Abstract
Genome-wide association studies (GWAS) have identified common variants for quantitative traits (insulin resistance and impaired insulin release) of type 2 diabetes (T2D) across different ethnics including China, but results were inconsistent. The study included 1654 subjects who were selected from the 2010–2012 China National Nutrition and Health Surveillance (CNNHS). Insulin resistance and impaired insulin release were assessed by homeostasis model assessment (HOMA). The study included 64 diabetes-related single nucleotide polymorphisms (SNPs), which were done using Mass ARRAY. A logistic regression model was employed to explore the associations of SNPs with insulin resistance and impaired insulin release by correcting for the confounders. The 5q11.2-rs4432842, RASGRP1-rs7403531, and SEC16B-rs574367 increased the risk of insulin resistance with OR = 1.23 (95% CI: 1.04–1.45, OR = 1.35 (95% CI: 1.13–1.62), OR = 1.34 (95% CI: 1.07–1.67), respectively, while MAEA-rs6815464 decreased the risk of insulin resistance (OR = 0.84, 95% CI: 0.71–1.00). CENTD2-rs1552224, TSPAN8-rs7961581 and ANK1-rs516946 was associated with increased risk of impaired insulin release with OR = 1.47 (95% CI: 1.09–1.99), OR = 1.25 (95% CI: 1.03–1.51), OR = 1.39 (95% CI: 1.07–1.81), respectively. Our findings would provide insight into the pathogenesis of individual SNPs and T2D.
Keywords: type 2 diabetes, insulin resistance, impaired insulin release, SNP, HOMA-IR, HOMA-β
1. Introduction
Type 2 diabetes (T2D) is a severe chronic non-communicable disease concerning public health in China [1]. It may cause multiple organ complications such as diabetic retinopathy, diabetic nephropathy, and peripheral neuropathy, which has become a burden not only for individual physiology and psychology but also for allocation of health resources [2]. The pathogenesis of T2D is complicated, although lifestyle is a great contributor to the T2D epidemic, genetic determinants can also affect the T2D susceptibility [3]. Individuals carrying risk alleles predisposed to T2D are mediated by insulin resistance or abnormal insulin secretion [4]. Insulin resistance [5] is one critical process to the development of T2D, which means the function of pancreatic β-cells is commonly normal and insulin secretion is sufficient, but the insulin receptors are insensitive to insulin levels and the ability to stimulate glucose utilization is decreasing, or impairments in insulin signal transduction, resulting in a compensatory hyperinsulinemia [6]. Besides, insulin resistance is ordinarily related to obesity or other metabolic syndromes [7].
Insulin release decrease is another mechanism of T2D when pancreatic β-cells are impaired, and individuals are unable to utilize glucose which will cause high postprandial blood glucose. A majority of variants have been identified to associate with β-cell dysfunction [8], but evidence suggests that Asians might be more susceptible to insulin resistance than Europeans [9]. Genome-wide association studies (GWAS) for T2D have been widely carried out in different racial types, with the exception of T2D-associated genes, obesity-associated and birth weight-associated genes have also been found to relate to T2D, but results were inconsistent. For example, loci rs13266634 in gene SLC30A8 was associated with T2D in Russian and the Danish populations [10,11], while the link disappeared in African populations [12]. A common obesity-associated gene, FTO, has been confirmed to have an association with T2D in different countries, and several gene loci were associated with insulin resistance such as rs9939609 and rs1558902 [13,14]. Birth weight-associated loci rs6931514 in gene CDKAL1 was associated with reduced insulin secretion in Europeans [15], which suggested that the genetic variation was affected differently among races and geographic areas [2]. Insulin resistance and insulin release were assessed by homeostasis model assessment (HOMA) in multiple studies based on fasting plasma glucose (FPG) and fasting serum insulin (FSIN) [4]. Thus, our aim was to replicate the relationship that has been confirmed in other ethnic groups, further, to explore novel loci that have never been clearly associated with T2D-related quantitative traits in the Chinese population, providing the pathogenesis of T2D.
2. Materials and Methods
2.1. Study Subjects
The data were from the 2010–2012 China National Nutrition and Health Surveillance (CNNHS). The CNNHS was a nationally representative cross-sectional surveillance conducted by the National Institute for Nutrition and Health, Chinese Center for Disease Control and Prevention (NINH, China CDC), which covered 31 provinces, autonomous regions, and municipalities (except for Taiwan, Hong Kong, and Macao). Cluster random sampling with multi-stage stratification proportional to population size sampling design was used in recruiting subjects [16]. For the present study, subjects born in 1960, 1961, and 1963 were included, and those with a low call rate of variants (<50%), unqualified blood samples, failed DNA extraction, abnormal gene detection results, incomplete basic information, and those with T2D who took control measures were excluded. Finally, a total of 1654 subjects were included in the present study. The protocol was approved by the Ethics Committee of NINH, China CDC (No.2013-010). Written informed consent was obtained from each subject.
2.2. Assessment and Definition of Insulin Resistance and Impaired Insulin Release
Fasting venous blood samples were taken from each subject in the morning. Fasting plasma glucose (FPG) was measured using the glucose oxidize enzymatic method within 3 hours of blood collection. Fasting serum insulin (FSIN) was measured by radioimmunoassay. Insulin resistance and impaired insulin release were assessed by homeostasis model assessment of insulin resistance (HOMA-IR), homeostasis model assessment of beta cell function (HOMA-β), respectively, which was calculated by the following equation:
HOMA-IR = FPG × FSIN/22.5, HOMA-β = FSIN × 20/(FPG − 3.5) (with serum insulin in mU/L and plasma glucose in mmol/L) [4].
Insulin resistance was defined by exceeding the top quartile of HOMA-IR index value and impaired insulin release was defined under bottom quartile of HOMA-β index value in non-diabetic individuals [17].
2.3. Genotyping and SNP Selection
The genotype of 81 diabetes-related single nucleotide polymorphisms (SNPs) were detected by Mass ARRAY (Agena, San Diego, CA, USA). SNPs’ exclusion criteria: (1) call rates <80%; (2) deviating from Hardy–Weinberg equilibrium p < 0.001 (Table S1); (3) a minor allele frequency (MAF) of each SNP <5%. Finally, a total of 64 variants were involved in the present study, of which 22 SNPs had previously confirmed association with insulin resistance (Table S2), 27 loci were associated with insulin release (Table S3). The association of the left 15 loci with T2D had been indicated, however, whether these SNPs related to insulin release or insulin resistance were still unclear.
