Highlights
-
•
We introduce the SBDiEM approach for modeling, forecasting and nowcasting disease dynamics.
-
•
Our model can be adjusted to describe past outbreaks as well as COVID-19.
-
•
The novel methodology could have important implications for NHSs, international stakeholders and policy makers.
-
•
We aim at the improvement of outbreak control, vaccination and prevention strategies.
-
•
The model can be embedded into a global AI surveillance system for combating epidemic and pandemic outbreaks.
Keywords: Epidemiology, Outbreak analysis, COVID-19, Contagious dynamics, Stochastic models, Virus transmissibility
Abstract
A worldwide multi-scale interplay among a plethora of factors, ranging from micro-pathogens and individual or population interactions to macro-scale environmental, socio-economic and demographic conditions, entails the development of highly sophisticated mathematical models for robust representation of the contagious disease dynamics that would lead to the improvement of current outbreak control strategies and vaccination and prevention policies. Due to the complexity of the underlying interactions, both deterministic and stochastic epidemiological models are built upon incomplete information regarding the infectious network. Hence, rigorous mathematical epidemiology models can be utilized to combat epidemic outbreaks. We introduce a new spatiotemporal approach (SBDiEM) for modeling, forecasting and nowcasting infectious dynamics, particularly in light of recent efforts to establish a global surveillance network for combating pandemics with the use of artificial intelligence. This model can be adjusted to describe past outbreaks as well as COVID-19. Our novel methodology may have important implications for national health systems, international stakeholders and policy makers.
1. Introduction
The World Health Organization (WHO) reported on December 31, 2019 cases of pneumonia of undetected etiology in the city of Wuhan, Hubei Province in China. A novel coronavirus (CoViD-19) was identified as the source of the disease by the Chinese authorities on January 7, 2020. Eventually, the International Committee on Taxonomy of Viruses on 11 February, 2020 named the Severe Acute Respiratory Syndrome Coronavirus as SARS-CoV-2 [1]. Concerns on public health were dispersed on a global scale about potentially infected countries. The virus might have been generated by animal populations and transmitted via the Huanan wholesale market [2], [3], [4] albeit not proven, while clinical findings demonstrated that international spread was caused mainly by commercial air travel [4], [5], [6], [7]. The WHO declared SARS-CoV-2 a pandemic on March 11, 2020. Throughout the globe, huge efforts were in progress to limit the spread of the virus and find medications and vaccines. However, the scientific community could not fully comprehend the dynamics of the spread [8], [9], [10].
Several outbreaks of infectious diseases have occurred in the past with immense impact on public health. For instance, the Severe Acute Respiratory Syndrome (SARS) occurred in 2003, the swine flu in 2009 and the Middle East Respiratory Syndrome Coronavirus (MERS) in Saudi Arabia in 2012, which still survives at a sub-critical level causing some peaks [11], [12], [13], [14]. Additionally, the Ebola epidemic emerged between 2014 and 2016 and caused over 28,000 cases in West Africa [15]. Its temporal decline coincided with the outbreak of Zika virus in Brazil [16]. Consequently, the outbreak of severe pathogens such as the aforementioned, require global interdisciplinary efforts in order to decode key epidemiological features and their transmission dynamics, and develop possible control policies.
Insights from mathematical modelling can be extremely beneficial. Indeed, dealing with infectious diseases from a mathematical angle could reveal inherent patterns and underlying structures that govern outbreaks. Stakeholders utilize available data from current and previous outbreaks in order to forecast infection rates, identify how to restrict the spread of diseases, and eventually introduce vaccination policies that will be most effective. Epidemiology is essentially a biology discipline concerned with public health and as such, it can be heavily influenced by mathematical theory. Most phenomena observed at population level are often very complex and difficult to decode just by observing the characteristics of isolated individuals [17]. Statistical analyses of epidemiological data help to characterize, quantify and summarize the way diseases spread in host populations. Interestingly, mathematical models appear as efficient ways to explore and test various epidemiological hypotheses, mostly due to the existence of ethical and practical limitations when deducting experiments on living populations. Models provide conceptual results on e.g., the basic reproduction number, threshold effects or herd immunity. One additional element of epidemiological modeling is the link with data via statistical methods. Although simple epidemiological models are often used, viral and bacterial infections commonly require increased complexity. There are many models in the literature on single epidemics, endemic diseases and spatiotemporal disease dynamics. The aim is to develop robust public health policies in defining optimal vaccination strategies.
Our study presents for the first time a new stochastic mathematical model for describing infectious dynamics and tracking virus temporal transmissibility on 3-dimensional space (earth). This model can be adjusted to describe all past outbreaks as well as CoViD-19. As a matter of fact, it introduces a novel approach to mathematical modelling of infectious dynamics of any disease, and sets a starting point for conducting simulations, forecasting and nowcasting investigations based on real-world stereographic and spherical tracking on earth.
In short, a single epidemic outbreak as opposed to disease endemicity occurs in a time span short enough not to have the demographic changes perturbing the dynamics of contacts among individuals. The most popular mathematical model in this category is the Susceptible-Infected-Recovered (SIR) epidemic model, in which all individuals of a finite population interact in the same manner. Individuals at time t are susceptible (S), infected (I) or recovered (R). The final size of the epidemic will strongly depend upon the initial conditions of the number of susceptible and infected individuals as well as the infection parameter. The final size distribution of the simple SIR model in most cases is bimodal presenting two local maxima. This bimodal feature is caused by two likely scenarios; either the epidemic dies out quickly infecting few individuals, or it becomes long-lasting and substantial. However, stochasticity in the form of random walk transmission mechanisms related to spreading processes has never been explored in epidemiology widely [18], [19], [20]. For example, in computer science, some artificially created viruses propagate randomly by a plethora of online communication channels. To the best of our knowledge, we are the first to scrutinize extensively the role of random walks in epidemic spreading and provide the proper mathematical arsenal to model it robustly. Interestingly, random walk paths converge in distribution to Brownian motions [21]. In this work, we assume that a biological carrier of virus Y is at position X(t) at any given time t. We call this the inaugural contamination focal point on earth.
The path defined by its motion is considered infectious. Xt, t ≥ 0 is supposed to follow a Brownian motion on a 2-dimensional sphere S 2 of radius a, i.e the sphere in of dimension 3. We consider this a proxy for earth, spreading via spherical and stereographic coordinates. Next, using the Laplace-Beltrami operator we construct the Brownian motion infectious process on the 2-dimensional sphere, using spherical and stereographic coordinates as local coordinates. We evaluate explicitly certain quantities related to generated diffusion processes. In what follows, we compute the transition and transmission density for the Xt, t ≥ 0, and we derive the stochastic differential equations that govern the infectious disease dynamics for Xt, t ≥ 0 in those local coordinates. We continue with the calculation of expectations of outbreak exit times in time and space of specific domains, possessing certain symmetries. Moreover, the moment generating functions are produced. In mathematical terms, we derive the stochastic reflection principle on S 2 for the infectious disease transmission process. Reflection points can be extremely useful to calculate the distribution functions of certain temporal quantities related to the dynamics. Additionally, we evaluate boundary local times of first hitting of the outbreak for an epidemic or a hybrid endemic-epidemic model. Hence, biological carrier(s) of a virus (infectious individuals) are tracked at any given time on earth coordinates, and the path(s) defined by each infectious dynamical motion. In the following two chapters we present a thorough literature review and a state-of-the-art analysis in order to pose clearly our novel approach optimally among the various methodologies followed thus far.
