Abstract
Aims
Arrhythmogenic cardiomyopathy (ACM) is a myocardial disease caused mainly by mutations in genes encoding desmosome proteins ACM patients present with ventricular arrhythmias, cardiac dysfunction, sudden cardiac death, and a subset with fibro-fatty infiltration of the right ventricle predominantly. Endurance exercise is thought to exacerbate cardiac dysfunction and arrhythmias in ACM. The objective was to determine the effects of treadmill exercise on cardiac phenotype, including myocyte gene expression in myocyte-specific desmoplakin (Dsp) haplo-insufficient (Myh6-Cre:DspW/F) mice.
Methods and results
Three months old sex-matched wild-type (WT) and Myh6-Cre:DspW/F mice with normal cardiac function, as assessed by echocardiography, were randomized to regular activity or 60 min of daily treadmill exercise (5.5 kJ work per run). Cardiac myocyte gene expression, cardiac function, arrhythmias, and myocardial histology, including apoptosis, were analysed prior to and after 3 months of routine activity or treadmill exercise. Fifty-seven and 781 genes were differentially expressed in 3- and 6-month-old Myh6-Cre:DspW/F cardiac myocytes, compared to the corresponding WT myocytes, respectively. Genes encoding secreted proteins (secretome), including inhibitors of the canonical WNT pathway, were among the most up-regulated genes. The differentially expressed genes (DEGs) predicted activation of epithelial–mesenchymal transition (EMT) and inflammation, and suppression of oxidative phosphorylation pathways in the Myh6-Cre:DspW/F myocytes. Treadmill exercise restored transcript levels of two-third (492/781) of the DEGs and the corresponding dysregulated transcriptional and biological pathways, including EMT, inflammation, and secreted inhibitors of the canonical WNT. The changes were associated with reduced myocardial apoptosis and eccentric cardiac hypertrophy without changes in cardiac function.
Conclusion
Treadmill exercise restored transcript levels of the majority of dysregulated genes in cardiac myocytes, reduced myocardial apoptosis, and induced eccentric cardiac hypertrophy without affecting cardiac dysfunction in a mouse model of ACM. The findings suggest that treadmill exercise has potential beneficial effects in a subset of cardiac phenotypes in ACM.
Keywords: Arrhythmogenic cardiomyopathy, Exercise, Gene expression
Graphical Abstract
Graphical Abstract.
1. Introduction
Arrhythmogenic cardiomyopathy (ACM) is a primary disease of the myocardium caused mainly by mutations in genes encoding intercalated discs (IDs), particularly desmosome proteins.1,2 Patients with ACM typically present with ventricular arrhythmias, which often occur early and in the absence of discernible cardiac dysfunction.1,2 Early ventricular arrhythmias are distinct from late ventricular and supra-ventricular arrhythmias, which typically occur in conjunction with cardiac dysfunction, as in other forms of heart failure.1,2 A histological hallmark of the classic form of ACM is fibro-fatty replacement of the myocardium, which manifests predominantly but not exclusively in the right ventricle, particularly in a subset referred to as arrhythmogenic right ventricular cardiomyopathy. Cardiac dysfunction typically occurs late and is progressive, leading to refractory heart failure.1,2 ACM is an important cause of sudden cardiac death, particularly in athletes.3–5
Mutations in PKP2, DSP, DSG2, DSC2, and JUP genes, encoding desmosome proteins plakophilin 2, desmoplakin, desmoglein 2, desmocollin 2, and junction protein plakoglobin, respectively, are major causes of ACM.1,2 Mutations in several other genes are also implicated as causes of ACM.2,6 The causal genes in a subset of ACM, particularly, in the sporadic cases, remain to be identified.
Desmosomes, which are members of the IDs, are abundantly present in cardiac myocytes and at low levels in other cardiac cell types.7,8 Desmosomes are responsible for myocyte–myocyte attachment and maintaining mechanical integrity of the myocardium.9 They are also signalling hubs for mechano-sensing pathways, including the Hippo and the canonical WNT pathways, which are implicated in the pathogenesis of ACM.9–12
Observational studies in patients with ACM suggest that intense endurance exercise accentuates cardiac phenotype by promoting cardiac dysfunction and ventricular arrhythmias.1,13–15 Accordingly, patients with ACM and individuals carrying mutations in genes encoding desmosome proteins are considered at an increased risk of developing heart failure and cardiac arrhythmias upon exercising.13,16 Therefore, they are discouraged from participation in heavy physical activity and endurance training.17,18 The apparent deleterious effects of exercise in ACM are in contrast to the beneficial effects of exercise observed in various cardiac pathologies, including post-myocardial infarction cardiac remodelling.19–23 Experimental data in mouse models implicate increased mechanical stress due to haemodynamic overload as a mechanism for cardiac dysfunction in ACM.24,25
Partial delineation of the causal mutations in genes encoding desmosome proteins has offered an opportunity to investigate the molecular basis of the apparent deleterious effects of exercise in ACM.26 Experimental data in mouse models of ACM support the potential deleterious effects of sustained exercise on cardiac function. The deleterious effects of exercise presumably result from an increased mechanical stress due to increased haemodynamic load during exercise as reducing preload has been reported to attenuate the effects.24,25 Thus, the objective of the present study was to identify the molecular pathways that mediate the effects of treadmill exercise on cardiac phenotype in the Myh6-Cre:DspW/F mouse model of ACM. The Myh6-Cre:DspW/F mice, first published in 2006, do not show a discernible phenotype within the first couple of months after birth, as these mice exhibit a normal cardiac function and do not show cardiac arrhythmias or fibro-adiposis. By 6 months of age, the Myh6-Cre:DspW/F mice exhibit cardiac systolic dysfunction and mild myocardial fibrosis.10 The phenotype fully evolves within a year and is manifested as increased mortality, cardiac systolic dysfunction, inducible ventricular arrhythmias, and fibro-adiposis. The phenotype involves both ventricles and there are no discernible differences in cardiac systolic function between male and female mice.10
2. Methods
Detailed material and methods are provided as Supplementary material online.
2.1 Data sharing
RNA-sequencing (RNA-Seq) data have been submitted to GEO (GSE129962). Detailed material and methods are available in Supplementary material online.
2.2 Regulatory approvals
Animal studies were in accord with the NIH Guide for the Care and Use of Laboratory Animals published and approved by the Animal Care and Use Committee (AWC-18-0048).
2.3 Myh6-Cre:DspW/F mice
The phenotype in the Myh6-Cre:DspW/F mice has been published.10 Sequence of the oligonucleotide primers used for genotyping is provided in Supplementary material online, Table S1.
Table 1.
Echocardiographic parameters in 6-month-old wild type and Myh6-Cre:DspW/F mice in the routine activity or treadmill exercise group
| Wild type |
P-value |
Myh6-Cre:Dsp
W/F
|
P-value | P (genotype effect) | P (exercise effect) | P (genotype *exercise) | |||
|---|---|---|---|---|---|---|---|---|---|
| RA | Exercise | RA | Exercise | ||||||
| N | 10 | 9 | NA | 10 | 12 | NA | NA | NA | NA |
| Male/female | 4/6 | 4/5 | 1a | 4/6 | 6/6 | 0.639a | NA | NA | NA |
| Age (days) | 191.6 ± 15.7 | 188.4 ± 3.1 | 0.281 | 194.60 ± 18.92 | 187.00 ± 4.09 | 0.094 | 0.843 | 0.175 | 0.571 |
| Body weight (g) | 31.4 ± 5.0 | 29.4 ± 3.5 | 0.167 | 33.77 ± 6.26 | 29.40 ± 4.24 | 0.177 | 0.434 | 0.044 | 0.438 |
| HR (b.p.m.) | 494.3 ± 40.0 | 514.2 ± 23.4 | 0.105 | 524.39 ± 55.98 | 477.27 ± 36.63 | 0.048b | 0.801 | 0.291 | 0.013 |
| IVST-d (mm) | 0.74 ± 0.08 | 0.77 ± 0.06 | 0.196 | 0.67 ± 0.03 | 0.69 ± 0.11 | 0.688 | 0.003 | 0.355 | 0.788 |
| IVST-s (mm) | 1.02 ± 0.14 | 1.22 ± 0.23 | 0.021 | 0.94 ± 0.11 | 0.90 ± 0.14 | 0.231 | 0.0004 | 0.470 | 0.054 |
| LVPWT-d (mm) | 0.84 ± 0.09 | 0.83 ± 0.12 | 0.423 | 0.79 ± 0.11 | 0.75 ± 0.09 | 0.168 | 0.035 | 0.436 | 0.609 |
| LVPWT-s (mm) | 1.21 ± 0.09 | 1.30 ± 0.13 | 0.056 | 0.99 ± 0.18 | 1.00 ± 0.11 | 0.817b | <0.0001 | 0.285 | 0.346 |
| LVEDD (mm) | 3.42 ± 0.14 | 3.46 ± 0.26 | 0.626 | 3.76 ± 0.37 | 4.08 ± 0.44 | 0.047 | <0.0001 | 0.111 | 0.190 |
| LVEDDI (mm/g) | 0.11 ± 0.02 | 0.12 ± 0.01 | 0.153 | 0.11 ± 0.02 | 0.14 ± 0.03 | 0.018b | 0.066 | 0.0135 | 0.143 |
| LVESD (mm) | 2.13 ± 0.19 | 1.97 ± 0.28 | 0.077 | 2.68 ± 0.53 | 3.03 ± 0.51 | 0.114b | <0.0001 | 0.470 | 0.054 |
| FS (%) | 37.7 ± 6.0 | 43.1 ± 7.4 | 0.048 | 29.38 ± 7.67 | 25.93 ± 5.50 | 0.129b | <0.0001 | 0.639 | 0.039 |
| EF (%) | 68.4 ± 7.5 | 74.6 ± 8.3 | 0.053 | 56.35 ± 12.12 | 51.07 ± 9.67 | 0.129b | <0.0001 | 0.882 | 0.065 |
| LV mass (mg) | 72.9 ± 11.9 | 79.5 ± 10.5 | 0.111 | 66.62 ± 15.60 | 84.73 ± 12.53 | 0.003 | 0.889 | 0.004 | 0.161 |
| LVMI (mg/g) | 2.37 ± 0.50 | 2.72 ± 0.34 | 0.046 | 2.01 ± 0.51 | 2.90 ± 0.36 | 0.0001 | 0.509 | <0.0001 | 0.054 |
Genotype-dependent effects of exercise on echocardiographic indices were analysed by two-way analysis of variance.
