Abstract
Traditional drug discovery focuses on identifying direct inhibitors of target proteins. This typically relies on a measurable biochemical readout and accessible binding sites whose occupancy influences the function of the target protein. These requirements preclude many disease-causing proteins from being 'druggable' targets, and these proteins are categorized as 'undruggable'. The proteolysis-targeting chimera (PROTAC) technology provides powerful tools to degrade these undruggable targets and has become a promising approach for drug discovery. However, the PROTAC technology has some limitations, and emerging new degrader technologies may greatly broaden the spectrum of targets that could be selectively degraded by harnessing a second major degradation pathway in cells. We review key emerging technologies that exploit the lysosomal degradation pathway and discuss their potential applications and limitations.
Keywords: undruggable targets, protein degradation, ATTEC, AUTAC, LYTAC, PROTAC
Highlights
Selective degradation of pathogenic proteins is a promising strategy to drug 'undruggable' targets.
Established degrader technologies such as PROTAC are extremely promising but still have limitations.
The lysosome is a major degradation pathway utilized by cells to degrade extracellular and intracellular content.
New technologies such as LYTAC, AUTAC, and ATTEC may have potential applications in several diseases by exploiting the lysosome for targeted degradation.
Importance of Degrader Technologies for Drug Discovery
Human genetic studies have revealed a large number of novel protein targets that cause disease via toxic gain of function. The abnormal accumulation and toxicity of these pathogenic proteins may directly cause many diseases including neurodegenerative disorders and type 2 diabetes [1,2]. Traditional inhibitor identification-based drug discovery strategies are limited by the requirements of specific measurable functions of the target protein and accessible binding sites whose occupancy directly influences their function. Because of these requirements, almost all the disease-relevant scaffolding proteins, transcription factors, and other non-enzymatic proteins are essentially undruggable by the inhibitor approach.
One appealing idea to address this is to enhance the global protein quality control system such that these undruggable pathogenic proteins can be corrected or eliminated. For example, remodeling the proteostasis network by small molecules targeting molecular chaperones or inducing stress responses may reduce the aggregation and toxicity of multiple disease-linked proteins in animal models (reviewed in [3]). Furthermore, the induction of autophagy may provide a potential therapeutic strategy for many neurodegenerative diseases that are possibly caused by pathogenic proteins (reviewed in [4]). Although these approaches have potential in treating multiple diseases, they induce global cellular changes and thus may have extensive nonspecific effects or induce strong compensatory mechanisms that offset their beneficial effects.
A more specific strategy would be to selectively reduce the levels of the pathogenic proteins. One possible approach is gene therapy: gene silencing through nucleic acid-based RNA- or DNA-targeting reagents such as antisense oligonucleotides (ASOs) and genome-editing reagents, respectively [5,6] (Figure 1A). Although highly promising, this approach faces some key challenges. Because they are large biomolecules, the gene therapy reagents are difficult to deliver, especially for neurological disease [7], and they are also prohibitively expensivei. Small molecule-induced protein degradation is an attractive alternative approach to selectively reduce the levels of pathogenic proteins, and may fulfill huge unmet medical needs. Several new approaches have been established to achieve this goal, as discussed later.
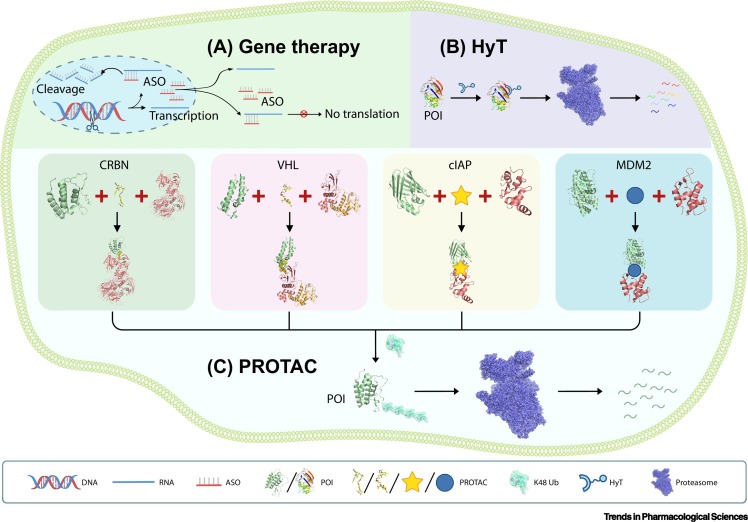
Figure 1.
Schematic Models of Established Strategies to Selectively Target a Protein of Interest (POI).