The rest of paper is organized as follows: Section 2 provides a brief literature review, past and recent, of mathematical epidemiology. Section 3 presents the state-of-the-art, and focal concepts and term definitions required to introduce our novel model. It also states which category the new model falls into, according to the official taxonomy of the various methodologies already utilized so far in the relevant literature. Next, section 4 exposes in detail the mathematical formulation of the model. Lastly, Section 5 discusses proposed policies and future paths of research, and concludes.
2. Literature review
The beginning of mathematical modeling in epidemiology dates back to 1766, when Bernoulli developed a mathematical model to analyze the mortality of smallpox in England [22]. Bernoulli used his model to show that inoculation against the virus would increase the life expectancy at birth by about three years. A revision of the main findings and a presentation of the criticism by D’Alembert, appears recently in Dietz and Heesterbeek [23]. Lambert in 1772 as well as Laplace in 1812 extended the Bernoulli model by incorporating age-dependent parameters [24], [25]. However, further systematic research was absent until the beginning of the twentieth century with the pioneering work of Ross in 1911, which is considered the inaugural study of modern mathematical epidemiology [26]. Ross used a set of equations to approximate the discrete-time dynamics of malaria via a mosquito-based pathogen transmission [27]. Importantly, the past century has witnessed the rapid emergence and development of substantial theories in epidemics. In 1927, Kermack and McKendrick [28] derived the celebrated threshold theorem, which is one of the key results in epidemiology. It predicts – depending on the transmission potential of the infection – the critical fraction of susceptibles in the population that must be exceeded if an epidemic is to occur. Kermack and McKendrick published three seminal papers, establishing what is called the deterministic compartmental epidemic modelling [29], [30], [31], wherein they addressed the mass–action incident in disease transmission cycles, assuming that the probability of infection of a susceptible is analogous to the number of its contacts with infected individuals. This deterministic representation was in line with the Law of Mass Action [32] introduced by Guldberg and Waage in 1864 and renders the basic most commonly used SIR model, which assumes homogeneous mixing of the contacts and conservation of the total population and low rates of interaction. MacDonald extended Ross’s model to explain in depth the transmission process of malaria. Utilizing modern computer power, the mathematical model for the dynamics and the control of mosquito-transmitted pathogens provided robust results in real-word applications. Overall, the family of models they introduced is known by now as Ross–MacDonald models [33]. Moreover, the classic work of Bartlett [34] examined models and data to explore the factors that determine disease persistence in large populations. Arguably, a landmark book on mathematical modelling of epidemiological systems was published by Bailey [35] and highlighted the importance of public health decision making [36]. Given the diversity of infectious diseases studied since the middle of the 1950s, an impressive variety of epidemiological models have been developed. In addition, we should highlight the 19th century works by Enko [37], [38], [39], who first published a probabilistic model for describing the epidemic of measles, yet in discrete time. This model is the precursor of the popular Reed-Frost chain binomial model introduced by Frost in 1928 in biostatistics’ lectures at Johns Hopkins University [40]. It assumes that the infection spreads from an infected to a susceptible individual via a discrete time Markov chain, and set the basis of contemporary stochastic epidemic modelling, on which we will also focus in our present work.
Moving to the 21st century, we mention some interesting works; Xing et al., [41] introduced a mathematical model on H7N9 influenza among migrant and resident birds, domestic poultry and humans in China. In this study they concluded that temperature seasonality might be a source of the disease, yet they suggested for the first time that controlling markets could help controlling outbreaks. Lee and Pietz [42] developed a mathematical model for Zika virus using logistic growth in human populations. Sun et al., [43] proposed a transmission model for cholera in China and observed that reducing the spread requires extensive immunization coverage of the population. Nishiura et al. [44] developed a Zika mathematical model which exhibited the same dynamics as dengue fever, and Khan et al. [45] introduced a model whereby a saturation function describes well the typhoid fever dynamics. Gui and Zhang [46], developed a modified SIR model demonstrating nonlinearities in recovery rates. Their model exhibited a backward bifurcation phenomenon, which in turn implied that a plain reduction of the reproduction number less than one, was not rendered sufficient to stop the disease spread. Li et al. [47] constructed a multi-group brucellosis model and found out that the best way to contain the disease is to avoid cross infection of animal populations. Moreover, Yu and Lin [48] identified complex dynamical behaviour in epidemiological models and particularly the existence of multiple limit cycle bifurcations using a predictor-prey model. Shi et al. [49] proposed an HIV model with a saturated reverse function to describe the dynamics of infected cells. Additionally, Bonyah et al. [50] developed a SIR model to study the dynamics of buruli ulcer and suggested policy measures to control the disease. Lastly, Zhang et al. [51] developed a model with a latent period of the disease wherein the person is not infectious with saturated incidence rates and treatment functions, called SEIR epidemic model.
3. State-of-the-art analysis and definitions
The SIR model is the basic one used for modelling epidemics. Kermack and McKendrick created the model in 1927 [29] in which they considered a fixed population with only three compartments, susceptible (S), infected (I) and recovered (R). There are a large number of modifications of the SIR model, including those that include births and deaths, the SIR without or with vital dynamics, a model where upon recovery there is no immunity called SIS and where immunity lasts for a short period of time, called SIRS model. Furthermore, a model where there is a latent period of the disease and where the person is not infectious is indentified as SEIS and SEIR respectively, or where infants can be born with immunity is named MSIR. Also, we mention the herd immunity model [52], [53].
Overall, the transmission mechanism from infective populations to susceptibles is not well-comprehended for many infectious diseases. Interactions in a population are very complex, hence it is extremely difficult to capture the large scale dynamics of disease spread without formal mathematical modeling. An epidemiological model uses microscopic effects - the role of an infectious individual - to forecast the macroscopic behavior of disease spread via a population.
Deterministic models do not incorporate any form of uncertainty and as such, they can be thought to account for the mean trend of a process, alone. On the other hand, stochastic models describe the mean trend as well as the variance structure of the underlying processes. Two basic types of stochasticity are commonly used: demographic and environmental. Within the context of demographic stochasticity, all individuals are subject to the same potential events with the exact same probabilities but differences in the fates of population individuals. Disease propagation in large populations obeys to the weak law of large numbers, thus effects of demographic stochasticity can be decreased significantly, and many times a deterministic model becomes more suitable. However, random events cannot be neglected and a stochastic model can be equally appropriate. Environmental stochasticity involves variations in the probability associated with an exogenous event. Model parameters of stochastic models are characterized by probability distributions, whilst for fixed parameter values deterministic models will always produce the same results, except when chaotic behaviour emerges.
In the classic SIR model it is assumed that the individuals leave the infectious class at a constant rate and even if this assumption seems most intuitive, it is not always the most realistic, regarding the duration individuals stay infective [54], [55], [56]. Usually, random variables describe the time of recovery since infection. For discrete random variables (e.g., number of individuals) it is easy to define a probability distribution, whilst for continuous variables the time of recovery since infection is modelled. Often, in this last category it is not possible to fix a probability as there is infinity of such times. Hence, we first define a cumulative distribution and then express a probability density function from this cumulative distribution. Infectious periods are exponentially distributed with a mean infectious duration, however as frequently real data does not back up this assumption, we rather use constant duration. To account for such more realistic distributions, the assumption that the probability of recovery does not depend on the time since infection, is often relaxed. Then, a common method of stages can be used to replace the infective compartment by a series of successive ones, each with an exponential distribution of the same parameter, leading to a total duration of the infectious period modelled by a gamma distribution [17].
