Abstract
Progestins are widely used for the treatment of gynecologic disorders and alone, or combined with an estrogen, are used as contraceptives. While their potencies, efficacies and side effects vary due to differences in structures, doses and routes of administration, little is known about their effects on the endometrial transcriptome in the presence or absence of estrogen. Herein, we assessed the transcriptome and pathways induced by progesterone (P4) and the three most commonly used synthetic progestins, medroxyprogesterone acetate (MPA), levonorgestrel (LNG), and norethindrone acetate (NETA), on human endometrial stromal fibroblasts (eSF), key players in endometrial physiology and reproductive success. While there were similar transcriptional responses, each progestin induced unique genes and biofunctions, consistent with their structural similarities to progesterone (P4 and MPA) or testosterone (LNG and NETA), involving cellular proliferation, migration and invasion. Addition of estradiol (E2) to each progestin influenced the number of differentially expressed genes and biofunctions in P4 and MPA, while LNG and NETA signatures were more independent of E2. Together, these data suggest different mechanisms of action for different progestins, with progestin-specific altered signatures when combined with E2. Further investigation is warranted for a personalized approach in different gynecologic disorders, for contraception, and minimizing side effects associated with their use.
Keywords: progestins, endometrial stromal fibroblasts, inflammation, angiogenesis, transcriptome
1. Introduction
Progestins, compounds with progestational activity, include naturally occurring progesterone (P4) and a variety of synthetic steroids [1,2]. They are widely used for contraception and the treatment of endometriosis, endometrial hyperplasia, and endometrial cancer, and are also used for postmenopausal hormone therapy [1,2,3,4,5,6]. Synthetic steroids that are structurally related to progesterone, testosterone, and spironolactone constitute the main classes of progestins. Progestins are key constituents of many contraceptives and either alone, or in combination with estrogens, are currently used by >660 million women globally [6,7,8]. The contraceptive effects of synthetic progestins result from a mimicry of the actions of progesterone, including inhibition of ovulation and thickening of cervical mucus [6,9]. Additionally, they can counteract estrogen-driven endometrial proliferation in endometriosis and have applications in postmenopausal hormone therapy and endometrial hyperplasia and cancer [1,3,10,11,12,13]. Recently, progestins have been implicated in the increased risk of HIV-1 acquisition, perhaps by modulating the integrity and cellular functions of the female reproductive tract and impact on immune functions, HIV-1 replication, and the vaginal microbiome [14,15]. However, “all progestins are not equal” [1], as they have different structures that alter their affinities for the progesterone nuclear receptor (PR), elicit unique intracellular signaling pathways and exhibit different potencies, metabolism, pharmacokinetics, efficacy, side effects and off-target effects [2,9]. Patient hormonal status and progestin dose, route of administration, formulation, combination with or without an estrogen, and duration of use also contribute to different effects locally within the female reproductive tract (e.g., histology, vascular integrity) [12,16], in other PR-expressing tissues, such as breast [2,6], and systemically [6,9].
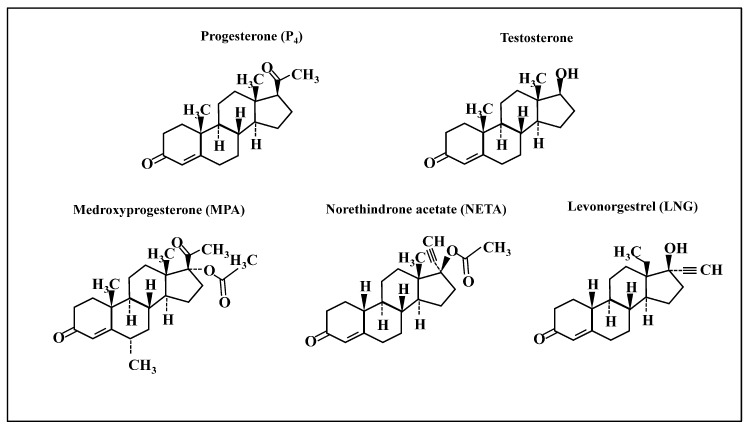
The study of transcriptional regulation within endometrial tissue and cells in response to progestins (with and without estrogens) has received limited focus [17,18,19,20], although they offer insights into understanding the molecular and functional impact of these steroids. The current study examined the transcriptome and related biological and functional pathways of human endometrial stromal fibroblasts (eSF) in vitro, in response to different progestins with and without estradiol (E2). This cell type is a central effector of endometrial physiology, homeostasis, and pathophysiology across a woman’s lifespan, including regulating endometrial cycling, receptivity to an implanting embryo, and generation of the decidua of pregnancy [21,22]. Additionally, eSF exhibit abnormalities in endometriosis and polycystic ovary syndrome, and they respond in situ during contraceptive and post-menopause hormone therapy. Thus, the study of this cell type has promise to provide insights into why some progestins have more efficacy, consequential side effects (e.g., breakthrough bleeding) [6,9] and susceptibility to HIV acquisition [15]. Given the plethora of progestins and their diverse bioactivities, herein, we focused on the most commonly formulated progestins in use today: medroxyprogesterone acetate (MPA, structurally similar to progesterone (P4)) and levonorgestrel (LNG) and norethindrone acetate (NETA)—structurally related to testosterone (Figure 1).
Figure 1.
Chemical structures of progestins used in the study. Chemical structures of progesterone, testosterone, medroxyprogesterone acetate (MPA, a progestin structurally related to progesterone), and two progestins structurally related to testosterone: norethindrone acetate (NETA) and levonorgestrel (LNG).
In the current study, we found that these progestins commonly display anti-inflammatory and pro-angiogenic profiles, altered effects on extracellular matrix integrity and exhibited distinct transcriptomic profiles depending on their subclass—i.e., structurally related to progesterone versus testosterone and within progestin sub-classes. Notably, addition of estradiol (E2) moderated some of the effects depending on the progestin, indicating that structural differences in the progestins are important in gene regulation and interactions with other steroid hormones in the endometrium.
2. Results
2.1. Progestins Structurally Related to Progesterone and Testosterone Induce Distinct Gene Expression Profiles
Gene expression profiles of eSF in response to each progestin versus vehicle control revealed that similar numbers of genes were differentially expressed in response to structurally related P4 and MPA; whereas, more than twice the number of genes were differentially expressed in response to LNG and NETA (Table 1). Moreover, LNG and NETA, structurally related to testosterone (Figure 1), affected similar numbers of differentially expressed genes (DEG) (Table 1 and Table 2). The top 30 DEG for each progestin treatment are presented in Table 2 (see Supplemental Table S1 for the full list of DEG for each progestin treatment).
Table 1.
Number of DEG in comparisons of each progestin vs. vehicle in the absence or presence of estradiol.
| Comparison | Up-Regulated DEG (FC ≥ 1.5) | Down-Regulated DEG (FC ≥ 1.5) | Total DEG | |
|---|---|---|---|---|
| Progestins without E2 | LNG vs. Vh | 249 | 304 | 553 |
| MPA vs. Vh | 122 | 129 | 251 | |
| NETA vs. Vh | 243 | 352 | 595 | |
| P4 vs. Vh | 55 | 61 | 116 | |
| E2 | E2 vs. Vh | 88 | 76 | 164 |
| Progestins + E2 | E2LNG vs. Vh | 233 | 338 | 571 |
| E2MPA vs. Vh | 259 | 350 | 609 | |
| E2NETA vs. Vh | 247 | 298 | 545 | |
| E2P4 vs. Vh | 208 | 314 | 522 |
E2: estradiol; MPA: medroxyprogesterone acetate; LNG: levonorgestrel; NETA: norethindrone acetate; FC: fold change; DEG: differentially expressed genes.
Table 2.
Select differentially expressed transcripts and genes in each progestin vs. vehicle without and with estradiol.
| P4 vs. Vh | E2P4 vs. Vh | MPA vs. Vh | E2MPA vs. Vh | LNG vs. Vh | E2LNG vs. Vh | NETA vs. Vh | E2NETA vs. Vh | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | FC | Gene Symbol | FC | Gene Symbol | FC | Gene Symbol | FC | Gene Symbol | FC | Gene Symbol | FC | Gene Symbol | FC | Gene Symbol | FC |
| SPARCL1 | 5.2 | SPARCL1 | 41.8 | SPARCL1 | 13.8 | SPARCL1 | 32.6 | SPARCL1 | 22.2 | SPARCL1 | 23.6 | SPARCL1 | 19.3 | SPARCL1 | 24.3 |
| SLC7A8 | 3.5 | SLC7A8 | 15.6 | SLC7A8 | 6.0 | FKBP5 | 11.1 | FKBP5 | 9.9 | FKBP5 | 11.2 | FKBP5 | 11.3 | FKBP5 | 11.1 |
| LCP1 | 3.4 | GREB1 | 15.0 | OMD | 5.1 | SLC7A8 | 10.4 | SLC7A8 | 9.4 | CNR1 | 8.8 | SLC7A8 | 9.3 | PARM1 | 9.1 |
| FKBP5 | 2.9 | LCP1 | 8.7 | FKBP5 | 5.0 | PARM1 | 10.2 | CNR1 | 8.3 | SLC7A8 | 8.7 | CNR1 | 8.9 | LCP1 | 8.0 |
