Dear Editor,
Liu et al. in this Journal recently reported a COVID-19 patient with prolonged viral shedding even after seroconversion.1 The patient's antibody to SARS-CoV-2 viral nucleocapsid (N) protein, as determined by western blotting using infected cell lysates, was firstly identified on the tenth day after symptom onset. However, detection of antibody to SARS-CoV-2 by western blot is time-consuming, labor-intensive, and expertise-dependent. Furthermore, data on antibodies to SARS-CoV-2 IgM or IgG were lacking.
Fourteen COVID-19 patients who were treated at six hospitals in Taiwan between January and March 2020 were enrolled in this study. The patients were classified into two groups: symptomatic and asymptomatic/mild symptoms. Patients designated in the symptomatic group had a fever for more than 3 days, obvious pneumonia patches on chest radiographs, and respiratory distress defined as oxygen saturation less than 95% or needing oxygen supply during hospitalization. Patients who did not meet the criteria of the symptomatic group were included in the asymptomatic/mild symptom group.
Samples from the respiratory tract for SARS-CoV-2 rRT-PCR testing included oropharyngeal and nasopharyngeal swabs, oral gargling, and sputum. All samples were submitted to virology laboratories validated and contracted by the Centers for Diseases Control and Prevention in Taiwan for testing.2 Three sets of primers and probes targeting SARS-COV-2 envelope (E), nucleocapsid (N), and RNA-dependent RNA polymerase (RdRp) genes were used. In addition to SARS-CoV-2 RT-PCR testing, all respiratory samples were simultaneously evaluated for the presence of Influenza A/B viruses using RT-PCR. When the results were negative for both SARS-CoV-2 and Influenza A/B viruses, an additional SARS-CoV-2 rRT-PCR test for another respiratory sample from the suspected COVID patient was performed.
The frequencies of antibody testing of the 14 COVID-19 patients were performed at the discretion of the attending physicians at each participating hospital. Anti-SARS-CoV-2 IgM and IgG antibodies in 33 serum samples collected from the 14 COVID-19 patients were determined using the ALLTEST 2019-nCoV IgG/IgM Rapid Test Cassette (Hangzhou ALLTEST Biotech Co., Ltd. Hangzhou, China).2 , 3 In addition, 28 control serum samples collected from 28 hospitalized patients (illness days 7-14), with respiratory tract infection but two negative results of SARS-CoV-2 rRT-PCR testing, were also evaluated to validate the performance of the assay. Among the 28 samples, three were known to be positive for either CMV-IgM and IgG, Chlamydophila pneumoniae IgG, or EBV VCA IgM. The ALLTEST 2019-nCoV IgG/IgM Rapid Test Cassette is a rapid lateral flow immunoassay (LFIA) that uses a recombinant SARS-CoV-2 N protein to detect both IgM and IgG antibodies. This in-vitro device was registered in Germany in March 2020 (DE/CA22/419-822.3-IVD; catalogue number INCP-402).3
Among the 14 patients with COVID-19, six belonged to the symptomatic group and eight belonged to the asymptomatic/mild symptom group. All the patients were hospitalized in isolation rooms within one of the six hospitals. The clinical characteristics of the patients are summarized in Table 1 . Age distribution of patients in both groups was similar, but more males tended to belong to the asymptomatic/mild symptom group. All patients in the symptomatic group acquired SARS-CoV-2 from abroad, including four in Wuhan, one in Macau, and one who had contact during their trip to Europe with a sick patient from Wuhan. Half of the patients in the asymptomatic/mild symptom group were infected locally in Taiwan, without any travel history. A few patients had underlying co-morbid diseases; one had diabetes mellitus and followed diet control, and another had HIV infection and was undergoing treatment with anti-retroviral agents (regimens not including protease inhibitor). All patients in the symptomatic group had fever, but only one patient in the asymptomatic/mild symptom group had fever.
Table 1.