Epidemic models presented above describe rapid outbreaks during which normally the host population is assumed to be in a constant state. For longer periods, deaths and births feed the population with new susceptibles, possibly allowing the disease to persist at a constant prevalence. This state renders an endemic state in the population [17]. In this case, we account for birth and death rate of the host population, whereby a good approximation is that the population size N = S + I + R is constant. When deterministic dynamics prevail a threshold on the value of the basic reproduction number exists. Conditions regarding this number guarantee the disease persistence, but in epidemic models such persistence can be dependent upon the magnitude of the stochastic fluctuations around the steady-state equilibrium. Furthermore, many times diseases are in an endemo-epidemic state. As endemic models exhibit damped oscillations which converge toward an endemic equilibrium, this equilibrium can be weakly stable with perturbations (intrinsic or extrinsic), which excite and sustain the inherent oscillation behaviour [57]. This behaviour is due to heterogeneity that is added temporally to the coefficient of transmission, spatially in the context of meta-populations, or by cohorts for age-structured models. Lastly, heterogeneity can be added statistically in case of stochastic versions. For example, a stochastic version of the endemic SIR model can utilize a Markov process, in which the future is independent of the past given the present, with a state space defined by the number of individuals in each of the three classes, and changes in the state space characterized by probabilistic transition events. And as future events are independent on past events, the time to the next event follows a negative exponential distribution.
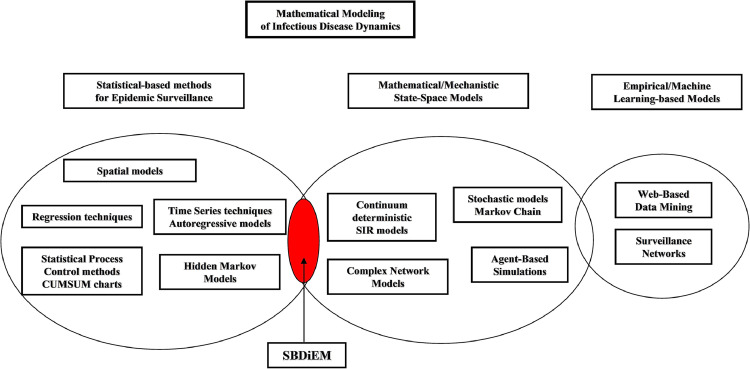
Over the years, a vast number of mathematical modeling approaches has been proposed, tackling the problem from different perspectives. The prevailing taxonomy proposed by Siettos and Russo [58] encompasses three general categories: (1) statistical methods of outbreaks and their identification of spatial patterns in real epidemics, (2) state-space models of the evolution of a “hypothetical” or on-going epidemic spread, and (3) machine learning methods, all utilized also for predictability purposes vis-à-vis an ongoing epidemic. In particular, the first category includes i) regression methods [59], [60], [61], [62], [63], [64], ii) times series analysis, namely ARIMA and seasonal ARIMA approaches [65], [66], [67], [68], iii) process control methods including cumulative sum (CUSUM) charts [69], [70], [71], [72], [73], [74] and exponentially weighted moving average (EWMA) methods [75], [76], as well as iv) Hidden Markov models (HMM) [77], [78]. The second category incorporates i) “continuum” models in the form of differential and/or (integro)-partial differential equations [79], [80], [81], [82], ii) discrete and continuous-time Markov-chain models [83], [84], [85], iii) complex network models which relax the hypotheses of the previous stochastic models that interactions among individuals are instantaneous and homogeneous [86], [87], [88], [89], [90], [91], and iv) Agent-based models [92], [93], [94], [95]. Lastly, the third category includes well-known machine learning approaches widely used in computer science, such as i) artificial neural networks [96], ii) web-based data mining [97], [98] and iii) surveillance networks [99], to name a few.
For the first time in the relevant literature, we introduce a new stochastic model laying in the intersection of categories (1) and (2), called “Stereographic Brownian Diffusion Epidemiology Model (SBDiEM)”. Fig. 1 presents a graphical overview of the models utilized so far, and the “positioning” of our novel approach for modelling infectious diseases.
Fig. 1.
Updated taxonomy of mathematical models for contagious diseases (source [58]). The new stochastic model lays in the intersection of categories (1) statistical methods and (2) state-space models of epidemic spreads.
4. Mathematical formulation
4.1. Preliminaries
4.1.1. The n-Sphere Sn
Definition 4.1
Let . The n-dimensional sphere Sn with center and radius a > 0 is (defined to be) the set of all points satisfying . Thus,
4.1.2. Stereographic projection coordinates
Definition 4.2
We consider to be the hyperplane given by . For convenience, we will let be coordinates on and be coordinates on . Let . The stereographic projection coordinates of Sn is the map given by
This map defines coordinates on Sn so that the point of Sn has coordinates where
The inverse map is given by
4.1.3. Spherical coordinates
The points of the 2-sphere with center at the origin and radius a may also be described in spherical coordinates in the following way: where 0 ≤ φ < 2π.
i.e.
where 0 ≤ θ < 2π and 0 ≤ φ ≤ π.
4.1.4. The Laplace-Beltrami operator
Definition 4.3
A C ∞ differentiable manifold of dimension n is a set M together with a family of one-to-one maps xα: Uα → M of open sets into M such that
- 1.
- 2.
For each pair α, β with we have that are open sets in and that are C ∞ differentiable maps.
- 3.
The family {Uα, xα} is maximal relative to conditions 1 and 2.
Each pair (xα, Uα) is called a coordinate chart on M. (For more details see [100])
Definition 4.4
A Cr function where M is a C ∞ differential manifold is a function f, such that is Cr for every cordinate chart (xα, Uα) on M.
Let be the Riemmanian metric tensor on a Riemmanian manifold M. This means that, in any coordinate chart on M, the length element can be computed by
Given local coordinates we can easily compute the matrix by the inner product
(see [100]). We denote by gij the elements of the inverse matrix .
Definition 4.5
The Laplace-Beltrami operator ΔM associated with the metric g is defined by
(4.1) where f is a Cr function on M.
In this work we are interested in the case where i.e., the 2 -dimensional sphere. We will denote the corresponding Laplace-Beltrami operator of S 2 by Δ2 or just Δ using the spherical coordinates. If i.e.
we have
i.e.,
and
Hence the Laplace-Beltrami operator of a smooth function f on S 2 is
| (4.2) |
where and Thus
or
| (4.3) |
In case where the function f is independent of θ the Laplace-Beltrami operator of f is
| (4.4) |
Generally the Laplace-Beltrami operator of a smooth function f on Sn is
| (4.5) |
where
| (4.6) |
If f is independent of , the Laplace Beltrami operator of f is
| (4.7) |
Using Stereographic projection coordinates, if i.e.
we have
Hence
Thus we have
Therefore, the Laplace Beltrami operator of a smooth function f on S 2, using Stereographic projection coordinates is
| (4.8) |
4.1.5. Brownian motion on a riemannian manifold
Definition 4.6
Let M be a Riemannian manifold (see definition 1.5) and Δ its corresponding Laplace-Beltrami operator. Any function P(t, x, y) on (0, ∞) × M × M satisfying the differential equation
(4.9) where Δx is Δ acting on the x-variables and the initial condition
(4.10) (where δx(y) is the delta mass at x ∈ M) is called a fundamental solution of the heat Eq. (4.9)on M.
The smallest positive fundamental solution of the heat Eqs. (4.9) and (4.10) is the heat kernel on M. It has been proved by J. Dodziak [101], that the heat kernel always exists, and is smooth in (t, x, y). Moreover the heat kernel possesses the following properties.
-
1.Symmetry in x, y, that is
-
2.The semigroup identity: For any s ∈ (0, t)
where dμ is the area measure element of M. In polar coordinates where and |g| is given by (4.6). -
3.The total mass inequality, i.e., for all t > 0 and x ∈ M
(4.11)
In case where M is compact and smooth, there is only one solution of (4.9) and (4.10) which is positive and satisfies
| (4.12) |
Definition 4.7
A process Xt, t ≥ 0 is a Markov process if for any t, s ≥ 0, the conditional distribution of given the information about the process up to time t, is the same as the conditional distribution of given Xt.
