Abstract
Sirtuins are NAD+-dependent deacylases that play crucial roles in the regulation of cellular metabolism, and as a result, are implicated in several diseases. The mitochondrial sirtuin Sirt4, for a long time considered as mainly a mono-ADP-ribosyltransferase, recently has shown a robust deacylase activity in addition to the already accepted substrate-dependent lipoamidase and deacetylase properties. Through these and likely other enzymatic and non-enzymatic activities, Sirt4 closely controls various metabolic events, and its dysregulation is linked to various aging-related disorders, including type 2 diabetes, cardiac hypertrophy, non-alcoholic fatty liver disease, obesity, and cancer. For its capability to inhibit glutamine catabolism and for the modulation of genome stability in cancer cells in response to different DNA-damaging conditions, Sirt4 is proposed as either a mitochondrial tumor suppressor or a tumor-promoting protein in a context-dependent manner. In addition to what is already known about the roles of Sirt4 in different biological settings, further studies are certainly still needed in order to validate this enzyme as a new potential target for various aging diseases.
Keywords: sirtuins, protein deacylation, mitochondria, metabolism, cancer
Introduction
Sirt4 is one of the three mitochondrial sirtuins and, despite that it was firstly described as a mono-ADP-ribosyltransferase, nowadays it has demonstrated a robust deacylase activity toward 3-hydroxy-3-methyl-glutarylated (HMG) lysine residues (1) as well as substrate-specific lipoamidase and deacetylase properties (2, 3). Sirt4 directly or indirectly regulates multiple mitochondrial functions closely connected to the progression of age-related diseases such as type 2 diabetes (T2D), neurodegeneration, and cancer (1–6).
Sirt4 Enzymatic Activities
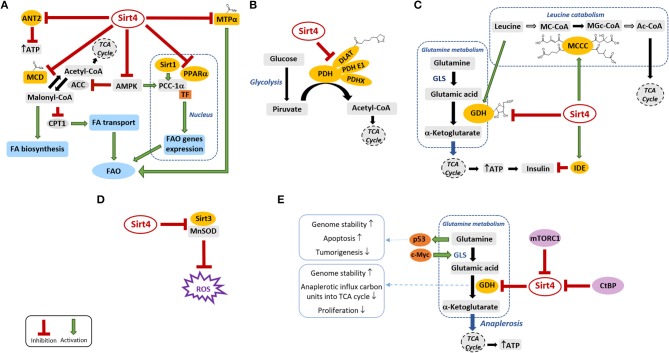
Sirt4 is distributed in both fetal and adult tissues, with higher expression levels in liver, heart, spleen, prostate, testis, kidney, ovary, white adipose tissue, and muscle, and it works as a “bridge protein” between mitochondrial metabolism and tumorigenesis (7). Several pieces of evidence showed that Sirt4 covers a crucial role when cells are subject to toxic stresses able to modulate the mitochondrial levels of the co-substrate NAD+ and/or the NAD+/NADH ratio (3–6). As a mono-ADP-ribosyltransferase, Sirt4 catalyzes the transfer of an ADP-ribosyl moiety from NAD+ to glutamate dehydrogenase (GDH), thus inhibiting its activity in mice pancreatic β-cells and blocking the anaplerotic influx of carbon units entering the tricarboxylic acids (TCA) cycle (8) (Figure 1). Beyond this capability to regulate energy and glutamine metabolism, an important Sirt4 activity is its lipoamidase-mediated inhibition of pyruvate dehydrogenase (PDH), the crucial multi-component enzymatic complex that modulates the entrance of acetyl-CoA deriving from glycolysis into the TCA cycle, identifying Sirt4 as a “guardian of cellular metabolism” (9). Despite the lack of deacetylase activity on histone peptides in vitro, an acetylome peptide microarray study showed a low but substrate-specific and reproducible deacetylase activity for Sirt4 against acetylation sites in, e.g., the mitochondrial proteins Stress-70, NAD(P) transhydrogenase, and Hsp60 (10). Moreover, Sirt4 was also shown to deacetylate and inhibit the activity of malonyl-CoA decarboxylase (MCD) in white adipose tissue and skeletal muscle, thereby regulating fatty acid oxidation (FAO) and biosynthesis processes (11) (Figure 1). Recently, the mitochondrial trifunctional protein α-subunit (MTPα) has also been identified as a substrate of Sirt4 deacetylation in hepatocytes, highlighting the capability of this enzyme to inhibit FAO also in the liver (12) (Figure 1).