2.4. Demographic Characteristic, Physical Activity and Dietary Measurement
Questionnaires were used to obtain information. Self-reported educational levels were categorized into three levels: primary school or lower, junior middle school, senior high school or above. According to the per capita annual income of urban and rural households in 2011, the family economic level was divided into three levels: low (<20,000 RMB), middle (20,000–40,000 RMB) and high (>40,000 RMB). Smoking was coded as “yes” if smoking during the past 30 days. Drinking alcohol was coded as “yes” if drinking any type of alcohol during the past 12 months. Physical exercise was coded as “yes” if attending leisure time physical exercise during the past 3 months. The sedentary time was defined as time spent sitting or lying in leisure time.
Height, weight, and waist circumference was measured by trained investigators under standard operation procedure. Body mass index (BMI) was calculated as weight in kilograms divided by height in meters squared (kg/m2).
The dietary intake was collected by food frequency questionnaire (FFQ) and three consecutive (two weekdays and one weekend day) 24-h recalls which were described elsewhere [16]. The whole cereal and beans intake level was divided into low (less than 50 g per day), medium (more than or equal to 50 g and less than or equal to 150 g per day), and high (more than 150 g per day). Livestock and poultry intake level was divided into low (less than 40 g per day), medium (more than or equal to 40 g and less than or equal to 75 g per day), and high (more than 75 g per day) based on the recommendations of Dietary Guideline for Chinese Residents (2016) [18].
2.5. Statistical Analysis
Data analyses were conducted using statistical analysis system (SAS) 9.4 software (SAS Institute, Cary, NC, USA). A Hardy–Weinberg equilibrium test was performed for each SNP of non-diabetic individuals by chi-square test [19]. Continuous variables were expressed as mean ± SD, and categorical variables were presented as frequency and percentage. T-tests and chi-square tests were performed for comparisons of continuous and categorical variables between males and females, respectively. A logistic regression model was utilized to validate the association of individual SNPs with T2D-related quantitative traits corrected for confounders, including gender, economical status, education levels, BMI, waist circumference, sedentary time, exercise, whole cereal and beans intake levels, livestock and poultry intake levels, drinking, and smoking. Two-tailed p < 0.05 regarded as statistical significance.
3. Results
3.1. Basic Characteristics of Study Subjects
Basic characteristics are shown in Table 1. The age of 1654 subjects (male 39.1%, female 60.9%) was 49.9 ± 1.5 years. The rate of impaired insulin release in males was 32.6%, which was higher than that in females (p < 0.05). There were no significant differences between male and female in the rate of insulin resistance. There were significant differences between males and females in education level, smoking, drinking, whole cereal and beans intake levels, livestock and poultry intake levels, BMI, waist circumference, and exercise.
Table 1.
Basic characteristics of 1654 subjects.
| Characteristics | Total | Male | Female | p Value |
|---|---|---|---|---|
| Total | 1654 | 647 (39.1%) | 1007 (60.9%) | |
| Age (year) | 49.9 ± 1.5 | 49.9±1.5 | 49.9 ± 1.5 | 0.9347 |
| Insulin resistance | 0.0950 | |||
| No | 1189 (71.9%) | 480 (74.2%) | 709 (70.4%) | |
| Yes | 465 (28.1%) | 167 (25.8%) | 298 (29.6%) | |
| Impaired insulin release | 0.0050 | |||
| No | 1179 (71.3%) | 436 (67.4%) | 743 (73.8%) | |
| Yes | 475 (28.7%) | 211 (32.6%) | 264 (26.2%) | |
| Education level | <0.0001 | |||
| Illiterate or primary school | 563 (34%) | 140 (21.6%) | 423 (42.0%) | |
| Middle school | 723 (43.7%) | 327 (50.5%) | 396 (39.3%) | |
| Senior school | 368 (22.2%) | 180 (27.8%) | 188 (18.7%) | |
| Economic status | 0.7453 | |||
| Low | 839 (52.7%) | 324 (52.1%) | 515 (53.1%) | |
| Middle | 633 (39.8%) | 254 (40.8%) | 379 (39.1%) | |
| High | 119 (7.5%) | 44 (7.1%) | 75 (7.7%) | |
| Smoking | <0.0001 | |||
| No | 1164 (70.5%) | 204 (31.6%) | 960 (95.4%) | |
| Yes | 487 (29.5%) | 441 (68.4%) | 46 (4.6%) | |
| Drinking | <0.0001 | |||
| Yes | 1121 (67.9%) | 250 (38.7%) | 871 (86.6%) | |
| No | 531 (32.1%) | 396 (61.3%) | 135 (13.4%) | |
| Whole cereal and beans intake levels | 0.0171 | |||
| Low | 1080 (86.5%) | 425 (88%) | 655 (85.5%) | |
| Medium | 133 (10.6%) | 39 (8.1%) | 94 (12.3%) | |
| High | 36 (2.9%) | 19 (3.9%) | 17 (2.2%) | |
| Livestock and poultry intake levels | 0.0003 | |||
| Low | 517 (41.4%) | 184 (38.1%) | 333 (43.5%) | |
| Medium | 284 (22.7%) | 93 (19.3%) | 191 (24.9%) | |
| High | 448 (35.9%) | 206 (42.7%) | 242 (31.6%) | |
| BMI (kg/m2) | 24.3 ± 3.4 | 24.0 ± 3.4 | 24.5 ± 3.4 | 0.002 |
| Waist circumference (cm) | 82.2 ± 10.0 | 83.7 ± 10.4 | 81.3 ± 9.6 | <0.0001 |
| Exercise | 0.0409 | |||
| No | 1500 (91.4%) | 600 (93.2%) | 900 (90.3%) | |
| Yes | 141 (8.6%) | 44 (6.8%) | 97 (9.7%) | |
| Sedentary time (h/d) | 2.7 ± 1.5 | 2.6 ± 1.4 | 2.7 ± 1.6 | 0.6235 |
Continuous variables were presented as mean ± SD, and categories variables were presented as N (%).
