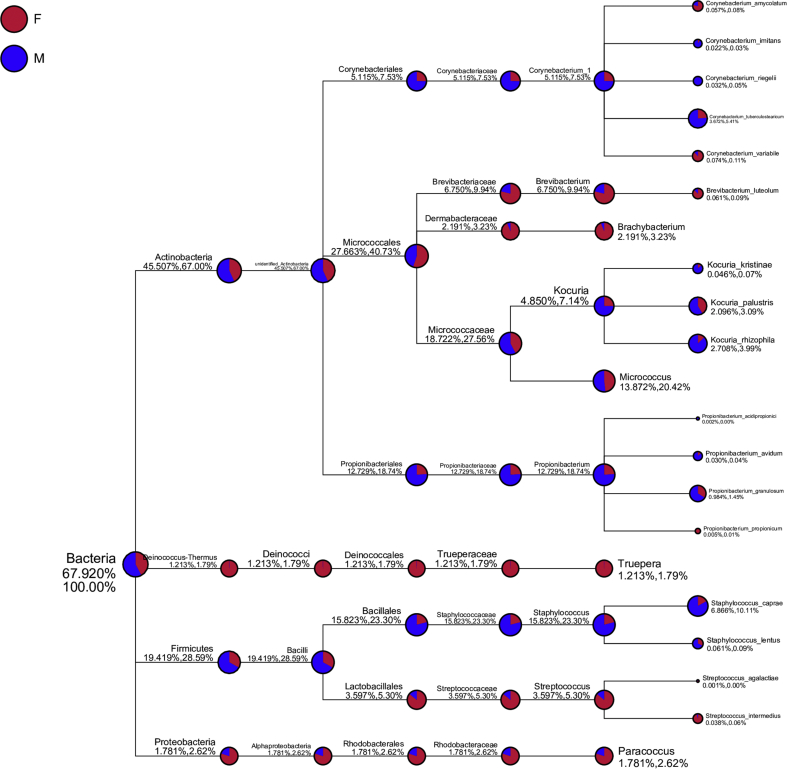
Figure 3.
Taxonomy tree of specific species from top 10 genus in high relative abundance. 21 bacterial species with highest relative abundance were identified based on top 10 genus Corynebacterium, Brevibacterium, Brachybacterium, Kocuria, Micrococcus, Propionibacterium, Truepera, Staphylococcus, Streptococcus, and Paracoccus. Relative abundance in female sample group (F) was marked as red, and male sample group (M) was marked as blue.
Table 1.
Characteristics of detected abundant species.
| Species | Characteristics | References |
|---|---|---|
| Corynebacterium amycolatum | Opportunistic pathogen, natural flora on human skin and mucous membranes, often found in immunosuppressed patients | [9, 10] |
| Corynebacterium imitans | Isolated from patients with suspected diphtheria | [11] |
| Corynebacterium riegelli | Isolated from female patients with urinary tract infections | [12] |
| Corynebacterium tuberculostearicum | Normal human skin microbiota, opportunistic pathogen | [13] |
| Corynebacterium variabile | Non-pathogenic, isolated from smear-ripened cheeses | [14] |
| Brevibacterium luteolum | Potential pathogenic (case report study), potential biosurfactant | [15] |
| Brachybacterium sp. | Isolated from gut microbiota of healthy infants | [16] |
| Kocuria kristinae | Non-pathogenic commensal can cause opportunistic infections | [17] |
| Kocuria palustris | Isolated from the rhizoplane of the narrow-leaved cattail | [18] |
| Kocuria rhizophila | Opportunistic pathogen | [19] |
| Micrococcus sp. | Possesses antimicrobial activity against food-borne pathogens | [20] |
| Propionibacterium acidipropionici | Probiotic microorganism modifies number of anaerobes and coliform caecal content | [21, 22] |
| Propionibacterium avidum | Cause of soft tissue infections, potential immunomodulator, treatment of neoplastic and infectious diseases, potential source of active metabolites | [23, 24, 25] |
| Propionibacterium granulosum | Immunomodulatory potential | [26] |
| Propionibacterium propionicum | Cause infections of the lacrimal apparatus | [27] |
| Truepera sp. | Radiation resistant species, possessing thermostable amylosucrase activity | [28, 29] |
| Staphylococcus caprae | Pathogen, causes bacteraemia and native bone infection | [30] |
| Staphylococcus lentus | Pathogen, causes peritonitis | [31] |
| Streptococcus agalactiae | Human pathogen that colonises the urogenital and/or the lower gastro-intestinal tract | [32] |
| Streptococcus intermedius | Pathogen which causes periodontitis and pyogenic infections in the brain and liver | [33] |
| Paracoccus sp. | Probiotic dietary supplement for livestock | [34] |
3.2. Bacterial strain identification
