Abstract
Renal cell carcinoma is the third type of urologic cancer and has a poor prognosis with 30% of metastatic patients at diagnosis. The antiangiogenics and targeted immunotherapies led to treatment remodeling emphasizing the role of the tumour microenvironment. However, long-term responses are rare with a high rate of resistance. New strategies are emerging to improve the efficacy and the emerging drugs are under evaluation in ongoing trials. With the different treatment options, there is an urgent need to identify biomarkers in order to predict the efficacy of drugs and to better stratify patients. Owing to the limitations of programmed death-ligand 1 (PD-L1), the most studied immunohistochemistry biomarkers, and of the tumor mutational burden, the identification of more reliable markers is an unmet need. New technologies could help in this purpose.
Keywords: immunotherapy, immune checkpoint inhibitors, renal cell carcinoma, PD-1, PD-L1, ongoing trials, biomarkers, emerging drugs
1. Introduction
Renal cell carcinoma is the third urological cancer, representing 3% of all cancers in women and 5% in men with an incidence of around 400,000 cases worldwide [1]. The prognosis is poor: 30% of patients are metastatic at diagnosis and almost 30% of the remaining patients will develop metastases detected during the follow-up [2].
Clear cell renal cell carcinomas (ccRCC) represent the vast majority of RCC (around 75%). The other histologies mainly encompass papillary (20%) and chromophobe RCC (5%). The other entities are very rare including translocation-associated RCC, medullar RCC and collecting duct carcinoma. The histological entities based on distinct pathological features present different molecular alterations. Indeed, ccRCC are hallmarked by a frequent alteration of the VHL gene, a tumour suppressor gene, leading to angiogenesis through the transcription of genes regulated by HIF such as VEGF [3,4,5,6,7].
Non ccRCC (nccRCC) represent a heterogeneous group with papillary, chromophobe RCC and translocation RCC, the most frequent entities. Papillary RCC (pRCC) include tumours with indolent outcome (type 1) and more aggressive tumours (type 2) [8]. Type 1 and type 2 pRCC commonly harbor frequent MET alterations. However, alterations of SETD2, CDKN2A, EGFR, NF2 and TERT have been described in type 2 and suggest the activation of MAP kinases pathway, cell cycle and deregulation of chromatin remodeling [9]. Chromophobe RCC (cRCC) are rarely metastatic and characterize mitochondrial alterations, frequently mutated p53 and activation of mTOR pathway [9]. Translocation RCC (tRCC) harbor gene fusions involve TFE3 and TFEB, members of the MiTF family [10]. These transcription factors have multiple partners, mainly involving messenger RNA splicing [11].
The other entities constitute less than 2% of renal tumours. The collecting duct carcinoma have been described as immunogenic tumours with high lymphocyte infiltration resultingfrom the upregulation of genes involved in T-cell activation and proliferation [12]. Renal medullary carcinoma present a frequent loss INI1 (SMARCB1) implicated in the chromatin remodeling complex [13]. Among familial RCC syndromes, patients with hereditary leiomyomatosis and renal cell carcinoma (HLRCC) syndrome harbor Fumarate Hydratase (FH) germline mutation and develop clinical aggressive tumours [14]. The FH mutation by inactivating the enzyme alters the function of the Krebs cycle.
The sarcomatoid component can be found in all the histologic subtypes and demonstrates an increased tumour mutation burden (TMB) with high frequency of p53, CDKN2A and NF2 mutations and also genes involved the chromatin remodeling such as ARID1A and BAP1 [15].
The treatment and management of metastatic RCC have radically changed over the past 20 years [16]. Initially, first-generation immunotherapy with cytokines: interleukins or interferon represented standard approaches but with poor results [17,18]. The development of tyrosine kinase inhibitors, mainly vascular endothelial growth factor (VEGF) receptor inhibitors, largely improved the prognosis of both progression free survival (PFS) and overall survival (OS) [19].
The emergence of immune checkpoint inhibitors (ICI) alone or in combination (anti-cytotoxic T-lymphocyte antigen-4 (CTLA4) and anti-programmed death 1 (PD-1)) showed interesting results [20,21]. Targeted immunotherapy is an alternative to antiangiogenics because ccRCC is also considered an immunogenic tumour with high numbers of immune cells such as tumour-infiltrating lymphocytes (TIL) [22,23,24]. Recent trials proposed antiangiogenics in association with targeted immunotherapy to overcome resistance emphasizing the role of the tumour microenvironment (TME) and this strategy is currently an option in first line treatment [25,26].
Mechanisms of resistance with ICI can be primary or innate and secondary or acquired [27]. They encompass neo-antigen loss, defect of antigen presentation, alternative immune checkpoints and defective interferon signalling. Interferon-γ is a major mechanism of resistance by enhancing programmed death-ligand 1 PD-L1 expression and inducing the expression of immune inhibitory molecules [28]. Other immune checkpoints such as TIM-3, LAG-3 and TIGIT play a role in the resistance by inhibiting antitumour immune response [29]. Novel therapeutic approaches try to overcome these mechanisms of resistance and are under evaluation in ongoing trials.
Identifying biomarkers is the key to better select treatments, reduce costs and improve survival in patients with metastatic kidney cancer. However, the limitations of the most studied biomarkers: PD-L1 immunohistochemistry and TMB make necessary the identification of robust markers. New technologies could help in this purpose.
In this comprehensive review, we will discuss: 1. the specificities of the TME in RCC, 2. the treatment update with the results of recent trials, 3. the emerging drugs used in ongoing trials, 4. the predictive biomarkers and 5. the novel technologies.
2. Specificities of the Tumour Microenvironment in Renal Cell Carcinoma
2.1. Vascular Component
Angiogenesis has been described to play an important role in the progression of RCC and leads to the recruitment of endothelial cells. Recent data suggest that endothelial cells in TME differ from normal endothelial cells [30]. Akino et al. identified aneuploidy in one third of endothelial cells freshly dissociated from RCC [31]. This warrants further investigation as it could impact the response to antiangiogenic drugs. Moreover, Edeline et al. demonstrated two different angiogenic phenotype described as mature and immature. These two patterns could coexist within the same tumour demonstrating the heterogeneity of the vascular component [32]. Dufies et al. demonstrated in experimental tumours that sunitinib stimulated the development of lymphatic vessels. Indeed, these vessels are crucial for the recruitment of immune cells [33].
2.2. Immune Component
The expression of PD-L1 is widely represented in RCC suggesting the important role of PD-1/PD-L1 checkpoint with the aberrant expression of tumour cells. Indeed, PD-L1 expression was reported in 23% of ccRCC, 10% of pRCC, 5.6% of cRCC, 30% of t RCC and 20% in collecting duct carcinoma [34,35]. An amplification of 9p24.1, locus of PD-L1, was recently identify in RCC with sarcomatoid component leading to PD-L1 constitutive expression [36].
The immune compartment mainly include T cells, NK cells, B cells, macrophages and dendritic cells with complex interactions. Recently, Chevrier and colleagues used mass cytometry to compile an atlas of immune cells from 73 RCC identifying 22 T cell and 17 tumour-associated macrophage phenotypes with distinct immune composition between tumours [37]. Some macrophages such as M-11 or M-13 were associated with a worse prognosis and could constitute new targets. The phenotype of T cells (CD8+) regarding the expression of immune checkpoints (PD-1, LAG-3, Tim-3) identified the immune-regulated profile as more aggressive in a cohort of 40 RCC [38]. The role of B cells in the TME is unclear with both anti- and pro-tumoral effects. In ccRCC, the density of B cells identified by immunohistochemistry was associated with poor prognosis [39]. Moreover, the B cell signature was correlated with poor prognosis in The Cancer Genome Atlas (TCGA) ccRCC cohort. B cells could exert pro-tumoral functions through different mechanisms such as secretion of immune-regulatory cytokines that affect T cells and macrophages.
To understand the immunomodulation of nivolumab, Chouieri et al. explored the morphological and molecular changes of mRCC (n = 91) at screening and on treatment [40]. Immunohistochemical analysis revealed an increase in CD3+, CD8+ and CD4+ lymphocytes. No consistent change was observe in the expression of PD-L1 in tumour cells. Transcriptional analysis identified the up-regulation of genes stimulated by interferon γ with high levels of related chemokine in peripheral blood. A study case reported a histological complete response after nivolumab in metastatic ccRCC [41]. Only fibrotic changes and CD8+ lymphocytes were detected after treatment.