3.2. Association between Individual SNP and Insulin Resistance
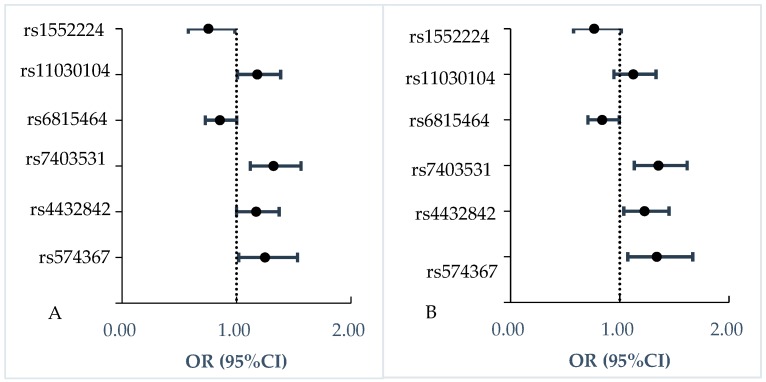
As shown in Table 2 and Figure 1, three SNPs were associated with increased insulin resistance risk with adjusting for covariates, including 5q11.2-rs4432842 (OR = 1.23, 95% CI: 1.04–1.45), RASGRP1-rs7403531 (OR = 1.35, 95% CI: 1.13–1.62), SEC16B-rs574367 (OR = 1.34, 95% CI: 1.07–1.67). Rs11030104 in gene BDNF (OR = 1.18, 95% CI: 1.01–1.39) with insulin resistance was found, which disappeared after adjusting for covariates (p = 0.184).
Table 2.
Association between each SNP and quantitative traits of T2D.
| Identified Association to | Loci | Chr | Reported Gene | Risk/Other Allele | Insulin Resistance | Impaired Insulin Release | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 1 | Model 2 | |||||||||
| OR (95%CI) | p Value | OR (95%CI) | p Value | OR (95%CI) | p Value | OR (95%CI) | p Value | |||||
| T2D | rs340874 | 1 | PROX1 | C/T | 0.97(0.83,1.14) | 0.718 | 0.96(0.81,1.13) | 0.595 | 0.89(0.76,1.04) | 0.137 | 0.90(0.76,1.05) | 0.187 |
| rs243021 | 2 | BCL11A | A/G | 1.08(0.92,1.27) | 0.362 | 1.07(0.89,1.27) | 0.486 | 0.94(0.80,1.10) | 0.436 | 0.94(0.80,1.11) | 0.483 | |
| rs2943641 | 2 | IRS1 | C/T | 1.20(0.89,1.62) | 0.226 | 1.26(0.92,1.73) | 0.157 | 1.00(0.75,1.32) | 0.985 | 0.98(0.74,1.31) | 0.911 | |
| rs3923113 | 2 | GRB14 | A/C | 1.03(0.83,1.30) | 0.770 | 1.02(0.80,1.30) | 0.852 | 1.16(0.92,1.46) | 0.210 | 1.15(0.91,1.45) | 0.247 | |
| rs7593730 | 2 | RBMS1, ITGB6 | C/T | 1.08(0.87,1.33) | 0.509 | 1.08(0.86,1.35) | 0.532 | 0.91(0.74,1.12) | 0.375 | 0.91(0.74,1.13) | 0.399 | |
| rs780094 | 2 | GCKR | C/T | 1.09(0.93,1.28) | 0.284 | 1.10(0.93,1.30) | 0.293 | 1.00(0.85,1.17) | 0.964 | 0.97(0.83,1.14) | 0.726 | |
| rs1470579 | 3 | IGF2BP2 | C/A | 0.98(0.82,1.18) | 0.854 | 1.04(0.86,1.25) | 0.716 | 1.04(0.87,1.24) | 0.646 | 1.04(0.86,1.24) | 0.708 | |
| rs16861329 | 3 | ST6GAL1 | T/C | 0.84(0.70,1.02) | 0.081 | 0.89(0.72,1.09) | 0.246 | 0.86(0.71,1.04) | 0.118 | 0.85(0.70,1.03) | 0.103 | |
| rs4607103 | 3 | ADAMTS9 | C/T | 1.13(0.97,1.33) | 0.123 | 1.15(0.97,1.37) | 0.099 | 0.87(0.74,1.01) | 0.067 | 0.87(0.74,1.02) | 0.083 | |
| rs4858889 | 3 | SCAP | A/G | 1.01(0.81,1.26) | 0.908 | 1.07(0.84,1.35) | 0.587 | 0.93(0.75,1.16) | 0.539 | 0.90(0.72,1.13) | 0.358 | |
| rs7612463 | 3 | UBE2E2 | C/A | 0.89(0.73,1.07) | 0.205 | 0.86(0.70,1.05) | 0.142 | 1.04(0.86,1.25) | 0.723 | 1.06(0.87,1.29) | 0.561 | |
| rs831571 | 3 | PSMD6 | C/T | 1.10(0.93,1.29) | 0.256 | 1.11(0.93,1.32) | 0.262 | 1.02(0.87,1.20) | 0.775 | 1.01(0.85,1.19) | 0.956 | |
| rs6815464 | 4 | MAEA | C/G | 0.85(0.73,1.00) | 0.053 | 0.84(0.71,1.00) | 0.044 | 1.12(0.96,1.32) | 0.153 | 1.15(0.98,1.35) | 0.098 | |
| rs459193 | 5 | ANKRD55 | G/A | 1.02(0.88,1.19) | 0.780 | 1.07(0.91,1.26) | 0.434 | 1.02(0.87,1.18) | 0.854 | 1.01(0.87,1.19) | 0.865 | |
| rs10946398 | 6 | CDKAL1 | C/A | 0.96(0.81,1.13) | 0.592 | 0.93(0.79,1.11) | 0.430 | 1.01(0.86,1.18) | 0.921 | 1.03(0.88,1.22) | 0.707 | |
| rs1535500 | 6 | KCNK16 | T/G | 0.93(0.80,1.09) | 0.390 | 0.91(0.77,1.07) | 0.239 | 1.06(0.91,1.23) | 0.492 | 1.06(0.90,1.24) | 0.486 | |
| rs9470794 | 6 | ZFAND3 | C/T | 0.97(0.82,1.14) | 0.691 | 1.00(0.84,1.19) | 0.973 | 1.10(0.93,1.29) | 0.256 | 1.08(0.92,1.28) | 0.343 | |
| rs2191349 | 7 | DGKB, TMEM195 | T/G | 1.07(0.91,1.25) | 0.419 | 1.04(0.88,1.23) | 0.648 | 0.93(0.79,1.08) | 0.329 | 0.91(0.77,1.06) | 0.225 | |
| rs4607517 | 7 | GCK | A/G | 1.09(0.91,1.31) | 0.354 | 1.06(0.87,1.29) | 0.550 | 0.96(0.79,1.15) | 0.629 | 0.97(0.81,1.18) | 0.783 | |
| rs864745 | 7 | JAZF1 | T/C | 1.03(0.85,1.24) | 0.783 | 1.01(0.82,1.23) | 0.951 | 0.92(0.76,1.11) | 0.367 | 0.95(0.79,1.15) | 0.600 | |