To identify the cultivable bacteria, 20 selected bacterial isolates from the eight volunteers were further phenotypically analysed by colony morphology and Gram staining. Parameters for morphological identification, such as pigment, shape, haemolysis, elevation, margin and surface characteristics, were considered for selection (see Supplementary file 1). The purity of individual species was confirmed by sub-culturing single colonies on new agar plates two times. All selected colonies were Gram-positive. This finding is supported by a previous study [35] confirming that the most prevalent bacteria on the skin were Gram-positive, which included various species of the genera Staphylococcus, Propionibacterium and Corynebacterium. Low and inconsistent attainment of Gram-negative bacteria in the culture method is predictably caused by the transient nature of such species [35]. Gram-negative bacterial species are believed to not include commensal skin microbiota because of the associations with environmental factors, such as the high humidity and climate of Depok City, Indonesia. However, Gram-negative bacteria are found on dry areas of the skin [4].
3.3. Molecular identification of bacterial isolates
DNA of selected bacterial isolates was extracted and analysed by 16S rDNA sequencing for molecular identification. As shown by the phylogenetic tree, the dominant isolates belonged to the phyla Firmicutes and Actinobacteria. This finding was in agreement with the NGS results and was supported by microscopic observation of morphology. S. hominis, S. warneri and M. luteus are potential probiotic strains [36]. These bacteria can be exploited either individually or as a bacterial cocktail for the development of microbial therapeutics for tropical diseases of the skin.
4. Discussion
As the first-line defensive barrier, the human skin is occupied by a diversity of bacteria that are potential pathogens or probiotics. Our study employed swabbing as sample collection method, as it was shown not to significantly differ with other methods i.e scrape and punch, without being invasive to the subjects [2]. The results of the present study showed that the dominant bacteria on the skin samples were members of the phyla Firmicutes and Actinobacteria (Figure 4). Bacterial composition of the skin microbiota differs depending on location, constitution, appendages, and topographical variability [37]. In this result, abundance of Staphylococcus and Propionibacterium reflected facial skin site as sebaceous. Age and sex also plays important role in variability of skin microbiota compositions, as described in skin microbiota survey on Chinese, Finnish, and Brazilian populations [38, 39, 40]. Diseased skin in particular, e.g atopic dermatitis (AD), leprosy, and hidradenitis suppurativa (HS) conditions are characterized by apparent changes in skin microbiota composition, such as underrepresentation of Firmicutes and increase in Proteobacteria for leprosy patients and appearance of genus Porphyromonas and Peptoniphilus in HS patients [39, 41, 42]. Unique microbial activities and communities on different populations may serve valuable information to target diseases based on predictive models, or utilized beneficial health purposes [43, 44]. Of the most abundant species from the top 10 genera, five bacterial strains that are prospective candidates for probiotics/postbiotics for use as skin microbiome therapeutics were Micrococcus sp., Propionibacterium acidipropionici, Propionibacterium granulosum, Propionibacterium avidum and Paracoccus sp, while others are considered as opportunistic pathogens.