3. Treatment Update in Renal Cell Carcinoma
3.1. First-Line Treatment
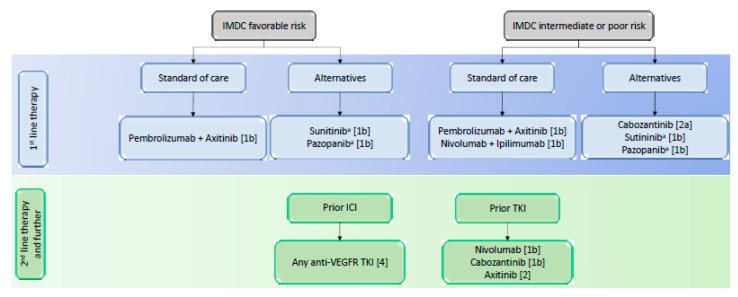
The ICI are the new backbone in the therapeutic landscape of renal cancer alongside tyrosine kinase inhibitors, Figure 1. Innovative combinations of ICI or ICI with TKI are now part of the treatment strategy and based on the results of recently published phase III trials in first line setting, Table 1 [20,25,26,42].
Figure 1.
European Association of Urology Guidelines on Renal Cell Carcinoma. IMDC = International Metastatic Renal Cell CarcinomaDatabase Consortium; OS = overall survival; Oxford level of evidence: [1b] = based on one randomised controlled phase 3 trial; [2a] = based on one randomised controlled phase 2 trial; [2b] = subgroup analysis of a randomised controlled phase 3 trial; [4] = expert opinion. a = No OS benefit proven.
Table 1.
Comparison of pivotal phase III clinical trials with available results evaluating immune checkpoints inhibitors.
| Study Name | Tested Drugs | Comparison | Phase | Histology | Therapy Setting | OS (HR, 95% CI, p) |
Median PFS (HR, 95% CI) |
ORR (%) |
CR (%) |
Grade 3 (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Javelin Renal 101 | avelumab + axitinib | sunitinib | III | ccRCC | First line | 12-mo: 86% vs. 83% (0.78; 0.55–1.08; p = 0.14) |
13.8 vs. 7.2 mo (0.61) |
55.2 vs. 25,5 | 3.4 vs. 1.8 | 71.2 vs. 71.5 |
| Keynote 426 | pembrolizumab + axitinib | sunitinib | III | ccRCC | First line | 12-mo: 90% vs. 78% (0.53; 0.38–0.74; p < 0.0001) |
15.1 vs. 11.1 mo (0.69; 0.57–0.84) |
59.3 vs. 35.7 | 5.8 vs. 1.9 | 75.8 vs. 70.6 |
| CheckMate 214 | nivolumab + ipilimumab | sunitinib | III | ccRCC | First line | 30-mo : 60% vs, 47% (0.66; 0.54–0.80; p < 0.0001) |
11.6 vs. 8.4 mo (0.82; 0.64–1,05) |
42 vs. 29 | 9 vs. 1 | 47 vs. 64 |
| Immotion 151 | atezolizumab + bevacizumab | sunitinib | III | ccRCC | First line | 24 mo : 63% vs. 60% (0.93; 0.76–1.14; p = 0·4751) |
11.2 vs. 7.7 mo (0.74; 0.57–0.96) |
43 vs. 25 | 9 vs. 4 | 40 vs. 54 |
Abbreviations: OS = overall survival; HR = Hazard Ratio; PFS = progression-free survival; ORR = objective response rate; CR = complete response; ccRCC: clear cell renal cell carcinoma, mo = months.
The Checkmate 214 trial confirmed the benefit of the nivolumab and ipilimumab association versus sunitinib in first line metastatic ccRCC among patients with intermediate or poor prognostic risk according to the International Metastatic Database Consortium (IMDC) risk model [20]. The objective response rate (ORR) was 42% versus 27%, and the complete response rate (CRR) was 11% versus 1% (p < 0.001). No differences were observed in terms of PFS. A recent update showed an interesting increase in OS after 30 months of follow-up in favor of nivolumab (anti-PD-1) plus ipilimumab (anti-CTLA4) combination (60% vs. 47%; HR: 0.66 ; CI 0.54 to 0.80 ; p < 0.0001) [43].
Recent updates also highlighted the benefit of ICI and TKI in combination. In Keynote 426, Pembrolizumab (anti-PD-1) plus axitinib showed a benefit in terms of OS at 12 month follow-up (90% vs. 78% ; HR: 0.53 ; CI: 0.38–0.74 ; p < 0.0001) leading to Food and Drug Administration (FDA) approval of the association in first line [26]. The Javelin 101 Renal comparing avelumab (anti-PD-L1) plus axitinib versus sunitinib in the PD-L1 positive population, defined as > 1% of positive immune cells staining within the tumour area, demonstrated a longer PFS, 13.8 months versus 8.4 months (HR = 0.69; 95% CI, 0.56 to 0.84; p < 0.001) and an improvement of the ORR (55.2% vs. 25%) [25]. After a 12 month follow-up, OS was not significantly different between the two arms. Similarly, the IMmotion 151 trial explored the atezolizumab (anti-PD-L1) and bevacizumab association regarding PD-L1 expression by immunohistochemistry (IHC) with a 1% cut-off on tumour-infiltrating immune cells [42]. The PFS was 11.2 months versus 8.4 months (HR = 0.74; 98.5% CI, 0.57 to 0.96; p = 0.02), but there was no statistical difference in OS at 24 months follow-up.
Among the recent phase III trials released, the distribution of patients in IMDC differed [20,25,26,42]. For instance, Checkmate 214 trial focused on intermediate- and poor-risk populations. The geography of recruitment may have an effect as the Keynote 426 trial had a large proportion of patients from outside the USA and Western Europe. Finally, RR and PFS are not strictly comparable. Moreover, follow-up remains short and it is difficult to formally conclude at this stage.
Considering the differences among trial populations and the lack of mature OS, the choice of first line therapy can be challenging. To date, guidelines for treatment decision rely on the IMDC risk model to stratify patients with untreated mRCC or previously treated with first-line targeted therapies [16,44]. However, the interest of this classification remains unclear with the advent of ICI combination therapies and patient characteristics stay major indicators in the absence of prospectively validated biomarkers. Indeed, a significant number of comorbidities could influence the treatment choice and should be confronted by the safety profile of ICI and TKI.
3.2. Second-Line Treatment
The choice of treatment in the second-line setting and beyond depends on the therapy previously received. In case of progression after antiangiogenics, the Checkmate 025 trial comparing nivolumab and everolimus showed an improvement of median OS in the nivolumab arm (25 vs. 19.6 months; HR = 0.73; 98.5% CI, 0.57 to 0.93; p = 0.0018) [45]. Notably, PD-L1 expression by IHC was associated with a poorer survival, but was not predictive of nivolumab efficacy. After first line ICI, any anti-angiogenic drug was recommended by the European Association of Urology with a low level of relevance [16]. However, due to the release of recent results, there is no prospectively validated data on the best therapeutic option after ICI and TKI combination in the front-line setting.
3.3. Adjuvant treatment
Finally, several ongoing phase III trials are studying the efficacy of ICI alone (atezolizumab, Pembrolizumab, nivolumab and tremelimumab) or in combination (nivolumab + ipilimumab) in the challenging peri-operative and adjuvant settings. Unsurprisingly, there is no combination of ICI plus TKI under evaluation as S-TRAC trial was the only one to show a benefit of adjuvant sunitinib in locally advanced high-risk ccRCC [46].
3.4. Non-Clear Cell Renal Cell Carcinoma
Unlike ccRCC, the management of nccRCC remains unclear and clinical trials are preferred [47]. Indeed, few prospective data for nccRCC treatment are available and trials have shown a lower efficacy of anti-angiogenic therapy compared to ccRCC with an impact on the prognosis of both PFS and OS [48]. In a recent retrospective trial including 41 patients, nivolumab seemed to demonstrate its efficacy with a favourable safety profile [49]. Indeed, the ORR was 20% and the median PFS was 3.5 months (95% CI; 1.9–5.0 months). Median OS was not reached, and the overall survival at the 10-month time point from the start of nivolumab treatment was 68%. Behind ccRCC, several trials evaluating ICI targeted drugs alone or in combination with TKI are currently recruiting in nccRCC without distinction of the entities, as shown in Table 2 and Table 3. Interestingly, the combination of durvalumab and savolitinib (MET TKI) is specifically evaluated in pRCC. Likewise, nivolumab plus axitinib is studied in tRCC. Of note, ongoing trials using emerging drugs or vaccinal strategies include de facto nccRCC under the non-restrictive RCC inclusion criteria, Table 4.
Table 2.