| GPX3 | 2.9 | OMD | 7.1 | THSD7A | 3.8 | LCP1 | 9.3 | LCP1 | 7.9 | PARM1 | 8.6 | PARM1 | 7.9 | CNR1 | 7.9 |
| IL1R1 | 2.7 | FKBP5 | 7.1 | IL1R1 | 3.5 | OMD | 8.9 | PARM1 | 6.7 | LCP1 | 8.1 | LCP1 | 7.9 | SLC7A8 | 7.8 |
| OMD | 2.6 | CNR1 | 6.7 | LCP1 | 3.4 | DKK1 | 8.1 | OMD | 6.5 | OMD | 7.1 | MAOB | 7.7 | OMD | 6.7 |
| DKK1 | 2.4 | GPX3 | 6.6 | GPX3 | 3.1 | CNR1 | 8.1 | MAOB | 6.5 | MAOB | 6.8 | OMD | 7.6 | MAOB | 6.1 |
| MT-TA | 2.3 | THSD7A | 6.0 | CNR1 | 3.0 | MAOB | 6.9 | GREB1 | 5.8 | DKK1 | 6.3 | DKK1 | 6.1 | DKK1 | 5.7 |
| THSD7A | 2.2 | MAOB | 5.6 | CRISPLD2 | 2.9 | CRYAB | 5.3 | DKK1 | 5.5 | GREB1 | 5.1 | CRYAB | 5.4 | ENPEP | 5.6 |
| LPAR1 | 2.1 | DKK1 | 5.6 | LAMA2 | 2.8 | ULK4 | 5.2 | ULK4 | 4.8 | CRYAB | 5.0 | ENPEP | 4.6 | PLCL1 | 4.9 |
| SPSB1 | 2.1 | IL1R1 | 4.9 | CRYAB | 2.7 | ENPEP | 5.1 | CRYAB | 4.7 | IL1R1 | 4.7 | IL1R1 | 4.5 | ULK4 | 4.9 |
| SEMA3C | 2.0 | C10orf10 | 4.8 | LPAR1 | 2.6 | THSD7A | 5.0 | IL1R1 | 4.4 | THSD7A | 4.4 | CRISPLD2 | 4.3 | CRYAB | 4.8 |
| SERPINE1 | 2.0 | ABHD5 | 4.7 | MAOB | 2.6 | PLCL1 | 4.7 | PLCL1 | 4.2 | LPAR1 | 4.3 | ALDH1A3 | 4.2 | ALDH1A3 | 4.7 |
| APOD | 2.0 | IMPA2 | 4.6 | ABCC9 | 2.5 | GREB1 | 4.6 | LPAR1 | 4.1 | ALDH1A3 | 4.3 | PLCL1 | 4.2 | ITPR1 | 4.4 |
| TOX | −1.8 | HSD17B2 | −3.8 | ETV1 | −2.7 | DACH1 | −4.4 | TNFRSF19 | −4.4 | DACH1 | −4.2 | TNFRSF19 | −4.3 | CLDN11 | −4.2 |
| ARHGAP29 | −1.8 | LYPD1 | −4.1 | NDNF | −2.7 | CD34 | −4.4 | CHRM2 | −4.4 | MXRA5 | −4.5 | CD34 | −4.3 | CD34 | −4.2 |
| CXCL12 | −1.9 | NCKAP5 | −4.2 | FJX1 | −2.8 | TOX | −4.5 | CD34 | −4.4 | TGFBI | −4.5 | F2RL2 | −4.5 | TNFRSF19 | −4.4 |
| DACH1 | −2.0 | CHRM2 | −4.2 | HPSE2 | −3.0 | TGFBI | −5.1 | LYPD1 | −4.8 | TNFRSF19 | −4.6 | TOX | −4.7 | LYPD1 | −4.6 |
| GUCY1A3 | −2.0 | FGF7 | −4.4 | HSD17B2 | −3.1 | LYPD1 | −5.4 | PRELP | −5.1 | LYPD1 | −4.7 | TGFBI | −4.8 | TOX | −4.7 |
| TNFRSF19 | −2.0 | CD34 | −4.5 | NCKAP5 | −3.1 | GBP4 | −5.6 | GBP4 | −5.2 | NCKAP5 | −6.1 | FJX1 | −4.9 | GBP4 | −4.7 |
| ITGA8 | −2.2 | EGR2 | −4.8 | CHRM2 | −3.2 | CCL2 | −6.0 | TGFBI | −5.2 | PRELP | −6.1 | NDNF | −4.9 | NCKAP5 | −5.1 |
| CHRM2 | −2.3 | MXRA5 | −5.0 | GBP4 | −3.2 | NCKAP5 | −6.3 | NCKAP5 | −5.2 | ETV1 | −6.2 | NCKAP5 | −5.0 | CCL2 | −5.1 |
| ETV1 | −2.3 | NDNF | −5.0 | SFRP1 | −3.4 | PRELP | −6.3 | NDNF | −5.6 | CCL2 | −6.2 | GBP4 | −5.1 | NDNF | −5.4 |
| LYPD1 | −2.4 | TGFBI | −5.4 | CST1 | −3.4 | NDNF | −6.6 | FJX1 | −5.9 | GBP4 | −6.4 | PRELP | −5.7 | PRELP | −5.4 |
| GBP4 | −2.4 | ETV1 | −5.7 | LYPD1 | −3.5 | SFRP1 | −6.8 | CCL2 | −6.4 | NDNF | −6.5 | CCL2 | −6.2 | SFRP1 | −5.5 |
| NDNF | −2.5 | GBP4 | −6.3 | MMP3 | −3.6 | FJX1 | −7.7 | ETV1 | −6.5 | FJX1 | −6.8 | ETV1 | −6.4 | ETV1 | −6.0 |
| KRT19 | −2.6 | CXCL12 | −7.2 | KRT19 | −3.7 | ETV1 | −8.0 | SFRP1 | −6.8 | SFRP1 | −6.8 | SFRP1 | −6.4 | FJX1 | −6.3 |
| SFRP1 | −2.6 | CCL2 | −7.4 | CD34 | −3.8 | HSD17B2 | −8.6 | HSD17B2 | −7.1 | MMP3 | −8.1 | MMP3 | −6.5 | MMP3 | −6.8 |
| CD34 | −2.9 | SFRP1 | −8.4 | CCL2 | −4.8 | MMP3 | −9.0 | MMP3 | −7.5 | HSD17B2 | −9.5 | HSD17B2 | −7.0 | HSD17B2 | −7.7 |
E2: estradiol; P4: progesterone; MPA: medroxyprogesterone acetate; LNG: levonorgestrel; NETA: norethindrone acetate; Vh: vehicle; FC: fold change.
Notably, in the response of eSF to all progestins, regardless of type, there were several classical progesterone-regulated genes, including secreted protein acidic and enriched in cysteine like 1 (SPARCL1), solute carrier family 7 member 8 (SLC7A8), olfactomedin (OMD), dikkopf 1 (DKK1), forkhead binding protein 5 (FKBP5), and interleukin 1 receptor (IL-1R) (Table 2) [23]. However, each progestin also differentially regulated unique genes compared to other progestins (full list in Supplemental Table S1), which were further altered in the presence of E2 (see below and Table 2).
The unique and common molecular functions of each progestin effect on eSF were analyzed by Ingenuity Pathway Analysis® (IPA) (Table 3).
Table 3.
Common and unique molecular functions of progesterone and testosterone structurally related progestins in the absence or presence of E2.
| Progestins vs. Vh & Predicted Activation | Progestins + E2 vs. Vh & Predicted Activation | E2 vs. Vh & Predicted Activation | ||||
|---|---|---|---|---|---|---|
| Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | |
| P4 | Angiogenesis | ↓ | Cell movement of epithelial cell lines | ↓ | Colony formation | ↓ |
| Binding of endothelial cells | ↓ | Cell movement of tumor cell lines | ↓ | Proliferation of smooth muscle cells | ↓ | |
| Cell viability | ↓ | Chemotaxis | ↓ | Inflammatory response | ↓ | |
| Cell viability of tumor cell lines + | ↓ | Colony formation of tumor cells | ↓ | Cell movement of blood cells | ↓ | |
| Endothelial cell development | ↓ | Cytostasis of tumor cell lines | ↓ | Cell movement of endothelial cells | ↓ | |
| Proliferation of endothelial cells | ↓ | Differentiation of fibroblasts ++ | ↓ | Migration of endothelial cells | ↓ | |
| Secretion of lipid | ↑ | Formation of skin | ↓ | Leukocyte migration | ↓ | |
| Growth of connective tissue ++ | ↓ | Response of tumor cell lines | ↓ | |||
| Homing of cells | ↓ | Quantity of Ca2 + | ↓ | |||
| Import of D-glucose | ↑ | |||||
| Internalization of carbohydrate | ↑ | |||||
| Invasion of cells ++ | ↓ | |||||
| Invasion of tumor cell lines ++ | ↓ | |||||
| Migration of breast cancer cell lines ++ | ↓ | |||||
| Migration of cells | ↓ | |||||
| Migration of colorectal cancer cell lines | ↓ | |||||
| Migration of tumor cell lines ++ | ↓ | |||||
| Proliferation of connective tissue cells | ↓ | |||||
| Proliferation of lung cancer cell lines | ↓ | |||||
| Proliferation of smooth muscle cells | ↓ | |||||
| Transcription | ↓ | |||||
| MPA | Cell viability of tumor cell lines + | ↓ | Cell movement of carcinoma cell lines | ↓ | ||
| Differentiation of fibroblasts | ↓ | Cell movement of sarcoma cell lines | ↓ | |||
| Growth of tumor | ↓ | Colony formation | ↓ | |||
| Import of D-glucose | ↑ | Colony formation of cells | ↓ | |||
| Migration of sarcoma cell lines | ↓ | Differentiation of fibroblasts ++ | ↓ | |||
| Migration of tumor cells | ↑ | Growth of connective tissue ++ | ↓ | |||
| Non-melanoma solid tumor | ↓ | Invasion of cells ++ | ↓ | |||
| Sphere formation of tumor cell lines | ↓ | Invasion of tumor cell lines ++ | ↓ | |||
| Migration of breast cancer cell lines ++ | ↓ | |||||
| Migration of sarcoma cell lines | ↓ | |||||
| Migration of tumor cell lines ++ | ↓ | |||||
| Sphere formation of tumor cell lines | ↓ | |||||
| LNG | Adhesion of lymphoma cell lines | ↓ | Activation of DNA endogenous promoter | ↓ | ||
| Apoptosis | ↑ | Apoptosis | ↑ | |||
| Binding of lymphoma cell lines * | ↓ | Binding of lymphoma cell lines ** | ↓ | |||
| Cell death | ↑ | Cell death ** | ↑ | |||
| Cell death of tumor cell lines | ↑ | Cell death of tumor cell lines | ↑ | |||
| Cell movement of carcinoma cell lines * | ↓ | Cell movement of carcinoma cell lines ** | ↓ | |||
| Cell movement of epithelial cell lines * | ↓ | Cell movement of sarcoma cell lines ** | ↓ | |||
| Cell movement of sarcoma cell lines * | ↓ | Cell movement of tumor cell lines ** | ↓ | |||
| Cell movement of tumor cell lines * | ↓ | Cell proliferation of carcinoma cell lines ** | ↓ | |||
| Cell proliferation of carcinoma cell lines * | ↓ | Cell viability of tumor cell lines ** | ↓ | |||
| Cell viability of tumor cell lines * | ↓ | Chemotaxis ** | ↓ | |||
| Chemotaxis * | ↓ | Colony formation ** | ↓ | |||
| Colony formation * | ↓ | Colony formation of cells ** | ↓ | |||
| Colony formation of cells * | ↓ | Colony formation of tumor cells ** | ↓ | |||
| Colony formation of tumor cell lines | ↓ | Differentiation of fibroblasts ** | ↓ | |||
| Colony formation of tumor cells * | ↓ | Growth of connective tissue ** | ↓ | |||
| Differentiation of fibroblasts * | ↓ | Growth of malignant tumor ** | ↓ | |||
| Growth of connective tissue * | ↓ | Growth of tumor ** | ↓ | |||
| Growth of malignant tumor * | ↓ | Homing of cells ** | ↓ | |||
| Growth of tumor * | ↓ | Invasion of cells ** | ↓ | |||
| Homing of cells * | ↓ | Invasion of tumor cell lines ** | ↓ | |||
| Invasion of cells * | ↓ | Microtubule dynamics | ↓ | |||
| Invasion of tumor cell lines * | ↓ | Migration of breast cancer cell lines ** | ↓ | |||
| Migration of breast cancer cell lines * | ↓ | Migration of cells ** | ↓ | |||
| Migration of cells * | ↓ | Migration of sarcoma cell lines ** | ↓ | |||
| Migration of colorectal cancer cell lines | ↓ | Migration of tumor cell lines ** | ↓ | |||