(A) Schematic illustration of DNA- and RNA-targeting technologies such as genome editing (represented by a pair of scissors cutting the target DNA) or antisense oligonucleotides (ASOs) that induce the target RNA degradation and protein translation inhibition. (B) Schematic illustration of the hydrophobic tagging (HyT) technology, which adds a hydrophobic tag onto POIs to induce their degradation via the proteasome, independently of E3 ligases and ubiquitination. (C) Schematic illustration of the four major PROTAC systems utilizing the indicated E3 ligase subunits. The PROTAC molecules bring the POIs in proximity to the corresponding E3 ligases to facilitate POI K48 polyubiquitination (Ub), leading to their subsequent degradation via the proteasome. All molecules were drawn based on public PDB files: CRBN-PROTAC-BRD4 (PDB: 6BN7); VHL-PROTAC-BRD4 (PDB: 5T35); cIAP1 (PDB: 3UW4), CRABP2 (PDB: 3CBS); MDM2 (PDB: 4HF2), AR (PDB: 1XOW); Ub (PDB: 5GOI); HyT POI DHFR (PDB: 5UII); proteasome (PDB: 4CR2). The PDB files of the cIAP and MDM2 PROTAC complexes were unavailable, and their PROTAC molecules therefore are represented by cartoon shapes.
Small Molecule-Induced Selective Protein Degradation
Small molecules are able to induce selective degradation of the target protein of interest (POI) by adding a tag that is recognized by the degradation machinery. In 2011–2012, Crews and colleagues reported small-molecule hydrophobic tagging (HyT)-induced degradation of POIs [8,9]. HyT was achieved by molecules consisting of a hydrophobic fragment and a ligand binding to the POI. Thus, the HyT molecules bind to the POI and add a hydrophobic fragment to it, leading to recognition and recruitment of heat-shock proteins followed by proteasomal degradation [8,9]. A similar concept was illustrated by another study that used a small molecule consisting of a hydrophobic group tert-butyl carbamate-protected arginine (Boc3Arg) attached to the dihydrofolate reductase (DHFR) non-covalent binding ligand trimethoprim (TMP) to degrade DHFR [10]. The Boc3Arg-TMP molecules may recycle during the degradation process, increasing its efficacy [10]. Hedstrom and colleagues synthesized chimeric molecules by linking other POI recognition ligands to Boc3Arg and also found that they can target POIs to the 20S proteasome for degradation directly without the requirement for ATP or involvement of ubiquitination pathways [11] (Figure 1).
Although HyT is an interesting concept, it has not been widely used and its clinical application may be limited by Boc3Arg off-target effects [12]. Currently, the prevailing approach for small molecule-induced protein degradation is the proteolysis-targeting chimera (PROTAC) technology, which provides a powerful strategy to degrade undruggable POIs and has been extensively reviewed [13., 14., 15., 16., 17.]. PROTAC molecules tether the POI to a specific E3 ligase subunit to enhance POI ubiquitination, leading to its subsequent degradation by the proteasome (Figure 1).
In 2001, Crews and colleagues provided the proof-of-concept evidence for PROTAC [18]. They synthesized a chimeric large molecule named Protac-1 to recruit MetAP-2 to the Skp1–Cullin–F box complex, leading to ubiquitination and degradation of MetAP-2 in a dose-dependent manner. However, the MetAP-2 covalently bound moiety was a peptide, hindering further applications because its large molecule size leads to poor cellular penetration. In 2008, a cell-permeable PROTAC molecule linking the non-steroidal androgen receptor ligand (SARM) and the MDM2 ligand nutlin with a polyethylene glycol (PEG)-based linker was synthesized [19]. This PROTAC molecule can induce the ubiquitination of the androgen receptor and its subsequent degradation by the proteasome. This PROTAC system is based on MDM2, the major E3 ligase that ubiquitinates the tumor-suppressor protein p53 [20]. The nutlins family consists of small molecules that bind to the p53-binding pocket of MDM2 and thus competitively inhibit the interaction between p53 and MDM2 [21], inducing possible off-target effects. The nutlins were further improved to RG7112 [22] and RG7388 [23] that showed superior potency and selectivity. In 2010, a new PROTAC system using the cellular inhibitor of apoptosis protein 1 (cIAP1) ubiquitin ligase was established. Hashimoto and colleagues verified that a hybrid molecule coupling the cIAP1 ligand methyl bestatin and a CRABP ligand can induce degradation of CRABP-II in cells via the proteasomal pathway [24]. In 2015, the group of Brader established another important PROTAC system based on cereblon (CRBN) [25]. CRBN was initially identified as the primary mediator of the teratogenic activity of thalidomide by Handa and colleagues [26]. Immunomodulatory drugs (IMiDs) including thalidomide and its derivatives such as lenalidomide and pomalidomide were later utilized to treat multiple myeloma [27]. These IMiDs were further reported to interact with the CRBN–DNA damage binding protein 1 (DDB1) complex [28]. Shortly thereafter a series of PROTAC molecules consisting of a CRBN-binding IMiD connected to small-molecule ligands for different POIs were designed and their degrader capability was verified [25,29,30]. For example, JQ1–penalidomide chimeric compounds can selectively degrade BRD4 protein via CRBN [25]. Another PROTAC system, the VHL-PROTAC system based on a HIF1-α-derived peptide, was initially introduced in 2004 [31]. In 2015, replacement of the VHL-targeting peptide by hydroxyproline-like small molecules significantly improved the affinity and specificity of the VHL-PROTAC system [32], greatly expanding its applications. Currently, although there are >600 known human E3 ligases, the established PROTAC systems rely on fewer than ten, of which the most widely used are CRBN [28], Von Hippel Lindau (VHL) [31], cIAP [24], and MDM2 [19], discussed earlier.