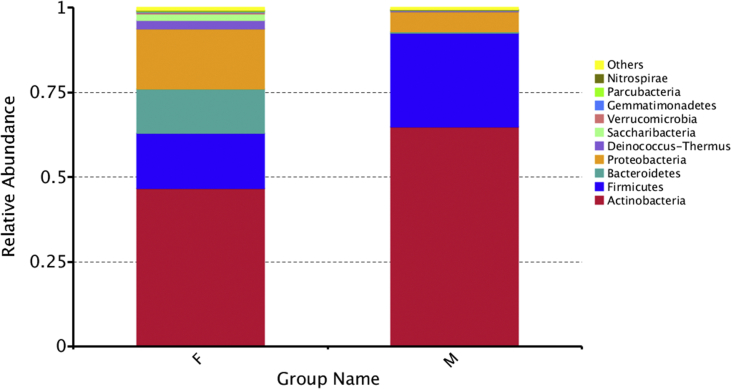
Figure 4.
Relative abundance at phyla level of (F) female and (M) male subjects.
According to Akbar et al. [19], Micrococcus luteus has antibacterial activities against the foodborne pathogens Salmonella typhimurium, Listeria monocytogenes, Escherichia coli, Klebsiella pneumoniae and Staphylococcus aureus. M. luteus is especially potent against S. typhimurium and produces essential enzymes, including protease, lipase and phytase [20]. Propionibacterium sp. found in dairy products are known for their probiotic activities. In particular, P. acidipropionici has received attention for its capability to modify the composition and metabolic activities of the intestinal microflora [22]. Recent studies, however, have reported that, other than dairy propionibacteria, cutaneous bacteria have potential health benefits [24]. The commensal skin residents P. avidum and P. granulosum are potent immunomodulators that were found to stimulate granulopoiesis in an in vivo study of swine [45]. Furthermore, P. avidum improved the quality of life of colorectal carcinoma patients by immunostimulation against cells with a cytotoxic phenotype [26]. Paracoccus sp. were initially used as probiotics for livestock because of the high contents of docosahexaenoic acid and eicosapentaenoic acid, which have potential to be further developed for skin therapeutics because of recent advancements in the attenuation of cutaneous inflammation by polyunsaturated fatty acids, which regulate cytokine synthesis and activities to promote wound healing [46].
In addition to NGS, culturing followed by 16S rDNA sequencing was performed to isolate potential probiotic strains. In agreement with the NGS results, Actinobacteria and Firmicutes were the dominant phyla. This result corresponds to another study executed among small Brazilian population which showed Firmicutes is the most dominant bacteria population presented on the skin, followed by other phylum such as Proteobacteria, Actinobacteria, and Bacteroidetes [39]. This finding also emphasize that geographical setting may affect the represented phyla, as Indonesia and Brazil have almost similar climate.
However, several strains have not been successfully isolated, namely, Brevibacterium, Brachybacterium, Propionibacterium, Paracoccus and Truepera. In particular, it is difficult to detect Propionibacterium as a member of the skin microbiota owing to its fastidious growth in cultivation medium, the aerotolerant anaerobic characteristics resulting in difficulties with the swabbing method, high subjectivity during colony selection and inappropriate growth conditions. Furthermore, the 16S rDNA regions can also affect the representation of several genera [47]. In most cases, sequencing of the V1–V3 region generates the best 16S-based profile. However, these regions were unable to precisely classify genera when applied for OTU-based methods. On the other hand, sequencing of the V4 region was used in the Earth Microbiome Project [48], but underrepresents Propionibacterium acnes.
From the cultivated bacteria, the species identified as probiotics included Staphylococcus hominis and Staphylococcus warneri, which secrete the antimicrobial peptide bacteriocin. This peptide can be developed as an active substance for skin microbiome therapeutics and cosmetic applications. Bacteriocin from commensal microbiota has been used in peptide-based cocktails for the treatment of infections and prophylaxis purposes [49]. S. hominis produces hominisin, a potent bacteriocin that inhibits the activities of S. aureus strains [36]. S. warneri is a commensal bacterium on the skin that rarely causes disease in humans [50]. S. warneri is also shown to produce molecules with antimicrobial activities, namely, the bacteriocins Nukacin ISK-1 and Warnericin RK, which was the first antibacterial peptide with activity against Legionella. Coagulase negative staphylococci (CNS) strains Staphylococcus hominis and Staphylococus warneri have been assessed for their probiotic and safety properties, in which they exhibited potent antioxidative properties, and non-pathogenic traits proven by negative results of haemolytic, DNase, and gelatinase tests [51]. Hofer et al. [52] reported that M. luteus can produce DNA-repair enzymes that can be used to prevent ultraviolet-light-induced activation of photo neoantigens on the skin. Series of characterization also showed that M. luteus is regarded as potential topical probiotic for its inhibitory against S. aureus and ability to prevent cutaneous membrane infection ex vivo [53]. It is reported that the application of probiotic bacteria may alter the microbiota composition in the skin of healthy individuals [8]. Emergence of concerns regarding probiotic administration in safety and stability aspects has encouraged development of postbiotics−non-viable, soluble factors produced by probiotics−to achieve effective therapeutic goals and avoid problems related to administration of viable bacteria [54]. The use of postbiotics-non-viable is also considered due to the instability and the difficulties in controlling the probiotic growth within cosmetic formulations. In the preparation, postbiotics can be recovered from probiotics using cell disruption methods including heat-killing, enzymatic treatments, solvent extraction, and ultrasonication [55].