Demographic data, underlying medical conditions, clinical manifestations, imaging studies, and laboratory findings from 14 hospitalized COVID-19 patients
| Symptomatic group (n = 6) | Asymptomatic/mild symptom group (n = 8) | |
|---|---|---|
| Media age, years (range) | 52 (45-73) | 50 (30-88) |
| Number of males (%) | 2 (33.3) | 5 (62.5) |
| Exposure history | ||
| Exposure abroad | 6 (100) | 4 (50) |
| Local transmission in Taiwan | 0 (0) | 4 (50) |
| Comorbid conditions, any (%) | 0 | 3 (37.5) |
| Diabetes | 0 | 1 |
| Cardiovascular diseases | 0 | 0 |
| Chronic obstructive pulmonary disease | 0 | 0 |
| Malignancy | 0 | 0 |
| Others | 0 | 1 (HIV) |
| Fever | 6 (100) | 1 (12.5) |
| Media duration of fever, days (range) | 6 (3-9) | 4 (4) |
| Increase C-reactive protein, >1 mg/L (%) | 5 (83.3) | 1 (12.5) |
| Leukocytopenia, <1,000/μL (%) | 4 (66.7) | 1 (12.5) |
| Pneumonia on chest radiographs | 6 (100) | 1 (12.5) |
| Pneumonia on chest computed tomography | 5/5a (100) | 2/7a (28.6) |
| Antibacterial agent(s) use | 5 (83.3) | 2 (25) |
| Antiviral agent use | 5 (83.3) | 1 (12.5) |
| Oseltamivir | 5 (83.3) | 1 (12.5) |
| Lopinavir/ritonavir | 2 (33.3) | 0 (0) |
| Hydroxychloroquine use | 2 (33.3) | 0 (0) |
| Steroid use | 0 (0) | 2 (25) |
Chest computed tomography was not performed in one patient in each group
Patients in the symptomatic group had higher proportions of abnormal laboratory data and data from imaging studies than those in the asymptomatic/mild symptom group (Table 1). Consequently, there were more patients in the symptomatic group prescribed with antibacterial agents, anti-viral agents, and even hydroxychloroquine. Eleven patients were discharged from the hospitals under relatively stable conditions, while three were still hospitalized. None of the patients developed respiratory failure and a need for ventilator support during hospitalization. Moreover, there were no mortalities observed in this study.
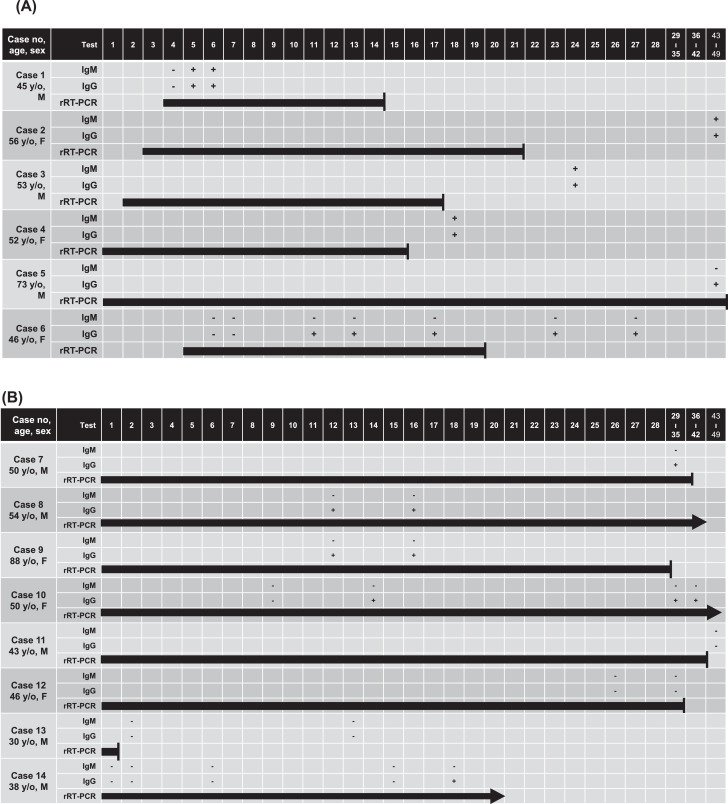
Of the six patients in the symptomatic group, all had positive anti-SARS-CoV-2 IgG and four had positive anti-SARS-CoV-2 IgM responses (Fig. 1 A). The duration of positive rRT-PCR results ranged from 12 to 46 days. Patients with positive anti-SARS-CoV-2 IgM results seemed to have a short duration of viral shedding (Fig. 1A). For the eight patients in the asymptomatic/mild symptom group, none had positive anti-SARS-CoV-2 IgM results and three (cases 11-13) had negative anti-SARS-CoV-2 IgG results (Fig. 1B). The last day of anti-SARS-CoV-2 IgM/IgG testing after the notification of positive rRT-PCR for these three cases was >42 days in case 11, >28 days in case 12, and 13 days in case 13. In case 13, a 30-year-old man was tested with rRT-PCR assay for SARS-CoV-2 due to travel exposure but did not have any related symptoms. His rRT-PCR showed a positive result only on day 1 (cycle threshold [ct] value, 33), but was negative in the three subsequent rRT-PCR tests with an interval of 48 hours. Except case 13, the duration of the presence of SARS-CoV-2 RNA by positive rRT-PCR was generally longer in the asymptomatic/mild symptom group than the symptomatic group.