One desirable property of PROTACs is that they are catalytically involved in multiple rounds of target protein degradation [29,32,33]. Therefore, the degradation induced by PROTACs is sub-stoichiometric – one PROTAC molecule is able to induce the ubiquitination and degradation of multiple POI molecules. The catalytic nature of PROTACs provides potential strong degradation power at relatively low concentration, and it can be desirable to introduce temporal and spatial control. Recent studies have developed controllable PROTACs, such as photoswitchable azobenzene-PROTACs (Azo-PROTACs) and photo-caged PROTACs (pc-PROTACs) [34,35]. Azo-PROTACs are light-controlled small-molecule tools for protein knockdown in cells. The light-induced configuration change can switch to the active state to induce protein degradation, and this can be reversed by light exposure in intact cells [34]. For pc-PROTACs, photo-removable blocking groups were added to a degrader of Brd4, and the resulting molecule showed potent degradation activity in live cells only after light irradiation [35].
Although the PROTAC field is booming, several potential limitations have been identified: PROTAC molecules often have high molecular weight (>800 Da) and a relatively high polar surface area, likely leading to low solubility, poor cell permeability, and low oral bioavailability and blood–brain barrier penetration. The effects of PROTAC are dependent on specific E3 ligase subunits and thus are influenced by their expression, limiting their application to particular cell types. The dependence on specific E3 ligase subunits may also cause cancer cell resistance following chronic PROTAC treatment [36]. Finally, PROTAC molecules induce POI degradation via the proteasome, which has limited ability to degrade proteins with expanded repeat sequences, protein aggregates, and other non-protein molecules [37].
Emerging New Concepts of Small Molecule-Induced Protein Degradation
As discussed earlier, although the PROTAC technology is extremely promising, its dependence on the expression of specific E3 ligases and the proteasomal pathway may limit its potential applications for particular cell types or proteasome-resistant proteins. Encouragingly, novel concepts have emerged very recently regarding the lysosomal degradation pathway – a second major pathway of degradation that is independent of the proteasome [38]. The lysosomal degradation pathway includes the endosome/lysosome pathway and the autophagy pathway (reviewed in [39,40]). The endosome/lysosome pathway involves sequential processing by several membrane-bound intracellular compartments: the endocytosed material is incorporated and proceeds through early endosomes, endosome carrier vesicles, late endosomes, and the lysosome for subsequent hydrolysis [39]. The autophagy pathway starts with an isolated membrane structure called a phagophore, which is derived from lipid bilayer with lipidated LC3 proteins [41]. This phagophore expands to engulf intracellular cargoes (autophagy substrates), including proteins and other biomolecules or even organelles, thereby sequestering them in a double-membrane vesicle called the autophagosome [40]. The loaded autophagosome matures through fusion with the lysosome, leading to cargo degradation. Thus, both the endosome/lysosome pathway and the autophagy pathway are able to degrade target material, and new concepts have been developed to harness each of these pathways for selective degradation.
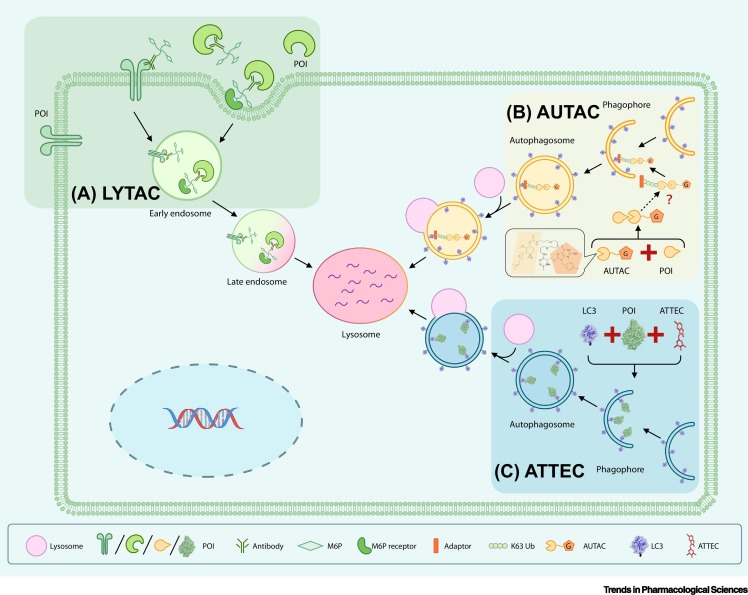
The LYTAC (lysosome targeting chimera) technology is a promising strategy to exploit the endosome/lysosome pathway to degrade POIs, currently published as a preprint paper without peer review [42]. Excitingly, LYTAC molecules may work on extracellular proteins as well as on membrane-bound proteins such as EGFR [42]. These types of proteins are typically resistant to PROTAC, which mainly targets intracellular proteins. A LYTAC molecule consists of an antibody to a specific POI, and a 20- or 90-mer of mannose-6-phosphate (M6P) is covalently attached to this antibody. M6P is a glycan that targets proteins for degradation by binding to the cation-independent M6P receptor (CI-M6PR) [42]. Thus, M6P-conjugated antibodies, namely the LYTAC molecules, are recognized by this endogenous system for lysosomal degradation of glycosylated proteins, and bring glycans together with the POI to the lysosome for degradation. In some cases the M6P-conjugated antibodies can be replaced by CI-M6PR polypeptide ligands conjugated to a small molecule that binds to the POI. The authors confirmed that a polypeptide with multiple ligands for CI-M6PR attached to biotin (a high-affinity avidin-binding molecule) targets the extracellular NeutrAvidin protein to endosomes and lysosomes [42]. The advantage of the LYTAC technology is that it enables degradation of both extracellular and membrane-associated POIs, and it utilizes a ubiquitously expressed endogenous degradation pathway. The major limitation is that LYTAC molecules are relatively large, and thus lose the desired properties of small-molecule drugs. The antibody or polypeptide nature of the molecules may also induce immune responses [43].