Figure 1.
Sirt4 and its substrates in the mitochondrial metabolic pathways. Sirt4 modulates directly or indirectly the activity of several targets (depicted in yellow) that play key roles in various metabolic processes. Green arrows and red lines indicate the promotion/activation and suppression/inhibition of a specific activity, respectively. (A) Sirt4 modulation of lipid metabolism. (B) Sirt4 inhibition of PDH through lipoamidase activity. (C) Sirt4 modulation of insulin secretion and sensitivity. (D) Sirt4 effects on mitochondrial oxidative stress. (E) Sirt4 and cancer. ACC, acetyl-CoA carboxylase; Ac-CoA, acetyl coenzyme A; AMPK, AMP-activated protein kinase; ANT2, adenine nucleotide translocator 2; ATP, adenosine triphosphate; CPT1, carnitine palmitoyl-transferase 1; CtBP, C-terminal binding protein; DLAT, dihydrolipoyllysine acetyltransferase; FA, fatty acids; FAO, fatty acids oxidation; GDH, glutamate dehydrogenase; GLS, glutaminase; IDE, insulin degrading enzyme; MCCC, 3-methylcrotonyl-CoA carboxylase; MC-CoA, 3-methylcrotonyl-CoA; MGc-CoA, 3-methylglutaconyl-CoA; MCD, malonyl-CoA decarboxylase; MnSOD, manganese-dependent superoxide dismutase; mTORC1, mammalian target of rapamycin complex 1; MTPα, mitochondrial trifunctional protein α; PGC-1α, peroxisome proliferator-activated receptor gamma co-activator 1α; PDH, pyruvate dehydrogenase; PPARα, peroxisome proliferator-activated receptors α; ROS, reactive oxygen species; TCA, tri-carboxylic acid; TF, transcription factor.
In line with its little to no-detectable ADP-ribosyltransferase and deacetylase activities in vitro (13, 14), the difficulty to purify the enzyme and the consequent impossibility to set up an efficient enzymatic assay represented for years the main obstacles in the investigation of Sirt4 functions and are the principal reasons for the absence of specific Sirt4 modulators up to now. In 2017, we reported for Sirt4 an efficient deacylase activity toward HMG-modified lysine residues of peptides and proteins and established a fluorescence-based Fluor de Lys-like assay for the screening of large numbers of potential modulators (1). We further solved the crystal structure of Sirt4 from Xenopus tropicalis in complex with ADP-ribose, revealing two main isoform-specific features: an extra-active site entry channel connected to the protein surface and a large, dynamic loop reaching the active site, which may contribute to substrate binding and regulate active site dynamics, thus explaining the broad acyl selectivity of Sirt4 (1).
Indeed in the same year Anderson et al. showed that Sirt4 removes HMG and related acyl moieties such as glutaryl (G), 3-methylglutaryl (MG), and 3-methylglutaconyl (MGc) from the lysine residues of various substrates both in vivo and in vitro (15). Among them, an important metabolic substrate is the enzyme complex methylcrotonyl-CoA carboxylase (MCC) that catalyzes the conversion of 3-methylcrotonyl-CoA (MC) into MGc-CoA and plays a crucial role in leucine catabolism. Important metabolic intermediates in this process are also HMG-CoA and MG-CoA (16) that, together with G-CoA, MGc-CoA, and other acyl-CoA, seem able to spontaneously acylate and destabilize the MCC itself, reducing leucine flux through the branched chain amino acid catabolic pathway (15) (Figure 1). In this context, the broad-spectrum deacylase activity of Sirt4 seems to play a key role to stimulate leucine catabolism in vivo and to indirectly inhibit amino acid-stimulated insulin secretion (AASIS) (15).