| Migration of prostate cancer cell lines * | ↓ | Necrosis ** | ↑ | |||
| Migration of sarcoma cell lines * | ↓ | Organization of cytoplasm | ↓ | |||
| Migration of tumor cell lines * | ↓ | Organization of cytoskeleton | ↓ | |||
| Necrosis | ↑ | Proliferation of connective tissue cells ** | ↓ | |||
| Phosphorylation of L-tyrosine | ↓ | Proliferation of lung cancer cell lines | ↓ | |||
| Proliferation of cancer cells | ↓ | Proliferation of smooth muscle cells | ↓ | |||
| Proliferation of connective tissue cells * | ↓ | ↓ | ↓ | |||
| Proliferation of smooth muscle cells * | ↓ | Sphere formation of tumor cell lines ** | ↓ | |||
| Proliferation of tumor cells * | ↓ | Transcription | ↓ | |||
| Rearrangement of cytoskeleton * | ↓ | Transcription of RNA | ↓ | |||
| Sphere formation of tumor cell lines * | ↓ | |||||
| NETA | Binding of lymphoma cell lines * | ↓ | Adhesion of lymphoma cell lines | ↓ | ||
| Cell movement of carcinoma cell lines * | ↓ | Binding of lymphoma cell lines ** | ↓ | |||
| Cell movement of epithelial cell lines * | ↓ | Cell death ** | ↑ | |||
| Cell movement of muscle cells | ↓ | Cell movement of carcinoma cell lines ** | ↓ | |||
| Cell movement of sarcoma cell lines * | ↓ | Cell movement of epithelial cell lines | ↓ | |||
| Cell movement of smooth muscle cells | ↓ | Cell movement of sarcoma cell lines ** | ↓ | |||
| Cell movement of tumor cell lines * | ↓ | Cell movement of smooth muscle cells | ↓ | |||
| Cell proliferation of carcinoma cell lines * | ↓ | Cell movement of tumor cell lines ** | ↓ | |||
| Cell survival | ↓ | Cell proliferation of carcinoma cell lines ** | ↓ | |||
| Cell viability | ↓ | Cell viability of tumor cell lines ** | ↓ | |||
| Cell viability of tumor cell lines * | ↓ | Chemotaxis ** | ↓ | |||
| Chemotaxis * | ↓ | Colony formation ** | ↓ | |||
| Colony formation * | ↓ | Colony formation of cells ** | ↓ | |||
| Colony formation of cells * | ↓ | Colony formation of tumor cell lines | ↓ | |||
| Colony formation of tumor cells * | ↓ | Colony formation of tumor cells ** | ↓ | |||
| Cytostasis of tumor cell lines | ↓ | Differentiation of fibroblasts ** | ↓ | |||
| Differentiation of fibroblasts * | ↓ | Growth of connective tissue ** | ↓ | |||
| Growth of connective tissue * | ↓ | Growth of malignant tumor ** | ↓ | |||
| Growth of malignant tumor * | ↓ | Growth of tumor ** | ↓ | |||
| Growth of tumor * | ↓ | Homing of cells ** | ↓ | |||
| Homing of cells * | ↓ | Invasion of cells ** | ↓ | |||
| Import of D-glucose | ↑ | Invasion of tumor cell lines ** | ↓ | |||
| Invasion of cells * | ↓ | Migration of breast cancer cell lines ** | ↓ | |||
| Invasion of tumor cell lines * | ↓ | Migration of cells ** | ↓ | |||
| Microtubule dynamics | ↓ | Migration of sarcoma cell lines ** | ↓ | |||
| Migration of breast cancer cell lines * | ↓ | Migration of tumor cell lines ** | ↓ | |||
| Migration of carcinoma cell lines | ↓ | Necrosis ** | ↑ | |||
| Migration of cells * | ↓ | Proliferation of cancer cells | ↓ | |||
| Migration of prostate cancer cell lines * | ↓ | Proliferation of connective tissue cells ** | ↓ | |||
| Migration of sarcoma cell lines * | ↓ | Proliferation of tumor cells ** | ↓ | |||
| Migration of smooth muscle cells | ↓ | Sphere formation of tumor cell lines ** | ↓ | |||
| Migration of tumor cell lines * | ↓ | |||||
| Migration of vascular smooth muscle cells | ↓ | |||||
| Proliferation of connective tissue cells * | ↓ | |||||
| Proliferation of lung cancer cell lines | ↓ | |||||
| Proliferation of smooth muscle cells * | ↓ | |||||
| Proliferation of tumor cells * | ↓ | |||||
| Rearrangement of cytoskeleton * | ↓ | |||||
| Sphere formation of tumor cell lines * | ↓ | |||||
| Transcription | ↓ | |||||
| Transcription of RNA | ↓ | |||||
E2: estradiol; P4: progesterone; MPA: medroxyprogesterone acetate; LNG: levonorgestrel; NETA: norethindrone acetate; Vh: vehicle; ↓, Decreased; ↑, Increased; +: Common between P4 and MPA; *: Common between LNG and NETA; ++: Common between P4+E2 and MPA+E2; **: Common between LNG+E2 and NETA+E2; Bold: Common in each progestin without or with E2.
The eSF response to P4 revealed that six molecular functions were decreased compared to vehicle control, including angiogenesis, endothelial cell development and proliferation, and cell viability and the secretion of lipid were increased. In response to MPA, cell viability, fibroblast differentiation, and tumor growth were decreased. Only one common function, decreased cell viability, was shared between P4 and MPA treatments of eSF (Table 3). When eSF cells were treated with progestins structurally similar to testosterone, there were considerably more altered molecular functions than in the P4 and MPA treated groups (Table 3). With LNG treatment of eSF, 37 molecular functions were regulated, including increased apoptosis and cell death and decreased cell movement, proliferation, migration, growth, and colony formation. Similar results were observed with NETA, wherein 41 molecular functions were regulated, including decreased cell movement, viability, survival, growth, invasion, proliferation, and migration. When comparing LNG and NETA treatment groups, 28 common molecular functions were observed, including decreased cell movement, cell migration, cell proliferation and cell viability, among others (Table 3).
2.2. Estrogen Enhances Progestin-Specific Effects on Gene Expression
The combined treatment of E2 with progestins resulted in higher numbers of differentially expressed genes compared to progestins alone (except for NETA), especially in the group structurally similar to progesterone (Table 1). Supplemental Table S1 and Table 2 contain the full gene lists and the top 15 up- and down-regulated genes in all groups, respectively. Of the 116 DEG in eSF treated with P4 versus vehicle control, 112 DEG (96.5%) were also differentially expressed in E2+P4 versus control. Of the 251 DEG expressed in eSF treated with MPA, 224 DEG (89.2%) were also expressed in the E2+MPA treatment group. In contrast, LNG and NETA exhibited similar effects (to each other) on changes in gene expression in the presence or absence of E2 (Table 1 and Table 2). Of the 553 DEG in eSF treated with LNG versus vehicle control, 511 DEG (92.4%) were also expressed in response to E2+LNG versus control. Of the 595 DEG resulting from NETA treatment, 502 DEG (84.3%) were also expressed in the E2+NETA treatment group.
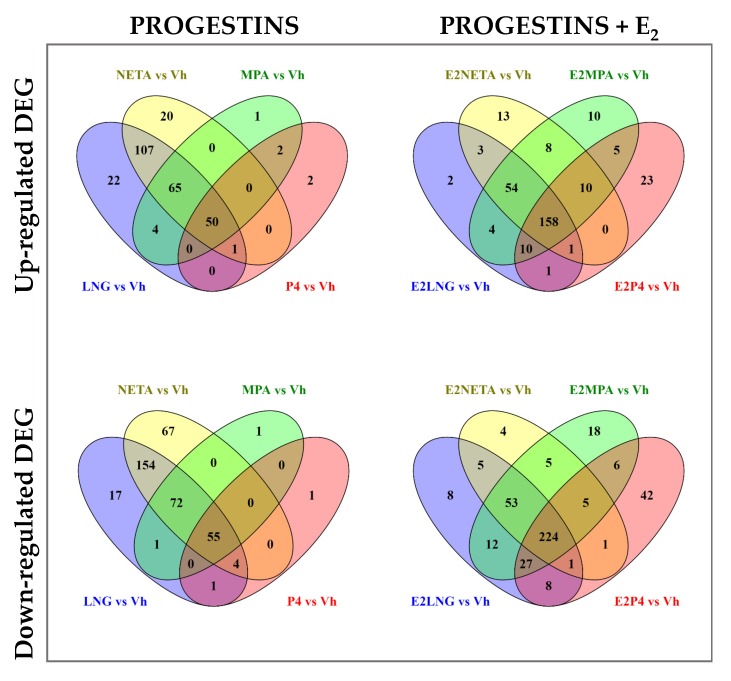
The Venn diagrams (Figure 2) show the number of upregulated and downregulated DEG in common and unique for each progestin treatment, with or without E2 versus vehicle control.
Figure 2.
Venn diagram of differentially expressed genes among the four progestins alone or in combination with E2. Upper panel shows Venn diagrams of up-regulated DEG for each progestin versus Vehicle alone (left panel), or in combination with E2 (right panel). Lower panel shows Venn diagrams of down-regulated genes for each progestin versus vehicle alone (left panel), or in combination with E2 (right panel). Fold change (FC) ≥ 1.5 and Benjamini-Hochberg adjusted p < 0.05.
The P4 and MPA treatments upregulated 52 common genes, with only 2 genes uniquely expressed in P4 alone and 1 in MPA alone. LNG and NETA resulted in the upregulation of 223 common genes, with 22 and 20 genes uniquely upregulated in LNG or NETA, respectively. Overall, there were 50 genes commonly upregulated by all four progestins. The number of upregulated genes increased with the addition of E2, particularly in P4 and MPA, with 23 uniquely upregulated genes in the P4 and 10 genes in MPA. Altogether, there were 158 genes in common among all four progestins when combined with E2 (vs. 50 genes in the absence of E2). Similarly, the majority of downregulated genes in P4 treatment were in common with MPA (55 genes). LNG and NETA induced more DEG than MPA and P4, with 285 downregulated genes in common between LNG and NETA. In addition, the addition of E2 increased the number of downregulated genes—particularly in P4 and MPA. These data demonstrated that the addition of E2 affected up- and down-regulation and increased the total number of common DEG in all four treatments to 224 genes, compared with 55 downregulated genes in progestins without the addition of E2. The lists of unique genes and pathways with and without E2 treatment are shown in Table 4 and Supplemental Table S1.
Table 4.