P (genotype effect) indicates the effects of genotype on the echocardiographic indices (independent of exercise).
P (exercise effect) indicates the effects of exercise on the echocardiographic indices (independent of the genotypes).
P (genotype*interaction) indicates interactive effects of genotypes and exercise on the echocardiographic indices.
BW, body weight; FS, fractional shortening; HR, heart rate; IVST, interventricular septum thickness; LVEDD, left ventricular end-diastolic diameter; LVEDDi, LVEDD indexed to the body weight; LVESD, left ventricular end-systolic diameter; LVM, left ventricular mass; LVMI, LVM indexed to the body weight; LVPWT, left ventricular posterior wall thickness; Myh6-Cre:DspW/F, cardiac myocyte specific heterozygous deletion of desmoplakin; RA, regular activity.
The Fisher’s exact test.
The Kruskal–Wallis.
2.4 Anaesthesia and euthanasia
Anaesthesia was induced with 4–5% inhaled isoflurane and was maintained at 2% isoflurane inhalation throughout the procedure. To isolate cardiac myocytes, anaesthesia was induced with one-time intraperitoneal injection of pentobarbital at 50 mg/kg dose. Mice were euthanized by 100% CO2 inhalation followed by cervical dislocation.
2.5 Exercise protocol
The study design is shown in Supplementary material online Figure S1. Three months old wild-type (WT) and Myh6-Cre:DspW/F mice with normal cardiac function were randomized to either regular activity or treadmill exercise. Mice were subjected to a 60-min treadmill run with a stepwise increase in slope and speed delivering 5.5 kJ work per day for a 30 g mouse (Supplementary material online,Table S2).
2.6 Isolation of adult mouse cardiac myocytes
Adult mouse cardiac myocytes were isolated as per a well-established protocol and as reported previously, with minor modifications.27,28 The perfusion method enables isolation of cardiac myocytes from non-myocyte cells and includes myocytes from both ventricles, which were used in these experiments.27
2.7 RNA-sequencing
Bulk RNA-Seq was performed, as published (N = 5 per group, matched for age and sex).11,28 Total RNA was extracted from isolated cardiac myocytes and samples with an RNA Integrity Number (RIN) of >8 were used to generate strand-specific sequencing libraries after depletion of rRNA. Sequencing was performed on the Illumina HiSeq 4000 instrument using the paired-end sample preparation chemistry.
Raw RNA sequencing reads were mapped to the mouse reference genome build 10 by Tophat2.29 Mapped reads were counted using the feature counts.30 Differentially expressed genes (DEGs) were identified using the edgeR package in R statistical programme. Significance level for the DEGs was set at q < 0.05. For comparisons across time points, data were normalized using the Remove Unwanted Variation (RUVr) method, as implemented in the R scientific analysis platform.31
2.8 Pathway analysis
Gene set enrichment analysis (GSEA, version 2.2.3) was performed on normalized count per million (CPM) or on ranked gene lists. Significance level was set at q < 0.05.32 Molecular signature database 3.0 curated gene sets for hallmark and canonical pathways were used for the analysis.
2.9 Identification of upstream regulators
Upstream regulators of the dysregulated genes were predicted using Ingenuity pathway analysis software. A Z score of ≥2 OR ≤−2 and q < 0.05 were considered significant.
2.10 Quantitative RT-PCR (qPCR)
Transcript levels of selected genes were quantified by qPCR.28,33–35 Target gene expression levels were normalized to Gapdh mRNA levels and compared using the ΔCT method. The list of TaqMan assays and oligonucleotide primers are provided in Supplementary material online, Table S1.
2.11 Echocardiography
M- and B-mode mouse echocardiography was performed in age- and sex-matched littermates using a Vevo 1100 ultrasound imaging system equipped with a 22-55 MHz MicroScan transducer, as published.8,11,28,33,34,36
2.12 Electrophysiological studies
Electrophysiological studies were performed using standard clinical pacing protocols.7,37 Ventricular tachycardia (VT) susceptibility was assessed by overdrive pacing and by delivering single, double, and triple extra-stimuli at the baseline and after intraperitoneal injection with 2 mg/kg isoproterenol or 2 mg/kg isoproterenol with 120 mg/kg caffeine. Cardiac rhythm was also monitored for 1 h in each mouse.
2.13 Gross morphology
Heart weight to body weight ratio was calculated and the mean values were compared among the groups.34
2.14 Quantification of myocardial fibrosis
Thin myocardial sections were stained for Picrosirius Red, and collagen volume fraction (CVF) was quantified, as published.7,8,28,33
2.15 Immunoblotting
Immunoblotting was performed using 50–100 µg of protein lysates28,38 Targeted proteins were detected using specific primary antibodies and the respective horseradish linked secondary antibodies, as listed in Supplementary material online, Table S1.
2.16 Immunofluorescence
Expression and localization of the proteins of interest were detected in thin myocardial frozen sections upon probing with the corresponding primary antibodies and incubation with the secondary antibody conjugates.7,28,34 Supplementary material online, Table S1 lists antibodies used in immunofluorescence studies.
2.17 TUNEL assay
Apoptosis was detected by terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay, using In-Situ Cell Death Detection Fluorescein Kit.10,28 The percentage of nuclei stained positive for TUNEL of a total of 12 000 to 20 000 cells was calculated in each heart and compared.
2.18 Wheat Germ Agglutinin staining
Wheat Germ Agglutinin staining was performed as described previously with minor modifications.39 The number of myocytes in each thin myocardial section was determined upon staining with an antibody against pericentriolar membrane protein (PCM1), which tags myocyte nuclei in the heart.40,41 At least 10 fields per section, 5 sections per mouse and 4–5 mice per genotype were analysed, representing about 12 000 to 20 000 cells per each mouse heart.
2.19 Secretome analysis
The list of mouse genes coding for secreted proteins (secretome) were obtained from the publicly available curated databases, which list 2332 genes.42,43 DEGs were compared with the secretome gene dataset to identify those encoding the secretome and assess its enrichment in the Myh6-Cre:DspW/F myocytes.
2.20 Statistical methods
Statistical analyses were performed either using GraphPad Prism 7 or STATA IC 15.1 and were as published.7,28,33 In brief, data were presented as mean ± standard deviation. Gaussian distribution of the data was determined using the Shapiro–Wilk normality test. Normally distributed data were compared using t-test between two groups and by analysis of variance (ANOVA) among multiple groups. The latter was followed by Bonferroni pairwise comparison test to compare differences between two specific groups. Data that departed from normality and non-parametric variables were compared by Mann–Whitney or Kruskal–Wallis test.
Statistical analyses of RNA-Seq data were as described above under each section. Genomic statistical results are depicted in each figure.
3. Results
3.1 Differentially expressed genes in cardiac myocytes isolated from Myh6-Cre:DspW/F mice
To identify DEGs, total RNA, extracted from cardiac myocytes isolated from the hearts of 3 or 6 months old WT and Myh6-Cre:DspW/F mice (N = 5 per group), was depleted from ribosomal RNA and analysed by RNA sequencing (RNA-Seq). The time point of 3 months was chosen, based on the previous data, to capture cardiac myocyte transcriptome prior to manifestation of major morphological, histological and functional phenotypes in the Myh6-Cre:DspW/F mice.10 The timepoint of 6 months enabled detection of early transcriptomic changes that coincide with the gradual evolution of cardiac phenotype in the Myh6-Cre:DspW/F mice.10
Analysis of transcripts of cardiac myocytes isolated from 3 months old WT and Myh6-Cre:DspW/F mice showed only 56 DEGs (out of 11 582 gene analysed by RNA-Seq), which were comprised of 21 up-regulated and 35 down-regulated genes with q < 0.05 (Supplementary material online, Figure S2). Serinc2 encoding serine incorporator 2, was the most up-regulated (17.8-fold, q = 0.001) and Ephbl4, encoding ephrin type-B receptor 1, was the most down-regulated (8.4-fold, q = 0.00002) genes in the Myh6-Cre:DspW/F cardiac myocytes. The ontology classification of the dysregulated gene is shown in Supplementary material online, Figure S2, albeit the number of DEGs was rather small to make a reliable prediction.