| rs972283 | 7 | KLF14 | G/A | 0.92(0.77,1.09) | 0.332 | 0.93(0.77,1.12) | 0.449 | 0.99(0.84,1.18) | 0.928 | 1.00(0.84,1.19) | 0.969 | |
| rs13266634 | 8 | SLC30A8 | C/T | 1.07(0.91,1.25) | 0.424 | 1.07(0.90,1.28) | 0.416 | 1.11(0.95,1.30) | 0.196 | 1.12(0.95,1.32) | 0.164 | |
| rs516946 | 8 | ANK1 | C/T | 1.15(0.90,1.48) | 0.271 | 1.27(0.97,1.65) | 0.083 | 1.42(1.10,1.83) | 0.008 | 1.39(1.07,1.81) | 0.014 | |
| rs896854 | 8 | TP53INP1 | T/C | 0.95(0.81,1.12) | 0.560 | 0.96(0.81,1.15) | 0.683 | 0.99(0.84,1.16) | 0.889 | 0.96(0.81,1.14) | 0.639 | |
| rs10811661 | 9 | CDKN2A, CDKN2B | T/C | 1.06(0.90,1.25) | 0.500 | 1.10(0.93,1.31) | 0.268 | 1.14(0.96,1.34) | 0.131 | 1.15(0.97,1.36) | 0.110 | |
| rs17584499 | 9 | PTPRD | T/C | 1.10(0.86,1.41) | 0.458 | 1.12(0.85,1.46) | 0.427 | 0.90(0.70,1.17) | 0.429 | 0.89(0.69,1.16) | 0.404 | |
| rs2796441 | 9 | TLE1 | G/A | 0.91(0.78,1.06) | 0.231 | 0.93(0.79,1.10) | 0.387 | 1.03(0.88,1.21) | 0.691 | 1.03(0.88,1.21) | 0.680 | |
| rs7041847 | 9 | GLIS3 | A/G | 0.95(0.81,1.11) | 0.526 | 0.98(0.83,1.16) | 0.812 | 0.96(0.82,1.13) | 0.627 | 0.93(0.80,1.10) | 0.400 | |
| rs10886471 | 10 | GRK5 | C/T | 1.03(0.85,1.25) | 0.748 | 1.07(0.87,1.31) | 0.538 | 1.17(0.96,1.42) | 0.118 | 1.20(0.99,1.47) | 0.068 | |
| rs10906115 | 10 | CDC123, CAMK1D | A/G | 0.91(0.77,1.07) | 0.262 | 0.97(0.81,1.16) | 0.747 | 1.13(0.96,1.33) | 0.139 | 1.10(0.93,1.30) | 0.260 | |
| rs11257655 | 10 | CDC123 | T/C | 0.90(0.77,1.06) | 0.194 | 0.95(0.80,1.13) | 0.538 | 1.12(0.96,1.31) | 0.159 | 1.08(0.92,1.27) | 0.366 | |
| rs5015480 | 10 | HHEX | C/T | 1.18(0.96,1.44) | 0.111 | 1.24(1.00,1.53) | 0.054 | 0.90(0.73,1.10) | 0.305 | 0.86(0.69,1.06) | 0.161 | |
| rs10830963 | 11 | MTNR1B | G/C | 0.99(0.84,1.16) | 0.876 | 1.06(0.89,1.25) | 0.524 | 1.09(0.93,1.27) | 0.304 | 1.08(0.92,1.27) | 0.371 | |
| rs1552224 | 11 | CENTD2 | A/C | 0.75(0.58,0.98) | 0.037 | 0.77(0.58,1.01) | 0.063 | 1.39(1.03,1.86) | 0.030 | 1.47(1.09,1.99) | 0.013 | |
| rs2237892 | 11 | KCNQ1 | C/T | 0.97(0.82,1.15) | 0.719 | 0.97(0.81,1.16) | 0.721 | 1.10(0.92,1.30) | 0.300 | 1.12(0.94,1.34) | 0.207 | |
| rs5215 | 11 | KCNJ11 | C/T | 0.93(0.79,1.09) | 0.362 | 0.90(0.76,1.07) | 0.225 | 1.13(0.97,1.33) | 0.123 | 1.15(0.97,1.35) | 0.100 | |
| rs10842994 | 12 | KLHDC5 | C/T | 1.09(0.89,1.34) | 0.386 | 1.12(0.91,1.39) | 0.293 | 0.95(0.78,1.15) | 0.577 | 0.95(0.77,1.16) | 0.606 | |
| rs7961581 | 12 | TSPAN8, LGR5 | C/T | 0.86(0.71,1.05) | 0.137 | 0.88(0.71,1.08) | 0.208 | 1.25(1.04,1.50) | 0.020 | 1.25(1.03,1.51) | 0.022 | |
| rs11634397 | 15 | ZFAND6 | G/A | 1.04(0.81,1.34) | 0.742 | 1.10(0.84,1.44) | 0.488 | 0.89(0.68,1.15) | 0.365 | 0.87(0.67,1.13) | 0.287 | |
| rs2028299 | 15 | AP3S2 | C/A | 1.07(0.89,1.29) | 0.472 | 1.09(0.90,1.32) | 0.391 | 1.19(0.99,1.42) | 0.065 | 1.17(0.97,1.41) | 0.100 | |
| rs7172432 | 15 | C2CD4A, C2CD4B | A/G | 1.16(0.98,1.36) | 0.082 | 1.13(0.95,1.35) | 0.156 | 1.15(0.98,1.35) | 0.092 | 1.12(0.95,1.33) | 0.166 | |
| rs7178572 | 15 | HMG20A | G/A | 1.06(0.90,1.25) | 0.480 | 1.09(0.92,1.30) | 0.331 | 1.11(0.94,1.30) | 0.219 | 1.09(0.92,1.28) | 0.337 | |
| rs7403531 | 15 | RASGRP1 | T/C | 1.32(1.12,1.56) | 0.001 | 1.35(1.13,1.62) | 0.001 | 0.95(0.81,1.13) | 0.565 | 0.93(0.78,1.10) | 0.373 | |
| rs1558902 | 16 | FTO | A/T | 1.09(0.87,1.38) | 0.454 | 1.05(0.82,1.34) | 0.72 | 1.08(0.86,1.36) | 0.507 | 1.08(0.86,1.37) | 0.504 | |
| rs7202877 | 16 | BCAR1 | T/G | 0.93(0.76,1.13) | 0.443 | 0.93(0.75,1.15) | 0.505 | 0.88(0.72,1.07) | 0.184 | 0.89(0.73,1.08) | 0.231 | |
| rs8050136 | 16 | FTO | A/C | 1.08(0.85,1.36) | 0.535 | 1.03(0.80,1.33) | 0.805 | 1.09(0.86,1.37) | 0.484 | 1.08(0.85,1.37) | 0.525 | |