Definition 4.8
The Brownian motion Xt, t ≥ 0, on a Riemannian manifold M is a Markov process with transition density function P(t, x, y) the heat kernel associated with the Laplace-Beltrami operator.
Remark 4.1
In the case where n ≥ 2, the transition density function P(t, x, y) of the Brownian motion Xt depends only on t and d(x, y), the distance between x and y. Thus in spherical coordinates it depends on t and the angle φ between x and y. Hence, the transition density function of the Brownian motion can be written as
(4.13) where p(t, φ) is the solution of
(4.14) and
(4.15) Here δ( · ) is the Dirac delta function on and An denotes the area of the n-dimensional sphere Sn with radius a. It is well known that [102]
(4.16) where Γ( · ) is the Gamma function. More precisely
(4.17)
(4.18)
Remark 4.2
The fact that Sn is a compact and smooth manifold implies that (4.14) and (4.15) has a unique positive solution which also satisfies
(4.19) Furthermore, as t → ∞, P(t, x, y) approaches the uniform density on Sn, i.e. P(t, x, y) → c, where
In the sequel for typographical convenience we will write Xt instead of {Xt}t ≥ 0.
4.2. Transition density function of Xt, t > 0
In this section we shall represent the transition density function p(t, φ) of the position X(t) of a biological carrier (infected individual) of virus Y at any given time t. For the next sections we suppose that the infected individual is at position X(t) at any given time t, namely the path defined by its motion is considered infectious. Xt, t ≥ 0 describes a Brownian motion on a 2-dimensional sphere S 2 of radius a. From the (4.14), (4.15) and (4.17) the transition density function p(t, φ) of Xt is the unique solution of
| (4.20) |
and
| (4.21) |
The solution of the diffusion equation
| (4.22) |
with initial condition
| (4.23) |
is given by the function
| (4.24) |
see[103]. Here is the associated Legendre polynomials of order zero, i.e.
| (4.25) |
This fact implies the following
Proposition 4.1
The transition density function of the Brownian motion Xt, t ≥ 0 on S 2 with radius a it is given by the function
(4.26)
Proof
First we prove that p(t, φ) satisfies the differential equation
We have that
where K(t, φ) is given by the (4.24), therefore
However from the (4.22)
(4.27) hence
i.e.
Furthermore p(t, φ) satisfies the
and if we set we imply that
Therefore
and this complete the proof. □
4.2.1. Stochastic differential equation of the brownian motion in local coordinates
We recall the following well-known fact
Theorem 4.1
Let
be such that is positive definite. If Xt is the Ito diffusion process
(4.28) then, its generator A is given by the formula
Conversely, the operator A given above is the generator of diffusion (4.28). For the proof see [104].
Case of spherical coordinates
The generator of Brownian motion on S 2 in spherical coordinates is
i.e.
Therefore, the Brownian motion on S 2 in spherical coordinates is the solution of the stochastic differential equation
where
Case of sterographic projection coordinates
Expressed in stereographic projection coordinates, the generator of Brownian motion on S 2 is
Hence, the Brownian motion on S 2 in stereographic projection coordinates is the solution of the stochastic differential equation
| (4.29) |
where
4.3. Expectations of exit times of X(t)
We recall some basic definitions.
Definition 4.9
A measurable space is said to be equipped with a filtration { t}, if for every t ≥ 0 { t} is a σ-algebra of subsets of Ω such that t and for every such that t 1 < t 2, we have that ⊂ . (i.e. { t} is an increasing family of sub σ-algebras of ).
Definition 4.10
Let us consider a measurable space equipped with a filtration { t}. A random variable T is a stopping time with respect to the filtration { t}, if for every t ≥ 0
Let Xt be the Brownian motion in Sn and D ⊂ Sn a domain. Then
is a stopping time with respect to t called the exit time on ∂D.
Proposition 4.2
Let φ0 ∈ [0, π) be fixed. We consider the set D in S 2, such that
Of course,
If Xt is the position of the biological carrier of the virus Y at a given time t starting at the point
and
then the expectation of T is given by
(4.30)
Proof
Based on [105],
we have the unique solution of the differential equation
(4.31) with boundary condition as
Here Δ2 is the Laplace-Beltrami operator on S 2. By symmetry of D, it follows that the expectation value of T is independent of θ. From (4.4) the differential Eq. (4.31) takes the form
(4.32) with boundary condition
(4.33) Set
hence from (4.32)
or
Thus
Therefore,
(4.34) However (see [104])
hence
Furthermore, we have
thus,
Consequently,
Thus
or
hence
Finally,
(4.35) □
Proposition 4.3
Let φ1, φ2 ∈ (0, π), such that φ1 < φ2, are both fixed. We consider the set D in S 2, such that
We have,
Let Xt be the position of the infectious individual Y is at a given time t starting at the point
and
then the expectation of T is given by
(4.36)
Proof
According to [105], E φ[t] satisfies the Poisson equation on D with Dirichlet boundary data. By uniqueness
is the unique solution of the differential Eq. (4.31), i.e.,
with boundary condition
Here Δ2 is the Laplace-Beltrami operator on S 2. By the symmetry of D, it follows that the expectation value of T is independent of θ. From (4.4) the differential Eq. (4.31) takes the form (4.32) with boundary condition
(4.37) Hence from (4.34)
However
i.e.
and
Thus
and
Consequently,
namely,
hence
from which we imply
(4.38) □
Proposition 4.4
We consider the 2-dimensional sphere S 2 of radius a. Let two circles pass through the North pole, such that in stereographic coordinates are represented by the parallel lines and where say b < c. We consider the set D in S 2, whose stereographic projection is
Of course
If Xt is the position of the carrier of virus Y at a given time t starting at the point A, where the stereogrpaphic projection coordinates of A are
and
then,
(4.39) where
(4.40) and
(4.41)
Proof
As we have seen the function
satisfies the differential equation
with boundary conditions
Here, Δ2 is the Laplace-Beltrami operator on S 2 expressed in stereographic projection coordinates. Hence, the differential equation takes the form
or
(4.42) However the function
satisfies the differential Eq. (4.42). Thus
where f(ξ 1, ξ 2) satisfies
with boundary conditions
and
If we take the transformation of variables and and set the function then ϕ(x, y) we satisfy
with boundary conditions
and
where Now let and i.e. Thus, if then
(4.43) Introducing the function It follows that ψ(u, v) satisfies
with boundary conditions
and
This is the standard Dirichlet boundary value problem for the half line, and it is well known that (see e.g. [106]) its solution is given by the Poisson integral formula for the half-plane:
or
where
Notice that Hence,
where u, v are given in (4.43). Therefore
i.e.
Finally
□
4.3.1. Hitting probabilities
Proposition 4.5
Let φ1, φ2 ∈ (0, π), such that φ1 < φ2, are both fixed. We consider the sets D 1, D 2 in S 2, such that
and
We have,
and
If Xt is the position of the infected (I) at a given time t starting at the point
and in case
and
then the probabilities
are given by
(4.44) and
(4.45)
Proof
It is known that (see [21]),
is the unique solution of the differential equation
(4.46) with boundary condition
Here Δn is the Laplace-Beltrami operator on S 2. By the symmetry of D, it follows that the probability value of is independent of θ. From (4.7) the differential Eq. (4.46) takes the form
(4.47) with boundary condition
(4.48) In we set
hence from (4.47)
or
Thus
i.e.