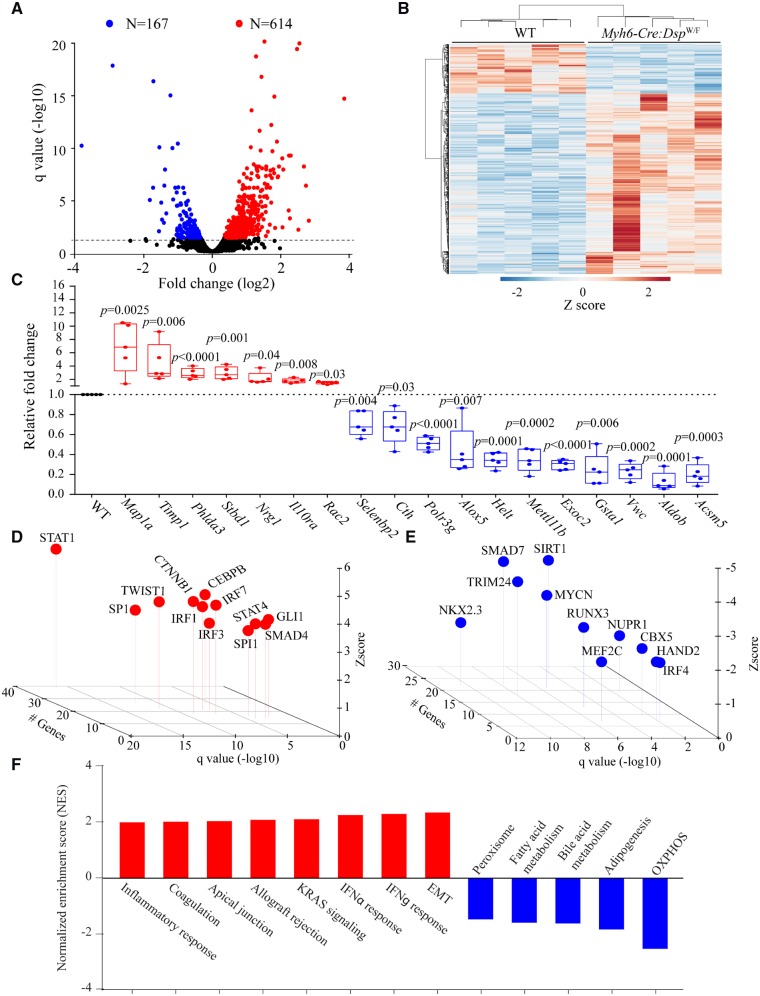
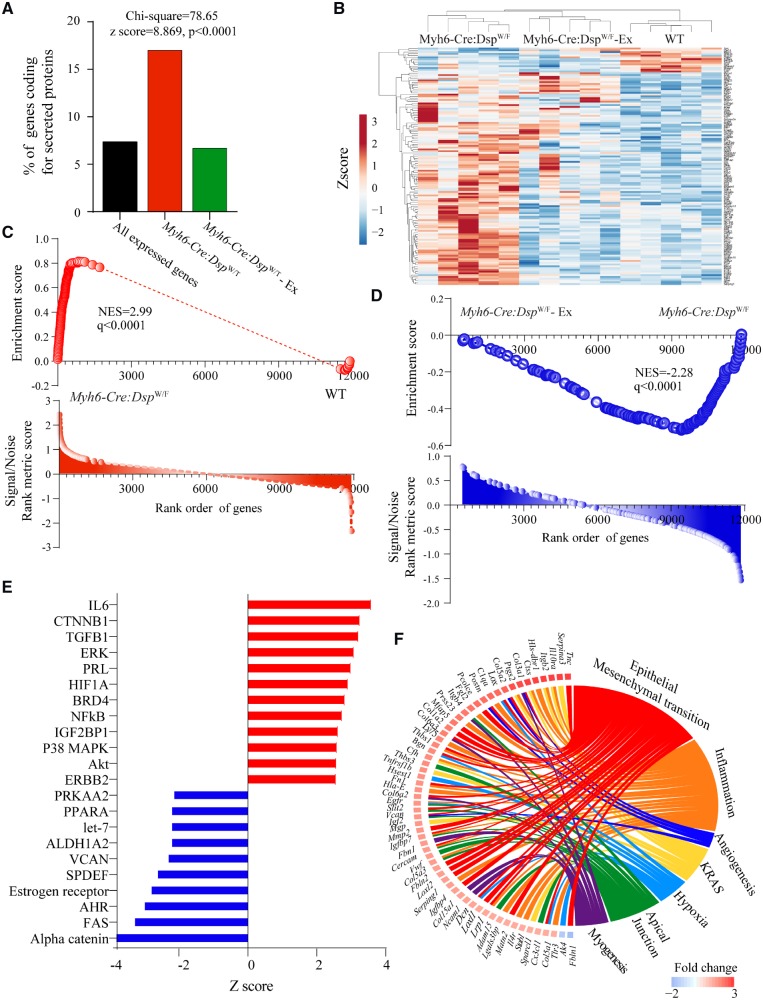
Transcriptome analysis of cardiac myocytes isolated from 6 months old WT and Myh6-Cre:DspW/F mice (N = 5 per group) was notable for differential expression of 781/11 122 genes (∼7% of gene analysed by RNA-Seq) at a q-value of <0.05 in the Myh6-Cre:DspW/F as compared to WT cardiac myocytes (Figure 1A). Transcript levels of 614 (5.5%) genes were increased, whereas those of 167 (1.5%) were suppressed (Figure 1A and B). Transcript levels of the top DEGs were quantified by qPCR and the results were in accord with the RNA-Seq findings, as all 18 genes tested by both methods showed changes in the same direction (Figure 1C and Supplementary material online, Table S3). Ingenuity pathway analysis of top differentially up-regulated transcripts were predicted to be targets of IRF1, 3, and 7 involved in inflammatory responses, canonical WNT signalling, and transforming growth factor β (TGFB) family (Figure 1D). In contrast, the down-regulated genes were targets of transcriptional factors regulating metabolism (such as SIRT1), negative regulator of extracellular matrix proteins (such as SMAD7), and chromatin modifiers (such as CBX5 and TRIM24) (Figure 1E). The corresponding predicted biological pathway pertained to activation of epithelial–mesenchymal transition (EMT), and inflammatory responses and suppression of oxidative phosphorylation (Figure 1F).
Figure 1.
Cardiac myocyte transcriptome in Myh6-Cre:DspW/F mice. (A) Volcano plot showing down-regulated and up-regulated genes in cardiac myocytes isolated from 6 months old Myh6-Cre:DspW/F as compared to wild type (WT) mice (N = 5 per group). (B) Heat map of the differentially expressed genes (DEGs). (C) Quantitative PCR validation of the top up-regulated and down-regulated genes in cardiac myocytes isolated from 6-month-old wild type and Myh6-Cre:DspW/F mice (N = 5). (D and E) Transcriptional factors (TFs) predicted to be activated (C) and suppressed (D) in Myh6-Cre:DspW/F myocytes. The X-axis represents −log10 q value, the Y-axis represents Z score, and the Z-axis represents number of target genes of the transcription factor present in the differentially expressed genes with q < 0.05. (F) Dysregulated biological pathways corresponding to the DEGs with q < 0.05 (Y-axis represents Z score). P-values were calculated by t-test for normally distributed data and Mann–Whitney test for those departing from normality.
3.2 Temporal evolution of cardiac transcriptome
Analysis of the cardiac myocyte transcriptome at two time points of 3 and 6 months of age enabled genotype-dependent assessment of changes in gene expression during the course of evolving cardiac phenotype. Comparing the transcriptome profiles of myocytes isolated from 3- and 6-month-old WT mice led to identification of 310 DEGs (q < 0.05), which was comprised of 251 up-regulated and 59 down-regulated genes (Supplementary material online, Figure S3). Pathway analysis did not identify significantly dysregulated biological pathways between 3- and 6-month-old myocytes in the WT mice (Supplementary material online, Figure S3).
The interval change in gene expression in myocytes isolated from 3- and 6-month-old Myh6-Cre:DspW/F mice was remarkable for 1643 DEGs (q < 0.05), which included 1405 up-regulated and 238 down-regulated genes (Supplementary material online, Figure S4). The five-fold higher number of DEGs between the two time points of 3 and 6 months in the Myh6-Cre:DspW/F myocytes, as compared to the WT myocytes, reflects the genotype effects and is in accord with the temporal onset of expression of histological and functional phenotypes in the Myh6-Cre:DspW/F mice.10 The up-regulated genes were identified as targets of STAT1, SMARCA4, HIF1, and TCF7L2 transcription factors, whereas the down-regulated genes were recognized as targets of SMAD7, TRIM24, and SIRT1 among others (Supplementary material online, Figure S4). Accordingly, genes involved in EMT, fibrosis, inflammatory response, and apoptosis were among the significantly up-regulated genes in 6 months compared to 3 months old Myh6-Cre:DspW/F cardiac myocytes (Supplementary material online, Figure S4). In contrast, genes involved in oxidative phosphorylation and fatty acid metabolism were significantly suppressed during this time interval.
To identify temporal changes in gene expression specific to the genotypes, the interval changes in gene expression between WT and Myh6-Cre:DspW/F myocytes were compared. The analyses identified 114/1643 (7%) DEGs that were common to both genotypes, of which 96 were up-regulated and 18 down-regulated (Supplementary material online, Figure S5). There were 1529 DEGs that were specific to Myh6-Cre:DspW/F myocytes, of which 1309 were up-regulated and 220 down-regulated (Supplementary material online, Figure S5). Altered transcriptional regulators and biological pathways predicted by the above sets of DEGs, were largely similar to those found when comparing the transcriptomes in 6 months old WT and Myh6-Cre:DspW/F myocytes (Figure 1 and Supplementary material online, Figure S5).
3.3 Morphological, histological, and functional phenotypes corresponding to transcriptomic changes
Consistent with the scant number of DEGs at 3 months of age, heart weight/body weight ratio, and CVF were similar between the two genotypes, as were the mean number of adipocytes, defined as cells containing fat droplets, in the heart and the myocyte cross-sectional area (Supplementary material online, Figure S6). However, the mean number of cells stained positive for the TUNEL assay in thin myocardial sections was increased modestly in the Myh6-Cre:DspW/F hearts (Supplementary material online, Figure S6). Cardiac size and function, as determined by echocardiography, were comparable between the two genotypes, except for a trend towards increased left ventricular end-systolic diameter (LVESD) in the Myh6-Cre:DspW/F as compared to WT mice (Supplementary material online,Table S4).