| rs4430796 | 17 | HNF1B | G/A | 0.99(0.83,1.17) | 0.884 | 0.96(0.80,1.16) | 0.676 | 0.98(0.83,1.16) | 0.824 | 0.98(0.82,1.17) | 0.825 | |
| rs12454712 | 18 | BCL2 | T/C | 1.02(0.87,1.19) | 0.852 | 1.00(0.85,1.18) | 0.966 | 1.00(0.86,1.17) | 0.997 | 0.98(0.84,1.15) | 0.815 | |
| rs12970134 | 18 | MC4R | G/A | 0.93(0.76,1.15) | 0.510 | 0.95(0.76,1.19) | 0.656 | 1.10(0.89,1.36) | 0.365 | 1.11(0.89,1.37) | 0.355 | |
| rs8090011 | 18 | LAMA1 | G/C | 0.92(0.77,1.09) | 0.339 | 0.94(0.78,1.13) | 0.476 | 1.10(0.93,1.32) | 0.276 | 1.11(0.93,1.33) | 0.249 | |
| rs10401969 | 19 | CILP2 | C/T | 1.03(0.79,1.36) | 0.826 | 1.00(0.74,1.33) | 0.977 | 0.99(0.75,1.30) | 0.937 | 0.98(0.74,1.30) | 0.907 | |
| rs3794991 | 19 | GATAD2A | T/C | 1.14(0.84,1.56) | 0.392 | 1.12(0.80,1.56) | 0.510 | 0.87(0.63,1.20) | 0.392 | 0.86(0.61,1.20) | 0.362 | |
| Birth weight | rs724577 | 4 | LCORL | C/A | 0.98(0.83,1.15) | 0.764 | 1.04(0.88,1.25) | 0.632 | 1.06(0.90,1.25) | 0.486 | 1.03(0.87,1.21) | 0.758 |
| rs4432842 | 5 | 5q11.2 | T/C | 1.17(1.00,1.37) | 0.050 | 1.23(1.04,1.45) | 0.017 | 1.03(0.88,1.21) | 0.701 | 1.04(0.89,1.22) | 0.609 | |
| rs6931514 | 6 | CDKAL1 | G/A | 1.00(0.85,1.19) | 0.976 | 1.01(0.85,1.21) | 0.882 | 0.93(0.79,1.10) | 0.390 | 0.95(0.80,1.13) | 0.562 | |
| rs1042725 | 12 | HMGA2 | T/C | 1.00(0.83,1.22) | 0.989 | 0.98(0.8,1.21) | 0.864 | 0.98(0.81,1.19) | 0.840 | 0.96(0.79,1.17) | 0.716 | |
| BMI | rs2568958 | 1 | 1p31 | A/G | 1.08(0.82,1.43) | 0.582 | 1.10(0.81,1.49) | 0.545 | 0.86(0.66,1.13) | 0.276 | 0.85(0.64,1.11) | 0.233 |
| rs574367 | 1 | SEC16B | G/T | 1.25(1.02,1.53) | 0.033 | 1.34(1.07,1.67) | 0.010 | 1.01(0.83,1.23) | 0.925 | 1.01(0.83,1.24) | 0.893 | |
| rs7561317 | 2 | TMEM18 | G/A | 0.95(0.73,1.24) | 0.725 | 0.98(0.74,1.3) | 0.898 | 1.07(0.81,1.39) | 0.645 | 1.11(0.84,1.46) | 0.468 | |
| rs16892496 | 8 | TRHR | C/A | 0.98(0.84,1.14) | 0.783 | 1.00(0.85,1.18) | 0.968 | 0.91(0.78,1.06) | 0.224 | 0.90(0.77,1.05) | 0.179 | |
| rs7832552 | 8 | TRHR | T/C | 0.96(0.82,1.12) | 0.585 | 0.98(0.83,1.15) | 0.782 | 0.90(0.78,1.05) | 0.193 | 0.89(0.76,1.04) | 0.142 | |
| rs11030104 | 11 | BDNF, BDNF-AS | A/G | 1.18(1.01,1.39) | 0.041 | 1.12(0.95,1.33) | 0.184 | 0.96(0.82,1.12) | 0.582 | 0.95(0.81,1.11) | 0.515 | |
| rs6265 | 11 | BDNF, BDNF-AS | C/T | 1.14(0.97,1.34) | 0.108 | 1.10(0.93,1.31) | 0.263 | 0.95(0.81,1.11) | 0.524 | 0.94(0.80,1.11) | 0.467 | |
| rs9939609 | 16 | FTO | A/T | 1.02(0.81,1.30) | 0.846 | 0.97(0.75,1.25) | 0.803 | 1.06(0.84,1.35) | 0.612 | 1.06(0.83,1.35) | 0.641 | |
Model 1 was logistic regression model to analyze the association between individual SNP and quantitative traits without adjusting covariates. Model 2 was logistic regression model to analyses the association between individual SNP and quantitative traits with adjusting continuous covariates, BMI, waist circumference, and sedentary time, and categorical covariates, gender, economical status, education levels, exercise, whole cereal and beans intake levels, livestock and poultry intake levels, drinking, and smoking.
Figure 1.
Association of six SNPs with insulin resistance. (A) Model 1 was logistic regression model to analyze the association of individual SNP with quantitative traits without adjusting for any covariates. (B) Model 2 was logistic regression model to analyze the association of individual SNP with quantitative traits adjusting for gender, economical status, education levels, BMI, waist circumference, sedentary time, exercise, whole cereal and beans intake levels, livestock and poultry intake levels, drinking, and smoking.