(4.49) However,
hence
and
Thus
Consequently,
or
(4.50) Moreover,
hence
(4.51) □
Proposition 4.6
We consider the 2-dimensional sphere S 2 of radius a. Let two circles pass through the North Pole, such that in stereographic coordinates are represented by the parallel lines and where with b < c. Next consider the sets D 1, D 2 in S 2, for which the stereographic projections are
and
Of course,
and
Let Xt be the position of the carrier of virus Y at a given time t starting at the point A, and the stereographic projection coordinates of A are
If
and
then
(4.52)
Proof
It is known that (see [21]) the function
is the unique solution of the differential equation
(4.53) with boundary condition
(4.54) Here, Δ2, is the Laplace-Beltrami operator on S 2 expressed in the stereographic projection coordinates. Hence from (4.8) the differential Eq. (4.53) takes the form
or
(4.55) From (4.54) and (4.55) we see easily that
Therefore,
□
4.3.2. Moment generating functions
Proposition 4.7
Let be fixed. We consider the set D in S 2, such that
Then,
If Xt is the infectious position at a given time t starting at the point
and
then the expectation of is given by
(4.56) where ν is such that and Pν( · ) is the Legendre function
where the multiple-valued function is to be determined in such a way that for it is equal to (the principal value of) zν (which is, in particular, real for positive z and real ν).
Proof
If where λ 1 is the first Dirichlet eigenvalue of D ⊂ S 2, then
it satisfies the differential equation
(4.57) with boundary condition
(4.58) Here Δ2 is the Laplace-Beltrami operator on S 2. By the symmetry of D, it follows that the expectation of is independent of θ. Hence u is independent of θ. From (4.4) the differential equation (4.57) takes the form
i.e.
(4.59) If we set
then
and (4.59) transforms to
or
This is Legendre’s differential equation. However, u(φ) is bounded for all and . Therefore (see [106]), the solution of (4.59) is
i.e.
where ν is such that □
4.4. Reflection principle
Theorem 4.2
Let Xt be the position of the infectious carrier of virus Y at a given time t starting at the point
where
If
then
(4.60)
Proof
(4.61) However, if Xt ∉ D then of course T < t.
Thus,
(4.62) On the other hand, if we set
then by the strong Markov property of Xt
but if then Xt ∉ D. Hence,
or
(4.63) Therefore from (4.61)–(4.63) we obtain that
□
4.4.1. Applications of the reflection principle
The reflection principle can help to calculate the distribution functions of certain exit times.
Let Xt be the position of the infected individual at a given time t starting at the point N(0, 0) in spherical coordinates. If
then
i.e.
where p(t, φ) is the transition density function of the Brownian motion on S 2 of radius a. Hence from (4.26)
or
| (4.64) |
However for every
It is known that (see [106])
However, for every Thus
or
It is also known that for every
Thus, if n is even then If n is odd, i.e. then
i.e.
| (4.65) |
From (4.64) and (4.65) we get that
| (4.66) |
Furthermore, if S(0, π) namely the South Pole of S 2, then
Therefore
| (4.67) |
By using Theorem 3.2, if then
| (4.68) |
4.5. Local time estimation
Definition 4.11
Let φ1 ∈ [0, π]. We set
is a subset of S 2. The reflected Brownian motion in D 1 is the diffusion Yt whose generator is Δn in D 1 with Neuman boundary condition at ∂D 1.
Roughly speaking Yt behaves like Xt inside D 1 but when it reaches the boundary, it is reflected back in D 1.
Definition 4.12
Let a fixed open set D ⊂ Sn with boundary ∂D. If Yt is the reflected Brownian motion in D, and Dδ the domain
we define the boundary local time Lt of Yt, as
It can be shown that the limit exist in the L 2 sense.
4.5.1. Boundary local time until first hitting
Proposition 4.8
Let φ0, φ1 ∈ (0, π), such that φ0 < φ1, both fixed. We consider the sets D, Γ0 in S 2, such that
and
Let Yt be the reflected Brownian motion in Γ0 starting at the point
if
and Lt is the boundary local time of Yt, then,
(4.69) and
(4.70)
Proof
It is known that the function
satisfies the differential equation
with boundary condition
and
as long as the function z is positive (see [107]). Here Δ2 is the Laplace-Beltrami operator on S 2. By the symmetry of D it follows that EA[ exp (λLT) ] is independent of θ. From (4.2) the differential equation takes the form
(4.71) We have shown that the solution of (4.71) is
However,
and
Hence
and
Thus
However,
Therefore,
and
i.e
and
□
5. Discussion and conclusions
A worldwide multilevel interplay among a plethora of factors ranging from micro-pathogens and individual interactions to macro-scale environmental, socio-economic and demographic conditions, necessitate the development of highly sophisticated mathematical models for robust representation of contagious dynamics of infectious diseases that would lead to the establishment of effective control strategies and prevention policies.
Ethical and practical reasons defer from conducting enormous experiments in public health systems, hence mathematical models appear to be an efficient way to explore contagion dynamics. A key aspect of epidemiological models is their link to real data, which is of particular utility toward the design of vaccination policies. Two major vaccination strategies exist currently, i.e., the mass vaccination, which is most applied, and the recently developed pulse vaccination which is used in an increasing number of countries. However, most vaccination strategies are imperfect in the sense that they decrease the number of cases, without however eradicating the disease.
Public-health organizations in the world use the epidemiological models that fall in the three categories already presented in this work, to evaluate disease outbreak policies for epidemics. As we pointed out, many shortcomings exist for those models. All the models already used in the literature assume that the host population has constant size. However, this excludes diseases in exponentially growing populations as in most developing countries, or disease-induced mortality as childhood diseases in developing countries e.g., malaria. Modeling infectious dynamics in non-stationary host populations requires explicit modeling of the host population as well as of the disease per se. Models sometimes can be highly complicated in order to improve best fit to real data. Nonetheless, very complex models do not always perform optimally in real-world applications or in simulations. Real-world models allow for swift decision making, and suitable quantification of the spatiotemporal dynamics of an outbreak. Multidisciplinary research efforts are speeding up, integrating the advances in epidemiology, molecular biology, computational science and applied mathematics. Mathematical modeling allows better understanding of the transmission process of infectious diseases in space and time, by setting forth rigorously the proper assumptions, the variables, the equations and their parameters.
Due to the complexity of the underlying complex interactions, either deterministic or stochastic epidemiological models are built upon incomplete information about e.g., the basic reproduction number, threshold effects, intensity of spread, precise data of infected versus susceptible individuals, and other inaccuracies regarding the entire infectious network. Simulations or brute-force computational techniques have been implemented in that direction to provide approximate solutions with encouraging results. Nevertheless, some of the underlying generating processes of the outbreaks, such as the virus pathogenicity or variant social network topologies, ethnological characteristics and other quantities, may influence the spread of an outbreak. Simulations often prove to be inefficient for the systematic analysis of an emergent epidemic. New rigorous mathematical modeling methodologies, such as the one presented in this work for the first time, can be used to address inherent incomplete data structure and hidden nonlinear complex dynamics, with an aim to enhance forecastability in combating epidemic outbreaks.
In the present study we introduced a novel approach for surveillance and modeling of infectious disease dynamics, called SBDiEM. We explicitly described the mathematical framework underpinning the implementation and conceptualization of our new-age epidemiological model. Our goal is to contribute to the arsenal of models already developed so far. It can be of particular interest, in light of a recent intensive worldwide effort to speed up the establishment of a global surveillance network for combating pandemics of emergent and re-emergent infectious diseases. Toward this aim, mathematical modeling will play a major role in assessing, controlling and forecasting potential outbreaks. We have to better understand and model the impact of numerous variables on contagious dynamics, ranging from the microscopic host–pathogen level, to individual and population interactions, as well as macroscopic environmental, social, economic and demographic factors all over the world.