Unique molecular functions and genes of each progestin without or with addition of E2.
| Unique P4 | Unique P4+E2 | Unique MPA | Unique MPA+E2 | Unique LNG | Unique LNG+E2 | Unique NETA | Unique NETA+E2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | Molecular functions | z ≥ 2 | |
| Angiogenesis | ↓ | Formation of skin | ↓ | Migration of tumor cells | ↑ | Cell death | ↑ | Activation of endogenous promoter | ↓ | Cell movement of muscle cells | ↓ | |||||
| Binding of endothelial cells | ↓ | Import of D-glucose | ↑ | Non-melanoma solid tumor | ↓ | Necrosis | ↑ | Organization of cytoplasm | ↓ | Cell survival | ↓ | |||||
| Endothelial cell development | ↓ | Internalization of carbohydrate | ↑ | Phosphorylation of L-tyrosine | ↓ | Organization of cytoskeleton | ↓ | Migration of carcinoma cell lines | ↓ | |||||||
| Proliferation of endothelial cells | ↓ | Migration of smooth muscle cells | ↓ | |||||||||||||
| Secretion of lipid | ↑ | Migration of vascular smooth muscle cells | ↓ | |||||||||||||
| Proliferation of lung cancer cell lines | ↓ | |||||||||||||||
| Transcription | ↓ | |||||||||||||||
| Unique Up- and Down-Regulated Genes | ||||||||||||||||
| 2 loci exclusively in “P4 vs. Vh” | 23 loci exclusively in “E2P4 vs. Vh” | 1 loci exclusively in “MPA vs. Vh” | 10 loci exclusively in “E2MPA vs. Vh” | 22 loci exclusively in “LNG vs. Vh” | 2 loci exclusively in “E2LNG vs. Vh” | 20 loci exclusively in “NETA vs. Vh” | 13 loci exclusively in “E2NETA vs. Vh” | |||||||||
| Up-regulated Genes | ABCA1, ABCG1 | IRS1, PTS, ITGB1BP1, LAMA3, DPH3, MRAS, JMY, MFSD5, PPP2CB, CCT5, B3GALT4, DTNA, HSD17B11, TUBB2A, CNTN3, C20orf194, CORIN, PRKCDBP, KIF13A, UNG, SLC22A23, FMN1, POLD4 | CH25H | P2RY1, SETMAR, HSPB1, BLVRB, RN7SKP283, ZDHHC7, TMEM120A, CDC25B, TMED10, PDE1A | TCEAL1|TCEAL3, OAT, TCEAL6, CDC42SE2, FAM199X, TRIM63, STK3, REV3L, SIDT2, GADD45A, PDE8A, NXPE3, PGR, GM2A, ATL3, TUBB2A, MORF4L2, SETMAR, TCEAL4, ATL1, PXK, PRTG | ARHGAP10, SMPD2 | SNORD13, ABLIM1, GALNT10, TMEM120A, LARGE, IRS1, TBCB, BLVRB, CHST7, ALOX5AP, KCNJ8, HSPB1, TANGO2, ZNF438, ZNF438, CHCHD10, SLC35E3, ARL8A, COX17, AGPAT6, TIPARP | NXPE3, SIDT2, SLC38A11, AGPAT6, STX2, RHOQP2, BCAT2, SNTB2, YBX3,CHCHD10, ATL3, C9orf91, CST3 | ||||||||
| 1 loci exclusively in “P4 vs. Vh” | 42 loci exclusively in “E2P4 vs. Vh” | 1 loci exclusively in “MPA vs. Vh” | 18 loci exclusively in “E2MPA vs. Vh” | 17 loci exclusively in “LNG vs. Vh” | 8 loci exclusively in “E2LNG vs. Vh” | 67 loci exclusively in “NETA vs. Vh” | 4 loci exclusively in “E2NETA vs. Vh” | |||||||||
| Down-regulated Genes | ZNF704 | BIVM, ORC2, NPAT, CNOT6L|CNOT6LP1, NQO1, LRRC37A4P, CREB5, ZNF462, SMAD5, DLC1, GRIA3, SMC5, CACNA2D1, C14orf1, NUCKS1, NEFM, PLEKHA5 TNFRSF10B, CNN3, NEO1, CDK19, HNRNPA1, LPHN1, CDH11, MXRA5P1 MASP1, HNRNPA1, TFAM, HNRNPA1, PPP1CC, ZKSCAN8, COL6A3, TFDP2, TSPYL2, LRRC8C, FAM171B, TLE4, CH25H, TMEM97, SLC7A11, PCDH18 |
FAM46C | MACF1, FAM46C, CACNA1C, ABCG1, FHL3, NF1P3, PTMAP4, TMEM51, PMEPA1, ARL15, CNNM1, RPL22L1, BAZ2B, PTMAP4, ZNF33B, NES, TMEM25, CADM4 | NHSL2, FAM43A, ALDH1A1, PARD3, RASL11A, ANGPT1, PCYT1B, DEPDC7, NPY1R, CKS1B, MBIP, FIGNL2, CADM4, PHLDA1, WBP1L, LOXL2, ACSL4 | PCNXL4, GABPA, TNRC6B, PCMTD2, CDK2, SNRK, STRA6, CEP57 | IFI16, PTMAP4, CROT, TOP2B, SYDE2, IRF1, CNN2, PAK1, RHBDD2, FARP1, CDK2, PCNXL4, CNOT2, ZNF483, CNNM1, ATP1A1, TANC1, TNFRSF10B, MACF1, CCDC109B, PTPN14, TGFBRAP1, HMGN1P30, TRIM5, CREB5, TFAM, PTDSS1, PPP1CC, TERF1, HNRNPA1, HNRNPA1P10, RNU6-674P, DUSP7, CH25H, TSHZ1, METAP1, ARL15, HNRNPA1P7, TBC1D8, RUNX1T1, ESR1, SALL2, PNISR, DGKH, HNRNPA1P6, EPHA5, NEO1, CCDC125, BAZ2B, ZNF713, C21orf91, CEP57, ZNF721, PDGFD, ADCY4, ZNF286A, IL17RD, SMC5 GABPA, FZD6, RBFOX2, DTWD1, PCDH18, RNU6-1152P, ZKSCAN8, LINC00597, ZNF217 |
ZNF483, PTMA, PDE9A, TM7SF2 | ||||||||
E2: estradiol; P4: progesterone; MPA: medroxyprogesterone acetate; LNG: levonorgestrel; NETA: norethindrone acetate; Vh: vehicle; ↓, Decreased; ↑, Increased.
2.3. Molecular Biofunctions
The numbers of molecular and cellular functions of DEG were also increased when E2 was added to progestin treatments of eSF, mainly for progestins structurally related to progesterone (Table 3). Twenty-one molecular functions were uniquely affected when E2 was added to P4, compared to 7 molecular functions affected by P4 alone (Table 3). Notably, E2 increased biofunctions involving the internalization of carbohydrate and the import of D-glucose and decreased cell movement, migration, invasion, and tumor cell colony formation, differentiation of fibroblasts, and gene transcription. When E2 was combined with MPA, the cellular functions that decreased included cell movement, invasion and migration. Common molecular functions in the MPA treatment with and without E2 included decreased fibroblast differentiation, and cell migration. Six shared molecular functions were observed between P4+E2 and MPA+E2, including decreased fibroblast differentiation and cell growth, invasion and migration (Table 3). In contrast, addition of E2 to the progestins structurally related to testosterone resulted in fewer affected molecular functions than without E2 (Table 3). Of the 36 molecular functions affected in LNG+E2, 29 of them were common to LNG alone, including increased apoptosis and necrosis, and decreased cell growth, viability, migration and proliferation, and colony formation. In NETA+E2, there were 31 cellular and molecular functions altered, of which 26 were common to NETA alone. These included decreased cell movement, growth, invasion and migration, and colony formation (Table 3). Twenty six molecular functions were common between LNG+E2 and NETA+E2 treatments, including decreased fibroblast differentiation and cell growth, invasion and migration (Table 3). Notably, eSF treatment with E2 alone resulted in decreased molecular functions of colony formation, proliferation of smooth muscle cells, migration of endothelial cells and leukocytes, and the inflammatory response (Table 3).
The unique versus common molecular biofunctions in the absence or presence of E2 are indicated in Table 3, based on the structurally related progestin groups.
2.3.1. Unique Molecular Biofunctions
Molecular functions that were unique for each progestin, as well as the upregulated and downregulated genes in each function, in the absence and presence of E2, are presented in Table 4. Note that these were unique between progestin-alone groups (P4 vs. MPA vs. LNG vs. NETA) and unique between progestin plus E2 groups (P4+E2 vs. MPA+E2 vs. LNG+E2 vs. NETA+E2). Unique biofunctions for each progestin involved decreased angiogenesis and endothelial cell-related functions in P4, increased tumor cell migration and decreased tumor formation in MPA, increased cell death and necrosis in LNG, and decreased transcription, cell survival, vascular smooth muscle cell movement and migration in NETA. In the presence of E2, only P4+E2 and LNG+E2 (but not MPA+E2 and NETA+E2) showed unique biofunctions, which were different from those of unique P4 and LNG without E2 (Table 4).
2.3.2. Common Molecular Biofunctions
Table 5 reflects the molecular biofunctions commonly shared among all four progestin treatments with and without E2, as well as the genes involved in each biofunction based on progestin treatments. In the absence of E2 (progestin alone treatments), only one molecular function, decreased cell viability, was common among all progestin treatments (Table 5).
Table 5.
Common biofunctions and associated differentially expressed genes between progestins without and with addition of E2. E2: estradiol; P4: progesterone; MPA: medroxyprogesterone acetate; LNG: levonorgestrel; NETA: norethindrone acetate; Vh: vehicle; DEG: differentially expressed genes.
