Abstract
Introduction
Triggering receptor expressed on myeloid cells‐like transcript 2 gene (TREML2) is a newly identified AD susceptibility gene. Its missense variant rs3747742‐C substantially decreases AD risk in both Caucasians and Han Chinese, but the underlying mechanisms remain elusive. In the present study, to uncover the possible mechanisms by which TREML2 rs3747742‐C reduces AD risk, we investigated the possible relation of this variant with AD‐related brain structures using a cognitively normal elderly population from Alzheimer's Disease Neuroimaging Initiative (ADNI) database.
Methods
In total, 158 cognitively normal elders from ADNI database with complete data for brain structures and TREML2 rs3747742 genotype were included in this study. The association of TREML2 rs3747742 genotype with the structures of three cerebral cortices (entorhinal cortex, middle temporal gyrus, and parahippocampal gyrus), two subcortical regions (amygdala and hippocampus), and three subfields of hippocampus (CA1, CA2 + CA3, and CA4 + dentate gyrus) was investigated.
Results
A significant difference was noted in the volume of right CA1 subfield among three genotypes of TREML2 rs3747742 (p = .0364). In the multivariate analysis, TREML2 rs3747742‐C significantly increased right CA1 subfield volume after adjusting for age, gender, education years, APOE ε4 status, and intracranial volume under the recessive genetic model (Bonferroni corrected p = .003586).
Conclusion
The present study provides the first evidence that TREML2 rs3747742‐C carriers have larger volumes of hippocampal CA1 subfield in a cognitively normal elderly population. These findings imply that enhancement of brain reserve may contribute to the protection of TREML2 rs3747742‐C in AD susceptibility.
Keywords: ADNI, Alzheimer' s disease, CA1, TREML2
The present study provides the first evidence that TREML2 rs3747742‐C carriers have larger volumes of hippocampal CA1 subfield in a cognitively normal elderly population. These findings imply that enhancement of brain reserve may contribute to the protection of TREML2 rs3747742‐C in AD susceptibility.

1. INTRODUCTION
Alzheimer's disease (AD) is the most common form of dementia in the world, affecting 5% of the population over 65 years (Alzheimer's, 2016; Lane, Hardy, & Schott, 2018). The late‐onset AD accounts for the majority of all AD cases and is considered as a genetically complex disorder that caused by an interaction between environmental factors and susceptibility genes (Jiang, Yu, Tian, & Tan, 2013). Investigating the mechanisms by which variants of these susceptibility genes modify AD risk will provide valuable insights into the pathogenesis of this devastating disease.
Triggering receptor expressed on myeloid cells‐like transcript 2 gene (TREML2) is a newly identified AD susceptibility gene. Its missense variant rs3747742‐C is revealed to substantially reduce AD risk in both Caucasians and Han Chinese (Benitez et al., 2014; Jiang et al., 2017), but the underlying mechanisms are still elusive. Previous studies investigated the association of TREML2 rs3747742 with cerebrospinal fluid (CSF) biomarkers of AD and found that carriers of C allele had a decreased level of both hyperphosphorylated and total CSF tau (Benitez et al., 2013; Song et al., 2019), indicating that TREML2 rs3747742‐C may decrease AD risk by attenuating neurodegeneration process.
In the present study, to uncover the other possible mechanisms by which TREML2 rs3747742‐C reduces AD risk, we investigated the possible relation of this variant with AD‐related brain structures using a cognitively normal elderly population from Alzheimer's Disease Neuroimaging Initiative (ADNI) database. For the first time, we showed that TREML2 rs3747742‐C carriers have larger volumes of hippocampal CA1 subfield after adjusting for age, gender, education years, APOE ε4 status, and intracranial volume. These findings imply that enhancement of brain reserve may also contribute to the protection of TREML2 rs3747742‐C in AD susceptibility.
2. METHODS
2.1. About ADNI database and subject selection
All data used in this article were obtained from the ADNI database (http://adni.loni.usc.edu). The ADNI was launched in 2003 as a public‐private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer's disease (AD). For up‐to‐date information, see http://www.adni-info.org. In total, 158 cognitively normal elders from ADNI database with complete data for brain structures and TREML2 rs3747742 genotype were included in this study.