Clinical trials evaluating immune checkpoint inhibitors in association with tyrosine kinase inhibitors.
| NCT number | Targeting Agents | Comparison | Phase | Histology | Primary Endpoint | Therapy Setting | Status |
|---|---|---|---|---|---|---|---|
| NCT03260894 | pembrolizumab + epacadostat | sunitinib or pazopanib | III | ccRCC | ORR | First line | Active not recruiting |
| NCT02811861 | lenvatinib + everolimus or pembrolizumab | sunitinib | III | ccRCC | PFS | First line | Active not recruiting |
| NCT03141177 | nivolumab + cabozantinib | sunitinib | III | ccRCC | PFS | First line | Active not recruiting |
| NCT03793166 | nivolumab, ipilimumab, cabozantinib | nivolumab or nivolumab + cabozantinib | III | ccRCC | OS | First line | Recruiting |
| NCT03937219 | nivolumab + ipilimumab + cabozantinib | nivolumab + ipilimumab + placebo | III | ccRCC | DFS | First line | Recruiting |
| NCT03680521 | sitravatinib + nivolumab | _ | II | ccRCC | ORR | First line | Recruiting |
| NCT02960906 | nivolumab, ipilimumab, VEGFR-TKI | _ | II | ccRCC | ORR | First line | Recruiting |
| NCT03736330 | axitinib + pembrolizumab + D-CIK | _ | II | ccRCC | ORR | At least second line | Recruiting |
| NCT02819596 | savolitinib, durvalumab, tremelimumab | _ | II | ccRCC, pRCC | DLT, ORR | At least second line | Unknown |
| NCT02964078 | pembrolizumab + interleukin-2 | _ | II | ccRCC | ORR | Any | Active not recruiting |
| NCT03092856 | anti-OX40 agonist antibody + axitinib | _ | II | ccRCC | PFS | No standard anymore available | Recruiting |
| NCT02724878 | atezolizumab + bevacizumab | _ | II | nccRCC | ORR | Any | Active not recruiting |
| NCT03635892 | nivolumab + cabozantinib | _ | II | nccRCC | ORR | Any | Recruiting |
| NCT03595124 | nivolumab + axitinib | _ | II | tRCC | PFS | Any | Recruiting |
| NCT02493751 | avelumab + axitinib | _ | I/II | ccRCC | DLT | First line | Published |
| NCT02899078 | ibrutinib + nivolumab | _ | I/II | ccRCC, nccRCC | PFS | At least second line | Recruiting |
| NCT02348008 | pembrolizumab + bevacizumab | _ | I/II | ccRCC | Safety, efficacy | At least second line | Active not recruiting |
| NCT03172754 | nivolumab + axitinib | _ | I/II | ccRCC | Safety | At least second line | Recruiting |
| NCT03024437 | atezolizumab + entinostat + bevacizumab | atezolizumab + entinostat | I/II | ccRCC | Safety, ORR | At least second line | Recruiting |
| NCT02501096 | pembrolizumab + lenvatinib | _ | I/II | ccRCC | DLT, ORR | No standard anymore available | Active not recruiting |
| NCT01472081 | nivolumab + sunitinib or pazopanib | nivolumab | I | ccRCC | Safety | At least second line | Published |
| NCT03307785 | TSR-022 (anti-TIM3)/niraparib/TSR-042 (anti-PD1)/chemotherapy/bevacizumab | _ | I | RCC and others | Safety | At least second line | Active not recruiting |
| NCT03200587 | avelumab + cabozantinib | _ | Ib | ccRCC | Safety | Any | Recruiting |
Abbreviations: OS = overall survival; HR = Hazard Ratio; PFS = progression-free survival; ORR = objective response rate; CR = complete response; ccRCC = clear cell renal cell carcinoma; nccRCC = non clear cell renal cell carcinoma; DLT = dose-limiting toxicity; DFS = disease-free survival; ICI = immune checkpoint inhibitor; TKI = tyrosine kinase inhibitor; VEGFR: vascular endothelial growth factor receptor; _ = na; SBRT = stereotactic body radiation.
Table 3.
Clinical trials evaluating targeted immunotherapies alone or in combination.
| NCT Number | Targeting Agents | Comparison | Phase | Histology | Primary Endpoint | Therapy Setting | Status |
|---|---|---|---|---|---|---|---|
| NCT03729245 | NKTR-214 (IL2R agonist) + nivolumab | sunitinib or cabozantinib | III | ccRCC | ORR, OS | First line | Recruiting |
| NCT03873402 | nivolumab + ipilimumab | nivolumab | III | ccRCC | PFS, ORR | First line | Recruiting |
| NCT01668784 | nivolumab | everolimus | III | ccRCC | OS | At least second line | Published |
| NCT03055013 | nivolumab | observation | III | ccRCC | DFS | Peri-operative | Recruiting |
| NCT03024996 | atezolizumab | placebo | III | ccRCC | DFS | Adjuvant | Active not recruiting |
| NCT03138512 | nivolumab + ipilimumab | placebo | III | ccRCC | DFS | Adjuvant | Recruiting |
| NCT03142334 | pembrolizumab | placebo | III | ccRCC | DFS | Adjuvant | Active not recruiting |
| NCT03288532 | nivolumab, tremelimumab | _ | III | ccRCC | DFS | Adjuvant | Recruiting |
| NCT02996110 | BMS-986205 (IDO1 oral inhibitor) +/− nivolumab +/− ipilimumab | nivolumab +/− ipilimumab | II | ccRCC, nccRCC | ORR, PFS | First line | Recruiting |
| NCT03552380 | nivolumab + ipilimumab + entinostat | _ | II | ccRCC | Safety | At least second line | Recruiting |
| NCT03501381 | entinostat + IL2 | IL2 | II | ccRCC | PFS | At least second line | Recruiting |
| NCT03469713 | nivolumab + SBRT | _ | II | ccRCC | ORR | At least second line | Recruiting |
| NCT03177239 | nivolumab + ipilimumab | _ | II | nccRCC | ORR | Any | Active, not recruiting |
| NCT04262375 | oleclumab (anti-CD73 antagonist mAb) + durvalumab | _ | II | RCC and others | ORR, PFS | Any | Not yet recruiting |
| NCT03207867 | NIR178 (A2aR antagonist) + spartalizumab (anti-PD1) | _ | II | RCC and others | ORR | At least second line | Recruiting |
| NCT03693612 | GSK3359609 (anti-ICOS) + tremelimumab | chemotherapies | II | RCC and others | DLT | No standard anymore available | Recruiting |
| NCT03693612 | anti-ICOS + tremelimumab | _ | II | RCC and others | Safety, DLT | No standard anymore available | Recruiting |
| NCT01038778 | entinostat + aldesleukin | _ | I/II | ccRCC | Dose, ORR | At least second line | Active, not recruiting |
| NCT03308396 | durvalumab + guadecitabine | _ | I/II | ccRCC | Safe dose/ORR | At least second line | Recruiting |
| NCT02989714 | nivolumab + interleukin-2 | - | I/II | ccRCC | Safety | Third line | Active not recruiting |
| NCT02460224 | LAG525 (anti-LAG3) + spartalizumab (anti-PD1) | _ | I/II | RCC and others | DLT, ORR | At least second line | Active, not recruiting |
| NCT03652077 | anti-TIM3 | _ | I/II | RCC and others | Safety | No standard anymore available | Recruiting |
| NCT02608268 | MBG453 (anti-TIM3) + spartalizumab (anti-PD1) | spartalizumab anti-PD1 | I/II | RCC and others | Safety, ORR, DLT | No standard anymore available | Recruiting |
| NCT01968109 | relatlimab (anti-LAG3) + nivolumab | relatlimab | I/II | RCC and others | Safety, ORR, DLT | No standard anymore available | Recruiting |
| NCT03126110 | INCAGN01876 (anti-GITR) + nivolumab + ipilimumab | _ | I/II | RCC and others | Safety, tolerability | No standard anymore available | Recruiting |
| NCT02335918 | varlilumab (anti-CD27) + nivolumab | _ | I/II | RCC and others | DLT, ORR | At least second line | Completed |
| NCT02718066 | HBI-8000 (HDACi) + nivolumab | _ | I/II | RCC and others | RP2D | At least second line | Recruiting |
| NCT02890069 | spartalizumab + LCL16 (IAP inhibitor) + everolimus + panobinostat | _ | I/II | RCC and others | DLT | At least second line | Recruiting |
| NCT02771626 | CB-839 (glutaminase inhibitor) + nivolumab | _ | I/II | RCC and others | Safety, efficacy | At least second line | Recruiting |
| NCT02817633 | TSR-022 (anti-TIM3)/TSR-042 (anti-PD1)/TSR-033 (anti-LAG3) | _ | I | RCC and others | Safety, tolerability | No standard anymore available | Recruiting |
| NCT03119428 | OMP-31M32 (anti-TIGIT) + nivolumab | _ | I | RCC and others | DLT | No standard anymore available | Terminated |
| NCT00351949 | IMP321 (anti-LAG3) | _ | I | RCC and others | Safety, tolerability | No standard anymore available | Completed |
| NCT02386111 | varlilumab (anti-CD27) | _ | I | RCC and others | Safety, tolerability | At least second line | Terminated |
| NCT03343613 | LY3300054 (IDO1 inhibitor) + PD-L1 inhibitor | _ | I | RCC and others | DLT | No standard anymore available | Recruiting |
| NCT02812875 | CA-170 (VISTA antagonist) | _ | I | RCC and others | DLT | No standard anymore available | Active not recruiting |
| NCT02655822 | ciforadenant (A2aR antagonist) +/- atezolizumab | _ | I | RCC and others | DLT | At least second line | Recruiting |
| NCT04198766 | anti-OX40 + pembrolizumab | _ | I | RCC and others | Safety | No standard anymore available | Recruiting |
Abbreviations: OS = overall survival; HR = Hazard Ratio; PFS = progression-free survival; ORR = objective response rate; CR = complete response; ccRCC = clear cell renal cell carcinoma; nccRCC = non clear cell renal cell carcinoma; DLT = dose-limiting toxicity; DFS = disease-free survival.