Histological and functional phenotypes in the 6 months old Myh6-Cre:DspW/F mice were largely in accord with the published data.10 CVF and the mean number of TUNEL positive cells in the myocardium were increased in the Myh6-Cre:DspW/F as compared to WT hearts (Supplementary material online, Figure S7). Heart weight/body weight ratio, the mean number of adipocytes, and mean myocyte cross-sectional areas were not different between the two genotypes (Supplementary material online, Figure S7). However, the Myh6-Cre:DspW/F mice exhibited echocardiographic evidence of left ventricular dysfunction, including increased LVESD and reduced left ventricular fractional shortening (LVFS), as compared to the corresponding WT mice (Supplementary material online, Table S5). Functional and histological findings are consistent with early stages of evolution of cardiac phenotype in the Myh6-Cre-DspW/F mice.
3.4 Effects of treadmill exercise on cardiac myocyte transcripts
Three months old WT and Myh6-Cre:DspW/F mice with a normal cardiac function, as determined by echocardiography, were randomized to regular activity or one hour of daily treadmill exercise, delivering 5.5 kJ work per session, for 3 months (Supplementary material online, Figure S1, Table S2). All except one Myh6-Cre:DspW/F mouse completed the exercise protocol. One Myh6-Cre:DspW/F mouse stopped exercising after 2.5 months. Echocardiography at this time point showed severe left ventricular dysfunction. The mouse was euthanized for isolation of cardiac myocytes and RNA extraction. There was no death in either group during the 3 months of daily treadmill exercise.
Treadmill exercise led to differential gene expression in the WT as well as Myh6-Cre-DspW/F mice, as compared to the corresponding mice assigned to the regular activity group. In the WT mice, transcript levels of 2529 genes were altered in the treadmill exercise group, which pertained to 1390 up-regulated and 1139 down-regulated genes (Supplementary material online, Figure S8A and B, all q < 0.05). The up-regulated genes were identified as targets of MEF2C and SMARCB1 transcription factor, whereas the down-regulated genes were transcriptionally regulated by CREB1, NFKB1A, and STATs, amongst others (Supplementary material online, Figure S8C). The magnitude of the changes in the predicted dysregulated biological pathways in the WT mice were modest and pertained to those involved in protein secretion, mitotic spindle, and PI3K/AKT/mTOR signalling amongst the activated and inflammation, EMT, and oxidative phosphorylation among the most down-regulated pathways (Supplementary material online, Figure S8D).
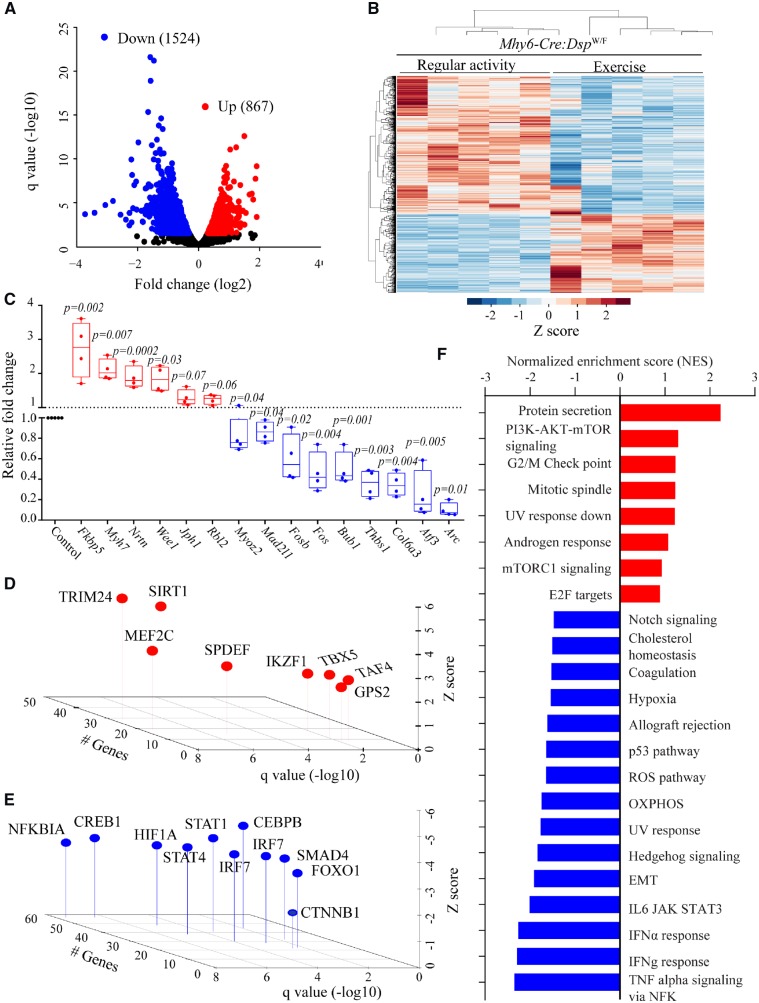
Treadmill exercise markedly affected transcriptome profile of Myh6-Cre-DspW/F cardiac myocytes, as noted by dysregulation of 2391 transcripts (q < 0.05) as compared to that in the regular activity group (Figure 2). The altered transcripts encompassed 867 up-regulated and 1524 down-regulated genes (Figure 2A and B). Transcript levels of selected DEGs involved in EMT and inflammation were also quantified by qPCR and the results were concordant with the RNA-Seq data for 12 out of 15 genes tested (Figure 2C and Supplementary material online,Table S6). Based on the DEGs, TRIM24, and SIRT1 were predicted to be among the top activated and NFKB1A, CREB1, STAT1, and CEBPB amongst the most suppressed transcriptional regulators (Figure 2D and E). GSEA predicted activation of pathways involved in protein secretion, PI3K/AKT/MTOR signalling, and mitotic spindle in the exercise group (Figure 2F). In contrast, gene sets involved in inflammation, EMT, oxidative phosphorylation, and oxidative stress were predicted to be suppressed (Figure 2F).
Figure 2.
Effects of treadmill exercise on cardiac myocyte transcriptome. (A) Volcano plot showing down-regulated and up-regulated genes in cardiac myocytes isolated from the hearts of Myh6-Cre:DspW/F mice in the treadmill exercise group as opposed to the regular activity group (N = 5 per group). (B) Heat map of DEGs between indicated groups showing clustering based on the genotypes. (C) Quantitative PCR data for top dysregulated genes in isolated cardiac myocytes from Myh6-Cre:DspW/F in the regular activity group (N = 5) and Myh6-Cre:DspW/F in the treadmill exercise group (N = 4). (D and E) TFs predicted to be activated (D) and suppressed (E) based on DEGs with a q < 0.05 in Myh6-Cre:DspW/F myocytes in the treadmill exercise group as compared to the regular activity group. The X-axis represents −log10 of q value, the Y-axis represents Z score, and the Z-axis represents number of genes predicted to be targets of each TF. (F) Dysregulated biological pathways corresponding to DEGs with q < 0.05. Pathways with q < 0.05 and normalized enrichment score, in the X-axis, are shown. P-values were calculated by t-test for normally distributed data and Mann–Whitney test for those departing from normality.
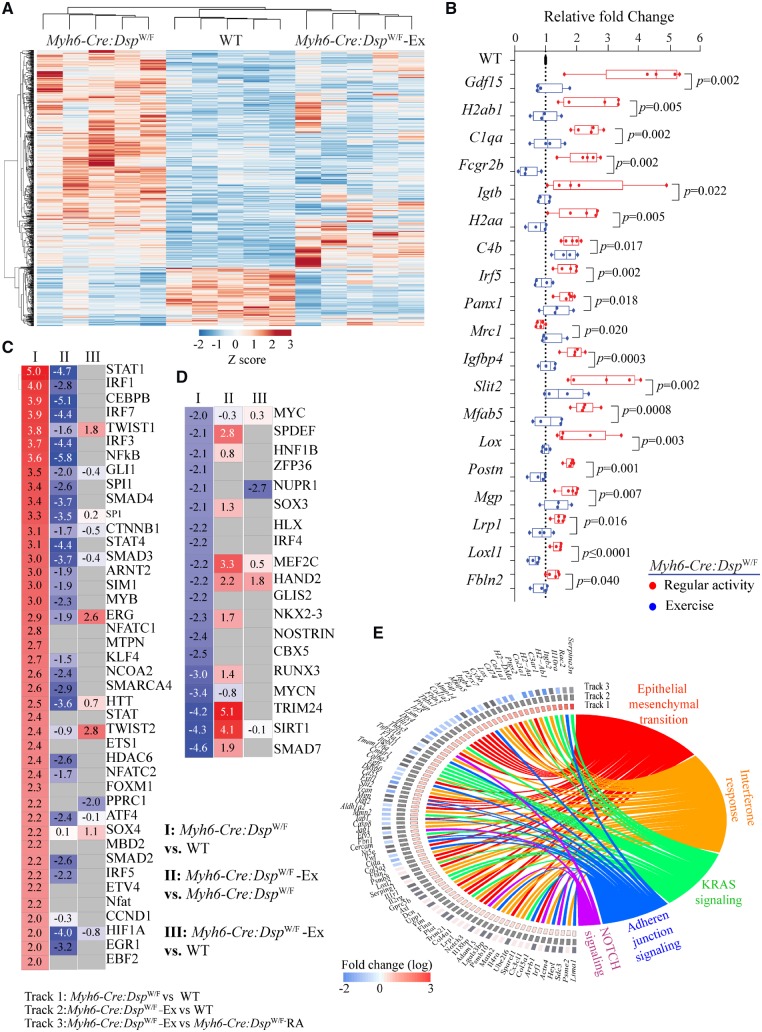
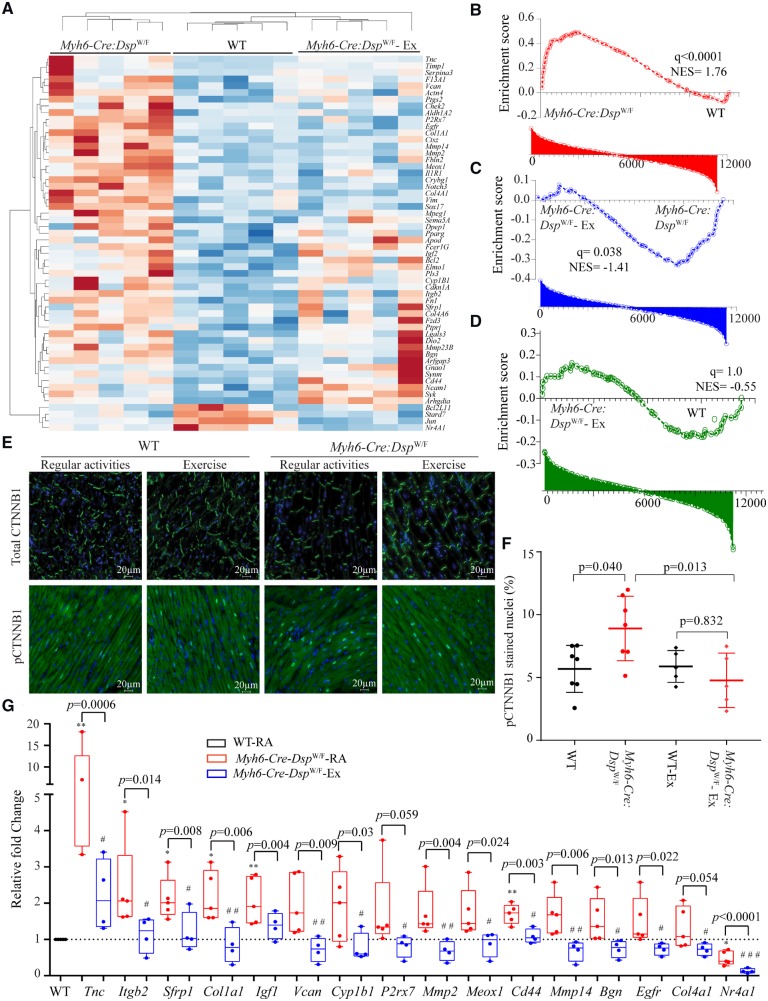
To compare global effects of exercise on cardiac myocyte transcripts, transcripts of WT, and Myh6-Cre-DspW/F myocytes in the regular activity and exercise groups were analysed by clustering. Exercise shifted the transcriptome profile of the Myh6-Cre-DspW/F cardiac myocytes towards that of the WT myocytes. Treadmill exercise rescued, defined as not being statistically different from the WT, the majority (492/781) of the DEGs in the Myh6-Cre-DspW/F myocytes (Figure 3A). One Myh6-Cre-DspW/F mouse that stopped exercising after 2.5 months due to severe cardiac dysfunction exhibited a transcriptome profile that was distinct from the WT myocytes (Figure 3A). This mouse had the signature of the molecular markers of heart failure, evidenced by increased Nppa, Nppb, Actc1, Myh7, and decreased Atp2a2 transcript levels, as would be expected. The other four Myh6-Cre-DspW/F mice in the exercise group had transcriptome profiles practically similar to those of the WT myocytes (Figure 3A). EMT and inflammation, which were among the top dysregulated pathways in 6 months old Myh6-Cre-DspW/F were rescued (partially for EMT) upon treadmill exercise, a finding which was also validated by qPCR of selected genes in these pathways (Figure 3B, Supplementary material online, Table S7). The predicted transcriptomic regulators also showed a near complete rescue in the Myh6-Cre-DspW/F cardiac myocytes in the exercise group with the notable exception of NUPR1, HAND2, and TWIST1, which remained significantly dysregulated in the Myh6-Cre-DspW/F myocytes (Figure 3C and D). The list of all dysregulated canonical upstream regulators of gene transcription along with rescue or the lack thereof upon treadmill exercise are shown in Figure 3F. A similar list that include all possible pairwise comparisons among the experimental groups is provided in Supplementary material online, Figure S9.