2.2. Ethical approval
As per ADNI protocols, all procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. More details can be found at adni.loni.usc.edu.
2.3. MRI for brain structures
MRI data of brain structures were extracted from ADNI website (TREML2 rs3747742 C/C genotype: 12 subjects, TREML2 rs3747742 C/T genotype: 71 subjects, TREML2 rs3747742 T/T genotype: 75 subjects). The details regarding image acquisition and process can be found at http://adni.loni.usc.edu/methods. In this study, three cerebral cortices (entorhinal cortex, middle temporal gyrus, and parahippocampal gyrus) and two subcortical structures (amygdala and hippocampus) that associated with AD were defined as ROIs. Regarding the subfields of hippocampus, three regions (CA1, CA2 + CA3, and CA4 + dentate gyrus) were selected as ROIs.
2.4. Statistical analysis
Statistical analyses were carried out by GraphPad Prism 6 (GraphPad Software), R 3.12 (http://www.r-project.org/) and PLINK 1.07 (http://pngu.mgh.harvard.edu/wpurcell/plink/). In univariate analysis, the association of TREML2 rs3747742 genotypes with AD‐related brain structures was investigated using one‐way ANOVA. In multivariate analysis, the association between TREML2 rs3747742 genotypes and AD‐related brain structures was analyzed using a multiple linear regression adjusting for age, gender, education years, APOE ε4 status, and intracranial volume under dominant and recessive genetic models. These two genetic models were defined as follows: (C/C + C/T) versus T/T for the dominant model, C/C versus (C/T + T/T) for the recessive model. Bonferroni correction was used for the adjustment of multiple comparisons. p < .05 was considered significant.
3. RESULTS
As indicated by Table S1 and Table S2, no difference was observed in the average thickness and volume of AD‐related cerebral cortices including bilateral entorhinal cortices, middle temporal gyri, and parahippocampal gyri among three genotypes of TREML2 rs3747742. Meanwhile, no difference was noted in the volume of subcortical structures including bilateral amygdalae and hippocampi among three genotypes of TREML2 rs3747742 (Table S3).
Regarding the hippocampal subfields, a significant difference was found in the volume of right CA1 subfield among three genotypes of TREML2 rs3747742 (p = .0364, see Table 1). We then investigated the association between TREML2 rs3747742 and right CA1 subfield volume using multiple linear regression adjusting for age, gender, education years, APOE ε4 status, and intracranial volume under dominant and recessive genetic models. As revealed by Figure 1, TREML2 rs3747742‐C significantly increased right CA1 subfield volume under the recessive genetic model after Bonferroni correction (Pc = 0.003586).
Table 1.
The influence of TREML2 rs3747742 genotypes on the volume of hippocampal subfields
| Hippocampal subfields | Genotypes | p value | ||
|---|---|---|---|---|
| C/C | C/T | T/T | ||
| CA1 subfield (Left, mm3) | 334.5 ± 62.15 (12) | 316.5 ± 42.16 (71) | 321.1 ± 43.77 (75) | .4191 |
| CA1 subfield (Right, mm3) | 358.7 ± 74.23 (12) | 327.4 ± 38.47 (71) | 322.1 ± 45.48 (75) | .0364 |
| CA2 + CA3 subfields (Left, mm3) | 909.4 ± 103.8 (12) | 911.5 ± 137.2 (71) | 903.9 ± 129.1 (75) | .9397 |
| CA2 + CA3 subfields (Right, mm3) | 981.3 ± 111.1 (12) | 968.5 ± 135.5 (71) | 947.4 ± 150.4 (75) | .5713 |
| CA4 + dentate gyrus subfields (Left, mm3) | 506.9 ± 56.2 (12) | 512.8 ± 71.32 (71) | 510.2 ± 73.27 (75) | .9541 |
| CA4 + dentate gyrus subfields (Right, mm3) | 535.9 ± 59.21 (12) | 539.2 ± 74.61 (71) | 524.9 ± 80.2 (75) | .5195 |
Data in this table are expressed as mean ± SD (number of subjects) and analyzed with one‐way ANOVA.