The LYTAC technology targets extracellular and/or membrane POIs to the endosome/lysosome pathway, and is not applicable to cytosolic proteins. However, these can be targets for autophagy, and autophagy-targeting degrader technologies are highly desirable for cytosolic POIs, especially those resistant to PROTAC molecules. Although there are several different types of autophagy, the term typically refers to macroautophagy, a multistep cellular process in which cargo proteins are engulfed into autophagosomes formed by intracellular double membranes that contain lipidated LC3 proteins [41]. Autophagy receptors such as SQSTM1/p62 may recognize Lys63 (K63) polyubiquitinated protein cargoes and shuttle them to autophagosomes for subsequent clearance [44], and this process is referred to as 'selective autophagy' [45,46]. Two independent new degrader technologies harnessing the autophagy pathway have been reported very recently.
The autophagy-targeting chimera (AUTAC) system has a similar design to the PROTAC technology [47]. Both AUTAC and PROTAC molecules function via ubiquitination. However, instead of tethering the POIs to an E3 ligase subunit and triggering K48 polyubiquitination, AUTAC molecules trigger K63 polyubiquitination, which is recognized by the selective autophagy pathway, leading to degradation of the target POI. An AUTAC molecule contains a degradation tag (guanine derivative) and a ligand of the POI to provide target specificity. The degradation tag mimics S-guanylation, a post-translational modification of Cys-cGMP adducts formed by treatment with 8-nitro-cGMP or a similar compound [48]. The authors suggest that S-guanylation is a standalone tag that marks the substrate protein for selective autophagy by inducing K63 polyubiquitination [47]. The authors reported encouraging results that AUTAC molecules are capable of degrading POIs as well as cellular organelles such as damaged mitochondria.
The design of AUTAC cleverly utilizes a process that defends cells against invading group A Streptococcus [49]. AUTAC molecules function through a complex multiple-step process that needs to be further clarified. In particular, the molecular mechanism mediating S-guanylation-triggered K63 ubiquitination of POIs needs to be elucidated, and the efficiency and potential off-target effects of this pathway need to be determined. This will clarify which proteins are required for the AUTAC system, how the AUTAC system is influenced by each mediator, and whether exploiting this pathway may cause changes to other cellular functions. In addition, whether AUTAC molecules influence the selective autophagy process per se and whether they are functional in vivo need to be investigated.
The concept of autophagosome-tethering compound (ATTEC) illustrates a more direct strategy to harness autophagy to degrade POIs [50,51]. In contrast to PROTAC and AUTAC, ATTEC molecules are independent of ubiquitination. Instead, ATTEC molecules tether the POI to the autophagosomes by direct binding to the POI and the key autophagosome protein LC3. A proof-of-concept study established a high-throughput screening strategy to identify compounds targeting the mutant HTT protein (mHTT), the Huntington’s disease (HD)-causing protein that has an expanded polyglutamine (polyQ) stretch [52]. The study demonstrated that these compounds can degrade mHTT both in cells and in vivo in animal models, and can rescue HD-relevant phenotypes [51]. The study also confirmed that these compounds can target mHTT to autophagsomes for subsequent degradation without influencing autophagy activity per se. By counter-screening for molecules binding to the wild-type HTT protein (wtHTT), the ATTEC molecules developed are allele-selective, in other words they degrade mHTT without influencing the level of wtHTT. In vitro experiments revealed that these compounds specifically interact with the expanded polyQ stretch, possibly by recognizing its unique structural features that differ from the short polyQ stretch [53,54]. The ATTEC molecules are also capable of degrading other disease-causing polyQ proteins, such as mutant ATXN3, which causes spinocerebellar ataxia type III [51]. Some of the ATTEC molecules are able to pass the blood–brain barrier and function at ~100 nM concentrations [51], providing encouraging entry points for drug discovery. By interacting with the autophagosome protein LC3 directly and bypassing the ubiquitination process, ATTEC molecules have great potential for degrading different types of cargoes, including autophagy-recognized non-protein cargoes such as DNA/RNA molecules, damaged organelles, etc., through a direct mechanism. ATTEC molecules do not influence global autophagy activity [51], but it remains to be elucidated whether AUTACs affect global autophagy. It is important not to perturb global autophagy to avoid non-specific degradation of functional proteins and organelles.