MAEA-rs6815464 was associated with decreased insulin resistance with correction for covariates, the OR was 0.84 (95% CI: 0.71–1.00). CENTD2-rs1552224 had an association with decreased insulin resistance (OR = 0.75, 95% CI: 0.58–0.98), while the link disappeared after controlling for covariates (p = 0.063).
3.3. Association between Individual SNP and Impaired Insulin Release
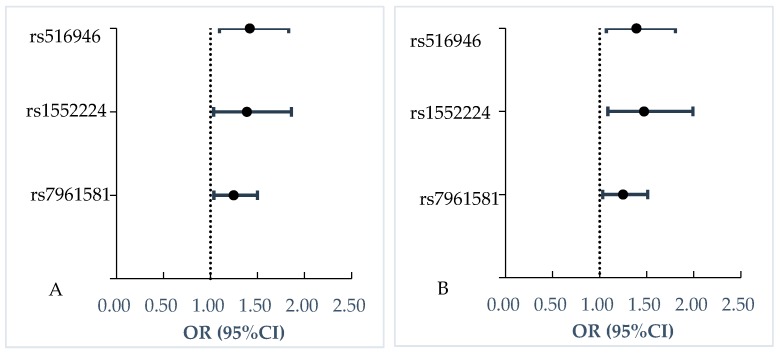
Association of CENTD2-rs1552224 (OR = 1.47, 95% CI: 1.09–1.99), TSPAN8-rs7961581 (OR = 1.25, 95% CI: 1.03–1.51) and rs516946 (OR = 1.39, 95% CI: 1.07–1.81) with increased impaired insulin release were significant without or with adjustment for covariates. (See Table 2 and Figure 2)
Figure 2.
Association of three SNPs with impaired insulin release. (A) Model 1 was logistic regression model to analyze the association of individual SNP with quantitative traits without adjusting for any covariates. (B) Model 2 was logistic regression model to analyze the association of individual SNP with quantitative traits adjusting for gender, economical status, education levels, BMI, waist circumference, sedentary time, exercise, whole cereal and beans intake levels, livestock and poultry intake levels, drinking, and smoking.
4. Discussion
Among the 64 susceptible loci we examined, RASGRP1-rs7403531, ANK1-rs516946, and SEC16B-rs574367 had associations with insulin resistance. HHEX-rs5015480, CENTD2-rs1552224, and TSPAN8-rs7961581 were associated with impaired insulin release in the current Chinese population.
Our study has replicated the association of rs7403531 with insulin resistance, which was consistent with a previous study which reported that rs7403531 had been associated with T2D and higher insulin in Chinese Hans [20]. However, Sakai et al. [21] found no significant association of rs7403531 with T2D. Insulin sensitivity is one of the predictors of diabetes [22], therefore, these findings indicated that carriers of rs7403531 may be predisposed to T2D by decreasing insulin sensitivity. Additionally, rs7403531 had the same OR for insulin resistance after correcting for BMI, suggesting its association with insulin resistance might not be mediated through obesity.
Most studies were conducted to explore the association of rs574367 with BMI, but seldom T2D. The G allele of SEC16B-rs574367 increased the risk for obesity in different ethnic populations [23]. SEC16B-rs574367 could affect the synthesis and transcription of lipase, thus inhibiting the decomposition of fat, leading to the occurrence of obesity [24]. Several obesity-associated loci including rs574367 constructed a BMI gene risk score (BMI-GRS), which had an enhanced effect on log (HOMA2-IR) in the age- and sex-adjusted model among Chinese adults [25]. Our study found rs574367 had the same risk for increased HOMA-IR. Further study should be conducted to confirm the relationship.
MAEA-rs6815464 (macrophage erythroblast attacher) was reported as a T2D risk variant in Asians [26,27]. In the Chinese population, no significant association was observed with insulin resistance or insulin release, measured by fasting-based homeostasis model assessment [28]. A genome-wide association study identified MAEA gene altered T2D risk through insulin secretion [29]. However, in our study, rs6815464 had an association with decreased insulin resistance after adjustment for confounding factors. Therefore, further functional characterization study is required to elucidate its role in the pathogenesis of T2D.
A Mendelian randomization study has shown that lowered birthweight 5q11.2-rs4432842 could increase susceptibility to T2D [30]. However, it remains unclear what its role in the pathogenesis of T2D is. Our study found 5q11.2-rs4432842 was associated with increased risk of insulin resistance.
The association of rs516946 with T2D has been confirmed by previous studies [31,32]. ANK1-rs516946 was associated with impaired insulin release by decreasing insulinogenic and disposition indexes in a Danish cohort study [32]. The risk C allele of rs516946 with a larger waist circumference was found in a Han Chinese population [31]. After adjusting for waist circumference, the risk C allele of ANK1-rs516946 was associated with increased impaired insulin release in our study.
CENTD2-rs1552224 could increase the risk of T2D through reducing insulin secretion [11]. CENTD2-rs1552224 associated with decreased glucose-stimulated insulin release, which increased 30-min plasma glucose values by 2.0% and reduced insulin release 30 min after an oral glucose load by 4.2% [33]. CENTD2-rs1552224 increased the risk of impaired insulin release after correction for covariates in our study.
TSPAN8-rs7961581 increased the risk of T2D in a previous study in Han Chinese [34]. A study in Denmark confirmed that the C allele of TSPAN8-rs7961581 associated with decreased levels of corrected insulin response, of AUC-insulin/AUC-glucose ratio, and of the insulinogenic index [35]. Our study has replicated the association of TSPAN8-rs7961581 with T2D, TSPAN8-rs7961581 increased the risk of impaired insulin release by 25%.
The present study had some limitations. Firstly, the method of HOMA was not the golden standard to assess insulin resistance and impaired insulin release. The standard intravenous methods (euglycemic insulin clamp technique) and comprehensive indices should be used in the Chinese population to validate the relationship in future studies. Secondly, the participants in the present study were about 50 years old and too young to get enough cases of T2D. The small numbers may partly explain why no significant associations of some SNPs with the risk of quantitative traits of T2D were found.