Fig. 1.
Dynamics of anti-SARS-CoV-2 IgG and IgM, and results of real‐time reverse transcriptase‐PCR (rRT-PCR) for 14 COVID-19 patients. (A) Six patients in the symptomatic group. (B) Eight patients in the asymptomatic/mild symptom group. The number in the columns indicates days after onset of symptoms or the time when COVID-19 was confirmed among asymptomatic patients. Arrows indicate patients who were still hospitalized and did not achieve negative conversion of rRT-PCR results at the end of the study.
In our study, the earliest detection of SARS-CoV-2 IgM was on day 5 in case 1 and the longest persistence was found on day 42 in case 2. For SARS-CoV-2 IgG, the earliest detection was on day 5 in case 1, and most cases persistently had positive SARS-CoV-2 IgG results after positive conversion. On the contrary, all 28 control serum samples, including the three samples with the presence of known non-SARS-CoV-2-viral antibodies, were negative for anti-SARS-CoV-2 IgM and IgG. The overall sensitivity and specificity of the rapid test kit based on the rRT-PCR results was 78.6% and 100%, respectively.
In our study, we demonstrated dynamics of anti-SARS-CoV-2 IgM or IgG among 14 patients with different clinical manifestations. The duration of positive rRT-PCR persistence was associated with antibody response and clinical manifestations. Patients with prominent symptoms and development of anti-SARS-CoV 2 IgM antibodies tended to have short viral shedding by rRT-PCR.
In China, another rapid LFIA using a recombinant antigen, the receptor-binding domain of the SARS-CoV-2 spike protein, to detect both SARS-CoV-2 IgM and IgG antibodies, was tested on 397 COVID-19 confirmed cases and 128 control cases.4 This assay showed a testing sensitivity of 88.7% and specificity of 90.3% for the diagnosis of COVID-19 (the presence of either IgG or IgM antibodies, or both). The turnaround time of IgG/IgM test is much less than that of rRT-PCR, which shortens the quarantine period and facilitates in efficacious use of hospital beds. However, the antibody test may also compromise the sensitivity and specificity of rRT-PCR. The timing, quality, and quantity of antibody response might be associated with clinical manifestations and disease course, and this therefore warrants further investigations.
The ALLTEST 2019-nCoV IgG/IgM Rapid Test Cassette is a rapid LFIA (turn-around time within 15 min) that uses a recombinant SARS-CoV-2 N protein to detect both IgM and IgG antibodies.3 In comparison with the results using conventional rRT-PCR (22 positive and 100 negative serum samples), the relative sensitivity, specificity, and accuracy rate of anti-SARS-CoV-2 IgG results from the ALLTEST 2019-nCoV IgG/IgM Rapid Test Cassette were claimed to be >99.9% (95% confidence interval (CI), 82.5%-100%), 98.0% (95% CI, 92.6%-99.9%), and 98.4% (95% CI, 93.9%-99.9%), respectively. However, the relative sensitivity, specificity, and accuracy rate for detecting anti-SARS-CoV-2 IgM antibodies were only 90.9% (95% CI, 71.0%-98.7%), 97.0% (95% CI: 91.8%-99.4%), and 95.9% (95% CI, 90.5%-98.5%), respectively.3 The recombinant SARS-CoV-2 N protein used in this assay shares homology with SARS-CoV and MERS-CoV but not common seasonal human coronaviruses (i.e., 229E, NL63, OC43, and HKU1). As a result, no cross-reaction with the four human coronaviruses was claimed by the manusfacturer.3 In a recent study conducted during the COVID-19 epidemic, 43 respiratory specimens collected from 43 suspected COVID-19 patients were subjected to rRT-PCR for SARS-CoV-2 and FilmArray respiratory panel (BioFire Diagnostics, bioMe´rieux, Utah, USA) assays Two were positive for SARS-CoV-2 rRT-PCR and another three were positive for coronavirus 229E/OC43; no co-infection with SARS-CoV-2 and other coronaviruses (HKU1, NL63, 229E, and OC43) were identified.5
In the present study, patients with symptoms and development of anti-SARS-CoV-2 IgM antibodies had a shorter duration of positive rRT-PCR result and were discharged with relatively stable conditions. Anti-SARS-CoV-2 IgG antibodies were absent in three patients (cases 11-13) in the asymptomatic/mild symptom group who had positive rRT-PCR results. In case 13, a false-positive rRT-PCR result was highly suspected by the attending physicians and the negative IgG result might support the clinicians’ suspicion. However, the presence of lower anti-SARS-CoV-2 IgG titers may have contributed to the negative anti-SARS-CoV-2 IgG results obtained by the rapid test. Further studies for detecting antibodies against SARS-CoV-2-specific proteins or neutralizing antibodies should be conducted to verify the performance of the rapid test kit.