Further studies will be necessary to further develop ATTEC. The compound compartments that interact with LC3 remain to be resolved for ATTEC molecules. Unlike PROTAC, LYTAC, and AUTAC, the ATTEC molecules targeting mHTT have very small sizes compared with the relatively large chimeric compounds with a linker between two separate chemical moieties that interact with the POI and the degradation machinery, respectively. Whether functional chimeric molecules can be developed by attaching the LC3-binding 'warhead' to POI binding compounds remains to be tested. On the other hand, these small compounds may have the advantage of having better drug properties.
In summary, in addition to the PROTAC technology and its further developments, at least three emerging new concepts of degrader technologies have been demonstrated recently (Figure 2 , Key Figure). Although each has its advantages and limitations (Table 1 ), they have greatly expanded the potential applications of degrader technologies and may open new avenues of research in the field of targeted degradation.
Figure 2.
Key Figure. Schematic Models of Emerging Degrader Technologies.
(A) LYTACs utilize a glycan tag to mark an extracellular protein of interest (POI) for intracellular lysosomal degradation following receptor-mediated internalization. Note that the LYTAC paper has not yet been peer reviewed and formally published. (B) AUTACs bind to the POI and add a degradation tag mimicking S-guanylation, a post-translational modification that triggers K63 polyubiquitination (Ub) of the POI. The POI is then recognized by the autophagy receptor SQSTM1/p62 and is recruited to the selective autophagy pathway for degradation. For the AUTAC molecule, the G pentagon represents the chemical moiety mimicking S-guanylation and the linked pacman shape represents the targeting recognition moiety. (C) ATTECs interact with both the POI and LC3, tethering the POI to the phagophores or autophagosomes for subsequent autophagic degradation.
Table 1.
Advantages and Limitations of PROTAC and Emerging Degrader Technologies
| Degrader technology | Degradation pathway | Potential targets | Advantages | Limitations | Refs |
|---|---|---|---|---|---|
| PROTAC | Proteasome pathway | Intracellular proteins | Well established with structural information; clear mechanisms of action; relatively high selectivity; catalytic and sub-stoichiometric |
E3-, ubiquitination-, and proteasome-dependent; generally undesirable pharmacokinetic profile; possible limitations of target spectrum |
[13., 14., 15., 16., 17.] |
| LYTAC | Endosome/lysosome pathway for degradation of glycosylated proteins | Extracellular proteins; transmembrane proteins | Applicable to extracellular and transmembrane proteins; independent of ubiquitination and proteasomal degradation |
Large molecular weight and poor permeability; possible induction of immune response in vivo | [42] |
| AUTAC | Selective macroautophagy pathway | Intracellular proteins; damaged organelles associated with specific proteins | Potentially a broad target spectrum; proteasome-independent; demonstrated ability to degrade mitochondria | Lack of key information of mechanisms of action; dependent on K63 ubiquitination; possible influence on selective autophagy |
[47] |
| ATTEC | Macroautophagy pathway | Intracellular proteins; non-protein autophagy substrates | Potentially a broad target spectrum; direct targeting to the degradation machinery; potentially effective in all cell types; low molecular weight |
The LC3-bound chemical moieties need to be solved; lack of studies on designed chimeras | [50,51] |
Potential Applications
The emerging new concepts of degrader technologies, especially novel developments that exploit the autophagy/lysosomal pathway, may greatly expand the landscape of the applications of degrader technologies. Although current studies have mainly focused on pathogenic proteins, these new concepts could also be explored for their ability to degrade pathogenic targets other than soluble proteins, such as protein aggregates, DNA/RNA molecules, peroxisomes, ribosomes, damaged mitochondria, or even microbial pathogens. In the following we briefly discuss possible future use of these strategies to degrade such targets as potential examples of the future applications of these technologies.
Many neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease, are associated with the formation of protein aggregates/oligomers by misfolded proteins [55,56]. Clearing these protein aggregates/oligomers without affecting their wild-type counterparts is likely a promising drug discovery paradigm. Given the poor ability of the proteasome to degrade these aggregates/oligomers [37], degraders exploiting autophagy may be a better strategy for targeting protein aggregates for selective degradation, at least in theory. In support, one of the ATTEC molecules reduced mutant HTT aggregates significantly [51]. Given that most of the relevant diseases are neurological, molecules with high lipid solubility and a molecular weight under 500 Da will be necessary because they are more likely to cross the blood–brain barrier via passive diffusion [57]. Thus, degraders with smaller molecular weights have greater potential as drug candidates for the treatment of neurodegeneration. Interestingly, Tau degraders were recently reported based on the PROTAC system [58]. The compounds efficiently degraded neurodegeneration-related soluble Tau protein, but Tau aggregates/tangles were not tested [58]. These Tau species might be degraded more efficiently by emerging lysosome-based technologies. Neurodegenerative disorders may also lead to altered lysosome-related functions. Taking Huntington’s disease as an example, there is robust evidence for expansion of endocytic and autophagic compartments [59., 60., 61., 62.] and reduced cargo/mHTT recognition [54,63., 64., 65.]. This further justifies enhancing the recognition of pathogenic proteins by degrader technologies such as ATTEC. Nevertheless, for diseases with severe lysosomal deficits, correcting lysosomal function per se is probably more important.