Table 4.
Vaccinal strategies and CAR T-cells trials.
| NCT Number | Targeting Agents | Comparison | Phase | Histology | Primary Endpoint | Therapy Setting | Status |
|---|---|---|---|---|---|---|---|
| NCT00458536 | dendritic cell tumor fusion vaccine + GM-CSF | _ | I/II | ccRCC, nccRCC | Safety | Any | Active not recruiting |
| NCT03633110 | GEN-009 Adjuvanted Vaccine + nivolumab + pembrolizumab | _ | I/II | RCC and others | Safety | At least second line | Recruiting |
| NCT00722228 | Autologous or Allogeneic tumor cells | _ | I/II | RCC and others | Safety, efficacy | At least second line | Recruiting |
| NCT03393936 | anti-ROR2 CAR-T or anti AXL CART-T | _ | I/II | ccRCC, nccRCC | Safety | No standard available | Recruiting |
| NCT02830724 | anti-CD70 CAR-T | _ | I/II | RCC and others | Safety | At least second line | Recruiting |
| NCT01218867 | anti-VEGFR2 CAR-T | _ | I/II | RCC and others | ORR | At least second line | Terminated |
| NCT03638206 | anti-C-MET CAR-T | _ | I/II | RCC and others | Safety | At least second line | Recruiting |
| NCT02950766 | neovax vaccine + ipilimumab | _ | I | ccRCC | DLT | At least second line | Recruiting |
| NCT00096629 | PSMA DNA vaccine | _ | I | ccRCC, nccRCC | Safety | Adjuvant | Completed |
| NCT03548467 | VB10.NEO vaccine +/- bempegaldesleukin (NKTR-214) | _ | I | RCC and others | Safety | At least second line | Recruiting |
| NCT03294083 | pexastimogene devacirepvec (Pexa-Vec) | _ | I | ccRCC, nccRCC | Safety, efficacy | At least second line | Recruiting |
| NCT03715985 | EVAX-01-CAF09b (peptide-based vaccine) +/- anti-PD1 or anti-PD-L1 | _ | I | RCC and others | Safety, efficacy | First line | Recruiting |
Abbreviations: OS = overall survival; HR = Hazard Ratio; PFS = progression-free survival; ORR = objective response rate; CR = complete response; ccRCC = clear cell renal cell carcinoma; nccRCC = non clear cell renal cell carcinoma; DLT = dose-limiting toxicity; DFS = disease-free survival; ICI = immune checkpoint inhibitor; TKI = tyrosine kinase inhibitor; VEGFR: vascular endothelial growth factor receptor; - = na; SBRT = stereotactic body radiation.
4. Emerging Drugs in Ongoing Trials Include Renal Cell Carcinoma
4.1. Inhibitory Immune Checkpoints
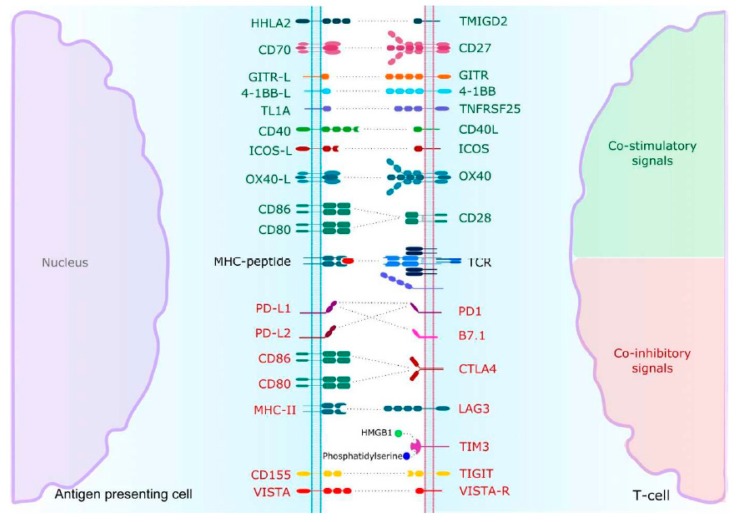
The anergy of T cell is commonly understood as the result of a sequential transduction of signals implicating immune checkpoints in the immune synapse, Figure 2. If CTLA4 and PD-1 are the most common and well-studied, others are emerging and could be implied in resistance to traditional ICI and are evaluated in clinical trials including RCC, as shown in Table 2 and Table 3.
Figure 2.
T-cell activation or inhibition is the result of an integrated and sequential intracellular signal cascade after MHC-peptide recognition by TCR. Abbreviations: GITR = Glucocorticoid-Induced TNF Receptor, ICOS = Inducible co-stimulator, OX40 = CD134, TCR = T-Cell Receptor, PD1 = Programmed Death 1, CTLA4 = Cytotoxic T-lymphocyte Antigen-4, LAG3 = Lymphocyte-associated gene 3, TIM3 = Tcell Immunoglobulin and Mucin domain-3, TIGIT = T-cell Immunoglobulin and ITIM domain, VISTA = Vdomain Immunoglobulin Suppressor of T-cell Activation.
Lymphocyte-associated gene 3 (LAG3) is a transmembrane protein mainly expressed in activated T and natural killer (NK) cells, as shown in Figure 2 [29]. LAG3 is located on T cells. It shares a structural homology with CD4 and binds its ligand, the major histocompatibility complex class II (MHCII) with higher affinity. Moreover, the LAG3 blockade leads to an increased production of interferon gamma (INFγ), tumour necrosis factor alpha (TNFα) and pro-inflammatory interleukins [50].
Other ICI are emerging. Among them, T cell immunoglobulin and mucin domain3 (TIM3) expressed on a wide variety of immune cells. TIM3 contributes to immune tolerance by inhibiting T cells activation, mostly by upregulation of apoptosis [51]. Interestingly, there could be a synergistic effect with PD-1-PD-L1 blockade, reversing T cell exhaustion and improving anti-tumoral immune response [52].
T cell immunoglobulin and ITIM domain (TIGIT) is mainly found on TIL and is immunosuppressive through disrupting interleukins production and antigen-presenting cell (APC) maturation [53]. The two ligands Nectin-2 and CD155 are expressed in various cellular types from tumour to immune cells. Similar results in terms of T cells exhaustion were observed in the B7-H3 pathway [54].
The v-domain immunoglobulin suppressor of T cell activation (VISTA) is predominantly expressed on myeloid-derived suppressive cells (MDSC) and APC and down-regulates T cells activation [55]. Remarkably, VISTA blockade seemed to inhibit regulatory T cell immunosuppressive functions [56].
4.2. Co-Activating Immune Checkpoints
Tumour-specific T cell-mediated immune response is balanced by both inhibitory and co-stimulatory signals. If ICI are mainly used to restore immune response, agonist drugs are developed to increase co-stimulatory signals and stimulate immune response, as shown in Figure 2. Like ICI, several co-activating immune checkpoints are used in ongoing trials, Table 2 and Table 3.
On T cells, CD28 is known to deliver an activating signal when binding CD80/86 after TCR recognition of major histocompatibility complex (MHC). Inducible co-stimulator (ICOS) mainly located on CD4+ T cells belongs to the immunoglobulin family as well as CD28 and produces inflammatory cytokines. Its intracytoplasmic structure has a strong affinity for phosphoinositide 3-kinase (PI3K) favoring the proliferation signal in lymphocytes [57].
The tumour necrosis factor (TNF) receptor superfamily is represented by a group of both soluble and transmembrane receptors involved in inflammation processes and able to bind a variety of ligands such as TNFα, TNFβ and OX40 ligand [58]. When binding its ligand, OX40 promotes T cells proliferation and survival, particularly CD4+ and CD8+ T cells, by upregulating pro-inflammatory cytokines and anti-apoptotic molecules. Other member TNF receptors such as CD40, CD27 and 4-1BB contribute to increased cytotoxic T cells mediated response through apoptosis or memory-cell differentiation. The glucocorticoid-induced TNF receptor (GITR), located on CD4+ and CD8+ T cells and predominantly on FoxP3+ regulatory T cells, is known to enhance immunity to tumours through the attenuation of the effector activity of immunosuppressive regulatory T cells [59].