Several opportunistic pathogens were identified in skin microbiome alongside the potential probiotic strains. From cultivation method, opportunistic pathogens such as Staphylococcus capitis, Staphylococcus saprophyticus, and Staphylococcus haemolyticus were isolated. Staphylococcus capitis were associated with endocarditis and meningitis in infants, while Staphylococcus saprophyticus is known as urinary tract infection (UTI)-causing bacteria [56, 57]. Staphylococcus haemolyticus is an infamous multidrug resistant strain commonly found in normal skin flora and may cause septicemia, peritonitis, otitis, and UTI in patients with underlying disease [58]. NGS analysis revealed Corynebacterium amycolatum, Corynebacterium tuberculostearicum, Brevibacterium luteolum, Kocuria rhizophila, Propionibacterium propionicum, and Staphylococcus lentus as opportunistic pathogens causing infections in various sites such as urinary tract, lacrimal apparatus, and peritoneum [9, 10, 13, 15, 19, 27, 31].
Although we do not demonstrate the probiotic/postbiotic properties of the bacterial isolates in this report, the more important thing is the information of bacterial composition from the NGS along with the bacteria strains from Indonesian young people's skin, as they are the potential target for skincare products. Thus, from this preliminary study, the development of bioactive products for skincare and for treatment of skin disorders/diseases with preferences to be marketed in the tropical countries is challenging. Hence, to expand this precious collection for further studies, first, we will focus on microbiome-based therapeutic with their holistic approach, such as the probiotic/postbiotic products.
An additional quantitative PCR method using species-specific primers can be carried out to further characterise the abundance of bacteria in skin microbiota samples [59]. Future studies are warranted to elucidate the metabolite composition and strain characteristics by genomic, proteomic, metabolomic approaches, and the potential of these isolated probiotics for use in skin microbiome therapeutics and skin care. Thus, the probiotic lysate can be implemented toward cell culture or other skin test models by comparing the microflora of healthy and diseased individuals to see the effect on the skin [60].
Nonetheless, for the development of skin microbiome therapeutics, the cause-effect relationships determination and the design of microbiome-based therapies that are able to achieve predictable outcomes on the microbial community and host health are the biggest challenges in skin microbiome research. In spite of their potential health impact, a full list of lifestyle factors capable of altering human microbiota remains incomplete, for example a large, and potentially unfeasible, number of interventional studies are needed to fully explore the rich diversity of human actions and behaviors [61]. Therefore, the interventions by design only test a small number of hypotheses [62].
5. Conclusions
In this study, we have successfully identified five potential probiotics through NGS profiling, namely Micrococcus sp., Propionibacterium acidipropionici, Propionibacterium granulosum, Propionibacterium avidum and Paracoccus sp. While, S. hominis, S. warneri and M. luteus were detected by performing 16S rRNA Sanger sequencing. Micrococcus genera was the only genera that was constantly detected among these two methods. There were differences in the probiotics variety detected by both NGS and sanger sequencing. However, the NGS profiling gives more significant results compared to conventional sanger sequencing. This study indicated that both NGS and sanger sequencing data are complemented one to each other to give insight towards bacteria compositions from human skin. The exploration of commensal bacteria from the human skin is a good step in the identification of probiotics as sources of active substances for use as pharmaceuticals for skin care and therapeutics.