Sirt4 Modulation of Lipid Metabolism
Sirt4 plays a pivotal role in the modulation of fatty acid metabolism in both skeletal muscle and white adipose tissue by deacetylating and inhibiting MCD (11) (Figure 1A). In its deacetylated form, MCD no longer catalyzes the conversion of malonyl-CoA into acetyl-CoA, thus causing malonyl-CoA accumulation. Malonyl-CoA serves at the same time as the chain-elongating unit for fatty acid biosynthesis and as an allosteric inhibitor of the fatty acid transporter carnitine palmitoyl-transferase (CPT1), responsible for the transfer of fatty acids from the cytosol to the mitochondrial matrix for β-oxidation. While nutritional rich conditions correlate with accumulated malonyl-CoA and the consequent arrest of the FAO process and increase of fat synthesis, during a fasted state, lower levels of malonyl-CoA have been registered and FAO is increased for energy production (17–19). This regulatory mechanism was confirmed in Sirt4 knock-out (KO) mice that showed raised FAO associated with increased exercise tolerance and resistance to diet-induced obesity (20–22). MTPα is a mitochondrial enzyme that catalyzes two steps of FAO and can be acetylated or ubiquitinated on the same three lysine residues. In hepatocytes, it was demonstrated that the Sirt4-mediated deacetylation of MTPα promotes its ubiquitination and proteasome-dependent degradation, thus contributing to the inhibition of FAO (12). Sirt4 expression is increased in livers of patients with non-alcoholic fatty liver disease (NAFLD) (12, 23). The evidence that decreased Sirt4 expression in mice livers is able to protect against NAFLD by inhibiting MTPα deacetylation and degradation suggested that the upregulated levels of Sirt4 observed in NAFLD patients could contribute to the onset of the disorder by decreasing MTPα-catalyzed FAO and, consequently, by promoting ectopic lipid accumulation (12). Sirt4 can also inhibit FAO in mice liver as well as hepatoma, kidney, and fibroblast cells by repressing the activity of peroxisome proliferator-activated receptor α (PPARα), a transcription factor that stimulates the expression of FAO genes. Indeed hepatocytes from Sirt4 KO mice showed an increased FAO rate, which was dependent on the interaction of Sirt1 with PPARα and on the Sirt1-dependent deacetylation and activation of the transcriptional co-activator peroxisome proliferator-activated receptor gamma co-activator 1-α (PGC-1α). These findings suggest that Sirt4 works in a retrograde signaling pathway from the mitochondria to the nucleus to decrease Sirt1 activity, likely by competing for NAD+ (17, 24) (Figure 1A).
AMP-activated protein kinase (AMPK) plays a crucial role in promoting FAO through the phosphorylation and the inhibition of acetyl-CoA carboxylase (ACC), the enzyme that, counteracting MCD, catalyzes the carboxylation of acetyl-CoA to provide malonyl-CoA. In fact, the consequently reduced malonyl-CoA levels correlate with an increased CPT1-mediated mitochondrial fatty acid uptake (17, 25, 26). In addition, AMPK is also known for its capability to act as a transcriptional co-activator of PGC-1α (27). In this context, it is noteworthy that Sirt4 KO mice livers presented elevated levels of activated AMPK, resulting in the inhibition of ACC, the reduction of malonyl-CoA, PGC1-α induction, and the final promotion of FAO (5) (Figure 1A).
Sirt4 as a Checkpoint Between Glycolysis and TCA Cycle
The mitochondrial multi-protein complex PDH (28) promotes the oxidative decarboxylation of pyruvate to provide acetyl-CoA, thus linking glycolysis and the TCA cycle. Sirt4 can remove lipoyl groups from the PDH subunit dihydrolipoyl lysine-residue acetyltransferase efficiently in a phosphorylation-independent manner, thereby repressing the enzymatic activity of the complex both in cells and in vivo (9). Since PDH activity indirectly modulates a large variety of downstream metabolic processes, the PDH regulator Sirt4 can be considered as one of the main regulators of cellular metabolism (Figure 1B).
Sirt4 Modulation of Insulin Secretion and Sensitivity
Insulin secretion by pancreatic β-cells can be promoted by amino acids or glucose (29). Amino acids are metabolized into TCA cycle intermediates to generate adenosine triphosphate (ATP) that finally stimulates insulin release. Glutamine is initially hydrolyzed by glutaminase (GLS) to glutamate, which is subsequently converted by GDH into the TCA cycle intermediate α-ketoglutarate (30). As mentioned, Sirt4 can catalyze the mono-ADP-ribosylation and inactivation of GDH in pancreatic β-cells (Figure 1C) and, through this mechanism, can contribute to the reduction of ATP levels and the repression of AASIS (8). Indeed pancreatic islets isolated from Sirt4 KO mice showed both increased GDH activity and circulating levels of insulin (8).
Sirt4 can modulate insulin secretion also by other mechanisms (3). Leucine is an allosteric activator of GDH, and Sirt4-promoted leucine catabolism via MCCC deacylation and activation will thus prevent GDH activation and result in decreased glutamine-stimulated insulin. Indeed Sirt4 KO mice displayed not only increased glutamine- and glucose-stimulated insulin secretion but also improved leucine-stimulated insulin release. Sirt4 can also interact with the insulin-degrading enzyme (IDE), thus reducing insulin secretion in response to glucose (29) (Figure 1C).