| Common Functions between Progesterone and Testosterone Structurally Related Progestins (Predicted Activation, z-score≥2) | Common DEG Progestins Related to Progesterone (P4/MPA) | Common DEG Progestins Related to Testosterone (LNG/NETA) | |
|---|---|---|---|
| −E2 | Cell viability of tumor cell lines (Decreased) | ABCA1, AK5, APOE, CCL2, CD200, DUSP6, FKBP5, GUCY1A3, INSR, KRT19, MAPK3K4, NTF3, SMAD3, TOX, ZBTB16 | AK5, AKAP13, APOE, ASAH1, BCL6, BDNF, BEX2, CASP1, CAV1, CCL2, CCND1, CD200, CDH2, CTGF, CXCL12, DNM1, DUSP6, FGFR1, FKBP5, GDF15, GUCY1A3, HMOX1, ID4, IL6, INPP5A, INSR, IRS2, JMJD1C, KIT, KRT19, LIMK2, MAP3K4, MCOLN1, NFAT5, NR1D1, NRP1, NTF3, PGRMC1, PIK3R1, PLAU, PLK2, POLB, PTPN11, PTPRK, SMAD3, TOX, TRIB2, UGCG, VEGFA, WEE1, ZBTB16 |
| +E2 | Differentiation of fibroblasts (Decreased) | CCL2, CD44, DKK1, F2RL1, IL6, PDE5A | CCL2, DKK1, F2RL1, IL6, PDE5A |
| Growth of connective tissue (Decreased) | CCL2, CCND1, CD44, CDH13, CTGF, CXCL12, FGF1, FGF7,, FGF9, FOXO1, GREM1, IGFBP4, IL6, PDE5A, PDGFD, PLAU, PTPRK, RUNX1T1, SFRP1, SMAD3, SPRY2, THRB, TMPO, WNT2 | CCL2, CCND1, CD83, CDH13, CTGF, CXCL12, FGF1, FGF7, FGF9, FGFR1, FOXO1, GREM1, IGFBP4, IL6, PDE5A, PDGFC, PLAU, PRDX4, PTPRK, RUNX1T1, SFRP1, SMAD3, SPRY2, THRB, TMPO, WNT2 | |
| Invasion of cells (Decreased) | ABLIM1, BDKRB1, BDNF, CAV1, CCL2, CCND1, CD44, CDH13, CDH2, CFH, CNR1, CTGF, TSB, CTSH, CTSL, CXCL12, DKK1, DOCK4, DRAM1, DUSP6, EFNB3, ELMO1, ETV1, ETV4, ETV5, FHL2, FUT8, GDF15, HMOX1, HTRA1, IL6, IRS2, JUNB, JUP, KIT, KRT19, LCP1, LIMA1, LIMK2, LPAR1, MGAT5, MMP16, MMP3, MSI2, NFAT5, NRP1, NRP2, PDLIM1, PIK3R1, PLAU, PTPRK, RECK, RGS4, SATB1, SEMA5A, SERPINE1, SFRP1, SMAD3, SPARCL1, SPRY2, SPSB1, TCF4, TFAP2C | ACSL4, BDKRB1, BDNF, CAV1, CCL2, CCND1, CDH13, CDH2, CFH, CNR1, CTGF, CTSB, CTSH, CTSL, CXCL12, DIAPH2, DKK1, DOCK4, DPYSL3, DRAM1, DUSP6, EFNB3, ELMO1, ESR1, ETV1, ETV4, ETV5, FGFR1, FHL2, FUT8, GDF15, HMOX1, HTRA1, ID4, IL6, IRS2, JUNB, JUP, KIT, KRT19, LCP1, LIMA1, LIMK2, LOXL2, LPAR1, MGAT5, MMP3, MSI2, NFAT5, NRP1, NRP2, PDLIM1, PGR, PIK3R1, PLAU, PTPRK, RECK, RGS4, SATB1, SEMA5A, SERPINE1, SFRP1, SMAD3, SPARCL1, SPRY2, SPSB1, TCF4, TFAP2C, TGFB3, TGFBI, TMPO, VEGFA, ZBTB16 | |
| Invasion of tumor cell lines (Decreased) | ABLIM1, BDKRB1, BDNF, CAV1, CCL2, CCND1, CD44, CDH13, CDH2, CFH, CNR1, CTGF, CTSB, CTSH, CXCL12, DKK1, DOCK4, DRAM1, DUSP6, EFNB3, ELMO1, ETV1, ETV4, ETV5, FHL2, FUT8, GDF15, HMOX1, HTRA1, IL6, IRS2, JUNB, KIT, KRT19, LCP1, LIMA1, LIMK2, LPAR1, MGAT5, MMP16, MMP3, MSI2, NFAT5, NRP1, NRP2, PDLIM1, PIK3R1, PLAU, PTPRK, RECK, SATB1, SEMA5A, SFRP1, SMAD3, SPARCL1, SPRY2, SPSB1, TCF4 | ACSL4, BDKRB1, BDNF, CAV1, CCL2, CCND1, CDH13, CDH2, CFH, CNR1, CTGF, CTSB, CTSH, CXCL12, DIAPH2, DKK1, DOCK4, DPYSL3, DRAM1, DUSP6, EFNB3, ELMO1, ESR1, ETV1, ETV4, ETV5, FGFR1, FHL2, FUT8, GDF15, HMOX1, HTRA1, ID4, IL6, IRS2, JUNB, KIT, KRT19, LCP1, LIMA1, LOXL2, LPAR1, MGAT5, MMP3, MSI2, NFAT5, NRP1, NRP2, PDLIM1, PGR, PIK3R1, PLAU, PTPRK, RECK, SATB1, SEMA5A, SFRP1, SMAD3, SPARCL1, SPRY2, SPSB1, TCF4, TFAP2C, TGFB3, TGFBI, TMPO, VEGFA | |
| Migration of breast cancer cell lines (Decreased) | CCL2, CCND1, CD44, CDH2, CTGF, CTSL, CXCL12, DUSP6, FGF1, FGF7, GAB1, IL6, ITGA6, KRT19, NRP1, PIK3R1, PLAU, SEMA3C, SERPINE1, SMAD3, TFAP2C, TGFB3, THRB, TNFRSF21 | ACSL4, CCL2, CCND1, CDH2, CTGF, CTSL, CXCL12, DUSP6, ESR1, FGF1, FGF7, GAB1, IL6, KRT19, NRP1, PGR, PIK3R1, PLAU, SEMA3C, SERPINE1, SLC16A4, SMAD3, TFAP2C, TGFB3, THRB, TNFRSF21, VEGFA | |
| Migration of tumor cell lines (Decreased) | BDKRB1, BDNF, CAV1, CCL2, CCND1, CD44, CDH13, CDH2, CTGF, CTSB, CTSH, CTSL, CXCL12, DUSP6, EFNB3, ELMO1, ETV4, ETV5, F2RL1, F3, FGF1, FGF7, FHL2, GAB1, GDF15, HMOX1, HTRA1, GFBP4, IL6, ITGA6, ITPR1, JUP, KDR, KIT, KRT19, LIMK2, LPAR1, LTBP2, MGAT5, MME, MMP19, MMP3, MYO10, NRP1, NRP2, NTF3, PDLIM1, PEAK1, PIK3R1, PLAU, PLCL1, PTGER4, PTPN11, PTPRK, RAP2A, RHOU, SEMA3C, SEMA5A, SERPINE1, SMAD3, SPARCL1, SPRY2, SPSB1, TCF4, TFAP2C, TGFB3, TGFBI, THRB, TMPO, TNFRSF21 | ACSL4, BDKRB1, BDNF, CAV1, CCL2, CCND1, CDH13, CDH2, CTGF, CTSB, CTSH, CTSL, CXCL12, DUSP6, EFNB3, ELMO1, ESR1, ETV4, ETV5, F2RL1, F3, FGF1, FGF7, FGFR1, FHL2, GAB1, GDF15, HMOX1, HTRA1, IGFBP4, IL6, ITPR1, JUP, KIT, KRT19, LPAR1, LTBP2, MGAT5, MME, MMP19, MMP3, MYO10, NRP1, NRP2, NTF3, PDLIM1, PEAK1, PGR, PIK3R1, PLAU, PLCL1, PTGER4, PTPN11, PTPRK, RAP2A, RHOU, SEMA3C, SEMA5A, SERPINE1, SH3PXD2B, SLC16A4, SMAD3, SPARCL1, SPRY2, SPSB1, TBX3, TCF4, TFAP2C, TGFB3, TGFBI, THRB, TMPO, TNFRSF21, VEGFA |
In contrast, with the addition of E2, six biological functions were common to all four treatments, including decreased fibroblast differentiation, growth of connective tissue, and the invasion and migration of cells and tumor cells (Table 5). Interestingly, the one common DEG in all biofunctions and in all groups, with or without E2, was the chemokine CCL2 (bolded in Table 5). Two DEG, CCL2 and IL-6, were common among all biofunctions and across all progestin treatment groups in the presence of E2.
2.4. Assessment of Secreted Protein Levels of the Two Differentially Expressed Genes Common to All Treatment Groups
The protein products of CCL2 and IL-6 genes were assessed because they were the only differentially expressed genes common to all treatment groups, including progestins alone or combined with E2. Moreover, vascular endothelia growth factor A (VEGFA) was assessed due to its important role in angiogenesis and as it was found to be downregulated in five of the eight studied conditions.
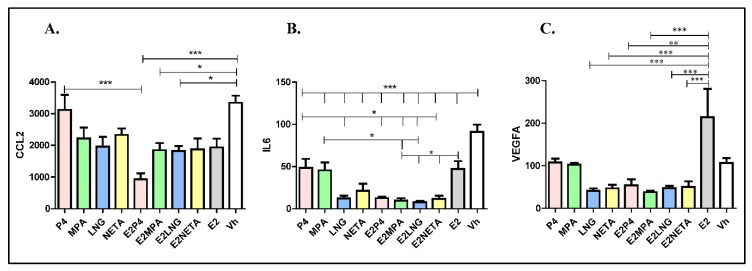
Concentrations of secreted CCL2 and IL-6 in media conditioned by eSF after progestin and progestin plus E2 treatments were determined (Figure 3).
Figure 3.
Concentrations of secreted CCL2, IL-6 and VEGFA in media conditioned by eSF after progestin and progestin plus E2 treatments. (A) Secreted CCL2. (B) Secreted IL-6. (C) Secreted VEGFA. In all cases, the figure shows the “amount” of secreted protein for each progestin and each progestin, plus E2 adjusted by cell number and total media. Pink-colored bars: progesterone (P4); green-colored bars: medroxyprogesterone acetate (MPA); blue-colored bars: levonorgestrel (LNG); yellow-colored bars: norethindrone acetate (NETA); gray-colored bars: E2 alone; white bars: vehicle. Error bars indicated SD; * p < 0.05, ** p < 0.01, *** p < 0.001.
All progestins, with or without E2, decreased secreted CCL2, IL-6 and VEGFA protein levels compared to vehicle control (Figure 3A), consistent with the gene expression data (Table 6).
Table 6.
Fold change of common DEG in all progestins in absence or presence of E2. E2: estradiol; P4: progesterone; MPA: medroxyprogesterone acetate; LNG: levonorgestrel; NETA: norethindrone acetate; Vh: vehicle.
| Progestins Related to Progesterone (P4/MPA) | Progestins Related to Testosterone (LNG/NETA) | |||||||
|---|---|---|---|---|---|---|---|---|
| P4 vs. Vh | E2P4 vs. Vh | MPA vs. Vh | E2MPA vs. Vh | LNG vs. Vh | E2LNG vs. Vh | NETA vs. Vh | E2NETA vs. Vh | |
| CCL2 | −1.55 | −7.44 | −4.77 | −6.04 | −6.41 | −6.25 | −6.16 | −5.08 |
| IL6 | −2.74 | −1.93 | −2.53 | −2.82 | −2.93 | −2.92 | −2.83 | |
| VEGFA | −1.76 | −1.68 | −1.78 | −1.74 | −1.51 | |||
The addition of E2 did not significantly alter the progestin inhibitory effect on CCL2 protein levels, except when combined with P4, resulting in a marked reduction of secreted CCL2 (Figure 3A). All progestins decreased IL-6 levels, with P4 and MPA having the least inhibitory effect (p < 0.05), and combined treatment with E2 further attenuated this effect (Figure 3B). E2 alone stimulates VEGFA, but progestins, alone or combined with E2, reduce its secretion (Figure 3C).
3. Discussion
The endometrium in natural cycles responds in a programmed fashion to E2 by induced cell proliferation, followed by P4-induced epithelial secretory transformation and stromal fibroblast decidualization, preparing for pregnancy. In non-conception cycles, it sheds and regenerates anew from epithelial and mesenchymal progenitors [24]. Normal endometrial homeostasis for growth, differentiation, desquamation, and regeneration revolves around appropriate cellular hormonal responses and paracrine interactions among the various cell types. Comprising this dynamic tissue are epithelial, endothelial, immune, vascular smooth muscle and stem cells, and stromal fibroblasts [25]. Progesterone promotes an epithelial-like phenotype of the latter, transforming them to master modulators of endometrial epithelial, vascular and immune function, acceptance of the conceptus, and controlled hemostasis during menses. Progestational agents share some, but not all, of native progesterone actions on eSF and are anticipated to have variable effects on this cell’s function in normal endometrial tissue and alternative effects not observed with P4 per se. Synthetic progestins are widely used for contraception, to treat endometriosis and endometrial cancer, and have been used in postmenopausal hormone therapy [1], and as a class, cause decidualization and atrophy of the endometrium [11,16]. A common side effect, often leading to their discontinuation, is abnormal uterine bleeding due to fragile endometrial vasculature [26] and overall altered signaling through the endometrial nuclear progesterone receptor (PR) [1,2]. The current study undertook, for the first time, an analysis of effects of synthetic progestins widely used in clinical practice on the human eSF transcriptional program, to identify each progestin’s effects and associated molecular functions relevant to normal and abnormal endometrial homeostasis. While we have identified the effects of these contraceptives on the eSF molecular phenotype, whether these differentially expressed genes are the direct or indirect targets of each progestins or reflect a transcriptional reprogramming in response to these hormone treatments are yet to be determined by time course and mechanistic analyses.