The significance of antibody response in COVID-19 is important, not only in the diagnosis but also prognosis. Specific antibodies, including IgG antibodies and neutralizing antibodies, are important for protecting the host from infection by blocking viral entry into host cells after viral infection.6 Contrarily, there are no vaccines or medications available for SARS-CoV-2 to date. The administration of convalescent sera from recovered individuals was proposed as an option for treatment to provide immediate immunity to susceptible or infected persons.7
Several limitations were found in our study. First, the sample size is small, as we included only 14 COVID-19 patients. Second, a further western blot assay for detecting the anti-SARS-CoV-2-specific protein IgG or IgM antibodies was not performed to validate the performance of the ALLTEST 2019-nCoV IgG/IgM Rapid Test and understand the inconsistency in the presence of anti-SARS-CoV-2 IgM and IgG. Third, cross reaction of serum specimens from the acute phase of different viral infections (e.g., influenza, respiratory syncytial virus, and rhinovirus.) in the IgM portion of this SARS-CoV-2 assay was not performed. Finally, different SARS-CoV-2 strains or genotypes may also interfere with disease outcome and antibody response; however, viral cultures for all patients in our study were not performed and genetic characteristics of SARS-CoV-2 strains were not investigated.8
In summary, we performed an anti-SARS-CoV-2 IgG/IgM test on 14 confirmed COVID-19 patients and 28 negative controls. Antibody response varied with different clinical manifestations and disease severity. Patients with symptoms and development of anti-SARS-CoV-2 IgM antibodies had a shorter duration of positive rRT-PCR result and no worsening clinical conditions compared to those without the presence of anti-SARS-CoV-2 IgM antibodies. The serological data can provide more information about epidemiological linkage and disease prognosis for the new emerging COVID-19 pandemic.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- 1.Liu W.D., Chang S.Y., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. 2020 Apr 10 doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee N.Y., Li C.W., Tsai H.P., Chen P.L., Syue L.S., Li M.C., Tsai C.S., Lo C.L., Hsueh P.R., Ko W.C. A case of COVID-19 and pneumonia returning from Macau in Taiwan: Clinical course and anti-SARS-CoV-2 IgG dynamic. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Package Insert. ALLTEST 2019-nCoV IgG/IgM Rapid Test Cassette, Hangzhou ALLTEST Biotech Co., Ltd. Hangzhou, China. https://www.siloambio-tech.com/en/product_1297294.html.
- 4.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsih W.H., Cheng M.Y., Ho M.W., Chou C.H., Lin P.C., Chi C.Y. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan. J Microbiol Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Y., Wang G., Shi X., Zhang H., Qiu Y., He Z. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2020;190:1119–1126. doi: 10.1086/423286. http://doi: 10.086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Xiong J., Bao L., Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;27:30141–30149. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J.T., Lin Y.Y., Chang S.Y., Yeh S.H., Hu B.H., Chen P.J. The role of phylogenetic analysis in clarifying the infection source of a COVID-19 patient. J Infect. 2020 Apr 8 doi: 10.1016/j.jinf.2020.03.031. S0163-4453(20)30159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]