Declarations
Author contribution statement
Amalia Sitti Khayyira, Aulia Elfa Rosdina: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Marina Ika Irianti: Analyzed and interpreted the data; Wrote the paper.
Amarila Malik: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by Research Grant PMDSU 2019 from The Ministry of Research, Technology, & Higher Education, Republic of Indonesia (NKB-3030/UN2.R3.1/HKP.05.00/2019) to Prof. Dr. Amarila Malik, and partially funded by The Ministry of Research, Technology, & Higher Education, Republic of Indonesia under WCU Program 2019, managed by Institut Teknologi Bandung.
Competing interest statement
The authors declare no conflict of interest.
Additional information
The following are the Supplementary data related to this article:
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018;16(3):143. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 2.Grice E.A., Kong H.H., Renaud G., Young A.C., Bouffard G.G., Blakesley R.W. A diversity profile of the human skin microbiota. Genome Res. 2008 doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice E.A., Segre J.A. The skin microbiome. Nat. Rev. Microbiol. 2011;9(4):244. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee S., Mitra R., Maitra A., Gupta S., Kumaran S., Chakrabortty A. Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci. Rep. 2016;6:36062. doi: 10.1038/srep36062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beri K. Skin microbiome & host immunity: applications in regenerative cosmetics & transdermal drug delivery. Future Sci. OA. 2018;4(6):FSO302. doi: 10.4155/fsoa-2017-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong A.C., Levy M. New approaches to microbiome-based therapies. mSystems. 2019;4(3) doi: 10.1128/mSystems.00122-19. e00122-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dore J., Blottiere H. The influence of diet on the gut microbiota and its consequences for health. Curr. Opin. Biotechnol. 2015;32:195–199. doi: 10.1016/j.copbio.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Paetzold B., Willis J.R., de Lima J.P., Knödlseder N., Brüggemann H., Quist S.R. Skin microbiome modulation induced by probiotic solutions. Microbiome. 2019;7(1):95. doi: 10.1186/s40168-019-0709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knox K.L., Holmes A.H. Nosocomial endocarditis caused by Corynebacterium amycolatum and other nondiphtheriae corynebacteria. Emerg. Infect. Dis. 2002;8(1):97. doi: 10.3201/eid0801.010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letek M., Ordóñez E., Fernández-Natal I., Gil J., Mateos L. Identification of the emerging skin pathogen Corynebacterium amycolatum using PCR-amplification of the essential divIVA gene as a target. FEMS Microbiol. Lett. 2006;265(2):256–263. doi: 10.1111/j.1574-6968.2006.00492.x. [DOI] [PubMed] [Google Scholar]
- 11.Funke G., Efstratiou A., Kuklinska D., Hutson R.A., De Zoysa A., Engler K.H. Corynebacterium imitans sp. nov. isolated from patients with suspected diphtheria. J. Clin. Microbiol. 1997;35(8):1978–1983. doi: 10.1128/jcm.35.8.1978-1983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funke G., Lawson P.A., Collins M.D. Corynebacterium riegelii sp. nov., an unusual species isolated from female patients with urinary tract infections. J. Clin. Microbiol. 1998;36(3):624–627. doi: 10.1128/jcm.36.3.624-627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abreu N.A., Nagalingam N.A., Song Y., Roediger F.C., Pletcher S.D., Goldberg A.N. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci. Transl. Med. 2012;4(151) doi: 10.1126/scitranslmed.3003783. 151ra24-ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schröder J., Maus I., Trost E., Tauch A. Complete genome sequence of Corynebacterium variabile DSM 44702 isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavor generation. BMC Genom. 2011;12(1):545. doi: 10.1186/1471-2164-12-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unás J.H., de Alexandria Santos D., Azevedo E.B., Nitschke M. Brevibacterium luteolum biosurfactant: production and structural characterization. Biocatal. Agric. Biotechnol. 2018;13:160–167. [Google Scholar]