3.1. Distinct Progestin-Induced Transcriptomes
In our comparison of the effects of four different and widely used progestins (P4, MPA, LNG and NETA) on the eSF transcriptome alone and in combination with E2, we found distinct differences between progestins structurally similar to progesterone (P4 and MPA) and those structurally similar to testosterone (NETA and LNG). As anticipated, the gene expression signatures of P4 and MPA treatments of eSF were more similar to each other and were different from signatures elicited by LNG and NETA, which were similar to each other, but also had unique transcriptomic patterns.
In the response of eSF to all progestins, regardless of type, several classical progesterone-regulated genes were upregulated, including SPARCL1, SLC7AB, OMD, DKK1, FKBP5, and IL-1R and down-regulated were CCL2, IL-6, transforming growth factor β1 (TGFβ1), matrix metalloproteinase-3 (MMP3), and 17β-hydroxysteroid dehydrogenase 2 (17βHSD2) [23]. These genes are regulated via P4:PR (ligand:receptor) binding with PR-mediated signaling, and the stimulation or silencing of gene expression. That all four progestins had similar effects on transcription of classically P4-regulated genes underscores the importance of PR in signaling by all progestins studied. However, the various progestins stimulated and repressed unique genes, as well. The unique genes differentially regulated by each of these progestins involved vastly different biofunctions that could potentially have distinct effects on the endometrial function and progestin-induced endometrial changes. For example, P4 uniquely affects angiogenesis and endothelial cell development and proliferation, while unique genes in MPA affect cell migration, and unique LNG genes affect cell death and necrosis, with NETA affecting a wide range of biofunctions. This is likely due to different PR activation by each ligand leading to altered gene transcription, perhaps their binding to other steroid hormone receptors, for which these progestins have measurable affinity (see below), by altering the chromatin per se, and/or by non-genomic pathways [27]. An example of the latter is the progestin R5020 that at picomolar concentrations promotes the proliferation of rat endometrial stromal cells via ERK1-2 and AKT activation mediated by PR interaction with ER, resulting in the PR regulation of gene expression, independent of hormone receptor binding to specific genomic targets [28].
3.2. Estrogen Effect on Progestin-Induced Transcriptome
The addition of E2 considerably affected eSF gene expression profiles in response to P4 and MPA; whereas, when E2 was combined with LNG and NETA, only a modest effect on their gene expression signature profiles was observed. We note that, in this study, we have assessed the effects of estradiol and not ethinyl E2, a commonly used synthetic estrogen in oral contraceptives. This is because the focus of the current study is on progestins and their effects in the absence/presence of E2 priming/stimulation, due to the known effects of E2 on biological responses to progestins. We have used estradiol as prototype, which addresses this question without potential pharmacological confounders beyond physiologic estrogenic signaling. The potentially divergent actions of different estrogenic compounds warrant a different experimental design.
Diverse molecular mechanisms may contribute to the distinct transcriptional responses to the various progestins observed in the current study. Even subtle changes in progesterone structure or synthetic progestins with progestational activity and varied binding affinities for PR and other steroid hormone receptors (see below), and in the presence of E2 binding to its cognate nuclear receptor and by non-genomic pathways, can result in the altered regulation of gene expression.
The effects observed herein likely derive from the unique structures of progestins with their additional chemical groups and conformations (Figure 1), that confer different binding affinities for cognate nuclear receptors [1]. For example, MPA, NETA and LNG have a greater affinity for PR than P4 (relative binding affinities of 115, and 150 and 50, respectively), and none of the synthetic progestins studied herein have an appreciable affinity for ER [29]. The binding affinity to the androgen receptor (AR) was reported to be higher for LNG and NETA than P4 [29]. However, in a steroid-receptor-deficient COS-1 cell line, with transiently expressing human AR; MPA, NETA, and P4 had a similar affinity to 5α-dihydrotestosterone (DHT) in binding to AR. However, NETA and MPA differed in inducing classical and AR-selective promoters [30]. Interestingly, it has been suggested that MPA displays greater glucocorticoid receptor (GR) agonist potency than P4 and NETA [31]. Moreover, MPA has immunosuppressive effects mediated by GR, as demonstrated by MPA repressing expression of IL-2 mRNA in normal human lymphocytes through GR [32].
It is important to note that the number of genomic regions containing exclusive hormone-specific response elements across the genome is limited, and many areas of the genome contain several response elements [33], which may explain how the combinatorial recruitment of ER with PR could alter target gene expression. Furthermore, progesterone-derived progestins binding to PR, MR and GR indicate that these progestins may use different mechanistic pathways from those of progestins structurally similar to testosterone, and subsequently have distinct differential combinatorial effects with ER.
3.3. Chemokines, Cytokines, and Angiogenic Factors
IL-6 and CCL2, encoding major pro-inflammatory cytokines, were the only two genes in common in biofunctions of eSF treated with all progestins in the presence of E2. CCL2 (also called monocyte chemoattractant protein-1 (MCP-1)) is a potent chemoattractant for monocytes and T cells [34], with lesser effects on basophils [35] and natural killer cells [36]. The CCL2 gene is located in chromosome 17 and has a promoter region containing binding sites for AP-1 and NFKB, and is a major product of macrophages and other cell types [37]. Of note, Arici et al. reported an anti-inflammatory phenotype produced in eSF, when P4 or MPA were added to these cells in vitro, specifically with regard to down-regulation of the CCL2 protein and mRNA [34]. Importantly, the effect of MPA on CCL2 expression was reversed by the PR antagonist, RU486, demonstrating the PR-mediated regulation of this chemokine (25). In the current study, in addition to P4 and MPA, LNG and NETA, progestins structurally related to testosterone also down-regulated CCL2, in the absence or presence of E2. Of note, CCL2 is highly expressed in peri-menstrual endometrium, when E2 levels are low but is down-regulated at ovulation, when E2 levels are high [38]. This regulation is consistent with the results obtained herein, demonstrating that its down-regulation is mediated by PR and ER.
Down-regulation of CCL2 and IL-6 in eSF treated with progestins and E2 suggests an anti-inflammatory environment in human endometrium. It is noteworthy that the downregulation of these two pro-inflammatory cytokines with all different progestin treatments is consistent with the known anti-inflammatory effects of progestins on human endometrium in vivo [11,16,39]. Early implantation is characterized by high levels of the pro-inflammatory T helper cells and cytokines such as TNFα, IL-6, and IL-8 [40,41], and the upregulation of pro-inflammatory cytokines positively correlates with IVF pregnancy outcomes [42]. Cytokines and chemokines such as IL-6 attract human trophoblast cells and therefore affect successful implantation [42], and the balance of pro- and anti-inflammatory cytokines and chemokines is required for proper endometrial tissue growth and remodeling [42,43]. Thus, downregulation of these cytokines has the potential to contribute to a sub-optimal environment for implantation and endometrial homeostasis.
CCL2 activates STAT1 signaling [44], and in mammals, the JAK/STAT signaling pathway is the principal pleiotropic cascade to transduce a wide array of cytokines and growth factors affecting pathways for cell differentiation, proliferation, migration and apoptosis [45]. Since the CCL2 receptor, CCR2, is present in human vascular smooth muscle cells (VSMCs) [46] and since STAT1 signaling induces VSMC proliferation [46], lowered CCL2 expression in eSF in response to progestins may contribute to the decreased VSMC proliferation involved in endometrial blood vessel formation in the proliferative phase, their growth and maintenance in luteal phase endometrium and potentially vessel fragility—a known side effect causing breakthrough bleeding in women using these progestins [47,48].
Moreover, VEGFA mRNA and protein (Table 6, Figure 3C) were decreased in eSF treated with progestins. By extrapolation, progestins could inhibit blood vessel formation in endometrium, leading to an inhospitable environment for embryo implantation and breakthrough bleeding, the latter of which is commonly associated with this class of drug [6,9]. Breakthrough bleeding in long term progestin-only contraceptives users is due to a complex interplay of progestin effects on eSF and thrombin, tissue factor, IL-8, and VEGF, resulting in aberrant angiogenesis and the enhanced fragility of endometrial blood vessels [39]. While the current findings are consistent with the potential involvement of some of these progestin-regulated molecules in these processes, the precise mechanisms underlying the role of eSF in breakthrough bleeding in women on progestin-only preparations, as well as those with combined estrogen-progestin preparations, await further study. Moreover, whether these findings have implications for disorders with abnormal progesterone signaling, such as the endometrium of women with endometriosis [49] and polycystic ovarian syndrome [50] remains to be determined.
3.4. Strengths and Limitations of This Study
This is the first study to assess the effects of four commonly used progestins, alone or in combination with E2 on the genome-wide transcriptome of normal human endometrial stromal fibroblasts, adjusting for biopotency. The data support common and unique responses of this cell type to progesterone and the other progestins studied herein in the absence or presence of estradiol that are relevant to endometrial effects of these steroids widely used by women across the lifespan. The data provide insights into potential mechanisms underlying breakthrough bleeding and other side effects commonly encountered among women taking these medications. Of note, the current study was an in vitro analysis, and while progestin biopotencies were similar, local effects in vivo can be modulated by numerous confounders, including paracrine interactions among endometrial cellular components as well as different bioavailability, metabolism, and pharmacodynamics of these steroids, ultimately leading to composite effects at the cellular level as well as systemically. A limitation of our study is that the effects of progestins on other cell types of endometrium are yet to be determined. We aim to investigate the effect of these contraceptives in endometrial epithelial and immune cells, to gain better insights into their effects on human endometrium. Another limitation is that the concentrations used for the progestins were determined by dose-response experiments, to identify concentrations that elicit cellular responses. These are not similar to physiological concentrations and remain to be determined in women using these contraceptives.
In conclusion, in vitro progestin effects on eSF alone or in combination with E2, differ from one progestin to another, and those structurally similar to progesterone and testosterone more closely align with their respective group effects on eSF gene expression. Despite these differences, there were many genes and pathways shared among the different progestins. All four progestins (alone or in combination with E2) decreased the expression of CCL2, prominently involved in immuno-regulatory and inflammatory processes. The results overall indicate that progestins have an anti-inflammatory effect, enhanced by E2 on the eSF cell, contributing to an anti-inflammatory environment in the endometrium. Notably, eSF in women with endometriosis and polycystic ovarian syndrome show an abnormal response to P4, and whether different progestins display varied responses in these populations warrants further analysis, potentially leading to more personalized progestin therapies for women.