Figure 3.
Rescue of cardiac myocyte transcriptome, TFs and biological pathways in Myh6-Cre:DspW/F mice in the exercise group. (A) Heat map of DEGs in Myh6-Cre:DspW/F mice assigned to the regular activity, WT mice, and Myh6-Cre:DspW/F mice assigned to treadmill exercise are shown. The map shows clustering of the transcripts in Myh6-Cre:DspW/F myocytes in the exercise group with those in the WT myocytes. (B) Transcript levels of selected DEGs corresponding to panel A, as determined by qPCR (one way ANOVA P-values are shown). The panel shows normalization of the transcript levels, similar to levels in the WT, in the Myh6-Cre:DspW/F in the treadmill exercise group. (C and D) Heat maps and Z scores showing rescue and normalization of all activated (C) and suppressed (D) TFs in Myh6-Cre:DspW/F myocytes upon treadmill exercise. Pairwise comparisons are shown; I: Myh6-Cre:DspW/F regular activity (RA) vs. WT; II: Myh6-Cre:DspW/F—treadmill exercise (Ex) vs. Myh6-Cre:DspW/F—RA; III: Myh6-Cre:DspW/F—Ex vs. WT. (E) Circos map showing rescue of DEGs and their corresponding enriched Hallmark pathways.
To identify exercise-induced genotype-specific changes in cardiac myocyte transcripts, DEGs in the WT and Myh6-Cre-DspW/F myocytes were compared. Accordingly, 447 exercise-induced up-regulated genes were common to both genotype, 693 genes were specific to Myh6-Cre-DspW/F, and 421 to the WT myocytes (Supplementary material online, Figure S10). The corresponding numbers for exercise-induced down-regulated genes were 658, 733, and 867, respectively (Supplementary material online, Figure S10). Exercise-induced DEGs predicted activation of TRIM24, SIRT1, and TAF4 transcription factors and suppression of STAT1 and IRF3 transcriptional regulator in the Myh6-Cre-DspW/F myocytes (Supplementary material online, Figure S10). In the WT myocytes, ERG was predicted to be up-regulated, whereas SREBF1 and IKZF1 were predicted as the suppressed transcription factors upon exercise. The corresponding predicted Hallmark biological pathways were notable for increased oxidative phosphorylation in Myh6-Cre-DspW/F myocytes in the exercise group exclusively and suppression of inflammation (IFNG and NFKB) upon exercise both in the Myh6-Cre-DspW/F and WT myocytes (Supplementary material online, Figure S10).
Pertinent to the pathways previously implicated in ACM, treadmill exercise reversed transcript levels of the majority of the canonical WNT signalling target genes, as indicated in the heat map and GSEA plots shown in Figure 4A–D.10 In accord with normalization of the transcript levels, the number of cells expressing phospho β-catenin, the co-effector of the canonical WNT pathway, was decreased significantly in the exercise group (Figure 4E and F). Consistent with the RNA-Seq and immunofluorescence data, exercise normalized transcript levels of majority of the canonical WNT target genes in Myh6-Cre-DspW/F myocytes, as determined by qPCR (Figure 4G, Supplementary material online, Table S8).
Figure 4.
Rescue of the canonical WNT pathway in Myh6-Cre:DspW/F mice upon treadmill exercise. (A) Heat map of selected known canonical WNT target gene transcripts in Myh6-Cre:DspW/F myocytes and their clustering with the transcripts in WT myocytes with exercise. (B–D) Pairwise gene set enrichment analysis plots and the corresponding q values, showing enrichment of transcript levels of the canonical WNT pathway target genes and the effects of exercise. (B) Myh6-Cre:DspW/F vs. WT myocytes; (C) Myh6-Cre:DspW/F—regular activity vs. Myh6-Cre:DspW/F—exercise; (D) Myh6-Cre:DspW/F—exercise vs. WT myocytes. (E) Representative immunofluorescence panels showing expression and subcellular localization of total and phospho-β catenin (Ser33/37/Thr41) in the experimental groups (N = 7 in WT and Myh6-Cre:DspW/F regular activity groups and N = 5 mice for WT and Myh6-Cre:DspW/F - exercise groups). (F) Quantitative data showing number of nuclei expressing phospho-β catenin. (G) Quantitative PCR depicting transcript levels of selected canonical WNT pathway target genes in myocytes isolated from WT (N = 5, set at 1 for normalization), Myh6-Cre:DspW/F—regular activity (N = 5), and Myh6-Cre:DspW/F—exercise groups (N = 4). The P-values were calculated by ANOVA. *P < 0.05, **P < 0.01 for pairwise comparison between WT and Myh6-Cre:DspW/F—regular activity groups, and #P < 0.05 and ##P < 0.01, respectively, between Myh6-Cre:DspW/F mice in the regular activity and exercise groups.
Notable among the DEGs in the Myh6-Cre-DspW/F myocytes was over-representation of genes whose protein products are known to be secreted (secretome). Accordingly, genes coding for secreted proteins comprised about 17.2% (135/782) of the DEGs, whereas genes encoding secretome comprised only 7.5% (893/11, 904) of the genes that were analysed by RNA-Seq (Figure 5A–D). In accord with the reversal of dysregulated transcripts with exercise in the Myh6-Cre-DspW/F myocytes, exercise rescued dysregulated secretome, as transcript levels of 118/135 (87%) of DEGs coding for secreted proteins were normalized (Figure 5A–D). Up-regulated genes encoding secretome were predicted to be targets of IL6, CTNNB1, TGFB1, and ERK, among others, whereas down-regulated genes were predicted to be targets of alpha-catenin, FAS, AHR, among others (Figure 5E). Consistent with the data presented in Figure 3, the rescued biological pathways pertaining to secretome included EMT and inflammation (Figure 5F).
Figure 5.
Normalization of transcript levels of genes encoding secreted proteins (secretome). (A) Bar graph illustrates the number of genes encoding secreted proteins in the experimental groups. (B) Heat map of secretome transcripts showing clustering of the transcripts in the Myh6-Cre:DspW/F—exercise with the WT myocytes, as opposed to Myh6-Cre:DspW/F—regular activity myocytes. (C and D) Gene set enrichment analysis plots showing enrichment of the secretome transcripts in the Myh6-Cre:DspW/F myocytes in the regular activity group and its reversal in the exercise group. (E) Predicted activated and suppressed TFs corresponding to dysregulated secretome, based on DEGs with q < 0.05 and Z score of >2. (F) Circos map of differentially expressed secretome (q < 0.05) and corresponding biological pathways in Myh6-Cre:DspW/F myocytes. Pathways were analysed by GSEA for hallmark signature and those with q < 0.05 are presented.