RNA molecules can be directly recognized by a lysosomal membrane protein, LAMP2C, that loads them into lysosomes for degradation [66]. Further studies revealed that a putative RNA transporter, SIDT2, mediates RNA translocation across the lysosomal membrane [67,68]. Thus, it may be possible to screen for high-affinity binding molecules for LAMP2C or SIDT2 and attach them to antisense oligonucleotides or small molecules specifically binding to the target RNA. These chimeric molecules could selectively degrade the target RNA and thus downregulate the expression of encoded pathogenic proteins. This may provide a novel degrader technology for RNA, in addition to the recent nuclease-targeting RNA degrader technology developed by the group of Disney [69]. For DNA molecules, clearance of cytosolic DNA molecules under pathological conditions is highly desired. Cyclic GMP-AMP (cGAMP) synthase (cGAS) detects infections or tissue damage by binding to microbial or self DNA in the cytoplasm [70]. Upon DNA binding, cGAS produces cGAMP that binds to and activates the adaptor protein STING [71], which induces LC3 lipidation and activates autophagy [72]. cGAMP-induced autophagy is crucial for the clearance of DNA and viruses in the cytosol [72]. This provides a promising possibility to use autophagy-targeting degrader technologies to enhance the clearance of these DNA molecules and their relevant pathogens or damaged cellular components. Because cytosolic DNA cannot be ubiquitinated, ATTEC is probably the most suitable degrader technology. For example, molecules binding to the target DNA could be attached to an LC3-binding chemical moiety, and these chimeric ATTEC molecules may tether the DNA to LC3 for subsequent autophagic degradation.
Bacterial and viral infections present a severe global threat to healthcare. Despite the enormous success of antibiotics, new therapeutic approaches will be vital because of the rise of antibiotic-resistant infections and the dearth of new antibiotics in the pipeline [73]. Selective autophagy is the major intracellular pathway that targets intracellular pathogens to lysosomes for degradation, a process referred to as xenophagy [74]. Thus, autophagy/lysosome degrader technologies such as ATTEC or AUTAC could be used to target key proteins of the pathogen, leading to enhanced clearance via xenophagy. This approach may provide synergistic effects with xenophagy enhancement approaches that have been recently studied for the control of bacterial infection [75]. Another potential strategy is to target microbial DNA/RNA for autophagic degradation by ATTEC as discussed earlier. The pathogen COVID-19 responsible for the recent pneumonia outbreak could also be targeted by this strategy [76] (see Outstanding Questions). However, many pathogens have evolved ways to subvert or exploit this defense mechanism, minimizing the actual effectiveness of xenophagy in innate immunity [73,77]. A better understanding of the complex relationship between autophagy and microbial pathogens will provide better opportunities to combine novel degrader technologies with xenophagy enhancement to achieve optimal therapeutic effects.
Outstanding Questions.
How can we improve the overall pharmacokinetic profile of PROTAC molecules?
What is the overall specificity of emerging degrader technologies, including LYTAC, AUTAC, and ATTEC?
How can we reduce the size of LYTAC molecules so that they become small-molecule compounds?
What is the molecular mechanism by which S-guanylation causes K63 ubiquitination?
Does AUTAC influence selective autophagy per se or other cellular pathways?
What is the atomic level structure of compound–protein interfaces between ATTEC and LC3?
Are chimeric ATTEC molecules constructed by attaching an LC3-interacting chemical moiety to a small-molecule ligand of the target protein effective degraders?
Are LYTAC, AUTAC, and ATTEC catalytic or non-catalytic?
Can we utilize the emerging new concepts of degrader technologies to degrade non-protein targets?
Can we utilize ATTEC or AUTAC to target COVID-19 by degrading its key protein or RNA?
Alt-text: Outstanding Questions
Concluding Remarks and Future Perspectives
Emerging new concepts of degrader technologies harness the relatively non-specific lysosomal degradation pathway for selective degradation of POIs. These greatly expand the landscape of therapeutic targets and provide exciting novel approaches of potential therapeutic benefit. LYTACs utilize a glycan tag to mark extracellular POIs for intracellular lysosomal degradation following receptor-mediated internalization. AUTACs cleverly exploit small-molecule mimics of S-guanylation to selectively tag POIs with K63 ubiquitination and induce their lysosomal degradation via selective autophagy. ATTECs exploit lipidated LC3 to directly tether polyQ proteins to growing autophagosomes to facilitate their selective removal via autophagy. Nevertheless, crucial concerns remain to be addressed before these strategies can mature into established technologies.