As a path for future research, we intend to conduct simulations, and empirical analyses based on real-time spatiotemporal datasets, in case of past outbreaks of infectious diseases as well as for COVID-19. Furthermore, we plan to convey an extensive comparative evaluation investigation of SBDiEM vis-à-vis the three major categories set forth by the taxonomy of Siettos and Russo [58], and more specifically versus (1) statistical methods for epidemic surveillance, (2) state-space models of epidemic spread and (3) machine learning methods. In this way, the forecasting and nowcasting capabilities of the new model will be thoroughly explored. We also intend to investigate embedding the proposed analytical model into integrated artificial intelligence systems in the near future.
Our novel methodology apart from offering a much better understanding of the complex and heterogeneous infectious disease dynamics could enhance predictability of epidemic outbreaks as well as have potentially important implications for national health systems, stakeholders and international policy makers.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Stelios Bekiros, Email: stelios.bekiros@eui.eu.
Dimitra Kouloumpou, Email: dimkouloumpou@hna.gr.
References
- 1.OWorld Health Organization. Coronavirus. world health organization, cited january 19. 2020. Available:https://www.who.int/health-topics/coronavirus.
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U., Khan K. Pneumonia of unknown etiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med. 2020 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao S., Musa S.S., Lin Q., Ran J., Yang G., Wang W. Estimating the unreported number of novel coronavirus (2019-nCoV) cases in China in the first half of january 2020: a data-driven modelling analysis of the early outbreak. J Clin Med. 2020 doi: 10.3390/jcm9020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cauchemez S., Donnelly C.A., Reed C., Ghani A.C., Fraser C., Kent C.K. Household transmission of 2009 pandemic influenza a (H1N1) virus in the United States. N Engl J Med. 2009;361:2619–2627. doi: 10.1056/NEJMoa0905498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cauchemez S., Fraser C., Van K.M.D., Donnelly C.A., Riley S., Rambaut A. Middle east respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cauchemez S., Nouvellet P., Cori A., Jombart T., Garske T., Clapham H. Unraveling the drivers of MERS-CoV transmission. Proc Natl Acad Sci USA. 2016;113:9081–9908. doi: 10.1073/pnas.1519235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley S., Fraser C., Donnelly C.A., Ghani A.C., Abu-Raddad L.J., Hedley A.J. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 15.WHO Ebola virus disease in West Africa — the first 9 months of the epidemic and forward projections. N Engl J Med. 2014;371:1481–1495. doi: 10.1056/NEJMoa1411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L.M., Guimarães K.P. The brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson R.M., May R.M. Vol. 28. Oxford University Press; Oxford: 1992. Infectious diseases of humans: dynamics and control. [Google Scholar]
- 18.Noh J.D., Rieger H. Random Walks on Complex Networks. Phys Rev Lett. 2004;92:118701. doi: 10.1103/PhysRevLett.92.118701. [DOI] [PubMed] [Google Scholar]
- 19.Draief M., Massouli L. Cambridge University Press; 2010. Epidemics and rumours in complex networks. [Google Scholar]
- 20.Bonaventura M., Nicosia V., Latora V. Characteristic times of biased random walks on complex networks. Phys Rev E. 2014;89:012803. doi: 10.1103/PhysRevE.89.012803. [DOI] [PubMed] [Google Scholar]
- 21.Klebaner F.C. Imperial College Press; Melbourne: 2004. Introduction to Stochastic Calculus with Applications. [Google Scholar]; Continuous Trading,’ Stoch. Process Appl., 11, 215–260
- 22.Bernoulli D. Essai d’une nouvelle analyse de la mortalité caus ée par la petite vérole. Mém Math Phys Acad R Sci Paris. 1766;1:1–45. [Google Scholar]
- 23.Dietz K., Heesterbeek J.A.P. Daniel bernoulli’s epidemiological model revisited. Math Biosci. 2002;180:1–21. doi: 10.1016/S0025-5564(02)00122-0. [DOI] [PubMed] [Google Scholar]; PMID:12387913
- 24.Lambert J.H. Die toedlichkeit der kinderblattern. beytrage zum gebrauche der mathematik und deren anwendung. Buchhand Realschule. 1772;3:568. [Google Scholar]
- 25.Laplace P.S. Courcier; Paris: 1812. Theorie analytique des probabilites. [Google Scholar]
- 26.Ross R. John Murray; London: 1911. The prevention of malaria; pp. 651–686. [Google Scholar]
- 27.Smith D.L., Battle K.E., Hay S.I., Barker C.M., Scott T.W., McKenzie F.E. Ross-macdonald, and a theory for the dynamics and control of mosquito-transmitted pathogens. PLoS Pathog. 2012;8:e1002588. doi: 10.1371/journal.ppat.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:22496640
- 28.Kermack W.O., McKendrick A.G. A contribution to the mathematical theory of epidemics. Proc R Soc Lond. 1927;A115:700–721. [Google Scholar]
- 29.Kermack W.O., McKendrick A.G. Contribution to the mathematical theory of epidemics. Proc R Soc Lond A Contain Pap Math Phys Character. 1927;115:700–721. doi: 10.1098/rspa.1927.0118. [DOI] [Google Scholar]
- 30.Kermack W.O., McKendrick A.G. Vol. 138. 1932. Contributions to the mathematical theory of epidemics, part II; pp. 55–83. [DOI] [Google Scholar]
- 31.Kermack W.O., McKendrick A.G. Vol. 141. 1933. Contributions to the mathematical theory of epidemics, part III; pp. 94–112. [DOI] [Google Scholar]
- 32.Guldberg C.M., Waage P. C. M. Forhandlinger: Videnskabs-Selskabet Christiana; 1864. Studies concerning affinity; p. 111. [Google Scholar]
- 33.Blower S., Bernoulli D. An attempt at a new analysis of the mortality caused by smallpox and of the advantages of inoculation to prevent it 1766. Rev Med Virol. 2004;14:275–288. doi: 10.1002/rmv.443. [DOI] [PubMed] [Google Scholar]; PMID:15334536
- 34.Bartlett M.S. The critical community size for measles in the United States. J R Stat Soc. 1960;123:37–44. [Google Scholar]
- 35.Bailey N.J.T. Griffin, London; 1957. The mathematical theory of infectious diseases and its application. [Google Scholar]
- 36.Anderson R.M., May R.M. Dynamics and control. Oxford University Press; Oxford: 1991. Infectious diseases of humans. [Google Scholar]
- 37.Enko P.D. Vrach. St. Petersburg. 1889. On the course of epidemics of some infectious diseases; pp. 1008–1010. [Google Scholar]; 1039–1042, 1061–1063
- 38.Enko P.D. On the course of epidemics of some infectious diseases. Int J Epidemiol. 1989;18:749–755. doi: 10.1093/ije/18.4.749. [DOI] [PubMed] [Google Scholar]; PMID:2695472
- 39.Dietz K. The first epidemic model: a historical note on P.D. Enko Aust J Stat. 1988;30A:56–65. doi: 10.1111/j.1467-842X.1988.tb00464.x. [DOI] [Google Scholar]
- 40.Frost W.H. Some conceptions of epidemics in general by wade hampton frost. Am J Epidemiol. 1976;103:141–151. doi: 10.1093/oxfordjournals.aje.a112212. [DOI] [PubMed] [Google Scholar]; PMID:766618
- 41.Xing Y., Song L., Sun G.Q., Jin Z., Zhang J. Assessing reappearance factors OF H7N9 avian influenza in China. Appl Math Comput. 2017;309:192–204. doi: 10.1016/j.amc.2017.04.007. [DOI] [Google Scholar]
- 42.Lee E.K., Liu Y., Pietz F.H. Vol. 2016. 2016. A compartmental model for zika virus with dynamic human and vector populations. AMIA; pp. 743–752. [PMC free article] [PubMed] [Google Scholar]; PMID: 28269870
- 43.Sun G.Q., Xie J.H., Huang S.H., Jin Z., Li M.T., Liu L. Transmission dynamics of cholera: mathematical modeling and control strategies. Commun Nonlinear Sci Numer Simul. 2017;45:235–244. doi: 10.1016/j.cnsns.2016.10.007. [DOI] [Google Scholar]
- 44.Nishiura H., Kinoshita R., Mizumoto K., Yasuda Y., Nah K. Transmission potential of Zika virus infection in the south pacific. Int J Infect Dis. 2016;45:95–97. doi: 10.1016/j.ijid.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 45.Khan M.A., Parvez M., Islam S., Khan I., Shafie S., Gul T. Mathematical analysis of typhoid model with saturated incidence rate. Adv Stud Biol. 2015;7:65–78. doi: 10.12988/asb.2015.41059. [DOI] [Google Scholar]