Sirt4 might also promote insulin release by increasing the ATP levels as a result of the deacylation and the inhibition of the uncoupling ADP/ATP carrier protein adenine nucleotide translocator 2 (31) (Figure 1C). Due to its many different activities in inhibiting insulin secretion, Sirt4 has been linked to the emergence of T2D, which was also confirmed by studies in Sirt4 KO mice that quickly develop hyperinsulinemia, insulin resistance, and glucose intolerance (15). Furthermore, Sirt4 protects podocytes against reactive oxygen species (ROS) accumulation and apoptosis under hyperglycemic conditions and thus has a protective role against diabetic nephropathy, one of the worst complications associated with diabetes (32).
Sirt4 and Mitochondrial Oxidative Stress
ROS are by-products of oxidative metabolism, mainly derived from oxidative phosphorylation and enzymatic reactions (α-glycerophosphate dehydrogenase, NADPH-oxidase, mono-aminoxidase, etc.) in the mitochondria. Since moderate levels of ROS have physiological roles in stress responses and signal transduction, both too high and excessively low ROS levels have significant pathogenic roles (6).
Sirt4 is involved in the regulation of ROS production in the mitochondria, very likely through a scaffolding function (19). Sirt3, another mitochondrial deacetylase, deacetylates, and activates the anti-oxidant enzyme manganese superoxide dismutase (MnSOD) to decrease the mitochondrial ROS levels. Sirt4 can compete with MnSOD for binding to Sirt3, thus sequestering Sirt3, and indirectly inhibiting Sirt3-mediated MnSOD activation (Figure 1D). This action, which seems non-enzymatic because the catalytically inactive mutant of Sirt4 H161Y is also able to block the Sirt3–MnSOD interaction, induces an increase of ROS levels and oxidative stress in the mitochondria of heart muscle cells and promotes cardiac hypertrophy (19). However, as mentioned, the overexpression of Sirt4 can prevent podocyte apoptosis induced by glucose with concomitantly increased mitochondrial membrane potential and reduced ROS production (32). These findings reveal that the effects of Sirt4 on mitochondrial ROS levels are context dependent.
Sirt4 as a Context-Dependent Tumor Suppressor and Oncoprotein
Most studies on the involvement of Sirt4 in tumor biology highlight this enzyme as a mitochondrion-localized tumor suppressor due to its crucial regulatory role of mitochondrial metabolism during tumorigenesis (5). Sirt4 mRNA levels are reduced in many human cancers, such as lung, pancreatic, ovarian, gastric, colorectal, prostate, renal, liver, and endometrial cancer as well as hematological tumors (33–45) (Table 1). Lower levels of Sirt4 protein expression in tumor tissues are often associated with worse pathological grading and reduced survival in cancer patients, and Sirt4 KO mice display an increased incidence of spontaneous tumors (2, 33, 36, 37, 44, 45, 49).
Table 1.
Roles of Sirt4 in tumorigenesis.
| Phenotype | Cancer types | Role of Sirt4 | References |
|---|---|---|---|
| Sirt4 downregulation | Acute myeloid leukemia, Burkitt lymphoma, and lung, colorectal, gastric, liver, lung, endometrial, pancreatic, ovarian, prostate, and renal cancers | Cancer suppression: inhibition of tumor proliferation, invasion, and migration | (4, 33–35, 37–46) |
| Sirt4 over-expression | Esophageal cancer, hepatocarcinoma (HepG2 cells) | Cancer promotion: enhanced cancer cell survival rate in “extreme” DNA-damaging conditios | (4, 47, 48) |
While quiescent cells exploit the TCA cycle to obtain energy from glucose, proliferating cells mainly use it as a carbon source for lipogenesis through the mitochondrial efflux of citric acid. This efflux needs to be replaced by an influx of TCA cycle intermediates, known as anaplerosis, and glutamine is the main source for TCA anaplerosis in proliferating cells (50) (Figure 1E). Therefore, glutamine catabolism plays a crucial role for the proliferation of tumor cells by replenishing TCA cycle intermediates to support increased growth and by generating ammonia, which neutralizes the acidic metabolites usually produced because of the increased glycolysis in tumor cells (Figure 1E). In addition, glutamine by itself promotes the activation of the tumor suppressor p53, which is responsible for apoptosis and tumor regression induction (51) (Figure 1E). For these reasons, Sirt4 repression of mitochondrial glutamine catabolism by the inhibition of GDH activity seems to contribute significantly to its function as a tumor suppressor.