5. Conclusions
In summary, we have replicated the association of seven SNPs and quantitative traits of T2D among the Chinese population, of which 5q11.2-rs4432842, RASGRP1-rs7403531, and SEC16B-rs574367 were associated with increased insulin resistance, MAEA-rs6815464 was associated with decreased insulin resistance, and ANK1- rs516946, CENTD2-rs1552224, and TSPAN8-rs7961581 were associated with increased impaired insulin release. Our results would provide insight into the pathogenesis of individual SNP and T2D.
Acknowledgments
We would like to thank all the participants who took part in this study and the staff of 31 provinces at the 2010–2012 China Nutrition and Health Surveillance.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-4601/17/7/2439/s1, Table S1: Hardy–Weinberg equilibrium test of T2D-associated loci, Table S2: Overview of investigated genetic variants associated with insulin resistance, Table S3: Overview of investigated genetic variants associated with insulin release.
Author Contributions
Data curation, F.Y., H.L.; formal analysis, F.Y., H.L.; methodology, F.Y., H.L.; project administration, A.L.; supervision, A.L.; writing—original draft, F.Y., H.L.; writing—review and editing, F.Y., H.L., C.S., H.F., R.W., Y.Z., W.G. and A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (No. 81372990). The China National Nutrition and Health Survey 2010–2012 (CNHS 2010–2012) was funded by (Major program for health care reform from National Health and Family Planning Commission of the People’s Republic of China), grant number (20120212), which was the source of design for the study and collection, analysis, and interpretation of data and writing of the manuscript.
Conflicts of Interest
All authors declare no conflict of interest.
References
- 1.Yang W.U., Lu J.M., Weng J.P., Jia W.P., Ji L.N., Xiao J.Z., Shan Z.Y., Liu J., Tian H.M., Ji Q.H., et al. China National Diabetes and Metabolic Disorders Study Group: Prevalence of diabetes among men and women in China. N. Engl. J. Med. 2010;362:1090–1101. doi: 10.1056/NEJMoa0908292. [DOI] [PubMed] [Google Scholar]
- 2.Chen M.J., Zhang X.L., Fang Q.X., Wang T.T., Li T.T., Qiao H. Three single nucleotide polymorphisms associated with type 2 diabetes mellitus in a Chinese population. Exp. Ther. Med. 2017;13:121–126. doi: 10.3892/etm.2016.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu C., Zhang R., Wang C.G., Wang J., Ma X.J., Hou X.H., Lu J.Y., Yu W.H., Jiang F., Bao Y.Q., et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 Are Associated with Glucose Metabolism in the Chinese. PLoS ONE. 2010;5:e15542. doi: 10.1371/journal.pone.0015542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong X., Zhang X.L., Xing X.Y., Zhang B., Hong J., Yang W.Y. The Association of Type 2 Diabetes Loci Identified in Genome-Wide Association Studies with Metabolic Syndrome and Its Components in a Chinese Population with Type 2 Diabetes. PLoS ONE. 2015;10:e143607. doi: 10.1371/journal.pone.0143607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marullo L., El-Sayed Moustafa J.S., Prokopenko I. Insights into the Genetic Susceptibility to Type 2 Diabetes from Genome-Wide Association Studies of Glycaemic Traits. Curr. Diabetes Rep. 2014;14:551. doi: 10.1007/s11892-014-0551-8. [DOI] [PubMed] [Google Scholar]
- 6.Gobato A.O., Vasques A.C., Zambon M.P., Barros Filho Ade A., Hessel G. Metabolic syndrome and insulin resistance in obese adolescents. Rev. Paul. Pediatr. 2014;32:55–62. doi: 10.1590/S0103-05822014000100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stumvoll M., Goldstein B.J., van Haeften T.W. Type 2 diabetes: Principles of pathogenesis and therapy. Lancet. 2005;365:1333–1346. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe R.M. The Genetics of Insulin Resistance: Where’s Waldo? Curr. Diabetes Rep. 2010;10:476–484. doi: 10.1007/s11892-010-0143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan J.C., Malik V., Jia W., Kadowaki T., Yajnik C.S., Yoon K.H., Hu F.B. Diabetes in Asia, Epidemiology, Risk Factors, and Pathophysiology. JAMA. 2009;20:2129–2140. doi: 10.1001/jama.2009.726. [DOI] [PubMed] [Google Scholar]
- 10.Nikitin A.G., Potapov V.Y., Brovkina O.I., Koksharova E.O., Khodyrev D.S., Philippov Y.I., Michurova M.S., Shamkhalova M.S., Vikulova O.K., Smetanina S.A., et al. Association of polymorphic markers of genes FTO, KCNJ11, CDKAL1, SLC30A8, and CDKN2B with type 2 diabetes mellitus in the Russian population. PEERJ. 2017;5:e3414. doi: 10.7717/peerj.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinthorsdottir V., Thorleifsson G., Reynisdottir I., Benediktsson R., Jonsdottir T., Walters G.B., Styrkarsdottir U., Gretarsdottir S., Emilsson V., Ghosh S., et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 12.Cheng L.Q., Zhang D.M., Zhou L.N., Zhao J., Chen B. Association between SLC30A8 rs13266634 Polymorphism and Type 2 Diabetes Risk: A Meta-Analysis. Med. Sci. Monit. 2015;21:2178–2189. doi: 10.12659/MSM.894052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoshi A., Bajestani M.K., Shakeri H., Goodarzi G., Azizi F. Association of Omentin rs2274907 and FTO rs9939609 gene polymorphisms with insulin resistance in Iranian individuals with newly diagnosed type 2 diabetes. Lipids Health Dis. 2019;18:142. doi: 10.1186/s12944-019-1085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M., Yoshida T., Bin W., Fukuo K., Kazumi T. FTO, Abdominal Adiposity, Fasting Hyperglycemia Associated with Elevated HbA1c in Japanese Middle-Aged Women. J. Atheroscler. Thromb. 2012;19:633–642. doi: 10.5551/jat.11940. [DOI] [PubMed] [Google Scholar]