- 46.Gui H.L., Yong X.Z. Dynamic behaviors of a modified SIR model in epidemic diseases using nonlinear incidence and recovery rates. PLoS One. 2017;12(4):e017578. doi: 10.1371/journal.pone.0175789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M.G., Sun G.Q., Wu Y.F., Zhang J., Jin Z. Transmission dynamics of a multi-group brucellosis model with mixed cross infection in public farm. Appl Math Comput. 2014;237:582–594. doi: 10.1016/j.amc.2014.03.094. [DOI] [Google Scholar]
- 48.Yua P., Linb W. Complex dynamics in biological systems arising from multiple limit cycle bifurcation. J Biol Dyn. 2016;10(1):263–285. doi: 10.1080/17513758.2016.1166270. [DOI] [PubMed] [Google Scholar]
- 49.Shi X., Li G., Zhou X., Song X. Analysis of a differential equation model of HIV infection of CD4 + T-cells with saturated reverse function. Turk J Math. 2011;35:649–666. [Google Scholar]
- 50.Bonyah E., Dontwi I., Nyabadza T. A theoretical model for the transmission dynamics of the Buruli ulcer with saturated treatment. Comput Math Meth Med. 2014;2014:1–13. doi: 10.1155/2014/576039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J., Jia J., Song X. Analysis of an SEIR epidemic model with saturated incidence and saturated treatment function. Sci World J. 2014;2014:1–11. doi: 10.1155/2014/910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daley D.J., Gani J. Cambridge University Press; NY: 2005. Epidemic modeling: an introduction. [Google Scholar]
- 53.Brauer F., Castillo-Chávez C. Springer; NY: 2001. Mathematical models in population biology and epidemiology. [Google Scholar]
- 54.Keeling M.J., Grenfell B.T. Understanding the persistence of measles: reconciling theory, simulation and observation. Proc R Soc Lond. 2002;B269:335–343. doi: 10.1098/rspb.2001.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lloyd A.L. Destabilization of epidemic models with the inclusion of realistic distributions of infectious periods. Proc R Soc Lond. 2001;B268:985–993. doi: 10.1098/rspb.2001.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wearing H.J., Rohani P., Keeling M.J. Appropriate models for the management of infectious diseases. Public Libr Sci – Med. 2005;2(7):e174. doi: 10.1371/journal.pmed.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grossman Z. Oscillatory phenomena in a model of infectious diseases. Theor Popul Biol. 1980;18:204–243. doi: 10.1016/0040-5809(80)90050-7. [DOI] [PubMed] [Google Scholar]
- 58.Siettos C., Russo L. Mathematical modelling of infectious disease dynamics. Virulence. 2013;4(4):295–306. doi: 10.4161/viru.24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Serfling R.E. Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Rep. 1963;78:494–506. doi: 10.2307/4591848. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:19316455
- 60.Stroup D.F., Thacker S.B., Herndon J.L. Application of multiple time series analysis to the estimation of pneumonia and influenza mortality by age 1962-1983. Stat Med. 1988;7:1045–1059. doi: 10.1002/sim.4780071006. [DOI] [PubMed] [Google Scholar]; PMID:3264612
- 61.Costagliola D., Flahault A., Galinec D., Garnerin P., Menares J., Valleron A.J. When is the epidemic warning cut-off point exceeded? Eur J Epidemiol. 1994;10:475–476. doi: 10.1007/BF01719680. [DOI] [PubMed] [Google Scholar]; PMID:7843360
- 62.Greenland S. Handbook of epidemiology. Part II. 2005. Regression methods for epidemiologic analysis; pp. 625–691. [DOI] [Google Scholar]
- 63.Pelat C., Boëlle P.Y., Cowling B.J., Carrat F., Flahault A., Ansart S. Online detection and quantification of epidemics. BMC Med Inform Decis Mak. 2007;7:29. doi: 10.1186/1472-6947-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:17937786
- 64.Dafni U.G., Tsiodras S., Panagiotakos D., Gkolfinopoulou K., Kouvatseas G., Tsourti Z. Algorithm for statistical detection of peaks-syndromic surveillance system for the athens 2004 olympic games. MMWR Morb Mortal Wkly Rep. 2004;24(Suppl):86–94. [PubMed] [Google Scholar]
- 65.Choi K., Thacker S.B. An evaluation of influenza mortality surveillance, 1962-1979. i. time series forecasts of expected pneumonia and influenza deaths. Am J Epidemiol. 1981;113:215–226. doi: 10.1093/oxfordjournals.aje.a113090. [DOI] [PubMed] [Google Scholar]; PMID:6258426
- 66.Abeku T.A., de Vlas S.J., Borsboom G., Teklehaimanot A., Kebede A., Olana D. Forecasting malaria incidence from historical morbidity patterns in epidemicprone areas of ethiopia: a simple seasonal adjustment method performs best. Trop Med Int Health. 2002;7:851–857. doi: 10.1046/j.1365-3156.2002.00924.x. [DOI] [PubMed] [Google Scholar]; PMID:12358620
- 67.Soebiyanto R.P., Adimi F., Kiang R.K. Modeling and predicting seasonal influenza transmission in warm regions using climatological parameters. PLoS One. 2010;5:e9450. doi: 10.1371/journal.pone.0009450. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:20209164
- 68.Nunes B., Viboud C., Machado A., Ringholz C., Rebelode-Andrade H., Nogueira P. Excess mortality associated with influenza epidemics in portugal, 1980 to 2004. PLoS One. 2011;6:e20661. doi: 10.1371/journal.pone.0020661. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:21713040
- 69.Page E.S. Continuous inspection scheme. Biometrika. 1954;41:100–115. [Google Scholar]
- 70.Raubertas R.F. An analysis of disease surveillance data that uses the geographic locations of the reporting units. Stat Med. 1989;8:267–271. doi: 10.1002/sim.4780080306. [DOI] [PubMed] [Google Scholar]; Discussion 279-81; PMID:2711060
- 71.Cowling B.J., Wong I.O.L., Ho L.M., Riley S., Leung G.M. Methods for monitoring influenza surveillance data. Int J Epidemiol. 2006;35:1314–1321. doi: 10.1093/ije/dyl162. [DOI] [PubMed] [Google Scholar]; PMID:16926216
- 72.Höhle M., Paul M. Count data regression charts for the monitoring of surveillance time series. Comput Stat Data Anal. 2008;52:4357–4368. doi: 10.1016/j.csda.2008.02.015. [DOI] [Google Scholar]
- 73.Watkins R.E., Eagleson S., Veenendaal B., Wright G., Plant A.J. Applying cusum-based methods for the detection of outbreaks of ross river virus disease in western australia. BMC Med Inform Decis Mak. 2008;8:37. doi: 10.1186/1472-6947-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:18700044
- 74.Spanos A., Theocharis G., Karageorgopoulos D.E., Peppas G., Fouskakis D., Falagas M.E. Surveillance of community outbreaks of respiratory tract infections based on house-call visits in the metropolitan area of athens, greece. PLoS One. 2012;7:e40310. doi: 10.1371/journal.pone.0040310. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:22905091
- 75.Roberts S.W. Control chart tests based on geometric moving averages. Technomearics. 1959;1:239–250. doi: 10.1080/00401706.1959.10489860. [DOI] [Google Scholar]
- 76.Elbert Y., Burkom H.S. Development and evaluation of a data-adaptive alerting algorithm for univariate temporal biosurveillance data. Stat Med. 2009;28:3226–3248. doi: 10.1002/sim.3708. [DOI] [PubMed] [Google Scholar]; PMID:19725023
- 77.Le Strat Y., Carrat F. Monitoring epidemiologic surveillance data using hidden markov models. Stat Med. 1999;18:3463–3478. doi: 10.1002/(sici)1097-0258(19991230)18:24<3463::aid-sim409>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]; PMID:10611619
- 78.Rath T.M., Carreras M., Sebastiani P. Springer; Berlin: 2003. Automated detection of influenza epidemics with hidden Markov models. [Google Scholar]
- 79.Anderson R.M., May R.M. Population biology of infectious diseases: Part i. Nature. 1979;280:361–367. doi: 10.1038/280361a0. [DOI] [PubMed] [Google Scholar]; PMID:460412
- 80.Brauder F., Castillo-Chavez C. Springer-Verlag; New York: 2001. Mathematical models in population biology and epidemiology. [Google Scholar]
- 81.Murray J.D. Springer-Verlag; New York: 2002. Mathematical biology II. [Google Scholar]
- 82.Feng Z., Dieckmann U., Levin S., editors. Disease evolution: models, concepts and data analysis. AMS; 2006.