Many studies have supported this notion, providing mechanistic insights into the relation between glutamine metabolism and Sirt4 dysregulation during tumorigenesis (3). c-Myc is an oncogenic transcription factor that can increase the expression of GLS via the repression of specific microRNAs (miR-23a and miR-23b) and consequently stimulates glutamine metabolism in many c-Myc-driven cancers (38), which typically show marked glutamine dependence (52, 53). Sirt4 can inhibit the proliferation of c-Myc-induced Burkitt lymphoma by inhibiting GDH activity, thereby preventing increased glutamine metabolism and sensitizing it to glucose depletion (38) (Figure 1E). Both the transcriptional regulator C-terminal binding protein (CtBP) and the mammalian target of rapamycin complex 1 (mTORC1) (54) were discovered to promote glutamine metabolism and tumor proliferation in various cancer contexts by repressing the expression of Sirt4 directly (55) and indirectly (54), respectively (Figure 1E).
In addition to the regulation of mitochondrial glutamine catabolism, Sirt4 also has the capability to modulate other cancer-related cellular features such as cell cycle progression, apoptosis, invasion, and metastatic potential. It is noteworthy that these processes, in many cases, are somehow linked to the effects on glutamine metabolism (56). Sirt4 expression is upregulated by different types of DNA damage. It can prevent cell cycle progression following DNA damage (37), and Sirt4-mediated inhibition of glutamine anaplerosis can be crucial for a productive cell cycle arrest upon DNA damage (56) and for the proper implementation of the cellular DNA damage repair program (6, 37). Moreover, in colorectal cancer (CRC) cells, Sirt4 overexpression induces anti-proliferative effects and increases the sensitivity to the drug 5-fluorouracil (46). Sirt4 can also display pro-apoptotic effects in lung cancer cells by inhibiting mitochondrial fission, which is known to prevent apoptosis and enhance cancer cell growth (44). Sirt4 is also able to prevent the epithelial–mesenchymal transition (EMT) in CRC cells by upregulating the expression of E-cadherin, which promotes cell–cell adhesion and consequently inhibits tumor invasion and metastasis (35) (Table 1). It is noteworthy that this upregulation seems to correlate with the Sirt4-mediated inhibition of glutamine metabolism since α-ketoglutarate, the product of the GDH-catalyzed reaction inhibited by Sirt4, decreases E-cadherin expression (35).
However, a few recent studies have demonstrated that Sirt4 can also be upregulated in some tumors and can exert oncogenic properties by regulating the stress responses of cancer cells and preparing them to gain selective advantages, including resistance to anti-tumor treatments (2, 4). Indeed Lai and coworkers showed that the Sirt4 levels in esophageal squamous cell carcinoma (ESCC) tissues from Chinese patients were higher than those in adjacent esophageal normal tissues and that the levels of the protein correlate inversely with the mean survival time of ESCC patients (48) (Table 1). In another report, Sirt4 overexpression in HepG2 cells correlated with reduced levels of cleaved caspase-3, thus reducing apoptotic cell death (anti-apoptotic effect) and increasing the survival and the cellular clone formation rate of tumor cells in response to different DNA-damaging conditions such as cisplatin, radiation, and UV irradiation, and Sirt4 loss sensitized cells to drug treatment (47) (Table 1). Therefore, as far as it concerns the modulation of genome stability, Sirt4 plays a dual role in inhibiting and promoting tumor formation. In fact, while in non-cancer cells Sirt4 protects against the accumulation of DNA damage and promotes DNA repair by acting as a mitochondrial tumor suppressor, it seems to play a different role in cancer cells depending on the external conditions. In “normal” conditions, Sirt4 can inhibit tumor proliferation via the repression of the mitochondrial glutamine catabolism and the subsequent reduction of tumor metabolism. In contrast, in “extreme” DNA-damaging conditions, including chemotherapy, Sirt4 can protect tumor cells, allowing them to escape apoptotic death and, potentially, to acquire more mutations and become more aggressive (4) (Table 1).
It is well-known that gene products known for their tumor-suppressive role can act as oncoproteins in relation to the specific (epi)genetic context, tumor type, tissue or organ microenvironment, stage of tumor development, and external stimuli. Further studies thus are necessary to define the role of Sirt4 and the exact mechanisms involved in the balance between anti-stress (anti-apoptosis) and tumor inhibition (pro-apoptosis) under different pathophysiological and pharmacological conditions.