- 16.Alou M.T., Cadoret F., Brah S., Diallo A., Sokhna C., Mehrej V. ‘Khelaifiella massiliensis’,‘Niameybacter massiliensis’,‘Brachybacterium massiliense’,‘Enterobacter timonensis’,‘Massilibacillus massiliensis’, new bacterial species and genera isolated from the gut microbiota of healthy infants. New Microb. New Infect. 2017;19:1–7. doi: 10.1016/j.nmni.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tewari R., Dudeja M., Das A.K., Nandy S. Kocuria kristinae in catheter associated urinary tract infection: a case report. J. Clin. Diagn. Res.: J. Clin. Diagn. Res. 2013;7(8):1692. doi: 10.7860/JCDR/2013/6077.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovács G., Burghardt J., Pradella S., Schumann P., Stackebrandt E., Màrialigeti K. Kocuria palustris sp. nov. and Kocuria rhizophila sp. nov., isolated from the rhizoplane of the narrow-leaved cattail (Typha angustifolia) Int. J. Syst. Evol. Microbiol. 1999;49(1):167–173. doi: 10.1099/00207713-49-1-167. [DOI] [PubMed] [Google Scholar]
- 19.Pękala A., Paździor E., Antychowicz J., Bernad A., Głowacka H., Więcek B. Kocuria rhizophila and Micrococcus luteus as emerging opportunist pathogens in brown trout (Salmo trutta Linnaeus, 1758) and rainbow trout (Oncorhynchus mykiss Walbaum, 1792) Aquaculture. 2018;486:285–289. [Google Scholar]
- 20.Akbar A., Sitara U., Ali I., Muhammad N., Khan S.A. Isolation and characterization of biotechnologically potent Micrococcus luteus strain from environment. Pakistan J. Zool. 2014;46(4) [Google Scholar]
- 21.Zárate G., De Ambrosini V.M., Chaia A.P., González S. Some factors affecting the adherence of probiotic Propionibacterium acidipropionici CRL 1198 to intestinal epithelial cells. Can. J. Microbiol. 2002;48(5):449–457. doi: 10.1139/w02-036. [DOI] [PubMed] [Google Scholar]
- 22.Chaia A.P., Zarate G., Oliver G. The probiotic properties of propionibacteria. Lait. 1999;79(1):175–185. [Google Scholar]
- 23.Wildeman P., Brüggemann H., Scholz C.F., Leimbach A., Söderquist B. Propionibacterium avidum as an etiological agent of prosthetic hip joint infection. PloS One. 2016;11(6) doi: 10.1371/journal.pone.0158164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zárate G. IntechOpen; 2012. Dairy Propionibacteria: Less Conventional Probiotics to Improve the Human and Animal health. Probiotic in Animals. [Google Scholar]
- 25.Piwowarek K., Lipińska E., Hać-Szymańczuk E., Kieliszek M., Ścibisz I. Propionibacterium spp.—source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 2018;102(2):515–538. doi: 10.1007/s00253-017-8616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isenberg J., Stoffel B., Wolters U., Beuth J., Stützer H., Ko H. Immunostimulation by propionibacteria--effects on immune status and antineoplastic treatment. Anticancer Res. 1995;15(5B):2363–2368. [PubMed] [Google Scholar]
- 27.Brazier J.S., Hall V. Propionibacterium propionicum and infections of the lacrimal apparatus. Clin. Infect. Dis. 1993;17(5):892–893. doi: 10.1093/clinids/17.5.892. [DOI] [PubMed] [Google Scholar]
- 28.Albuquerque L., Simoes C., Nobre M.F., Pino N.M., Battista J.R., Silva M.T. Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2005;247(2):161–169. doi: 10.1016/j.femsle.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X., Tian Y., Xu W., Bai Y., Zhang T., Mu W. Biochemical characterization of a highly thermostable amylosucrase from Truepera radiovictrix DSM 17093. Int. J. Biol. Macromol. 2018;116:744–752. doi: 10.1016/j.ijbiomac.2018.05.096. [DOI] [PubMed] [Google Scholar]