- 83.Severo N. Generalizations of some stochastic epidemic models. Math Biosci. 1969;4:395–402. doi: 10.1016/0025-5564(69)90019-4. [DOI] [Google Scholar]
- 84.Prosperi M.C.F., D’Autilia R., Incardona F., De Luca A., Zazzi M., Ulivi G. Stochastic modelling of genotypic drug-resistance for human immunodeficiency virus towards long-term combination therapy optimization. Bioinformatics. 2009;25:1040–1047. doi: 10.1093/bioinformatics/btn568. [DOI] [PubMed] [Google Scholar]; PMID:18977781
- 85.Vardavas R., Blower S. The emergence of HIV transmitted resistance in botswana: ”when will the WHO detection threshold be exceeded?”. PLoS One. 2007;2:e152. doi: 10.1371/journal.pone.0000152. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:17225857
- 86.Newman M.E.J. The structure and function of networks. SIAM Rev. 2003;45:167–256. doi: 10.1137/S003614450342480. [DOI] [Google Scholar]
- 87.Barabasi A.L. Statistical mechanics of complex networks. Rev Mod Phys. 2002;74:47–97. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- 88.Boccaletti S., Latora V., Moreno Y., Chavez M., Hwang D.U. Complex networks: structure and dynamics. Phys Rep. 2006;424:175–308. doi: 10.1016/j.physrep.2005.10.009. [DOI] [Google Scholar]
- 89.Keeling M.J., Danon L., Vernon M.C., House T.A. Individual identity and movement networks for disease metapopulations. Proc Natl Acad Sci USA. 2010;107:8866–8870. doi: 10.1073/pnas.1000416107. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:20421468
- 90.Keeling M.J., Eames K.T. Networks and epidemic models. J R Soc Interface. 2005;2:295–307. doi: 10.1098/rsif.2005.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:16849187
- 91.Read J.M., Eames K.T.F., Edmunds W.J. Dynamic social networks and the implications for the spread of infectious disease. J R Soc Interface. 2008;5:1001–1007. doi: 10.1098/rsif.2008.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:18319209
- 92.Eubank S., Guclu H., Kumar V.S., Marathe M.V., Srinivasan A., Toroczkai Z. Modelling disease outbreaks in realistic urban social networks. Nature. 2004;429:180–184. doi: 10.1038/nature02541. [DOI] [PubMed] [Google Scholar]; PMID:15141212
- 93.Ferguson N.M., Cummings D.A., Cauchemez S., Fraser C., Riley S., Meeyai A. Strategies for containing an emerging influenza pandemic in southeast asia. Nature. 2005;437:209–214. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]; PMID:16079797
- 94.Burke D.S., Epstein J.M., Cummings D.A., Parker J.I., Cline K.C., Singa R.M. Individual-based computational modeling of smallpox epidemic control strategies. Acad Emerg Med. 2006;13:1142–1149. doi: 10.1111/j.1553-2712.2006.tb01638.x. [DOI] [PubMed] [Google Scholar]; PMID:17085740
- 95.Balcan D., Colizza V., Gonçalves B., Hu H., Ramasco J.J., Vespignani A. Multiscale mobility networks and the spatial spreading of infectious diseases. Proc Natl Acad Sci USA. 2009;106:21484–21489. doi: 10.1073/pnas.0906910106. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:20018697
- 96.Bai Y., Jin Z. Prediction of SARS epidemic by BP neural networks with online prediction strategy. Chaos Solitons Fractals. 2005;26:559–569. doi: 10.1016/j.chaos.2005.01.064. [DOI] [Google Scholar]
- 97.Ginsberg J., Mohebbi M.H., Patel R.S., Brammer L., Smolinski M.S., Brilliant L. Detecting influenza epidemics using search engine query data. Nature. 2009;457:1012–1014. doi: 10.1038/nature07634. [DOI] [PubMed] [Google Scholar]; PMID:19020500
- 98.Hulth A., Rydevik G., Linde A. Web queries as a source for syndromic surveillance. PLoS One. 2009;4:e4378. doi: 10.1371/journal.pone.0004378. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:19197389
- 99.Chan E.H., Sahai V., Conrad C., Brownstein J.S. Using web search query data to monitor dengue epidemics: a new model for neglected tropical disease surveillance. PLoS Negl Trop Dis. 2011;5:e1206. doi: 10.1371/journal.pntd.0001206. [DOI] [PMC free article] [PubMed] [Google Scholar]; PMID:21647308
- 100.Do Carmo M.P. Prentice-Hall, Inc.; Upper Saddle River, New Jersey: 1976. Differential geometry of curves and surfaces. [Google Scholar]
- 101.Dodziuk J. Maximum principle for parabolic inequalities and the heat flow on open manifolds. Indiana Oniv Math J. 1983;32(5):115–142. [Google Scholar]
- 102.John F. Springer; USA: 1982. Partial differential equations. [Google Scholar]
- 103.Chung M.K. Proceedings of IEEE International symposium on biomedical imaging (ISBI) 2006. Heat kernel smoothing on unit sphere; pp. 992–995. [Google Scholar]
- 104.Oksendal B. Springer-Verlag; 1995. Stochastic differential equations. [Google Scholar]
- 105.Dynkin E.B. Vol. 2. Springer; Berlin: 1965. Markov processes. [Google Scholar]
- 106.Spiegel M.R. Schaum’s Outline Series. 1999. Complex variables with an introduction to conformal mapping and its applications. [Google Scholar]
- 107.Papanicolaou V.G. The probabilistic solution of the third boundary value problem for second order elliptic equations. Probab Theory Relat Fields. 1990;87:27–77. [Google Scholar]