Conclusion
In the last few years, many studies about Sirt4 have only started to shed light on the multiple enzymatic activities and the biological functions of this, for a long time, enigmatic sirtuin. It is now evident that several Sirt4 actions and substrates are tissue specific, and it is likely that additional enzymatic and non-enzymatic activities and substrates still have to be identified. For example, while Sirt4 associates with different biotin-dependent carboxylases, including MCCC, and is able to hydrolyze various lysine-biotinylated peptides in vitro, the capability to remove biotinyl groups from substrate proteins in vivo has not been investigated yet (3). In humans, Sirt4 is a key regulator of several mitochondrial metabolic processes (Figure 1) including fatty acid and branched-chain amino acid metabolism, carbon entry into the TCA cycle, electron transport and ROS production, insulin secretion and sensitivity, apoptosis, ATP homeostasis, and redox state responding to nutrient condition and exercise. Moreover, Sirt4 has been indicated as a mitochondrion-localized tumor-suppressor protein that, like other sirtuins, may also act as an oncoprotein depending on the specific tumor type, stage, and overall biological context (Table 1). A better mechanistic understanding of how Sirt4 contributes to tumorigenesis, and more generally, to age-related diseases is certainly necessary to provide new insights for the potential targeting of this enzyme for therapeutic scopes.
Author Contributions
DT, CS, AM, and DR contributed to conception, manuscript writing, and proofreading. DT and DR searched the literature and prepared the figure about the biological roles of Sirt4. DR supervised and coordinated the whole writing work. All the authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank our lab members for the fruitful discussions and help with the editing.
Footnotes
Funding. We are grateful for the funding agencies for supporting work on Sirt4 - and mention financial support by Dr. Robert Pfleger foundation (to CS), PRIN 2016 (prot. 20152TE5PK) (to AM), Ricerca Finalizzata 2013 PE-2013-02355271 (to AM), AIRC 2016 (no. 19162) (to AM), Progetto di Ateneo Sapienza 2017 no. RM11715C7CA6CE53 (to DR), and NIH (no. R01GM114306) (to AM).
References
- 1.Pannek M, Simic Z, Fuszard M, Meleshin M, Rotili D, Mai A, et al. Crystal structures of the mitochondrial deacylase sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nat Commun. (2017) 8:1513. 10.1038/s41467-017-01701-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gertz M, Steegborn C. Using mitochondrial sirtuins as drug targets: disease implications and available compounds. Cell Mol Life Sci. (2016) 73:2871–96. 10.1007/s00018-016-2180-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betsinger CN, Cristea IM. Mitochondrial function, metabolic regulation, and human disease viewed through the prism of sirtuin 4. (SIRT4) functions. J Proteome Res. (2019) 18:1929–38. 10.1021/acs.jproteome.9b00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang G, Zhu G. Sirtuin-4. (SIRT4), a therapeutic target with oncogenic and tumor-suppressive activity in cancer. Onco Targets Ther. (2018) 11:3395–400. 10.2147/OTT.S157724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min Z, Gao J, Yu Y. The roles of mitochondrial SIRT4 in cellular metabolism. Front Endocrinol. (2018) 9:783. 10.3389/fendo.2018.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han Y, Zhou S, Coetzee S, Chen A. SIRT4 and its roles in energy and redox metabolism in health, disease and during exercise. Front Physiol. (2019) 10:1006. 10.3389/fphys.2019.01006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Yan Y, Principe DR, Zou X, Vassilopoulos A, Gius D. SIRT3 and SIRT4 are mitochondrial tumor suppressor proteins that connect mitochondrial metabolism and carcinogenesis. Cancer Metab. (2014) 2:1–15. 10.1186/2049-3002-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. (2006) 126:941–54. 10.1016/j.cell.2006.06.057 [DOI] [PubMed] [Google Scholar]
- 9.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. (2014) 159:1615–25. 10.1016/j.cell.2014.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, et al. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun. (2013) 4:2327. 10.1038/ncomms3327 [DOI] [PubMed] [Google Scholar]