The specificity and potential off-target effects need to be clarified (see Outstanding Questions). Proteomic studies have been performed for ATTEC in cellular and tissue samples [51], although such studies for LYTAC and AUTAC are currently missing. In addition to proteomic studies measuring steady-state protein levels, researchers may consider investigating whether the compounds interact with proteins other than their designated targets. Pull-down of compound-binding proteins in cell or tissue lysates and proteomic analysis may reveal potential off-target binding partners. In addition, whether these technologies are catalytic and function substoichiometrically needs to be clarified (see Outstanding Questions). In theory, AUTAC and ATTEC could be catalytic because the small molecules involved may be recycled after lysosomal degradation of the target proteins, but this needs to be further tested and validated. LYTAC molecules are probably non-catalytic because of their large biomolecular size, leading to less efficient but more controllable degradation compared with catalytic technologies.
Specific concerns remain to be addressed for each of these emerging technologies (see Outstanding Questions). The large LYTAC molecules need to be miniaturized for easier delivery and prevention of immune responses. To achieve this, small molecules binding to CI-M6PR may be identified by screening or structure-based design. The CI-M6PR-binding small molecule may then replace the long M6P chain or CI-M6PR polypeptide ligand to reduce the size of LYTACs. For AUTAC, the mechanism of action remains unclear, especially concerning how S-guanylation induces K63 polyubiquitination. This could potentially be addressed by interactome studies for proteins recognizing this modification, or by biochemical screening for modifiers of S-guanylation-induced K63 polyubiquitination. The potential influence of AUTAC on autophagy also needs to be tested. Compared with LYTAC and AUTAC studies, the biological effects of ATTEC molecules have been investigated relatively thoroughly. However, atomic level structural information for the compound–protein interfaces is crucial for ATTEC and remains to be elucidated. Chimeric ATTEC molecules also need to be designed and tested.
Although the emerging degrader technologies are in their infancy and have key concerns that need to be addressed, they have unprecedented potential. With appropriate research and development, they may greatly expand the target spectrum of degrader technologies and develop into exciting new avenues of research.
Acknowledgments
The authors would like to thank the National Natural Science Foundation of China (81925012, 81870990, 31961130379) and the Newton Advanced Fellowship from Academy of Medical Sciences (NAF\R1\191045) for funding support.
Resource
iPicchi, A. The cost of Biogen’s new drug: $750,000 per patient; www.cbsnews.com/news/the-cost-of-biogens-new-drug-spinraza-750000-per-patient/.
References
- 1.Walker L.C., LeVine H. The cerebral proteopathies: neurodegenerative disorders of protein conformation and assembly. Mol. Neurobiol. 2000;21:83–95. doi: 10.1385/MN:21:1-2:083. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Sala A.J. Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 2017;216:1231–1241. doi: 10.1083/jcb.201612111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Djajadikerta A. Autophagy induction as a therapeutic strategy for neurodegenerative diseases. J. Mol. Biol. 2019 doi: 10.1016/j.jmb.2019.12.035. Published online December 27, 2019. http://dx.doi.org/j.jmb.2019.1012.1035. [DOI] [PubMed] [Google Scholar]
- 5.Schoch K.M., Miller T.M. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94:1056–1070. doi: 10.1016/j.neuron.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeder M.L., Gersbach C.A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evers M.M. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv. Drug Deliv. Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Neklesa T.K. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tae H.S. Identification of hydrophobic tags for the degradation of stabilized proteins. ChemBioChem. 2012;13:538–541. doi: 10.1002/cbic.201100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long M.J. Inhibitor mediated protein degradation. Chem. Biol. 2012;19:629–637. doi: 10.1016/j.chembiol.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y. Boc3Arg-linked ligands induce degradation by localizing target proteins to the 20S proteasome. ACS Chem. Biol. 2016;11:3328–3337. doi: 10.1021/acschembio.6b00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffey R.T. Ubiquilin-mediated small molecule inhibition of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 2016;291:5221–5233. doi: 10.1074/jbc.M115.691584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konstantinidou M. PROTACs – a game-changing technology. Expert Opin. Drug Discovery. 2019;14:1255–1268. doi: 10.1080/17460441.2019.1659242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schapira M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 2019;18:949–963. doi: 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- 15.Sun X. PROTACs: great opportunities for academia and industry. Signal Transd. Target. Ther. 2019;4:64. doi: 10.1038/s41392-019-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burslem G.M., Crews C.M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y. Degradation of proteins by PROTACs and other strategies. Acta Pharm. Sin. B. 2020;10:207–238. doi: 10.1016/j.apsb.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto K.M. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneekloth A.R. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuchs S.Y. Mdm2 association with p53 targets its ubiquitination. Oncogene. 1998;17:2543–2547. doi: 10.1038/sj.onc.1202200. [DOI] [PubMed] [Google Scholar]
- 21.Vassilev L.T. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 22.Vu B. Discovery of RG7112: a small-molecule MDM2 Inhibitor in clinical development. ACS Med. Chem. Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Q. Discovery of RG7388, a potent and selective p53–MDM2 inhibitor in clinical development. J. Med. Chem. 2013;56:5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 24.Itoh Y. Protein knockdown using methyl bestatin-ligand hybrid molecules: design and synthesis of inducers of ubiquitination-mediated degradation of cellular retinoic acid-binding proteins. J. Am. Chem. Soc. 2010;132:5820–5826. doi: 10.1021/ja100691p. [DOI] [PubMed] [Google Scholar]