- 15.Horikoshi M., Yaghootka H., Mook-Kanamori D.O., Sovio U., Taal H.R., Hennig B.J., Bradfield J.P., St Pourcain B., Evans D.M., Charoen P., et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao L.Y., Ma G.S., Piao J.H., Zhang J., Yu D.M., He Y.N., Huo J.S., Hu X.Q., Yang Z.Y., Yang X.G. Scheme of the 2010–2012 Chinese nutrition and health surveillance. Chin. J. Preval. Med. 2016;50:204–207. doi: 10.3760/cma.j.issn.0253-9624.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Liu R., Zheng Z., Du J., Christoffel K.K., Liu X. Racial Disparity in the Associations of Cotinine with Insulin Secretion: Data from the National Health and Nutrition Examination Survey, 2007–2012. PLoS ONE. 2016;11:e167260. doi: 10.1371/journal.pone.0167260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Chinese Nutrition Society . Dietary Guidelines for Chinese Residents (2016) People’s Medical Publishing House; Beijing, China: 2016. pp. 3–88. [Google Scholar]
- 19.Nielsen D.M., Ehm M.G., Weir B.S. Detecting Marker-Disease Association by Testing for Hardy-Weinberg Disequilibrium at a Marker Locus. Am. J. Hum. Genet. 1998;63:1531–1540. doi: 10.1086/302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H.X., Gan W., Lu L., Dong X., Han X.Y., Hu C., Yang Z., Sun L., Bao W., Li P.T., et al. A Genome-Wide Association Study Identifies GRK5 and RASGRP1 as Type 2 Diabetes Loci in Chinese Hans. Diabetes. 2013;62:291–298. doi: 10.2337/db12-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai K., Imamura M., Tanaka Y., Iwata M., Hirose H., Kaku K., Maegawa H., Hirotaka Watada H., Tobe K., Kashiwagi A., et al. Replication study of the association of rs7578597 in THADA, rs10886471 inGRK5, and rs7403531 inRASGRP1with susceptibility to type 2 diabetes among a Japanese population. Diabetol. Int. 2015;6:306–312. doi: 10.1007/s13340-015-0202-6. [DOI] [Google Scholar]
- 22.Liese A.D., Roach A.K., Sparks K.C., Marquart L., D’Agostino R.B., Jr., Mayer-Davis E.J. Whole-grain intake and insulin sensitivity: The Insulin Resistance Atherosclerosis Study. Am. J. Clin. Nutr. 2003;78:965–971. doi: 10.1093/ajcn/78.5.965. [DOI] [PubMed] [Google Scholar]
- 23.Hong J., Shi J., Qi L., Cui B., Gu W., Zhang Y., Li L., Xu M., Wang L., Zhai Y., et al. Genetic susceptibility, birth weight and obesity risk in young Chinese. Int. J. Obes. (Lond.) 2013;37:673–677. doi: 10.1038/ijo.2012.87. [DOI] [PubMed] [Google Scholar]
- 24.Fu L.W., Zhang M.X., Gao L.W., Mi J. Association between SEC16B polymorphisms and body mass index variation or risk of obesity: A Meta-analysis. Chin. J. Epidemiol. 2016;37:1288–1295. doi: 10.3760/cma.j.issn.0254-6450.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y., Xu M., Xie L., Wang T., Huang X., Lv X.F., Chen Y., Ding L., Lin L., Wang W.Q., et al. Obesity and peripheral arterial disease: A Mendelian Randomization analysis. Atherosclerosis. 2016;247:218–224. doi: 10.1016/j.atherosclerosis.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 26.Cho Y.S., Chen C.H., Hu C., Long J., Ong R.T., Sim X., Takeuchi F., Wu Y., Go M.J., Yamauchi T., et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in East Asians. Nat. Genet. 2012;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imamura M., Maeda S., Yamauchi T., Hara K., Yasuda K., Morizono T., Takahashi A., Horikoshi M., Nakamura M., Fujita H., et al. A single-nucleotide polymorphism in ANK1 is associated with susceptibility to type 2 diabetes in Japanese populations. Hum. Mol. Genet. 2012;21:3042–3049. doi: 10.1093/hmg/dds113. [DOI] [PubMed] [Google Scholar]
- 28.Xu K.F., Jiang L., Zhang M., Zheng X.Q., Gu Y., Wang Z.X., Cai Y., Dai H., Shi Y. Type 2 Diabetes Risk Allele UBE2E2 Is Associated with Decreased Glucose-Stimulated Insulin Release in Elderly Chinese Han Individuals. Medicine. 2016;95:e3604. doi: 10.1097/MD.0000000000003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood A.R., Jonsson A., Jackson A.U., Wang N., van Leewen N., Palmer N.D., Kobes S., Deelen J., Boquete-Vilarino L., Paananen J., et al. A Genome-Wide Association Study of IVGTT-Based Measures of First-Phase Insulin Secretion Refines the Underlying Physiology of Type 2 Diabetes Variants. Diabetes. 2017;66:2296–2309. doi: 10.2337/db16-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T., Huang T., Li Y., Zheng Y., Manson J.E., Hu F.B., Qi L. Low birthweight and risk of type 2 diabetes: A Mendelian randomisation study. Diabetologia. 2016;59:920–1927. doi: 10.1007/s00125-016-4019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun L., Zhang X., Wang T., Chen M., Qiao H. Association of ANK1 variants with new-onset type 2 diabetes in a Han Chinese population from northeast China. Exp. Ther. Med. 2017;14:3184–3190. doi: 10.3892/etm.2017.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harder M.N., Ribel-Madsen R., Justesen J.M., Sparsø T., Andersson E.A., Grarup N., Jørgensen T., Linneberg A., Hansen T., Pedersen O. Type 2 diabetes risk alleles near bcar1 and in ank1 associate with decreased β-cell function whereas risk alleles near ankrd55 and grb14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J. Clin. Endocrinol. Metab. 2013;98:E801–E806. doi: 10.1210/jc.2012-4169. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen T., Sparsø T., Grarup N., Jørgensen T., Pisinger C., Witte D.R. Type 2 diabetes risk allele near CENTD2 is associated with decreased glucose-stimulated insulin release. Diabetologia. 2011;54:1052–1056. doi: 10.1007/s00125-011-2054-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhou D.Z., Liu Y., Zhang D., Liu S.M., Yu L., Yang Y.F., Zhao T., Chen Z., Kan M.Y., Zhang Z.F., et al. Variations in/nearby genes coding for JAZF1, TSPAN8/LGR5 and HHEX-IDE and risk of type 2 diabetes in Han Chinese. J. Hum. Genet. 2010;55:810–815. doi: 10.1038/jhg.2010.117. [DOI] [PubMed] [Google Scholar]
- 35.Grarup N., Andersen G., Krarup N.T., Albrechtsen A., Schmitz O., Jørgensen T., Borch-Johnsen K., Hansen T., Pedersen O. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57:2534–2540. doi: 10.2337/db08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.