- 11.Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. (2013) 50:686–98. 10.1016/j.molcel.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo L, Zhou SR, Wei XB, Liu Y, Chang XX, Liu Y, et al. Acetylation of mitochondrial trifunctional protein alpha-subunit enhances its stability to promote fatty acid oxidation and is decreased in nonalcoholic fatty liver disease. Mol Cell Biol. (2016) 36:2553–67. 10.1128/MCB.00227-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. (2003) 11:437–44. 10.1016/S1097-2765(03)00038-8 [DOI] [PubMed] [Google Scholar]
- 14.Verdin E, Dequiedt F, Fischle W, Frye R, Marshall B, North B. Measurement of mammalian histone deacetylase activity. Methods Enzymol. (2004) 377:180–96. 10.1016/S0076-6879(03)77010-4 [DOI] [PubMed] [Google Scholar]
- 15.Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. (2017) 25:838–55 e815. 10.1016/j.cmet.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner GR, Bhatt DP, O'Connell TM, Thompson JW, Dubois LG, Backos DS, et al. A class of reactive acyl-coa species reveals the non-enzymatic origins of protein acylation. Cell Metab. (2017) 25:823–37 e828. 10.1016/j.cmet.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurent G, de Boer VC, Finley LW, Sweeney M, Lu H, Schug TT, et al. SIRT4 represses peroxisome proliferator-activated receptor alpha activity to suppress hepatic fat oxidation. Mol Cell Biol. (2013) 33:4552–61. 10.1128/MCB.00087-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugger H, Witt CN, Bode C. Mitochondrial sirtuins in the heart. Heart Fail Rev. (2016) 21:519–28. 10.1007/s10741-016-9570-7 [DOI] [PubMed] [Google Scholar]
- 19.Luo YX, Tang X, An XZ, Xie XM, Chen XF, Zhao X, et al. SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J. (2017) 38:1389–98. 10.1093/eurheartj/ehw138 [DOI] [PubMed] [Google Scholar]
- 20.George J, Ahmad N. Mitochondrial sirtuins in cancer: emerging roles and therapeutic potential. Cancer Res. (2016) 76:2500–6. 10.1158/0008-5472.CAN-15-2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choubey SK, Prabhu D, Nachiappan M, Biswal J, Jeyakanthan J. Molecular modeling, dynamics studies and density functional theory approaches to identify potential inhibitors of SIRT4 protein from homo sapiens: a novel target for the treatment of type 2 diabetes. J Biomol Struct Dyn. (2017) 35:3316–29. 10.1080/07391102.2016.1254117 [DOI] [PubMed] [Google Scholar]
- 22.Elkhwanky MS, Hakkola J. Extranuclear sirtuins and metabolic stress. Antioxid Redox Signal. (2018) 28:662–76. 10.1089/ars.2017.7270 [DOI] [PubMed] [Google Scholar]
- 23.Wu T, Liu YH, Fu YC, Liu XM, Zhou XH. Direct evidence of sirtuin downregulation in the liver of non-alcoholic fatty liver disease patients. Ann Clin Lab Sci. (2014) 44:410–8. [PubMed] [Google Scholar]
- 24.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha. (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci USA. (1999) 96:7473–8. 10.1073/pnas.96.13.7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alam N, Saggerson ED. Malonyl-CoA and the regulation of fatty acid oxidation in soleus muscle. Biochem J. (1998) 334:233–41. 10.1042/bj3340233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. (2008) 7:11–20. 10.1016/j.cmet.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Jiang L, Wang J, Li S, Yu Y, You J, et al. Abrogation of hepatic ATP-citrate lyase protects against fatty liver and ameliorates hyperglycemia in leptin receptor-deficient mice. Hepatology. (2009) 49:1166–75. 10.1002/hep.22774 [DOI] [PubMed] [Google Scholar]
- 28.Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ, Stoops JK. The remarkable structural and functional organization of the eukaryotic pyruvate dehydrogenase complexes. Proc Natl Acad Sci USA. (2001) 98:14802–7. 10.1073/pnas.011597698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Argmann C, Auwerx J. Insulin secretion: SIRT4 gets in on the act. Cell. (2006) 126:837–9. 10.1016/j.cell.2006.08.031 [DOI] [PubMed] [Google Scholar]
- 30.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. (2007) 282:33583–92. 10.1074/jbc.M705488200 [DOI] [PubMed] [Google Scholar]
- 31.Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, et al. SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging. (2013) 5:835–49. 10.18632/aging.100616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi JX, Wang QJ, Li H, Huang Q. SIRT4 overexpression protects against diabetic nephropathy by inhibiting podocyte apoptosis. Exp Ther Med. (2017) 13:342–8. 10.3892/etm.2016.3938 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. (2001) 98:13784–9. 10.1073/pnas.241500798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. (2005) 19:1751–9. 10.1038/sj.leu.2403910 [DOI] [PubMed] [Google Scholar]