4. Materials and Methods
4.1. Samples
This study was approved by the Committee on Human Research of the University of California San Francisco (UCSF) (IRB: 10-02786). Samples were collected from the UCSF/NIH Human Endometrial Tissue and DNA Bank after written informed consent. For these studies, endometrial stromal fibroblasts (eSF) were isolated from endometrial biopsies and established in primary culture, as previously described [51]. Briefly, five eutopic endometrial biopsies were collected from women without any gynecological abnormalities. Each tissue biopsy was digested separately using collagenase and the tissue digests were then filtered using a 40micron cell strainer to separate the dissociated eSF from endometrial glands and undigested tissue. The eSF from the filtered through fraction were then established in monolayer primary culture and grown in medium supplemented with 10% charcoal-stripped fetal bovine serum (FBS), as previously described [51]. Thus, a separate eSF primary culture was established from each of the five different patient biopsies. For the hormone treatment experiments, sub-confluent primary cultures (approximately 75% confluent) were harvested and eSF seeded onto 24-well plates at 105 cells/well and grown in 10% serum-supplemented medium as above until confluent. Confluent cultures were treated with the various steroid hormones in medium supplemented with 2% charcoal-stripped FBS, according to our previously described protocol [52]. In brief, cultures were pre-incubated in medium supplemented with 2% charcoal-stripped FBS without hormones for 24 h. After 24 h, the culture medium was replaced with fresh medium containing steroid hormones (E2, progestin, progestin plus E2) or vehicle control for 14 days, with media renewal every three days. Concentrations of E2 and P4 used were maximally effective concentrations, as shown before for eSF in vitro decidualization [51]. Other progestins were used at concentrations equipotent to that of P4, adjusted according to their reported transactivation biopotency relative to P4 [53]. Concentrations of progestins, alone or in combination with 10 nM E2, were: 1 µM P4, 54.7 nM LNG, 0.294 μM NETA, 6 nM MPA and ethanol vehicle control (Vh, (0.1% ethanol)). After 14 days of treatment, eSF conditioned media were collected for secreted cytokine analysis and cells were harvested by trypsinization, counted, and frozen at -80ºC until RNA extraction. Each sample was processed separately for RNA extraction and microarray, without pooling samples derived from different patient biopsies.
To ensure proper cellular responsiveness to P4, the conditioned media from each sample after 14 days of treatment was used for the analysis of IGFBP1 protein levels by ELISA, in duplicate and normalized to the cell count for each sample. IGFBP1 is a progesterone-responsive gene, and we found its increased protein level in all treatments, and the highest with E2+P4 treatment, as expected. See Supplementary Figure S1.
4.2. RNA Extraction and Whole Genome Microarrays
Cellular RNA was extracted using the NuceloSpin Tissue Kit (Marcherey-Nagel Inc, Bethlehem, PA), according to manufacturer’s recommendations and stored at -80ºC until use. RNA quality was assessed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA) and only samples with an RNA integrity number (RIN) higher than 7 were considered of high quality and used for further analysis by microarray. RNA samples were reverse transcribed to cDNA, followed by 2nd strand DNA generation and cDNA generation. For microarray analysis, all hormone treatments for each of the five cell lines were prepared according to Affymetrix (Santa Clara, CA) specifications and hybridized to the Affymetrix Exon Expression Chip HuGene1_0-st-v1 gene array at the UCSF Genome Core facility, as previously described [49].
4.3. Whole Genome Microarray Data Analysis
A gene expression microarray data analysis was conducted using GeneSpring GX software (Agilent Technologies). The data from the raw CEL files were normalized together by RMA. The normalized data were then used to identify differentially expressed gene between different comparisons across the five biological replicates using ANOVA. To correct for multiple comparisons, we used the Benjamini–Hochberg correction, and significance was determined at p-value < 0.05 and a fold change (FC)) > ± 1.5. Venn diagrams were used to assess common DEG among the four progestins, using Venny 2.1.0 software [54]. Subsequently, all DEG lists for all comparisons were analyzed using IPA software (Ingenuity LLC, Portola Valley, CA) [55], to identify common and unique pathways among the different progestins. Pathways with a z-score > ± 2.00 were considered to be significant.
4.4. Luminex Analysis
The validation of IL-6 (interleukin-6), CCL2 (C-C motif chemokine ligand 2 (monocyte chemotactic protein-1)) and VEGFA (vascular endothelia growth factor A) gene expression for secreted proteins was performed using multiplexed immunoassay analysis (Luminex, EMD Millipore, Burlington, MA, USA) of these analytes in eSF conditioned media in the various treatment groups. These analytes were selected because they were decreased in gene expression analyses and were common among all comparisons (IL-6 and CCL2) or importance in angiogenesis (VEGFA) (see Results). Conditioned media from treated eSF cells were centrifuged at 3000xg for five minutes to remove debris, and the supernatant was used for Luminex analysis, according to the manufacturer’s instructions. Briefly, conditioned media were incubated overnight in Luminex plates with antibody-coated fluorescent-dyed analyte-specific microspheres, and bound analytes were resuspended in sheath fluid and analyzed on a Bioplex bead sorter (Bio-Rad, Hercules, CA), adjusted by media volume and cell number. Statistical significance of concentration differences of analytes in specific comparisons was determined as p < 0.05 by ANOVA after Bonferroni’s multiple comparison correction, using GraphPad Prism V.5.
Acknowledgments
We acknowledge UCSF Genomics CORE for the performance of the microarray platform.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/7/2625/s1. Supplemental Table S1. Differentially expressed genes of each progestin vs. vehicle without and with addition of E2 (FC ≥ 1.5). Highlighted rows are the common genes between each progestin without or with addition of estradiol. E2: estradiol; pink: progesterone (P4); green: medroxyprogesterone acetate (MPA); blue: levonorgestrel (LNG); yellow: norethindrone acetate (NETA); FC: fold change. Supplemental Figure S1. IGFBP1 protein level in different treatments. Protein levels of IGFBP1, a progesterone target gene, was determined by ELISA in conditioned media of n=5 eSF cell lines grown in culture and normalized to cell number. For each sample, each assay was done in duplicate. Values for each progestin or progestin plus E2 is the measurements from 5 samples with error bars.
Author Contributions
Conceptualization, S.H., J.C.C., K.S.-M., L.C.G.; methodology, S.H., J.C.C., J.C.I., L.C.G.; sample procurement, K.C.V.; software, S.H., J.C.C., J.V.-J., S.B.; validation, J.C.C., S.B.; formal analysis, S.H., J.C.C., J.V.-J.; resources, L.C.G., K.S.-M., R.M.G.; original draft preparation, S.H., J.V.-J.; review and editing, S.H., J.V.-J., J.C.C., K.S.-M., R.M.G., J.C.I., L.C.G. All authors have read and agreed to the published version of the manuscript.
Funding
Supported by NIH/NIAID P01 grant # AI083050-05 (L.C.G., K.S.M., R.M.G.), and the National Institutes of Health Eunice Kennedy Shriver National Institute for Child Health and Human Development, Ruth L Kirschstein National Research Service Award # 1F32 HD074423-02 (J.C.C.).
Conflicts of Interest
The authors have nothing to disclose.
References
- 1.Stanczyk F.Z. All progestins are not created equal. Steroids. 2003;68:879–890. doi: 10.1016/j.steroids.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Stanczyk F.Z., Hapgood J.P., Winer S., Mishell D.R. Progestogens used in postmenopausal hormone therapy: Differences in their pharmacological properties, intracellular actions, and clinical effects. Endocr. Rev. 2013;34:171–208. doi: 10.1210/er.2012-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giudice L.C. Clinical Practice: Endometriosis. N. Engl. J. Med. 2010;362:2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J.J., Chapman-Davis E. Seminars in Reproductive Medicine. Volume 28. Thieme Medical Publishers; New York, NY, USA: 2017. Role of Progesterone in Endometrial Cancer; pp. 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gezer A., Oral E. Progestin therapy in endometriosis. Women’s Health. 2015;11:643–652. doi: 10.2217/whe.15.42. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber C.A., Barnhart K. Yen & Jaffe’s Reproductive Endocrinology: Physiology, Pathophysiology, and Clinical Management. 8th ed. W.B. Saunders; Philadelphia, PA, USA: 2018. Contraception; pp. 962–978. [Google Scholar]
- 7.Wilcox T., Hirshkowitz A. Types of combined oral contraceptives used by US women. Contraception. 2015;85:1–27. [Google Scholar]
- 8.Chen M.J., Kim C.R., Whitehouse K.C., Berry-Bibee E., Gaffield M.E. Development, updates, and future directions of the World Health Organization Selected Practice Recommendations for Contraceptive Use. Int. J. Gynecol. Obstet. 2017;136:113–119. doi: 10.1002/ijgo.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jameson J.L., De Groot L.J., de Kretser D.M., Giudice L.C., Grossman A.B., Melmed S., Potts J.T., Weir G.C., Glasier A. Endocrinol Adult Pediatr. Saunders; Philadelphia, PA, USA: 2016. Contraception; pp. 2297–2309. [Google Scholar]
- 10.Whitehead M.I., Townsend P.T., Pryse-Davies J., Ryder T.A., King R.J.B. Effects of Estrogens and Progestins on the Biochemistry and Morphology of the Postmenopausal Endometrium. N. Engl. J. Med. 1981;305:1599–1605. doi: 10.1056/NEJM198112313052701. [DOI] [PubMed] [Google Scholar]
- 11.Benagiano G., Pera A., Primiero F.M. The endometrium and hormonal contraceptives. Hum. Reprod. 2000;15:101–118. doi: 10.1093/humrep/15.suppl_1.101. [DOI] [PubMed] [Google Scholar]
- 12.Dinh A., Sriprasert I., Williams A.R., Archer D.F. A review of the endometrial histologic effects of progestins and progesterone receptor modulators in reproductive age women. Contraception. 2015;91:360–367. doi: 10.1016/j.contraception.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Epplein M., Reed S.D., Voigt L.F., Newton K.M., Holt V.L., Weiss N.S. Endometrial Hyperplasia Risk in Relation to Recent Use of Oral Contraceptives and Hormone Therapy. Ann. Epidemiol. 2009;19:1–7. doi: 10.1016/j.annepidem.2008.08.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polis C.B., Phillips S.J., Hillier S.L., Achilles S.L. Levonorgestrel in contraceptives and multipurpose prevention technologies: Does this progestin increase HIV risk or interact with antiretrovirals? AIDS. 2016;30:2571. doi: 10.1097/QAD.0000000000001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hapgood J.P., Kaushic C., Hel Z. Hormonal contraception and HIV-1 acquisition: Biological mechanisms. Endocr. Rev. 2018;39:36–78. doi: 10.1210/er.2017-00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deligdisch L. Hormonal pathology of the endometrium. Mod. Pathol. 2000;13:285–294. doi: 10.1038/modpathol.3880050. [DOI] [PubMed] [Google Scholar]