Finally, transcriptomes were analysed for genotype-by-exercise interactions. Only transcript levels of 65 genes were affected by the interactions (q < 0.05), which included 19 up-regulated and 46 down-regulated genes (Supplementary material online, Figure S9). The dysregulated genes were members of metabolic pathways, cytokine production, and cell–cell communication among others (Supplementary material online, Figure S11). However, the number of genes whose transcript levels were influenced by genotype-by-treatment interaction was too small to make firm conclusions.
3.5 Effects of exercise on cardiac function
All but one mice in the Myh6-Cre:DspW/F group completed the 3-month long daily exercise protocol. One Myh6-Cre:DspW/F mouse stopped exercising 2.5 months after the start of exercise protocol. Prior to exercise the mouse had a left ventricular end-diastolic diameter (LVEDD) of 3.5 mm, LVESD of 2.4 mm, and LVFS of 27%. After 2.5 months of exercise, the corresponding numbers were 4.4 mm, 3.6 mm, and 17%, indicating cardiac dilatation and dysfunction.
To determine effects of treadmill exercise on cardiac size and function, echocardiographic indices were compared in 6 months old Myh6-Cre:DspW/F mice assigned to the routine activity or treadmill exercise groups. As shown in Table 1, treadmill exercise was associated with a modest increase in left ventricular mass, LVFS, and systolic septal thickness in the WT mice. It was associated with a slower heart rate and eccentric cardiac hypertrophy in the Myh6-Cre:DspW/F mice, the latter was indicated by an increased LVEDD (indexed to body weight) and increased left ventricular mass. However, treadmill exercise did not lead to further deterioration of cardiac systolic function, as left ventricular fractional shortening and ejection fraction were similar between Myh6-Cre:DspW/F mice in the routine activity and treadmill exercise groups (Table 1). Effects of treadmill exercise on genotype-dependent evolution of cardiac phenotype in the WT and Myh6-Cre:DspW/F mice were also analysed (exercise-by-genotype interaction analysis). Genotype has the most prominent effects on indices of cardiac size and function, whereas exercise had more pronounced effect on left ventricular mass indexed to the body weight. There were modest genotype-by-exercise interactions for heart rate and LVFS (Table 1).
3.6 Effects of exercise on electrophysiological parameters
Surface electrocardiogram and cardiac rhythm monitoring for an approximately 1 h did not show discernible differences between WT and Myh6-Cre:DspW/F mice prior to exercise. Cardiac rhythm was not monitored during treadmill exercise. Electrocardiographic and electrophysiological indices of cardiac conduction or susceptibility to VT, performed upon completion of 3 months of treadmill exercise, were not significantly different between the WT and Myh6-Cre:DspW/F mice at the baseline or after infusion of isoproterenol or a combination of isoproterenol and caffeine (N = 7 per group, Supplementary material online, Table S10).
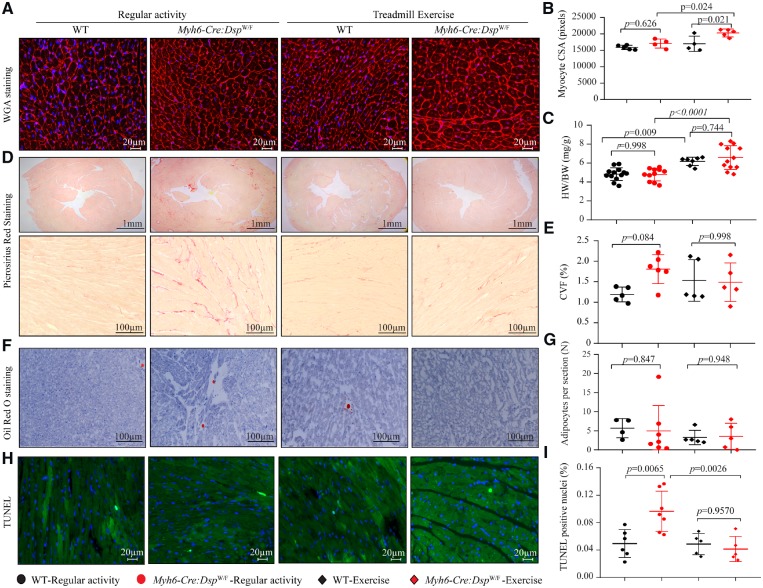
3.7 Effects of exercise on myocyte size, myocardial fibrosis, adipocytes, and apoptosis
To determine effects of treadmill exercise on histological phenotypes, thin myocardial sections (5 micron) were analysed for fibrosis, adipocyte accumulation, apoptosis, and myocyte size in the WT and Myh6-Cre:DspW/F mice subjected to 3 months of treadmill exercise (N = 4–7 mice per group). Treadmill exercise was associated with increased myocyte cross-sectional area in Myh6-Cre:DspW/F mice (Figure 6A and B) as well as increased heart weight/body weight ratio in the WT and Myh6-Cre:DspW/F mice (Figure 6C). However, there were no differences in the CVF among mice in the exercise and normal activity groups (Figure 6D and E). Likewise, the mean number of adipocytes in myocardial sections was unchanged (Figure 6F and G). In contrast, exercise was associated with a significant reduction in the number of TUNEL positive cells in the myocardium, a marker for apoptosis (Figure 6H and I). Immunofluorescence staining of thin myocardial sections and immunoblotting for the expression of selected ID proteins showed no distinct changes on localization of the ID proteins in either genotype in the normal activity and exercise groups (Supplementary material online, Figure S12).
Figure 6.
Effects of exercise on morphometric and histological phenotypes. (A) Wheat germ agglutinin (WGA) and DAPI co-stained thin myocardial sections. (B) Mean myocyte cross sectional area (N = 4–5 per group). (C) Heart weight/body weight ratio (N = 7–13 per group). (D) Low (upper) and high (lower) magnification fields of picrosirius red stained myocardial sections (N = 5–6 per group). (E) Quantitative data of collagen volume fraction (CVF, N = 5–6 per group). (F) Oil red O stained thin myocardial section (N = 4–7 per group). (G) Mean number of adipocytes per thin myocardial section (N = 4–7 per group). (H) Myocardial sections stained with TUNEL assay (N = 4–5 per group). (I) Quantitative data showing the percent of TUNEL positive cells in the myocardium (N = 4–5 per group). The P-values were calculated by ANOVA followed by Bonferroni pairwise comparison test.
4. Discussion
Conditional heterozygous deletion of Dsp specifically in mouse cardiac myocytes led to dysregulation of expression of about 800 genes, predicting activation of inflammation and EMT, and suppression of oxidative phosphorylation in cardiac myocytes. Treadmill exercise partially restored dysregulated gene expression, particularly those involved in inflammation, EMT, and oxidative phosphorylation. The anti-inflammatory effects of exercise on gene expression was evident in the WT as well as in the Myh6-Cre:DspW/F myocytes. Likewise, the effects of treadmill exercise on the transcript levels of genes in the EMT pathway pertained to WT as well as Myh6-Cre:DspW/F mice. Treadmill exercise also rescued transcript levels of the majority of the dysregulated genes encoding secreted proteins, including inhibitors of the canonical WNT pathway SFRP1, SFRP3, APOE, and LRP1, which are implicated in the pathogenesis of ACM. The inhibitors, upon secretion, would be expected to target myocyte (autocrine effects) as well as the non-myocyte cells (paracrine effects) in the myocardium as well as serve as potential biomarkers in ACM. Overall, the findings highlight activation of inflammation and EMT and suppression of oxidative phosphorylation in Myh6-Cre:DspW/F cardiac myocytes and their restoration by treadmill exercise, predicting salutary effects of exercise in a subset of phenotypes in ACM.
The study was designed with the primary focus on determining the effects of Dsp gene haplo-insufficiency and treadmill exercise on cardiac myocyte transcriptome. Conditional deletion of one copy of the Dsp gene specifically in cardiac myocytes led to dysregulation of expression of about 800 genes, which pertained to those involved in inflammation and oxidative phosphorylation. Treadmill exercise was associated with normalization of approximately two-third of the dysregulated transcripts in Myh6-Cre:DspW/F cardiac myocytes, including those in the inflammatory pathway and oxidative phosphorylation. However, despite the remarkable effect, the rescue was incomplete and genes which were targets of several important transcriptional regulators, such as SIRT1, NUPR1 and TWIST1, remained dysregulated. Treadmill exercise induced significant eccentric cardiac hypertrophy, as indicated by increased left ventricular mass and end-diastolic diameter, but did not lead to further deterioration of cardiac systolic function. Nevertheless, in accord with the incomplete transcriptomic rescue, cardiac systolic function remained suppressed in the Myh6-Cre:DspW/F mice assigned to treadmill exercise. Likewise, CVF, a measure of myocardial fibrosis, was not rescued, despite rescue of transcript levels of genes involved in EMT, including collagen genes, the findings which were also verified by qPCR. The latter disparity might reflect the higher sensitivity of the transcript analysis as opposed to the histological detection of fibrosis. Apoptosis, as determined by TUNEL assay, was rescued in the treadmill exercise group but its rescue was not associated with an improved cardiac function, likely reflective of the small number of cells undergoing apoptosis in the myocardium in the Myh6-Cre:DspW/F mice. Finally, treadmill exercise had no discernible effects on electrophysiological parameters, albeit the Myh6-Cre:DspW/F mice subjected to daily treadmill exercise were somewhat resistant to induction of ventricular arrhythmias. Likewise, no cardiac arrhythmia was detected during monitoring of cardiac rhythm for about an hour. Cardiac rhythm, however, was not monitored during exercise. Therefore, possible occurrence of cardiac arrhythmia during exercise cannot be excluded. The Myh6-Cre:DspW/F mice upon completion of the exercise protocol were 6 months old, which by design is in accord with the phenotypic expression of ventricular arrhythmias in young human patients. Notably, ventricular arrhythmias are typically the first manifestation of ACM in humans.1,14 Likewise, exercise-induced ventricular arrhythmias are typically encountered in young individuals with ACM.14 One may surmise that the low prevalence of ventricular arrhythmias in the Myh6-Cre:DspW/F mice reflects the inherent shortcoming of this particular myocyte-specific deletion of Dsp model in properly recapitulating the human phenotype of early ventricular arrhythmias. The present study does not address effects of treadmill exercise on cardiac arrhythmias in older mice.