Figure 1.
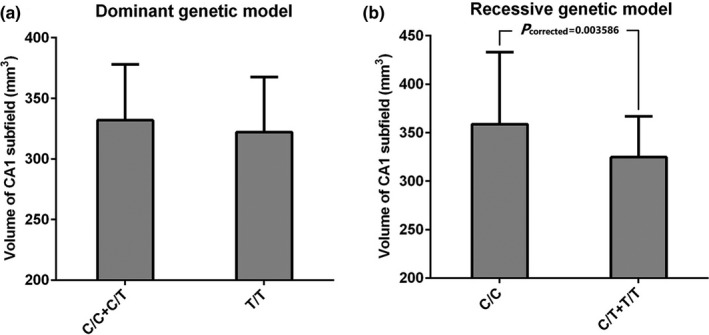
The influence of TREML2 rs3747742‐C on right CA1 subfield volume. (a) The association between TREML2 rs3747742‐C and right CA1 subfield volume after adjusting for age, gender, education years, APOE ε4 status, and intracranial volume under the dominant genetic model ([C/C + C/T] vs. T/T). (b) The association between TREML2 rs3747742‐C and right CA1 subfield volume after adjusting for age, gender, education years, APOE ε4 status, and intracranial volume under the recessive genetic model (C/C vs. [C/T + T/T])
4. DISCUSSION
As a newly identified susceptibility gene for AD, TREML2 locates on chromosome 6p21.1‐q15, a region showing a strong association with AD risk (Lambert et al., 2013). Previously, by mining Caucasian exome‐sequencing datasets, Benitez and colleagues revealed that TREML2 rs3747742‐C was associated with a lowered risk of AD (Benitez et al., 2014). Recently, we had confirmed this association in a large Han Chinese population including 992 AD patients and 1,358 healthy controls (Jiang et al., 2017), further supporting a protective role of TREML2 rs3747742‐C in AD susceptibility.
However, the underlying mechanisms by which TREML2 rs3747742‐C reduces AD risk remain largely unknown. Benitez et al. recently found that TREML2 rs3747742‐C carriers had a decreased level of CSF hyperphosphorylated tau (Benitez et al., 2013). Meanwhile, Song and colleagues revealed that TREML2 rs3747742‐C was related to a reduced CSF total tau level in AD patients after controlling for age, gender, education, and APOE ε4 status (Song et al., 2019). Since CSF tau levels reflect the degree of neurodegeneration (Frost, Gotz, & Feany, 2015), these findings implied that TREML2 rs3747742‐C might modify AD risk by attenuating neurodegeneration process. In the present study, we investigated the possible relation of TREML2 rs3747742‐C with AD‐related brain structures using a cognitively normal elderly population from ADNI database. For the first time, we revealed that TREML2 rs3747742‐C carriers had larger volumes of hippocampal CA1 subfield after adjusting for age, gender, education years, APOE ε4 status, and intracranial volume. The hippocampal CA1 subfield, also known as Sommer's sector, is required for the retrieval of spatial and contextual memory (Guerreiro et al., 2013; Thomas, 2015). Since CA1 subfield is particularly susceptible to cytotoxicity (Yang, Tian, Yang, & Zhang, 2010), a larger volume of this region might provide a better compensation for neuropathological damages during AD progression. Therefore, it seemed that enhancement of brain reserve might also contribute to the protection of TREML2 rs3747742‐C in AD susceptibility.