- 30.Hilliard C.A., El Masri J., Goto M. Staphylococcus caprae bacteraemia and native bone infection complicated by therapeutic failure and elevated MIC: a case report. JMM Case Rep. 2017;4(9) doi: 10.1099/jmmcr.0.005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivera M., Dominguez M.D., Mendiola N.R., Roso G.R., Quereda C. Staphylococcus lentus peritonitis: a case report. Perit. Dial. Int. 2014;34(4):469–470. doi: 10.3747/pdi.2012.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosini R., Margarit I. Biofilm formation by Streptococcus agalactiae: influence of environmental conditions and implicated virulence factors. Front. Cell. Infect. Microbiol. 2015;5:6. doi: 10.3389/fcimb.2015.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa N., Sekizuka T., Sugi Y., Kawakami N., Ogasawara Y., Kato K. Characterization of the pathogenicity of Streptococcus intermedius TYG1620 isolated from a human brain abscess based on the complete genome sequence with transcriptome analysis and transposon mutagenesis in a murine subcutaneous abscess model. Infect. Immun. 2017;85(2) doi: 10.1128/IAI.00886-16. e00886-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wanka K.M., Damerau T., Costas B., Krueger A., Schulz C., Wuertz S. Isolation and characterization of native probiotics for fish farming. BMC Microbiol. 2018;18(1):119. doi: 10.1186/s12866-018-1260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myles I.A., Reckhow J.D., Williams K.W., Sastalla I., Frank K.M., Datta S.K. A method for culturing Gram-negative skin microbiota. BMC Microbiol. 2016;16(1):60. doi: 10.1186/s12866-016-0684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung C., Kim B.G., Kim S., Joo H.S., Kim P. Probiotic potential of Staphylococcus hominis MBBL 2–9 as anti-Staphylococcus aureus agent isolated from the vaginal microbiota of a healthy woman. J. Appl. Microbiol. 2010;108(3):908–916. doi: 10.1111/j.1365-2672.2009.04485.x. [DOI] [PubMed] [Google Scholar]
- 37.Moissl-Eichinger C., Probst A.J., Birarda G., Auerbach A., Koskinen K., Wolf P. Human age and skin physiology shape diversity and abundance of Archaea on skin. Sci. Rep. 2017;7(1):4039. doi: 10.1038/s41598-017-04197-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying S., Zeng D.-N., Chi L., Tan Y., Galzote C., Cardona C. The influence of age and gender on skin-associated microbial communities in urban and rural human populations. PloS One. 2015;10(10) doi: 10.1371/journal.pone.0141842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva P.E., Reis M.P., Ávila M.P., Dias M.F., Costa P.S., Suhadolnik M.L. Insights into the skin microbiome dynamics of leprosy patients during multi-drug therapy and in healthy individuals from Brazil. Sci. Rep. 2018;8(1):8783. doi: 10.1038/s41598-018-27074-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehtimäki J., Karkman A., Laatikainen T., Paalanen L., Von Hertzen L., Haahtela T. Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Sci. Rep. 2017;7:45651. doi: 10.1038/srep45651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander H., Paller A., Traidl-Hoffmann C., Beck L., De Benedetto A., Dhar S. The role of bacterial skin infections in atopic dermatitis: expert statement and review from the International Eczema Council Skin Infection Group. Br. J. Dermatol. 2019 doi: 10.1111/bjd.18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ring H.C., Thorsen J., Saunte D.M., Lilje B., Bay L., Riis P.T. The follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls. JAMA Dermatol. 2017;153(9):897–905. doi: 10.1001/jamadermatol.2017.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knights D., Parfrey L.W., Zaneveld J., Lozupone C., Knight R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe. 2011;10(4):292–296. doi: 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBain A.J., O’Neill C.A., Amezquita A., Price L.J., Faust K., Tett A. Consumer safety considerations of skin and oral microbiome perturbation. Clin. Microbiol. Rev. 2019;32(4) doi: 10.1128/CMR.00051-19. e00051-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markowska-Daniel I., Pejsak Z., Szmigielski S., Jeljaszewicz J., Pulverer G. Stimulation of granulopoiesis in pregnant swine and their offspring by Propionibacterium avidum KP-40. Br. Vet. J. 1992;148(2):133–145. doi: 10.1016/0007-1935(92)90105-A. [DOI] [PubMed] [Google Scholar]