- 35.Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer. (2015) 113:492–9. 10.1038/bjc.2015.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi YL, Tsukasaki K, O'Neill MC, Yamada Y, Onimaru Y, Matsumoto K, et al. A genomic analysis of adult T-cell leukemia. Oncogene. (2007) 26:1245–55. 10.1038/sj.onc.1209898 [DOI] [PubMed] [Google Scholar]
- 37.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. (2013) 23:450–63. 10.1016/j.ccr.2013.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeong SM, Lee A, Lee J, Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. (2014) 289:4135–44. 10.1074/jbc.M113.525949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. (2014) 33:1609–20. 10.1038/onc.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang G, Cui F, Yu F, Lu H, Zhang M, Tang H, et al. Sirtuin-4. (SIRT4) is downregulated and associated with some clinicopathological features in gastric adenocarcinoma. Biomed Pharmacother. (2015) 72:135–9. 10.1016/j.biopha.2015.04.013 [DOI] [PubMed] [Google Scholar]
- 41.Bartosch C, Monteiro-Reis S, Almeida-Rios D, Vieira R, Castro A, Moutinho M, et al. Assessing sirtuin expression in endometrial carcinoma and non-neoplastic endometrium. Oncotarget. (2016) 7:1144–54. 10.18632/oncotarget.6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakahara Y, Yamasaki M, Sawada G, Miyazaki Y, Makino T, Takahashi T, et al. Downregulation of SIRT4 expression is associated with poor prognosis in esophageal squamous cell carcinoma. Oncology. (2016) 90:347–55. 10.1159/000445323 [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Lai X, Wu C, Tian Q, Lei T, Pan J, et al. Decreased SIRT4 protein levels in endometrioid adenocarcinoma tissues are associated with advanced AJCC stage. Cancer Biomark. (2017) 19:419–24. 10.3233/CBM-160419 [DOI] [PubMed] [Google Scholar]
- 44.Fu L, Dong Q, He J, Wang X, Xing J, Wang E, et al. SIRT4 inhibits malignancy progression of NSCLCs, through mitochondrial dynamics mediated by the ERK-Drp1 pathway. Oncogene. (2017) 36:2724–36. 10.1038/onc.2016.425 [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, Lin J, Feng S, Chen X, Huang H, Wang C, et al. SIRT4 inhibits the proliferation, migration, and invasion abilities of thyroid cancer cells by inhibiting glutamine metabolism. Onco Targets Ther. (2019) 12:2397–408. 10.2147/OTT.S189536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y, Wang G, Li X, Wang T, Weng M, Zhang Y. Knockout of SIRT4 decreases chemosensitivity to 5-FU in colorectal cancer cells. Oncol Lett. (2018) 16:1675–81. 10.3892/ol.2018.8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeong SM, Hwang S, Seong RH. SIRT4 regulates cancer cell survival and growth after stress. Biochem Biophys Res Commun. (2016) 470:251–6. 10.1016/j.bbrc.2016.01.078 [DOI] [PubMed] [Google Scholar]
- 48.Lai X, Yu Z, Chen X, Huang G. SIRT4 is upregulated in Chinese patients with esophageal cancer. Int J Clin Exp Pathol. (2016) 9:10543–9. [Google Scholar]
- 49.Shi Q, Liu T, Zhang X, Geng J, He X, Nu M, et al. Decreased sirtuin 4 expression is associated with poor prognosis in patients with invasive breast cancer. Oncol Lett. (2016) 12:2606–12. 10.3892/ol.2016.5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukey MJ, Wilson KF, Cerione RA. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med Chem. (2013) 5:1685–700. 10.4155/fmc.13.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim MH, Kim H. Oncogenes and tumor suppressors regulate glutamine metabolism in cancer cells. J Cancer Prev. (2013) 18:221–6. 10.15430/JCP.2013.18.3.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. (2009) 458:762–5. 10.1038/nature07823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci USA. (2011) 108:21069–74. 10.1073/pnas.1117500108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. (2013) 153:840–54. 10.1016/j.cell.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Zhou H, Wang Y, Cui G, Di LJ. CtBP maintains cancer cell growth and metabolic homeostasis via regulating SIRT4. Cell Death Dis. (2015) 6:e1620. 10.1038/cddis.2014.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong SM, Haigis MC. Sirtuins in cancer: a balancing act between genome stability and metabolism. Mol Cells. (2015) 38:750–8. 10.14348/molcells.2015.0167 [DOI] [PMC free article] [PubMed] [Google Scholar]