4.3. Metabolic Pathways
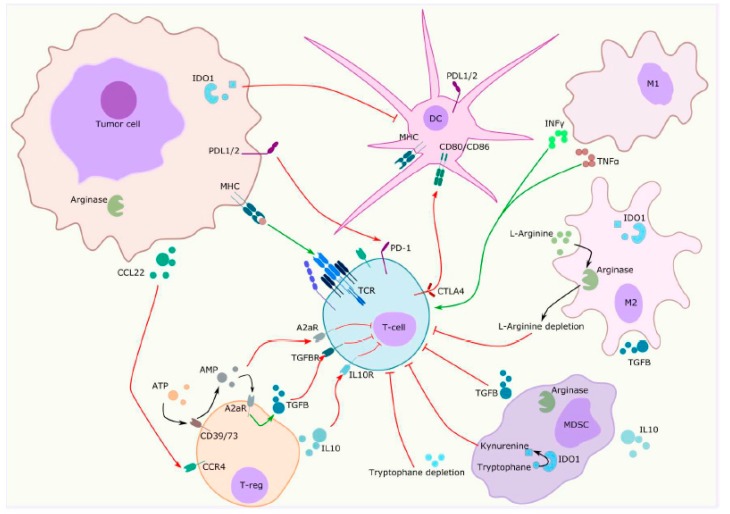
Metabolic changes influence the TME by providing immunosuppressive metabolites and favoring tumour growth in hypoxic conditions, as shown in Figure 3 [60]. Multiple enzymes have been identified as key regulators of cytotoxic T cells immune response and are under evaluation in clinical trials, as shown in Table 2 and Table 3.
Figure 3.
Tumor microenvironment is commonly defined as the co-existence of tumor cells interacting with resident and infiltrating host cells, secreted factors and extracellular matrix proteins. Among them, immunosuppressive cells are recruited in the tumor microenvironment by chemotaxis and are responsible for immunomodulatory cytokines production enhancement and decreased essential amino acids availability, resulting in favorable conditions for tumor growth. MDSC = Myeloïd-derived suppressive cells; T-cell = T lymphocyte; T-reg = Regulator lymphocyte; M2 = Type 2 macrophage; DC = dendritic cell; TGBß = tumor growth factor ß; A2aR = adenosine 2a receptor; ATP = adenosine triphosphate; AMP = adenosine monophosphate; IL10 = interleukin-10; MHC = major histocompatibility complex; PD1 = programmed death protein 1; PDL1 = programmed death ligand 1; IDO1 = indoleamine 2,3 dioxygenase 1; CTLA4 = cytotoxic T-lymphocyte antigen-4; CCL22 = C-C motif chemokine Ligand L22; CCR4 = C-C motif chemokine receptor 4; M1 = type 1 macrophage; INFγ = interferon γ; TNF α = tumor necrosis factor α.
Among them, indoleamine 2,3 dioxygenase 1 (IDO1), an intracellular enzyme, catalyses the conversion of tryptophan into kynurenine [61]. Although physiologically expressed in stromal and dendritic cells, it is overexpressed in MDSC and tumour cells. The tryptophan depletion induced by IDO1 expression leads to T cell exhaustion and apoptosis. Moreover, high concentration of kynurenine promotes immune-tolerant dendritic cells and regulatory T cell proliferation [62].
Adenosine is a purine base known to bind G-protein coupled adenosine receptors, upregulated in activated immune cells [63]. Adenosine 2a receptor (A2aR) triggers upregulation of adenylate cyclase activity leading to increased cyclic adenosine monophosphate (cAMP) concentration. It has a profound immunomodulatory effect on immune cells by various mechanisms including ZAP70 inhibition in TCR signalling, IL2 down-regulation, FoxP3 expression enhancement on regulatory T cells and TGFß marked secretion [64].
As previously reported, TME acquires immunosuppressive features by reprogramming metabolic processes. Arginine is an essential amino acid in immune and tumour cells demonstrating a high level of arginase [65]. Arginase inhibitors are currently evaluated in combination with several ICI, Table 3.
4.4. Other Strategies
Various innovative strategies in immunotherapy are applied for RCC. The histone conformation impacts the transcription and is regulated through phosphorylation, sumoylation, ubiquitination, acetylation and deacetylation [66]. Histone deacetylase (HDAC) inhibitors modify the chromatin accessibility and play a crucial role in cycle arrest and apoptosis and could eventually enhance tumour antigens release and indirectly improve antigen presentation by APC and T cells priming [67]. In RCC, entinostat, panobinostat and chidamide are under evaluation in combination with ICI or TKI, as shown in Table 2 and Table 3.
Several vaccinal strategies are tested in cancers with the common objective to upregulate tumour neo-antigens exposure to immune system, particularly by T cells priming phase improvement [68]. At this time, three broad vaccine types are under investigation including DNA/RNA-based, peptide-based and cell-based vaccines with encouraging results in RCC. Oncolytic viruses are interesting alternatives, designed to infect tumour cells and hijack cellular machinery to induce transgene expression.
The chimeric Antigen Receptor (CAR) T cells are T cells genetically engineered to produce an artificial T cell receptor that combines both antigen-binding and T cell activating functions. The use of CART cells is hampered in RCC by the tumour heterogeneity. CAR T cells are currently tested in renal cell carcinoma, targeting various antigens such as ROR2, AXL, CD70, VEGFR2, MET or CAIX, as seen in Table 3. Moreover, pre-clinical data suggest the rationale of combining CAR T cells with TKI or radiotherapy [69].
5. Predictive Biomarkers in RCC
5.1. Clinico-Biological Biomarkers
The IMDC risk model includes six clinical and biological variables (poor Karnofsky performance status, less than 1 year between the diagnosis and the treatment, low hemoglobin concentration, high platelet count, high neutrophil count and high serum calcium) and has been validated in ccRCC and nccRCC [70,71]. However, the use of this classification is limited in nccRCC because the group is heterogeneous and samples in each group are small. The guidelines are dichotomized between favorable- and intermediate- or poor-risk disease, as shown in Figure 1 [16]. For instance, the clinical outcome was better in patients with intermediate- or poor-risk disease under nivolumab plus ipilimumab [20]. On the contrary, patients with favorable-risk disease better answered to sunitinib. These results suggest a distinct underlying biology.
5.2. Immunohistochemical Biomarkers
The most studied immunohistochemical biomarker PD-L1 failed to demonstrate a predictive capability in metastatic RCC [20]. If it was demonstrated as a poor prognostic factor in both ccRCC and nccRCC; indeed its ability to discriminate between patients the good responders is questionable [35]. Limitations to the use of PD-L1 have been well described and encompass intra-tumoral heterogeneity, variability of cut-offs and heterogeneous expression between primary and metastatic sites among others [72].
The contradictory and unresolved issue about PD-L1 as a biomarker also reveals the complicated interactions between the tumour and immune response, as shown in Figure 3. A comprehensive immune phenotype including other immunosuppressive factors such as TGFβ or IDO-1 and a better characterization of immune cells could be another step forward in the search of predictive biomarkers. Interestingly, IDO-1 expression was higher in endothelial cells of responders to nivolumab in a cohort of 15 patients with metastatic RCC [73]. This results in a reduced influx of tryptophan in the surrounding tumour tissue leading to a decrease in tumour proliferation.
5.3. Transcriptomic Analysis
Beuselink and colleagues first performed clustering transcriptomic analysis in patients with metastatic ccRCC (n = 53) receiving first-line sunitinib [74,75]. Hakimi et al. performed the same analysis in ccRCC of patients included in COMPARZ trial receiving pazopanib or sunitinib (n = 453) and identified four similar clusters [75]. Cluster 3 had the best prognosis with high angiogenic gene expression being associated with a better outcome under antiangiogenic therapy and similar to ccrcc2 cluster reported by Beuselinck et al. [76]. Of note, PBRM1 mutation was frequently associated with angiogenic gene expression. Cluster 4 had a worse prognosis and was similar to ccrcc4 with upregulation of immune pathways. Interestingly, ccrcc4 was enriched in tumours with sarcomatoid differentiation with a frequent expression of PD-L1 [77]. Clusters 1 and 2 were intermediate clusters with a lower expression of angiogenic and immune genes. A consensus classification could emerge from these different studies as proposed in the bladder cancer [78].
McDermott and colleagues explored three gene expression signatures: angiogenesis, T-effector/IFN-γ response, and myeloid inflammatory genes in ccRCC (n = 263) from IMmotion 150 trial: a phase 2 trial before IMmotion 151 of atezolizumab (anti-PD-L1) alone or combined with bevacizumab (anti-VEGF) versus sunitinib [76]. Interestingly, high T effector/IFNγ signature was associated with a better outcome to atezolizumab plus bevacizumab and a high myeloid inflammation signature was associated with reduced survival in the atezolizumab alone arm.
5.4. Tumour Mutational Burden and Mismatch Repair Deficiency
The TMB is based on the total number of mutations per coding area of the tumour genome. High mutation burden favors the formation of neo-antigens that enhance the tumour immune response [79]. This biomarker initially failed to identify responders to immunotherapy in ccRCC but is still under evaluation [80,81]. However, Limitations of this marker may be due to technical requirements such as coverage, DNA amount and analysis time and lack of standardization.