The disparity in the transcriptomic rescue and failure to rescue fibrosis or cardiac function might also reflect the study design, which was devised to identify early cardiac myocyte transcriptomic changes during the course of evolution of the phenotype in the Myh6-Cre:DspW/F mice as opposed to studies in mice with a fully manifested phenotype. In addition, the study was performed in mice with conditional heterozygous deletion of the Dsp gene specifically in cardiac myocytes, which led to 31% reduction on Dsp transcript level, as opposed to the expected 50%, likely reflective of incomplete cre-mediated deletion as well as allelic compensation. As would be expected, the reduction is beyond the sensitivity of the western blot for detecting reduced DSP protein levels. While the approach enabled determination of cell-type specific contributions to the ACM phenotype, it only pertains to the human genotype of haplo-insufficiency and not missense mutations, which comprise the majority of the mutations in humans with ACM. In accord with a mild reduction in Dsp transcript levels, the phenotype in the Myh6-Cre:DspW/F mice evolves slowly and is relatively mild at 3 to 6 months of age, as evidenced by a modest increase in CVF and apoptosis, mild cardiac dysfunction, and absence of excess myocardial adipocytes. Likewise, early cardiac arrhythmias were absent in mice with myocyte-specific deletion of Dsp gene, as opposed to the phenotype in humans and deletion of Dsp gene in the cardiac conduction system.7 The observed phenotype in the young Myh6-Cre:DspW/F mice is in keeping with the previous data showing age-dependent penetrance of the Dsp-haploinsufficiency, predominantly presenting between 6 months to 2 years of age, including cardiac arrhythmias occurring in the context of left ventricular dysfunction.10
A number of studies have reported beneficial effects of exercise in humans as well as in various models of cardiac injury, particularly post-myocardial infarction but not in ACM models.19–23 The exercise protocol employed in the present study provided a gradual increase in the workload delivering a total of 5.5 kJ work per day for a 30 g mouse. The protocol, as compared to other exercise protocols used in mice, was more intense, however, it might not sufficiently emulate exercise physiology in human patients with ACM.20,21,23 The beneficial effects of treadmill exercise on cardiac transcripts are in accord with the existing literature on the beneficial effects of exercise on cardiac remodelling and regeneration.20–23 The reversal of the majority of the dysregulated gene in Dsp heterozygous myocytes, however, did not translate into beneficial effects on cardiac dilatation, dysfunction, or arrhythmias in the Myh6-Cre:DspW/F mice. The absence of beneficial effects of treadmill exercise in the present study might reflect model-specific effects, i.e. the lack of benefit or potential deleterious effects in ACM caused by defective desmosomes, as opposed to post-myocardial infarction, wherein the desmosomes are largely intact. The effects of treadmill exercise on adverse cardiac remodelling was evident in one Myh6-Cre:DspW/F mouse with a borderline cardiac function. Whether early implementation of an exercise programme prior to any discernible cardiac dysfunction could delay the onset or attenuate the phenotype is an untested hypothesis. Overall, the study design was best suited for serial and comparative analysis of cardiac myocyte gene expression, as opposed to detection of clinical phenotype, such as cardiac dysfunction, arrhythmias or death.
The findings of the present study demonstrate salutary effects of treadmill exercise on cardiac myocyte transcriptome. Nevertheless, despite the reversal of the majority of the dysregulated transcripts, the findings should not be interpreted to advocate to discard potential deleterious effects of heavy physical exercise on the risk of heart failure and cardiac arrhythmias in human patients with ACM.13–16,26,44 The findings provoke further interest in exploring potential beneficial effects of early and long-term supervised treadmill exercise in human patients with ACM.
Authors’ contributions
S.M.C. performed the molecular biology experiments, including isolation of myocytes, extraction of RNA, RNA-Sequencing, and qPCR. She also assisted in data analysis and preparation of the manuscript. J.H. supervised the exercise protocol and performed morphometric analysis of the histological data, including apoptosis, phospho-beta catenin nuclear localization, and myocyte cross-sectional area. S.F. performed immunofluorescence and immunoblotting experiments pertaining to desmosome proteins and the canonical WNT signalling. P.Y. performed histological evaluation of the myocardial phenotype, including quantification of fibrosis. J.K. supervised the exercise protocol. She also supervised the mouse colony. G.C. supervised exercise protocol and assisted in maintaining the mouse colony. M.J.R. performed bioinformatics analysis of the RNA-Seq data. C.C. supervised bioinformatics analysis of the RNA-Seq data and assisted with interpretation of the findings K.H. reviewed the manuscript and made critical revisions. Y.Y. reviewed the manuscript and made critical revisions. H.C.M. acquired data and gave experimental design input for the experiments summarized in Supplementary material online,Table S5. She also drafted portions of the methods section relevant to these experiments. X.W. supervised electrophysiological studies, interpreted the findings, and make revisions. P.G. assisted with data analysis and manuscript preparation and made critical revisions. A.J.M. developed the concept and designed the experiments and wrote the manuscript.
Conflict of interest: none declared.
Supplementary Material
Funding
This work was supported in part by the National Institutes of Health (NHLBI R01 HL088498 and 1R01HL132401 and 1S10OD018135-01A1), Leducq Foundation (14 CVD 03), the Ewing Halsell Foundation, and George and Mary Josephine Hamman Foundation. P.Y. was supported by the National Key R&D Program of China (2017YFC1307804) and the China Scholarship Council/CSC. J.H. was supported by the Department of Cardiovascular Medicine, the Second Affiliated Hospital of Nanchang University, Nanchang of Jiangxi, China. S.F. was sponsored by the National Nature Science Foundation of China (81570309) and the China Scholarship Council/CSC. X.W. was sponsored by National Institutes of Health (NHLBI grants HL089598, HL091947, HL117641 and HL147108).
Time for primary review: 22 days
Translational perspective
Patients with arrhythmogenic cardiomyopathy (ACM) are advised to avoid physical exercise, because of potential adverse outcomes. To gain insights into the molecular mechanisms, we performed a randomized study testing effects of treadmill exercise on cardiac phenotype and myocyte gene expression in a mouse model of ACM. Genes involved in epithelial-mesenchymal transition, inflammation, metabolism, and WNT pathways were most dysregulated in cardiac myocytes isolated from the ACM mice. Exercise restored two-third of the ∼800 differentially expressed genes, rescued the main dysregulated pathways, and induced eccentric cardiac hypertrophy without affecting cardiac function. The findings suggest treadmill exercise might have partial salutary phenotypic effects in ACM.