Triggering receptor expressed on myeloid cells‐like transcript 2 gene encodes a single‐pass type I membrane protein that belongs to the immunoglobulin superfamily (Klesney‐Tait, Turnbull, & Colonna, 2006). In the brain, TREML2 is mainly expressed by microglia, the immune cell within the central nervous system (Zheng et al., 2016). In a previous functional study, Zheng and colleagues showed that the expression of TREML2 on microglia was upregulated by oligomeric amyloid‐β stimulation (Zheng et al., 2016). More importantly, knockdown of TREML2 facilitated microglia proliferation and suppressed microglia‐mediated release of proinflammatory cytokines (Zheng et al., 2016). These findings implied that TREML2 might be a modulator of microglia during AD progression. As a missense variant, TREML2 rs3747742‐C leads to a change of amino acids at 144 residue (p.S144G) of TREML2, and functional studies are warranted to determine whether the protective effects of TREML2 rs3747742‐C against AD susceptibility are related to the alteration of TREML2 protein and microglia functions. However, according to the prediction of MutationTaster and PolyPhen‐2 software (Adzhubei, Jordan, & Sunyaev, 2013; Adzhubei et al., 2010; Schwarz, Cooper, Schuelke, & Seelow, 2014), p.S144G amino acid change was unlikely to affect the structure or functions of TREML2 protein. Therefore, it is also possible that the TREML2 rs3747742‐C is in LD with other nearby functional variants and thus protects against AD risk (Carrasquillo et al., 2017).
The main limitation of this study is the relatively small sample size of the cognitively normal elders in the ADNI dataset. Therefore, our findings should be further validated using a larger cohort in the future.
In conclusion, the present study provides the first evidence that TREML2 rs3747742‐C carriers have larger volumes of hippocampal CA1 subfield after adjusting for age, gender, education years, APOE ε4 status, and intracranial volume in a cognitively normal elderly population. These findings imply that enhancement of brain reserve may also contribute to the protection of TREML2 rs3747742‐C in AD susceptibility.
CONFLICT OF INTEREST
The authors confirm that this article has no conflict of interest.
AUTHORS' CONTRIBUTION
Si‐Yu Wang and Xiao Xue were involved in data analysis. Rui Duan and Peng‐Yu Gong were involved in data acquisition. Yan E and Ying‐Dong Zhang were involved in data interpretation. Teng Jiang was involved in manuscript preparation. The investigators within the ADNI contributed to the design and implementation of ADNI database.
INFORMED CONSENT
A written informed consent was obtained from each participant or the legal guardian. More details can be found at adni.loni.usc.edu.
Supporting information
ACKNOWLEDGMENT
Data collection and sharing for this project was funded by the ADNI (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. This work was also supported by National Natural Science Foundation of China (81974156), “Six Talent Summit” Foundation of Jiangsu Province (2016‐WSN‐180), Youth Medical Talent Program of Jiangsu Province (QNRC2016068), and Nanjing Medical Science and Technology Development Foundation for Distinguished Young Scholars (JQX17008). We deeply thank Dr. Hui‐Fu Wang for the assistance of data analysis.
Wang S‐Y, Xue X, Duan R, et al; For the Alzheimer’s Disease Neuroimaging Initiative . A TREML2 missense variant influences specific hippocampal subfield volumes in cognitively normal elderly subjects. Brain Behav. 2020;10:e01573 10.1002/brb3.1573
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
The peer review history for this article is available at https://publons.com/publon/10.1002/brb3.1573 [Correction added on 11 March 2020, after first online publication: Peer review history statement has been added.]
Contributor Information
Teng Jiang, Email: jt870918@163.com.