Even if RCC are not considered to be in the spectrum of hereditary non-polyposis colon cancer (HNPCC) or Lynch syndrome, loss of mismatch repair (MMR) proteins leading to microsatellite instability is frequently observed [82]. MMR deficient tumours demonstrate a higher rate of mutations but on specific genes. In RCC, data on the response to immunotherapy according to MMR status are also still ongoing.
5.5. Gut Microbiome
The gut microbiome, defined by the environmental conditions and the collection of microorganism and host genomes in an ecosystem, seems to influence the response to ICI. Indeed, the use of antibiotics could unfavorably impact the response. Indeed, Derosa et al. demonstrated a shorter survival and higher rates of progressive disease in patients with mRCC who received antibiotics within the month following the beginning of the treatment [83]. Indeed, antibiotics-related dysbiosis impacted the mirobiome and could decrease the activity of ICI. Conversely, they identified that some bacteria such as B. salyersiae or A. muciniphila could restore the efficacy of ICI.
6. Future Directions
6.1. Microenvironment Cell Population Counter
The microenvironment cell population counter (MCP counter) aims to assess the proportion of immune and stromal cells in the TME from transcriptomic data [84]. They classified tumours into four molecular TME subgroups: immune infiltration, T and NK lymphocytes, MCH1 expression and fibroblastic infiltration. This classification could be judiciously applied to previous transcriptomic data.
6.2. Single-Cell Technologies
The development and proper use of immunotherapies rely on the detailed understanding of tumour composition. However, this purpose is hampered by intratumour heterogeneity, which was demonstrated to be extensive in ccRCC [85]. With recent developments in single-cell technologies though, we can now circumvent and integrate intratumour heterogeneity into analyses in oncology by characterizing each individual cells within tumours [86]. In RCC, Kim and collaborators performed single-cell RNA sequencing to study the intratumour heterogeneity of paired primary RCC and lung metastasis [87]. A considerable variability between the primary and metastatic sites and among tumour cells was demonstrated by the activation of drug target pathways. This study encourages the use of single-cell RNA sequencing to better characterize cell populations and to favor the discovery of new biomarkers.
6.3. 3D Culture Models
The 3D culture models of ccRCC could represent good pre-clinical models based on tissue slices preserving the stromal components of the TME [88]. Already used for TKI targeted therapies, these models are increasingly being considered as potential platforms for monitoring immunotherapy responses [89]. Recent studies indeed reported an active immune TME after applying immunomodulatory molecules on tumour slice organotypic cultures, opening the way for their study on 3D ex vivo tumour models [90]. Interestingly, co-cultures of tumour-derived organoids were enriched with lymphocytes, leading Dijkstra and collaborators to recently demonstrate the generation of tumour-reactive T cells by co-culture of peripheral blood lymphocytes and colorectal cancer-derived organoids [91]. These precursor approaches will allow to learn more about immune escape mechanisms and determine associated predictive biomarkers.
6.4. Imaging
Novel methods of imaging are non-invasive and assess RCC at different time points. Bensch et al. demonstrated that positron emission tomography (PET) imaging was able to localize 89Zr-labeled atezolizumab to tumours expressing PD-L1 between primary and metastatic sites [92]. The uptake in tumours was heterogeneous and the clinical response better correlated with the pretreatment PET signal than with PD-L1 immunohistochemistry or T-effector gene expression signature. Moreover, CD8+ T cell infiltration was inferred from computed tomography [93].
6.5. Circulating Tumour Cells
The circulating tumour cells (CTC) is a non-invasive method isolating CTC from blood circulation [94]. The challenge is to detect them because of the rare expression of the usual marker epithelial cell adhesion molecule (EpCAM). This is due to the transdifferentiation of tumour cells through the epithelial mesenchymal transition [95]. The antibodies directed against membrane carbonic anhydrase 9 (CA9) and CD147 largely improved the detection of CTC from 17% with EpcAM to 97% of samples with CA9 and CD147 markers [96]. Other methods are based on RT-PCR targeting VHL gene alteration and size-based blood filtration associated with genetic and morphological analyses [97]. The CTC analyses could be used to select patients for clinical trials guided by biomarkers. The phenotype could evolve under treatment and be timely investigated to adapt the therapeutic strategy.
7. Conclusions
Renal cell carcinoma include distinct entities with specific molecular alterations. Among them, ccRCC, the most frequent, is particularly characterized by its angiogenic and immunogenic TME with complex interactions between stromal and immune cells. Like tumour cells, intratumour heterogeneity is present in the TME with a variable distribution and phenotype and consequently favours resistance to treatment.
Therapeutic options for patients with mRCC have expanded rapidly over the past decade with targeted immunotherapy being the new corner stone. Several emerging drugs are designed to enhance the antitumour immune response and are tested in ongoing trials.
With an increasing number of treatment options available, improved biomarkers are needed to better stratify patients and define the optimal selection of patients and the sequence of treatment to overcome resistance. Promising biomarkers such as gene expression signatures or gut microbiome are under evaluation. It is likely that the future of predictive biomarkers relies on the combination of different approaches reflecting the complexity of the TME.
We hope that, in the future, new technologies such as single-cell technologies contribute to unravel the intratumour heterogeneity, identify predictive biomarkers and discover new treatment targets.
Author Contributions
Conception and design: S.-F.K-.J., N.R.-L.; Collection and assembly of data: S.-F.K.-J., A.D., J.S., L.C., B.L., F.D., M.-A.B.-R., R.M., K.B., G.V.; Manuscript writing: A.D., J.S., S.-F.K.-J.; Final approval of the manuscript: all authors; Accountable for all aspects of the work: all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Ljungberg B., Campbell S.C., Choi H.Y., Jacqmin D., Lee J.E., Weikert S., Kiemeney L.A. The epidemiology of renal cell carcinoma. Eur. Urol. 2011;60:615–621. doi: 10.1016/j.eururo.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 3.Gossage L., Eisen T., Maher E.R. Vhl, the story of a tumour suppressor gene. Nat. Rev. Cancer. 2015;15:55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 4.Akaza H., Fukuyama T. Axitinib for the treatment of advanced renal cell carcinoma. Expert Opin. Pharmacother. 2014;15:283–297. doi: 10.1517/14656566.2014.868436. [DOI] [PubMed] [Google Scholar]
- 5.Motzer R.J., Hutson T.E., Cella D., Reeves J., Hawkins R., Guo J., Nathan P., Staehler M., de Souza P., Merchan J.R., et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N. Engl. J. Med. 2013;369:722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 6.Choueiri T.K., Escudier B., Powles T., Mainwaring P.N., Rini B.I., Donskov F., Hammers H., Hutson T.E., Lee J.L., Peltola K., et al. Cabozantinib versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015;373:1814–1823. doi: 10.1056/NEJMoa1510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rini B.I., Pal S.K., Escudier B.J., Atkins M.B., Hutson T.E., Porta C., Verzoni E., Needle M.N., McDermott D.F. Tivozanib versus sorafenib in patients with advanced renal cell carcinoma (tivo-3): A phase 3, multicentre, randomised, controlled, open-label study. Lancet Oncol. 2020;21:95–104. doi: 10.1016/S1470-2045(19)30735-1. [DOI] [PubMed] [Google Scholar]