References
- 1. Corrado D, Basso C, Judge DP.. Arrhythmogenic cardiomyopathy. Circ Res 2017;121:784–802. [DOI] [PubMed] [Google Scholar]
- 2. Gandjbakhch E, Redheuil A, Pousset F, Charron P, Frank R.. Clinical diagnosis, imaging, and genetics of arrhythmogenic right ventricular cardiomyopathy/dysplasia: JACC State-of-the-Art review. J Am Coll Cardiol 2018;72:784–804. [DOI] [PubMed] [Google Scholar]
- 3. Finocchiaro G, Papadakis M, Robertus JL, Dhutia H, Steriotis AK, Tome M, Mellor G, Merghani A, Malhotra A, Behr E, Sharma S, Sheppard MN.. Etiology of sudden death in sports: insights from a United Kingdom Regional Registry. J Am Coll Cardiol 2016;67:2108–2115. [DOI] [PubMed] [Google Scholar]
- 4. Thiene G, Nava A, Corrado D, Rossi L, Pennelli N.. Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med 1988;318:129–133. [DOI] [PubMed] [Google Scholar]
- 5. Corrado D, Basso C, Thiene G, McKenna WJ, Davies MJ, Fontaliran F, Nava A, Silvestri F, Blomstrom-Lundqvist C, Wlodarska EK, Fontaine G, Camerini F.. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: a multicenter study. J Am Coll Cardiol 1997;30:1512–1520. [DOI] [PubMed] [Google Scholar]
- 6. Hoorntje ET, Te Rijdt WP, James CA, Pilichou K, Basso C, Judge DP, Bezzina CR, van Tintelen JP.. Arrhythmogenic cardiomyopathy: pathology, genetics, and concepts in pathogenesis. Cardiovasc Res 2017;113:1521–1531. [DOI] [PubMed] [Google Scholar]
- 7. Karmouch J, Zhou QQ, Miyake CY, Lombardi R, Kretzschmar K, Bannier-Helaouet M, Clevers H, Wehrens XHT, Willerson JT, Marian AJ.. Distinct cellular basis for early cardiac arrhythmias, the cardinal manifestation of arrhythmogenic cardiomyopathy, and the skin phenotype of cardiocutaneous syndromes. Circ Res 2017;121:1346–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lombardi R, Chen SN, Ruggiero A, Gurha P, Czernuszewicz GZ, Willerson JT, Marian AJ.. Cardiac fibro-adipocyte progenitors express desmosome proteins and preferentially differentiate to adipocytes upon deletion of the desmoplakin gene. Circ Res 2016;119:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broussard JA, Getsios S, Green KJ.. Desmosome regulation and signaling in disease. Cell Tissue Res 2015;360:501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ.. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest 2006;116:2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ.. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ Res 2014;114:454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karaman R, Halder G.. Cell junctions in hippo signaling. Cold Spring Harb Perspect Biol 2018;10:a028753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. James CA, Bhonsale A, Tichnell C, Murray B, Russell SD, Tandri H, Tedford RJ, Judge DP, Calkins H.. Exercise increases age-related penetrance and arrhythmic risk in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. J Am Coll Cardiol 2013;62:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sawant AC, Calkins H.. Relationship between arrhythmogenic right ventricular dysplasia and exercise. Card Electrophysiol Clin 2015;7:195–206. [DOI] [PubMed] [Google Scholar]
- 15. Sawant AC, Bhonsale A, Te Riele AS, Tichnell C, Murray B, Russell SD, Tandri H, Tedford RJ, Judge DP, Calkins H, James CA.. Exercise has a disproportionate role in the pathogenesis of arrhythmogenic right ventricular dysplasia/cardiomyopathy in patients without desmosomal mutations. J Am Heart Assoc 2014;3:e001471.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perrin MJ, Angaran P, Laksman Z, Zhang H, Porepa LF, Rutberg J, James C, Krahn AD, Judge DP, Calkins H, Gollob MH.. Exercise testing in asymptomatic gene carriers exposes a latent electrical substrate of arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol 2013;62:1772–1779. [DOI] [PubMed] [Google Scholar]
- 17. Corrado D, Wichter T, Link MS, Hauer RN, Marchlinski FE, Anastasakis A, Bauce B, Basso C, Brunckhorst C, Tsatsopoulou A, Tandri H, Paul M, Schmied C, Pelliccia A, Duru F, Protonotarios N, Estes NM 3rd, McKenna WJ, Thiene G, Marcus FI, Calkins H.. Treatment of arrhythmogenic right ventricular cardiomyopathy/dysplasia: an international task force consensus statement. Circulation 2015;132:441–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maron BJ, Udelson JE, Bonow RO, Nishimura RA, Ackerman MJ, Estes NA 3rd, Cooper LT Jr, Link MS, Maron MS; American Heart Association Electrocardiography and Arrhythmias Committee of Council on Clinical Cardiology, Council on Cardiovascular Disease in Young, Council on Cardiovascular and Stroke Nursing, Council on Functional Genomics and Translational Biology, and American College of Cardiology. Eligibility and disqualification recommendations for competitive athletes with cardiovascular abnormalities: task force 3: hypertrophic cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy and other cardiomyopathies, and myocarditis: a scientific statement from the American Heart Association and American College of Cardiology. Circulation 2015;132:e273–e280. [DOI] [PubMed] [Google Scholar]
- 19. Anderson L, Oldridge N, Thompson DR, Zwisler AD, Rees K, Martin N, Taylor RS.. Exercise-based cardiac rehabilitation for coronary heart disease: cochrane systematic review and meta-analysis. J Am Coll Cardiol 2016;67:1–12. [DOI] [PubMed] [Google Scholar]
- 20. de Waard MC, van der Velden J, Bito V, Ozdemir S, Biesmans L, Boontje NM, Dekkers DH, Schoonderwoerd K, Schuurbiers HC, de Crom R, Stienen GJ, Sipido KR, Lamers JM, Duncker DJ.. Early exercise training normalizes myofilament function and attenuates left ventricular pump dysfunction in mice with a large myocardial infarction. Circ Res 2007;100:1079–1088. [DOI] [PubMed] [Google Scholar]
- 21. Qin R, Murakoshi N, Xu D, Tajiri K, Feng D, Stujanna EN, Yonebayashi S, Nakagawa Y, Shimano H, Nogami A, Koike A, Aonuma K, Ieda M.. Exercise training reduces ventricular arrhythmias through restoring calcium handling and sympathetic tone in myocardial infarction mice. Physiol Rep 2019;7:e13972.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonilla IM, Belevych AE, Sridhar A, Nishijima Y, Ho HT, He Q, Kukielka M, Terentyev D, Terentyeva R, Liu B, Long VP, Gyorke S, Carnes CA, Billman GE.. Endurance exercise training normalizes repolarization and calcium-handling abnormalities, preventing ventricular fibrillation in a model of sudden cardiac death. J Appl Physiol (1985) 2012;113:1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vujic A, Lerchenmuller C, Wu TD, Guillermier C, Rabolli CP, Gonzalez E, Senyo SE, Liu X, Guerquin-Kern JL, Steinhauser ML, Lee RT, Rosenzweig A.. Exercise induces new cardiomyocyte generation in the adult mammalian heart. Nat Commun 2018;9:1659.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirchhof P, Fabritz L, Zwiener M, Witt H, SchäFers M, Zellerhoff S, Paul M, Athai T, Hiller KH, Baba HA, Breithardt G, Ruiz P, Wichter T, Levkau B.. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation 2006;114:1799–1806. [DOI] [PubMed] [Google Scholar]
- 25. Fabritz L, Hoogendijk MG, Scicluna BP, van Amersfoorth SC, Fortmueller L, Wolf S, Laakmann S, Kreienkamp N, Piccini I, Breithardt G, Noppinger PR, Witt H, Ebnet K, Wichter T, Levkau B, Franke WW, Pieperhoff S, de Bakker JM, Coronel R, Kirchhof P.. Load-reducing therapy prevents development of arrhythmogenic right ventricular cardiomyopathy in plakoglobin-deficient mice. J Am Coll Cardiol 2011;57:740–750. [DOI] [PubMed] [Google Scholar]
- 26. La Gerche A, Rakhit DJ, Claessen G.. Exercise and the right ventricle: a potential Achilles' heel. Cardiovasc Res 2017;113:1499–1508. [DOI] [PubMed] [Google Scholar]
- 27. O'Connell TD, Rodrigo MC, Simpson PC.. Isolation and culture of adult mouse cardiac myocytes. Methods Mol Biol 2007;357:271–296. [DOI] [PubMed] [Google Scholar]
- 28. Auguste G, Gurha P, Lombardi R, Coarfa C, Willerson JT, Marian AJ.. Suppression of activated FOXO transcription factors in the heart prolongs survival in a mouse model of laminopathies. Circ Res 2018;122:678.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Risso D, Ngai J, Speed TP, Dudoit S.. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hochberg Y, Benjamini Y.. More powerful procedures for multiple significance testing. Stat Med 1990;9:811–818. [DOI] [PubMed] [Google Scholar]
- 33. Gurha P, Chen X, Lombardi R, Willerson JT, Marian AJ.. Knockdown of plakophilin 2 downregulates miR-184 through CpG hypermethylation and suppression of the E2F1 pathway and leads to enhanced adipogenesis in vitro. Circ Res 2016;119:731–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen SN, Lombardi R, Karmouch J, Tsai JY, Czernuszewicz GZ, Taylor M, Mestroni L, Coarfa C, Gurha P, Marian AJ.. DNA damage response/TP53 pathway is activated and contributes to the pathogenesis of dilated cardiomyopathy associated with lamin A/C mutations. Circ Res 2019;124:856.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marreddy Cheedipudi S, Matkovich SJ, Coarfa C, Hu X, Robertson MJ, Sweet ME, Taylor M, Mestroni L, Cleveland JC, Willerson JT, Gurha P, Marian AJ.. Genomic reorganization of lamin-associated domains in cardiac myocytes is associated with differential gene expression and DNA methylation in human dilated cardiomyopathy. Circ Res 2019;124:1198–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ruggiero A, Chen SN, Lombardi R, Rodriguez G, Marian AJ.. Pathogenesis of hypertrophic cardiomyopathy caused by myozenin 2 mutations is independent of calcineurin activity. Cardiovasc Res 2013;97:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Oort RJ, McCauley MD, Dixit SS, Pereira L, Yang Y, Respress JL, Wang Q, De Almeida AC, Skapura DG, Anderson ME, Bers DM, Wehrens XH.. Ryanodine receptor phosphorylation by calcium/calmodulin-dependent protein kinase II promotes life-threatening ventricular arrhythmias in mice with heart failure. Circulation 2010;122:2669–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lombardi R, da Graca Cabreira-Hansen M, Bell A, Fromm RR, Willerson JT, Marian AJ.. Nuclear plakoglobin is essential for differentiation of cardiac progenitor cells to adipocytes in arrhythmogenic right ventricular cardiomyopathy. Circ Res 2011;109:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen SN, Czernuszewicz G, Tan Y, Lombardi R, Jin J, Willerson JT, Marian AJ.. Human molecular genetic and functional studies identify TRIM63, encoding Muscle RING Finger Protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ Res 2012;111:907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bergmann O, Zdunek S, Alkass K, Druid H, Bernard S, Frisen J.. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res 2011;317:188–194. [DOI] [PubMed] [Google Scholar]
- 41. Bergmann O, Jovinge S.. Isolation of cardiomyocyte nuclei from post-mortem tissue. J Vis Exp 2012;65:e4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grimmond SM, Miranda KC, Yuan Z, Davis MJ, Hume DA, Yagi K, Tominaga N, Bono H, Hayashizaki Y, Okazaki Y, Teasdale RD, Group RG, Members GSL.. The mouse secretome: functional classification of the proteins secreted into the extracellular environment. Genome Res 2003;13:1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meinken J, Walker G, Cooper CR, Min XJ.. MetazSecKB: the human and animal secretome and subcellular proteome knowledgebase. Database (Oxford) 2015;2015:bav077.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martherus R, Jain R, Takagi K, Mendsaikhan U, Turdi S, Osinska H, James JF, Kramer K, Purevjav E, Towbin JA.. Accelerated cardiac remodeling in desmoplakin transgenic mice in response to endurance exercise is associated with perturbed Wnt/beta-catenin signaling. Am J Physiol Heart Circ Physiol 2016;310:H174–H187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.