- 46.Huang T.-H., Wang P.-W., Yang S.-C., Chou W.-L., Fang J.-Y. Cosmetic and therapeutic applications of fish oil’s fatty acids on the skin. Mar. Drugs. 2018;16(8):256. doi: 10.3390/md16080256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meisel J.S., Hannigan G.D., Tyldsley A.S., SanMiguel A.J., Hodkinson B.P., Zheng Q. Skin microbiome surveys are strongly influenced by experimental design. J. Invest. Dermatol. 2016;136(5):947–956. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilbert J.A., Jansson J.K., Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12(1):69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hols P., Ledesma-García L., Gabant P., Mignolet J. Mobilization of microbiota commensals and their bacteriocins for therapeutics. Trends Microbiol. 2019 doi: 10.1016/j.tim.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Ivić I., Karanović J., Pavičić-Ivelja M. Sepsis with multiple abscesses caused by staphylococcus warneri: a case report. Open Med. 2013;8(1):45–47. [Google Scholar]
- 51.Khusro A., Aarti C., Dusthackeer A., Agastian P. Anti-tubercular and probiotic properties of coagulase-negative staphylococci isolated from Koozh, a traditional fermented food of South India. Microb. Pathog. 2018;114:239–250. doi: 10.1016/j.micpath.2017.11.054. [DOI] [PubMed] [Google Scholar]
- 52.Hofer A., Legat F.J., Gruber-Wackernagel A., Quehenberger F., Wolf P. Topical liposomal DNA-repair enzymes in polymorphic light eruption. Photochem. Photobiol. Sci. 2011;10(7):1118–1128. doi: 10.1039/c1pp05009e. [DOI] [PubMed] [Google Scholar]
- 53.Sawers C. University of Otago; 2012. Factors Influencing the Probiotic Potential of an Inhibitor-Producing Micrococcus Luteus. [Google Scholar]
- 54.Cicenia A., Scirocco A., Carabotti M., Pallotta L., Marignani M., Severi C. Postbiotic activities of lactobacilli-derived factors. J. Clin. Gastroenterol. 2014;48:S18–S22. doi: 10.1097/MCG.0000000000000231. [DOI] [PubMed] [Google Scholar]
- 55.Aguilar-Toalá J., Garcia-Varela R., Garcia H., Mata-Haro V., González-Córdova A., Vallejo-Cordoba B. Postbiotics: an evolving term within the functional foods field. Trends Food Sci. Technol. 2018 [Google Scholar]
- 56.Cameron D., Jiang J.-H., Hassan K., Elbourne L., Tuck K., Paulsen I. Insights on virulence from the complete genome of Staphylococcus capitis. Front. Microbiol. 2015;6:980. doi: 10.3389/fmicb.2015.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Motwani B., Khayr W. Staphylococcus saprophyticus urinary tract infection in men: a case report and review of the literature. Infect. Dis. Clin. Pract. 2004;12(6):341–342. [Google Scholar]
- 58.Takeuchi F., Watanabe S., Baba T., Yuzawa H., Ito T., Morimoto Y. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 2005;187(21):7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonk F., Popp D., Harms H., Centler F. PCR-based quantification of taxa-specific abundances in microbial communities: quantifying and avoiding common pitfalls. J. Microbiol. Methods. 2018 doi: 10.1016/j.mimet.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 60.Saxena R., Sharma V. Medical and Health Genomics. Elsevier; 2016. A metagenomic insight into the human microbiome: its implications in health and disease; pp. 107–119. [Google Scholar]
- 61.Oh J., Byrd A.L., Park M., Kong H.H., Segre J.A., Program N.C.S. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.David L.A., Materna A.C., Friedman J., Campos-Baptista M.I., Blackburn M.C., Perrotta A. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilantho A., Deekaew P., Srisuttiyakorn C., Tongsima S., Somboonna N. Diversity of bacterial communities on the facial skin of different age-group Thai males. PeerJ. 2017;5 doi: 10.7717/peerj.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.