- 8.Moch H., Cubilla A.L., Humphrey P.A., Reuter V.E., Ulbright T.M. The 2016 who classification of tumours of the urinary system and male genital organs-part a: Renal, penile, and testicular tumours. Eur. Urol. 2016;70:93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research N., Linehan W.M., Spellman P.T., Ricketts C.J., Creighton C.J., Fei S.S., Davis C., Wheeler D.A., Murray B.A., Schmidt L., et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016;374:135–145. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argani P. Mit family translocation renal cell carcinoma. Semin. Diagn. Pathol. 2015;32:103–113. doi: 10.1053/j.semdp.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Malouf G.G., Monzon F.A., Couturier J., Molinie V., Escudier B., Camparo P., Su X., Yao H., Tamboli P., Lopez-Terrada D., et al. Genomic heterogeneity of translocation renal cell carcinoma. Clin. Cancer Res. 2013;19:4673–4684. doi: 10.1158/1078-0432.CCR-12-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malouf G.G., Comperat E., Yao H., Mouawad R., Lindner V., Rioux-Leclercq N., Verkarre V., Leroy X., Dainese L., Classe M., et al. Unique transcriptomic profile of collecting duct carcinomas relative to upper tract urothelial carcinomas and other kidney carcinomas. Sci. Rep. 2016;6:30988. doi: 10.1038/srep30988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlo M.I., Chaim J., Patil S., Kemel Y., Schram A.M., Woo K., Coskey D., Nanjangud G.J., Voss M.H., Feldman D.R., et al. Genomic characterization of renal medullary carcinoma and treatment outcomes. Clin. Genitourin. Cancer. 2017;15:e987–e994. doi: 10.1016/j.clgc.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.B., Brannon A.R., Toubaji A., Dudas M.E., Won H.H., Al-Ahmadie H.A., Fine S.W., Gopalan A., Frizzell N., Voss M.H., et al. Hereditary leiomyomatosis and renal cell carcinoma syndrome-associated renal cancer: Recognition of the syndrome by pathologic features and the utility of detecting aberrant succination by immunohistochemistry. Am. J. Surg. Pathol. 2014;38:627–637. doi: 10.1097/PAS.0000000000000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malouf G.G., Ali S.M., Wang K., Balasubramanian S., Ross J.S., Miller V.A., Stephens P.J., Khayat D., Pal S.K., Su X., et al. Genomic characterization of renal cell carcinoma with sarcomatoid dedifferentiation pinpoints recurrent genomic alterations. Eur. Urol. 2016;70:348–357. doi: 10.1016/j.eururo.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 16.Albiges L., Powles T., Staehler M., Bensalah K., Giles R.H., Hora M., Kuczyk M.A., Lam T.B., Ljungberg B., Marconi L., et al. Updated european association of urology guidelines on renal cell carcinoma: Immune checkpoint inhibition is the new backbone in first-line treatment of metastatic clear-cell renal cell carcinoma. Eur. Urol. 2019;76:151–156. doi: 10.1016/j.eururo.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Motzer R.J., Bacik J., Murphy B.A., Russo P., Mazumdar M. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J. Clin. Oncol. 2002;20:289–296. doi: 10.1200/JCO.20.1.289. [DOI] [PubMed] [Google Scholar]
- 18.Fyfe G., Fisher R.I., Rosenberg S.A., Sznol M., Parkinson D.R., Louie A.C. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. J. Clin. Oncol. 1995;13:688–696. doi: 10.1200/JCO.1995.13.3.688. [DOI] [PubMed] [Google Scholar]
- 19.Motzer R.J., Hutson T.E., Tomczak P., Michaelson M.D., Bukowski R.M., Rixe O., Oudard S., Negrier S., Szczylik C., Kim S.T., et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N. Engl. J. Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 20.Motzer R.J., Tannir N.M., McDermott D.F., Aren Frontera O., Melichar B., Choueiri T.K., Plimack E.R., Barthelemy P., Porta C., George S., et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motzer R.J., Escudier B., McDermott D.F., George S., Hammers H.J., Srinivas S., Tykodi S.S., Sosman J.A., Procopio G., Plimack E.R., et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leite K.R., Reis S.T., Junior J.P., Zerati M., Gomes Dde O., Camara-Lopes L.H., Srougi M. Pd-l1 expression in renal cell carcinoma clear cell type is related to unfavorable prognosis. Diagn. Pathol. 2015;10:189. doi: 10.1186/s13000-015-0414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson R.H., Dong H., Kwon E.D. Implications of b7-h1 expression in clear cell carcinoma of the kidney for prognostication and therapy. Clin. Cancer Res. 2007;13:709s–715s. doi: 10.1158/1078-0432.CCR-06-1868. [DOI] [PubMed] [Google Scholar]
- 24.Thompson R.H., Dong H., Lohse C.M., Leibovich B.C., Blute M.L., Cheville J.C., Kwon E.D. Pd-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin. Cancer Res. 2007;13:1757–1761. doi: 10.1158/1078-0432.CCR-06-2599. [DOI] [PubMed] [Google Scholar]
- 25.Motzer R.J., Penkov K., Haanen J., Rini B., Albiges L., Campbell M.T., Venugopal B., Kollmannsberger C., Negrier S., Uemura M., et al. Avelumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019;380:1103–1115. doi: 10.1056/NEJMoa1816047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rini B.I., Plimack E.R., Stus V., Gafanov R., Hawkins R., Nosov D., Pouliot F., Alekseev B., Soulieres D., Melichar B., et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N. Engl. J. Med. 2019;380:1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 27.Sharma P., Hu-Lieskovan S., Wargo J.A., Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 29.Anderson A.C., Joller N., Kuchroo V.K. Lag-3, tim-3, and tigit: Co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwe H., Pircher A., Van den Eynde K., et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 31.Akino T., Hida K., Hida Y., Tsuchiya K., Freedman D., Muraki C., Ohga N., Matsuda K., Akiyama K., Harabayashi T., et al. Cytogenetic abnormalities of tumor-associated endothelial cells in human malignant tumors. Am. J. Pathol. 2009;175:2657–2667. doi: 10.2353/ajpath.2009.090202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edeline J., Mottier S., Vigneau C., Jouan F., Perrin C., Zerrouki S., Fergelot P., Patard J.J., Rioux-Leclercq N. Description of 2 angiogenic phenotypes in clear cell renal cell carcinoma. Hum. Pathol. 2012;43:1982–1990. doi: 10.1016/j.humpath.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 33.Dufies M., Giuliano S., Ambrosetti D., Claren A., Ndiaye P.D., Mastri M., Moghrabi W., Cooley L.S., Ettaiche M., Chamorey E., et al. Sunitinib stimulates expression of vegfc by tumor cells and promotes lymphangiogenesis in clear cell renal cell carcinomas. Cancer Res. 2017;77:1212–1226. doi: 10.1158/0008-5472.CAN-16-3088. [DOI] [PubMed] [Google Scholar]
- 34.Thompson R.H., Kuntz S.M., Leibovich B.C., Dong H., Lohse C.M., Webster W.S., Sengupta S., Frank I., Parker A.S., Zincke H., et al. Tumor b7-h1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66:3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 35.Choueiri T.K., Fay A.P., Gray K.P., Callea M., Ho T.H., Albiges L., Bellmunt J., Song J., Carvo I., Lampron M., et al. Pd-l1 expression in nonclear-cell renal cell carcinoma. Ann. Oncol. 2014;25:2178–2184. doi: 10.1093/annonc/mdu445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta S., Cheville J. C., Jungbluth A. A., Zhang Y., Zhang L., Chen Y. B., Tickoo S. K., Fine S. W., Gopalan A., Al-Ahmadie H. A., et al. JAK2/PD-L1/PD-L2 (9p24.1) amplifications in renal cell carcinomas with sarcomatoid transformation: Implications for clinical management. Mod. Pathol. 2019;32:1344–1358. doi: 10.1038/s41379-019-0269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chevrier S., Levine J.H., Zanotelli V.R.T., Silina K., Schulz D., Bacac M., Ries C.H., Ailles L., Jewett M.A.S., Moch H., et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–749. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giraldo N.A., Becht E., Vano Y., Petitprez F., Lacroix L., Validire P., Sanchez-Salas R., Ingels A., Oudard S., Moatti A., et al. Tumor-infiltrating and peripheral blood t-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin. Cancer Res. 2017;23:4416–4428. doi: 10.1158/1078-0432.CCR-16-2848. [DOI] [PubMed] [Google Scholar]
- 39.Sjoberg E., Frodin M., Lovrot J., Mezheyeuski A., Johansson M., Harmenberg U., Egevad L., Sandstrom P., Ostman A. A minority-group of renal cell cancer patients with high infiltration of cd20 + b-cells is associated with poor prognosis. Br. J. Cancer. 2018;119:840–846. doi: 10.1038/s41416-018-0266-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choueiri T.K., Fishman M.N., Escudier B., McDermott D.F., Drake C.G., Kluger H., Stadler W.M., Perez-Gracia J.L., McNeel D.G., Curti B., et al. Immunomodulatory activity of nivolumab in metastatic renal cell carcinoma. Clin. Cancer Res. 2016;22:5461–5471. doi: 10.1158/1078-0432.CCR-15-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirotake S., Kaneko G., Nagata K., Oyama M., Nishimoto K. Histological complete response with nivolumab for renal cell carcinoma with multiple metastases: A case report. Mol. Clin. Oncol. 2019;10:244–248. doi: 10.3892/mco.2018.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rini B.I., Powles T., Atkins M.B., Escudier B., McDermott D.F., Suarez C., Bracarda S., Stadler W.M., Donskov F., Lee J.L., et al. Atezolizumab plus bevacizumab versus sunitinib in patients with previously untreated metastatic renal cell carcinoma (immotion151): A multicentre, open-label, phase 3, randomised controlled trial. Lancet. 2019;393:2404–2415. doi: 10.1016/S0140-6736(19)30723-8. [DOI] [PubMed] [Google Scholar]