Ying‐Dong Zhang, Email: zhangyingdong@aliyun.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Adzhubei, I. , Jordan, D. M. , & Sunyaev, S. R. (2013). Predicting functional effect of human missense mutations using PolyPhen‐2. Current Protocols in Human Genetics, 76, 7.20.1–7.20.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei, I. A. , Schmidt, S. , Peshkin, L. , Ramensky, V. E. , Gerasimova, A. , Bork, P. , … Sunyaev, S. R. (2010). A method and server for predicting damaging missense mutations. Nature Methods, 7(4), 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer's Association (2016). 2016 Alzheimer's disease facts and figures. Alzheimer's & Dementia: The Journal of the Alzheimer's Association, 12(4), 459–509. [DOI] [PubMed] [Google Scholar]
- Benitez, B. A. , Cooper, B. , Pastor, P. , Jin, S.‐C. , Lorenzo, E. , Cervantes, S. , & Cruchaga, C. (2013). TREM2 is associated with the risk of Alzheimer's disease in Spanish population. Neurobiology of Aging, 34(6), 1711.e15–1711.e17. 10.1016/j.neurobiolaging.2012.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez, B. A. , Jin, S. C. , Guerreiro, R. , Graham, R. , Lord, J. , Harold, D. , … Cruchaga, C. (2014). Missense variant in TREML2 protects against Alzheimer's disease. Neurobiology of Aging, 35(6), 1510.e19–1510.e26. 10.1016/j.neurobiolaging.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo, M. M. , Allen, M. , Burgess, J. D. , Wang, X. , Strickland, S. L. , Aryal, S. , … Ertekin‐Taner, N. (2017). A candidate regulatory variant at the TREM gene cluster associates with decreased Alzheimer's disease risk and increased TREML1 and TREM2 brain gene expression. Alzheimer's & Dementia: The Journal of the Alzheimer's Association, 13(6), 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost, B. , Gotz, J. , & Feany, M. B. (2015). Connecting the dots between tau dysfunction and neurodegeneration. Trends in Cell Biology, 25(1), 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro, R. , Wojtas, A. , Bras, J. , Carrasquillo, M. , Rogaeva, E. , Majounie, E. … Alzheimer Genetic Analysis Group (2013). TREM2 variants in Alzheimer's disease. The New England Journal of Medicine, 368(2), 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T. , Wan, Y. , Zhou, J.‐S. , Tan, M.‐S. , Huang, Q. , Zhu, X.‐C. , … Yu, J.‐T. (2017). A missense variant in TREML2 reduces risk of Alzheimer's Disease in a Han Chinese population. Molecular Neurobiology, 54(2), 977–982. [DOI] [PubMed] [Google Scholar]
- Jiang, T. , Yu, J. T. , Tian, Y. , & Tan, L. (2013). Epidemiology and etiology of Alzheimer's disease: From genetic to non‐ genetic factors. Current Alzheimer Research, 10(8), 852–867. [DOI] [PubMed] [Google Scholar]
- Klesney‐Tait, J. , Turnbull, I. R. , & Colonna, M. (2006). The TREM receptor family and signal integration. Nature Immunology, 7(12), 1266–1273. [DOI] [PubMed] [Google Scholar]
- Lambert, J. C. , Ibrahim‐Verbaas, C. A. , Harold, D. , Naj, A. C. , Sims, R. , Bellenguez, C. , … Bellenguez, C. (2013). Meta‐analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature Genetics, 45(12), 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, C. A. , Hardy, J. , & Schott, J. M. (2018). Alzheimer's disease. European Journal of Neurology: The Official Journal of the European Federation of Neurological Societies, 25(1), 59–70. [Google Scholar]
- Schwarz, J. M. , Cooper, D. N. , Schuelke, M. , & Seelow, D. (2014). MutationTaster2: Mutation prediction for the deep‐sequencing age. Nature Methods, 11(4), 361–362. [DOI] [PubMed] [Google Scholar]
- Song, Y. N. , Li, J. Q. , Tan, C. C. , Wang, H.‐F. , Tan, M.‐S. , Cao, X.‐P. , … The Alzheimer's Disease Neuroimaging Initiative (2019). TREML2 mutation mediate Alzheimer's Disease risk by altering neuronal degeneration. Frontiers in Neuroscience, 13, 455 10.3389/fnins.2019.00455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S. A. (2015). Neuromodulatory signaling in hippocampus‐dependent memory retrieval. Hippocampus, 25(4), 415–431. 10.1002/hipo.22394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. J. , Tian, Y. T. , Yang, Z. , & Zhang, T. (2010). Effect of melamine on potassium currents in rat hippocampal CA1 neurons. Toxicology in Vitro: An International Journal Published in Association with BIBRA, 24(2), 397–403. [DOI] [PubMed] [Google Scholar]
- Zheng, H. , Liu, C.‐C. , Atagi, Y. , Chen, X.‐F. , Jia, L. , Yang, L. , … Bu, G. (2016). Opposing roles of the triggering receptor expressed on myeloid cells 2 and triggering receptor expressed on myeloid cells‐like transcript 2 in microglia activation. Neurobiology of Aging, 42, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