- 17.Illouz S., Dales J.P., Sferlazzo K., Garcia S., Carpentier-Meunier S., Boubli L., Lavaut M.N., Charpin C. Effects of progestins of human proliferative endometrium: An in vitro model of potential clinical relevance. Int. J. Mol. Med. 2003;12:517–523. doi: 10.3892/ijmm.12.4.517. [DOI] [PubMed] [Google Scholar]
- 18.Jones R.L., Morison N.B., Hannan N.J., Critchley H.O.D., Salamonsen L.A. Chemokine expression is dysregulated in the endometrium of women using progestin-only contraceptives and correlates to elevated recruitment of distinct leukocyte populations. Hum. Reprod. 2005;20:2724–2735. doi: 10.1093/humrep/dei140. [DOI] [PubMed] [Google Scholar]
- 19.Goldfien G.A., Barragan F., Chen J., Takeda M., Irwin J.C., Perry J., Greenblatt R.M., Smith-Mccune K.K., Giudice L.C. Progestin-containing contraceptives alter expression of host defense-related genes of the endometrium and cervix. Reprod. Sci. 2015;22:814–828. doi: 10.1177/1933719114565035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuevas C.A., Tapia-Pizarro A., Salvatierra A.M., Munroe D.J., Velasquez L., Croxatto H.B. Effect of single post-ovulatory administration of mifepristone (RU486) on transcript profile during the receptive period in human endometrium. Reproduction. 2016;151:331–349. doi: 10.1530/REP-15-0458. [DOI] [PubMed] [Google Scholar]
- 21.Okada H., Tsuzuki T., Murata H. Decidualization of the human endometrium. Reprod. Med. Biol. 2018;17:220–227. doi: 10.1002/rmb2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinketova K., Mourdjeva M., Oreshkova T. Human Decidual Stromal Cells as a Component of the Implantation Niche and a Modulator of Maternal Immunity. J. Pregnancy. 2016;2016:1–17. doi: 10.1155/2016/8689436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aghajanova L., Velarde M., Giudice L. Altered Gene Expression Profiling in Endometrium: Evidence for Progesterone Resistance. Semin. Reprod. Med. 2010;28:051–058. doi: 10.1055/s-0029-1242994. [DOI] [PubMed] [Google Scholar]
- 24.Gargett C.E., Schwab K.E., Deane J.A. Endometrial stem/progenitor cells: The first 10 years. Hum. Reprod. Update. 2016;22:137–163. doi: 10.1093/humupd/dmv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner R.M., Slayden O.D. The Physiology of Reproduction. 2nd ed. Academic Press; Cambridge, MA, USA: 1994. Cyclic changes in the primate oviduct and endometrium; pp. 541–569. [Google Scholar]
- 26.Smith O.P.M., Critchley H.O.D. Progestogen only contraception and endometrial break through bleeding. Angiogenesis. 2005;8:117–126. doi: 10.1007/s10456-005-9003-z. [DOI] [PubMed] [Google Scholar]
- 27.Singh M., Su C., Ng S. Non-genomic mechanisms of progesterone action in the brain. Front. Neurosci. 2013;7:159. doi: 10.3389/fnins.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vallejo G., La Greca A.D., Tarifa-Reischle I.C., Mestre-Citrinovitz A.C., Ballaré C., Beato M., Saragüeta P. CDC2 mediates progestin initiated endometrial stromal cell proliferation: A PR signaling to gene expression independently of its binding to chromatin. PLoS ONE. 2014;9:e97311. doi: 10.1371/journal.pone.0097311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindler A.E., Campagnoli C., Druckmann R., Huber J., Pasqualini J.R., Schweppe K.W., Thijssen J.H.H. Classification and pharmacology of progestins. Maturitas. 2003;46:7–16. doi: 10.1016/j.maturitas.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Africander D.J., Storbeck K.H., Hapgood J.P. A comparative study of the androgenic properties of progesterone and the progestins, medroxyprogesterone acetate (MPA) and norethisterone acetate (NET-A) J. Steroid Biochem. Mol. Biol. 2014;143:404–415. doi: 10.1016/j.jsbmb.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Koubovec D., Ronacher K., Stubsrud E., Louw A., Hapgood J.P. Synthetic progestins used in HRT have different glucocorticoid agonist properties. Mol. Cell. Endocrinol. 2005;242:23–32. doi: 10.1016/j.mce.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Bamberger C.M., Else T., Bamberger A.M., Beil F.U., Schulte H.M. Dissociative glucocorticoid activity of medroxyprogesterone acetate in normal human lymphocytes. J. Clin. Endocrinol. Metab. 1999;84:4055–4061. doi: 10.1210/jc.84.11.4055. [DOI] [PubMed] [Google Scholar]
- 33.Yin P., Roqueiro D., Huang L., Owen J.K., Xie A., Navarro A., Monsivais D., Coon J.S., Kim J.J., Dai Y., et al. Genome-wide progesterone receptor binding: Cell type-specific and shared mechanisms in T47D breast cancer cells and primary leiomyoma cells. PLoS ONE. 2012;7:e29021. doi: 10.1371/journal.pone.0029021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arici A. Regulation of Monocyte Chemotactic Protein-1 Expression in Human Endometrial Stromal Cells by Estrogen and Progesterone. Biol. Reprod. 1999;61:85–90. doi: 10.1095/biolreprod61.1.85. [DOI] [PubMed] [Google Scholar]
- 35.Alam R., Lett-Brown M.A., Forsythe P.A., Anderson-Walters D.J., Kenamore C., Kormos C., Grant J.A. Monocyte chemotactic and activating factor is a potent histamine-releasing factor for basophils. J. Clin. Investig. 1992;89:723–728. doi: 10.1172/JCI115648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allavena P., Bianchi G., Zhou D., Van Damme J., Jílek P., Sozzani S., Mantovani A. Induction of natural killer cell migration by monocyte chemotactic protein−1, −2 and −3. Eur. J. Immunol. 1994;24:3233–3236. doi: 10.1002/eji.1830241249. [DOI] [PubMed] [Google Scholar]
- 37.Ueda A., Okuda K., Ohno S., Shirai A., Igarashi T., Matsunaga K., Fukushima J., Kawamoto S., Ishigatsubo Y., Okubo T. NF-kappa B and Sp1 regulate transcription of the human monocyte chemoattractant protein-1 gene. J. Immunol. 1994;153:2052–2063. [PubMed] [Google Scholar]
- 38.Arici A., MacDonald P.C., Caseya M.L. Regulation of monocyte chemotactic protein-1 gene expression in human endometrial cells in cultures. Mol. Cell. Endocrinol. 1995;107:189–197. doi: 10.1016/0303-7207(94)03442-V. [DOI] [PubMed] [Google Scholar]
- 39.Lockwood C.J., Krikun G., Hickey M., Huang J.S.F. Decidualized Human Endometrial Stromal Cells Mediate Hemostasis, Angiogenesis, and Abnormal Uterine Bleeding. Reprod. Sci. 2009;16:162–170. doi: 10.1177/1933719108325758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mor G., Cardenas I., Abrahams V., Guller S. Inflammation and pregnancy: The role of the immune system at the implantation site. Ann. N. Y. Acad. Sci. 2011;1221:80–87. doi: 10.1111/j.1749-6632.2010.05938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Sinderen M., Menkhorst E., Winship A., Cuman C., Dimitriadis E. Preimplantation Human Blastocyst-Endometrial Interactions: The Role of Inflammatory Mediators. Am. J. Reprod. Immunol. 2013;69:427–440. doi: 10.1111/aji.12038. [DOI] [PubMed] [Google Scholar]
- 42.Dekel N., Gnainsky Y., Granot I., Racicot K., Mor G., Dekel C.N. The Role of Inflammation for a Successful Implantation. Am. J. Reprod. Immunol. 2014;72:141–147. doi: 10.1111/aji.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagamatsu T., Schust D.J. The Contribution of Macrophages to Normal and Pathological Pregnancies. Am. J. Reprod. Immunol. 2010;63:460–471. doi: 10.1111/j.1600-0897.2010.00813.x. [DOI] [PubMed] [Google Scholar]
- 44.Hu X., Park-Min K.-H., Ho H.H., Ivashkiv L.B. IFN-γ-Primed Macrophages Exhibit Increased CCR2-Dependent Migration and Altered IFN-γ Responses Mediated by Stat1. J. Immunol. 2005;175:3637–3647. doi: 10.4049/jimmunol.175.6.3637. [DOI] [PubMed] [Google Scholar]
- 45.Hess A.P., Nayak N.R., Giudice L.C. Oviduct and Endometrium: Cyclic Changes in the Primate Oviduct and Endometrium. 3rd ed. Volume 3. Elsevier Inc.; Amsterdam, The Netherlands: 2006. [Google Scholar]
- 46.Hayes I.M., Jordan N.J., Towers S., Smith G., Paterson J.R., Earnshaw J.J., Roach A.G., Westwick J., Williams R.J. Human vascular smooth muscle cells express receptors for CC chemokines. Arterioscler. Thromb. Vasc. Biol. 1998;18:397–403. doi: 10.1161/01.ATV.18.3.397. [DOI] [PubMed] [Google Scholar]
- 47.Lash G.E., Innes B.A., Drury J.A., Robson S.C., Quenby S., Bulmer J.N. Localization of angiogenic growth factors and their receptors in the human endometrium throughout the menstrual cycle and in recurrent miscarriage. Hum. Reprod. 2012;27:183–195. doi: 10.1093/humrep/der376. [DOI] [PubMed] [Google Scholar]
- 48.Smith S.K. Regulation of angiogenesis in the endometrium. Trends Endocrinol. Metab. 2001;12:147–151. doi: 10.1016/S1043-2760(01)00379-4. [DOI] [PubMed] [Google Scholar]
- 49.Barragan F., Irwin J.C., Balayan S., Erikson D.W., Chen J.C., Houshdaran S., Piltonen T.T., Spitzer T.L.B., George A., Rabban J.T., et al. Human Endometrial Fibroblasts Derived from Mesenchymal Progenitors Inherit Progesterone Resistance and Acquire an Inflammatory Phenotype in the Endometrial Niche in Endometriosis. Biol. Reprod. 2016;94:118. doi: 10.1095/biolreprod.115.136010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., Feng Y., Lin J.F., Billig H., Shao R. Endometrial progesterone resistance and PCOS. J. Biomed. Sci. 2014;21:1–7. doi: 10.1186/1423-0127-21-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irwin J.C., Kirk D., King R.J.B., Quigley M.M., Gwatkin R.B.L. Hormonal regulation of human endometrial stromal cells in culture: An in vitro model for decidualization. Fertil. Steril. 1989;52:761–768. doi: 10.1016/S0015-0282(16)61028-2. [DOI] [PubMed] [Google Scholar]
- 52.Aghajanova L., Tatsumi K., Horcajadas J.A., Zamah A.M., Esteban F.J., Herndon C.N., Conti M., Giudice L.C. Unique transcriptome, pathways, and networks in the human endometrial fibroblast response to progesterone in endometriosis. Biol. Reprod. 2011;84:801–815. doi: 10.1095/biolreprod.110.086181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Attardi B.J., Koduri S., Hild S.A. Relative progestational and androgenic activity of four progestins used for male hormonal contraception assessed in vitro in relation to their ability to suppress LH secretion in the castrate male rat. Mol. Cell. Endocrinol. 2010;328:16–21. doi: 10.1016/j.mce.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Oliveros J.C. Venny 2.1.0. [(accessed on 20 March 2018)]; Available online: http://bioinfogp.cnb.csic.es/tools/venny/
- 55.QIAGEN Inc. [(accessed on 7 July 2018)]; Available online: https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.