- 25.Winter G.E. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito T. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 27.Chanan-Khan A.A. Pomalidomide: the new immunomodulatory agent for the treatment of multiple myeloma. Blood Cancer J. 2013;3 doi: 10.1038/bcj.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Girona A. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu J. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai A.C. Modular PROTAC design for the degradation of oncogenic BCR–ABL. Angew. Chem. Int. Ed. Engl. 2016;55:807–810. doi: 10.1002/anie.201507634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneekloth J.S., Jr. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- 32.Bondeson D.P. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015;11:611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson C.M. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin Y.H. Azo-PROTAC: novel light-controlled small-molecule tool for protein knockdown. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.1029b02058. Published online March 19, 2020. [DOI] [PubMed] [Google Scholar]
- 35.Xue G. Light-induced protein degradation with photocaged PROTACs. J. Am. Chem. Soc. 2019;141:18370–18374. doi: 10.1021/jacs.9b06422. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L. Acquired resistance to BET-PROTACs (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol. Cancer Ther. 2019;18:1302–1311. doi: 10.1158/1535-7163.MCT-18-1129. [DOI] [PubMed] [Google Scholar]
- 37.Wong E., Cuervo A.M. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tooze S.A. Endocytosis and autophagy: exploitation or cooperation? Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pillay C.S. Endolysosomal proteolysis and its regulation. Biochem. J. 2002;363:417–429. doi: 10.1042/0264-6021:3630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundu M., Thompson C.B. Autophagy: basic principles and relevance to disease. Annu. Rev. Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 41.Kabeya Y. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banik S. Lysosome targeting Chimeras (LYTACs) for the degradation of secreted and membrane proteins. ChemRxiv. 2019 doi: 10.26434/chemrxiv.7927061. Published online November 20, 2019. [DOI] [Google Scholar]
- 43.Van Regenmortel M.H. Antigenicity and immunogenicity of synthetic peptides. Biologicals. 2001;29:209–213. doi: 10.1006/biol.2001.0308. [DOI] [PubMed] [Google Scholar]
- 44.Kirkin V. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 45.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filimonenko M. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi D. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell. 2019;76:797–810. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Sawa T. Protein S-guanylation by the biological signal 8-nitroguanosine 3',5'-cyclic monophosphate. Nat. Chem. Biol. 2007;3:727–735. doi: 10.1038/nchembio.2007.33. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa I. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 50.Li Z. ATTEC: a potential new approach to target proteinopathies. Autophagy. 2020;16:185–187. doi: 10.1080/15548627.2019.1688556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019;575:203–209. doi: 10.1038/s41586-019-1722-1. [DOI] [PubMed] [Google Scholar]
- 52.Walker F.O. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 53.Sun X. Conformation-dependent recognition of mutant HTT (huntingtin) proteins by selective autophagy. Autophagy. 2017;13:2111–2112. doi: 10.1080/15548627.2017.1382783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu Y. A toxic mutant huntingtin species is resistant to selective autophagy. Nat. Chem. Biol. 2017;13:1152–1154. doi: 10.1038/nchembio.2461. [DOI] [PubMed] [Google Scholar]
- 55.Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 57.Pardridge W.M. Drug transport across the blood–brain barrier. J. Cereb. Blood Flow Metab. 2012;32:1959–1972. doi: 10.1038/jcbfm.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva M.C. Targeted degradation of aberrant tau in frontotemporal dementia patient-derived neuronal cell models. Elife. 2019;8 doi: 10.7554/eLife.45457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies S.W. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 60.Sapp E. Huntingtin localization in brains of normal and Huntington's disease patients. Ann. Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 61.Kegel K.B. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J. Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagata E. Autophagosome-like vacuole formation in Huntington's disease lymphoblasts. Neuroreport. 2004;15:1325–1328. doi: 10.1097/01.wnr.0000127073.66692.8f. [DOI] [PubMed] [Google Scholar]
- 63.Martinez-Vicente M. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ochaba J. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rui Y.N. Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujiwara Y. Discovery of a novel type of autophagy targeting RNA. Autophagy. 2013;9:403–409. doi: 10.4161/auto.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aizawa S. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy. 2016;12:565–578. doi: 10.1080/15548627.2016.1145325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hase K. Cytosolic domain of SIDT2 carries an arginine-rich motif that binds to RNA/DNA and is important for the direct transport of nucleic acids into lysosomes. Autophagy. 2020:1–15. doi: 10.1080/15548627.2020.1712109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costales M.G. Small-molecule targeted recruitment of a nuclease to cleave an oncogenic RNA in a mouse model of metastatic cancer. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2406–2411. doi: 10.1073/pnas.1914286117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun L. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gui X. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimmey J.M., Stallings C.L. Bacterial pathogens versus autophagy: implications for therapeutic interventions. Trends Mol. Med. 2016;22:1060–1076. doi: 10.1016/j.molmed.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dupont N. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009;6:137–149. doi: 10.1016/j.chom.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 75.Laing E.D. Enhanced autophagy contributes to reduced viral infection in black flying fox cells. Viruses. 2019;11:260. doi: 10.3390/v11030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu F. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogawa M. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]