- 43.Motzer R.J., Rini B.I., McDermott D.F., Aren Frontera O., Hammers H.J., Carducci M.A., Salman P., Escudier B., Beuselinck B., Amin A., et al. Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: Extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol. 2019;20:1370–1385. doi: 10.1016/S1470-2045(19)30413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lalani A.A., McGregor B.A., Albiges L., Choueiri T.K., Motzer R., Powles T., Wood C., Bex A. Systemic treatment of metastatic clear cell renal cell carcinoma in 2018: Current paradigms, use of immunotherapy, and future directions. Eur. Urol. 2019;75:100–110. doi: 10.1016/j.eururo.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Tomita Y., Fukasawa S., Shinohara N., Kitamura H., Oya M., Eto M., Tanabe K., Kimura G., Yonese J., Yao M., et al. Nivolumab versus everolimus in advanced renal cell carcinoma: Japanese subgroup analysis from the checkmate 025 study. Jpn. J. Clin. Oncol. 2017;47:639–646. doi: 10.1093/jjco/hyx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravaud A., Motzer R.J., Pandha H.S., George D.J., Pantuck A.J., Patel A., Chang Y.H., Escudier B., Donskov F., Magheli A., et al. Adjuvant sunitinib in high-risk renal-cell carcinoma after nephrectomy. N. Engl. J. Med. 2016;375:2246–2254. doi: 10.1056/NEJMoa1611406. [DOI] [PubMed] [Google Scholar]
- 47.Motzer R.J., Jonasch E., Michaelson M.D., Nandagopal L., Gore J.L., George S., Alva A., Haas N., Harrison M.R., Plimack E.R., et al. Nccn guidelines insights: Kidney cancer, version 2.2020. J. Natl. Compr. Cancer Netw. 2019;17:1278–1285. doi: 10.6004/jnccn.2019.0054. [DOI] [PubMed] [Google Scholar]
- 48.Vera-Badillo F.E., Templeton A.J., Duran I., Ocana A., de Gouveia P., Aneja P., Knox J.J., Tannock I.F., Escudier B., Amir E. Systemic therapy for non-clear cell renal cell carcinomas: A systematic review and meta-analysis. Eur. Urol. 2015;67:740–749. doi: 10.1016/j.eururo.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Koshkin V.S., Barata P.C., Zhang T., George D.J., Atkins M.B., Kelly W.J., Vogelzang N.J., Pal S.K., Hsu J., Appleman L.J., et al. Clinical activity of nivolumab in patients with non-clear cell renal cell carcinoma. J. Immunother. Cancer. 2018;6:9. doi: 10.1186/s40425-018-0319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benci J.L., Xu B., Qiu Y., Wu T.J., Dada H., Twyman-Saint Victor C., Cucolo L., Lee D.S.M., Pauken K.E., Huang A.C., et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167:1540–1554. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao X., Zhu Y., Li G., Huang H., Zhang G., Wang F., Sun J., Yang Q., Zhang X., Lu B. Tim-3 expression characterizes regulatory t cells in tumor tissues and is associated with lung cancer progression. PLoS ONE. 2012;7:e30676. doi: 10.1371/journal.pone.0030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakuishi K., Apetoh L., Sullivan J.M., Blazar B.R., Kuchroo V.K., Anderson A.C. Targeting tim-3 and pd-1 pathways to reverse t cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casado J.G., Pawelec G., Morgado S., Sanchez-Correa B., Delgado E., Gayoso I., Duran E., Solana R., Tarazona R. Expression of adhesion molecules and ligands for activating and costimulatory receptors involved in cell-mediated cytotoxicity in a large panel of human melanoma cell lines. Cancer Immunol. Immunother. 2009;58:1517–1526. doi: 10.1007/s00262-009-0682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inamura K., Amori G., Yuasa T., Yamamoto S., Yonese J., Ishikawa Y. Relationship of b7-h3 expression in tumor cells and tumor vasculature with foxp3+ regulatory t cells in renal cell carcinoma. Cancer Manag. Res. 2019;11:7021–7030. doi: 10.2147/CMAR.S209205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L., Rubinstein R., Lines J.L., Wasiuk A., Ahonen C., Guo Y., Lu L.F., Gondek D., Wang Y., Fava R.A., et al. Vista, a novel mouse ig superfamily ligand that negatively regulates t cell responses. J. Exp. Med. 2011;208:577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Mercier I., Chen W., Lines J.L., Day M., Li J., Sergent P., Noelle R.J., Wang L. Vista regulates the development of protective antitumor immunity. Cancer Res. 2014;74:1933–1944. doi: 10.1158/0008-5472.CAN-13-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fos C., Salles A., Lang V., Carrette F., Audebert S., Pastor S., Ghiotto M., Olive D., Bismuth G., Nunes J.A. Icos ligation recruits the p50alpha pi3k regulatory subunit to the immunological synapse. J. Immunol. 2008;181:1969–1977. doi: 10.4049/jimmunol.181.3.1969. [DOI] [PubMed] [Google Scholar]
- 58.Aspeslagh S., Postel-Vinay S., Rusakiewicz S., Soria J.C., Zitvogel L., Marabelle A. Rationale for anti-ox40 cancer immunotherapy. Eur. J. Cancer. 2016;52:50–66. doi: 10.1016/j.ejca.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 59.Shevach E.M., Stephens G.L. The gitr-gitrl interaction: Co-stimulation or contrasuppression of regulatory activity? Nat. Rev. Immunol. 2006;6:613–618. doi: 10.1038/nri1867. [DOI] [PubMed] [Google Scholar]
- 60.Ramapriyan R., Caetano M.S., Barsoumian H.B., Mafra A.C.P., Zambalde E.P., Menon H., Tsouko E., Welsh J.W., Cortez M.A. Altered cancer metabolism in mechanisms of immunotherapy resistance. Pharmacol. Ther. 2019;195:162–171. doi: 10.1016/j.pharmthera.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 61.Chen J.Y., Li C.F., Kuo C.C., Tsai K.K., Hou M.F., Hung W.C. Cancer/stroma interplay via cyclooxygenase-2 and indoleamine 2,3-dioxygenase promotes breast cancer progression. Breast Cancer Res. 2014;16:410. doi: 10.1186/s13058-014-0410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheong J.E., Sun L. Targeting the ido1/tdo2-kyn-ahr pathway for cancer immunotherapy—challenges and opportunities. Trends Pharmacol. Sci. 2018;39:307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Leone R.D., Emens L.A. Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer. 2018;6:57. doi: 10.1186/s40425-018-0360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Linnemann C., Schildberg F.A., Schurich A., Diehl L., Hegenbarth S.I., Endl E., Lacher S., Muller C.E., Frey J., Simeoni L., et al. Adenosine regulates cd8 t-cell priming by inhibition of membrane-proximal t-cell receptor signalling. Immunology. 2009;128:e728–e737. doi: 10.1111/j.1365-2567.2009.03075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ananieva E. Targeting amino acid metabolism in cancer growth and anti-tumor immune response. World J. Biol. Chem. 2015;6:281–289. doi: 10.4331/wjbc.v6.i4.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawson M.A., Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 67.Eckschlager T., Plch J., Stiborova M., Hrabeta J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18071414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pan R.Y., Chung W.H., Chu M.T., Chen S.J., Chen H.C., Zheng L., Hung S.I. Recent development and clinical application of cancer vaccine: Targeting neoantigens. J. Immunol. Res. 2018;2018:4325874. doi: 10.1155/2018/4325874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H., Ding J., Lu M., Liu H., Miao Y., Li L., Wang G., Zheng J., Pei D., Zhang Q. Caix-specific car-t cells and sunitinib show synergistic effects against metastatic renal cancer models. J. Immunother. 2020;43:16–28. doi: 10.1097/CJI.0000000000000301. [DOI] [PubMed] [Google Scholar]
- 70.Heng D.Y., Xie W., Regan M.M., Harshman L.C., Bjarnason G.A., Vaishampayan U.N., Mackenzie M., Wood L., Donskov F., Tan M.H., et al. External validation and comparison with other models of the international metastatic renal-cell carcinoma database consortium prognostic model: A population-based study. Lancet Oncol. 2013;14:141–148. doi: 10.1016/S1470-2045(12)70559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kroeger N., Klatte T., Chamie K., Rao P.N., Birkhauser F.D., Sonn G.A., Riss J., Kabbinavar F.F., Belldegrun A.S., Pantuck A.J. Deletions of chromosomes 3p and 14q molecularly subclassify clear cell renal cell carcinoma. Cancer. 2013;119:1547–1554. doi: 10.1002/cncr.27947. [DOI] [PubMed] [Google Scholar]
- 72.Kammerer-Jacquet S.F., Deleuze A., Saout J., Mathieu R., Laguerre B., Verhoest G., Dugay F., Belaud-Rotureau M.A., Bensalah K., Rioux-Leclercq N. Targeting the pd-1/pd-l1 pathway in renal cell carcinoma. Int. J. Mol. Sci. 2019;20:1692. doi: 10.3390/ijms20071692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seeber A., Klinglmair G., Fritz J., Steinkohl F., Zimmer K.C., Aigner F., Horninger W., Gastl G., Zelger B., Brunner A., et al. High ido-1 expression in tumor endothelial cells is associated with response to immunotherapy in metastatic renal cell carcinoma. Cancer Sci. 2018;109:1583–1591. doi: 10.1111/cas.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beuselinck B., Job S., Becht E., Karadimou A., Verkarre V., Couchy G., Giraldo N., Rioux-Leclercq N., Molinie V., Sibony M., et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin. Cancer Res. 2015;21:1329–1339. doi: 10.1158/1078-0432.CCR-14-1128. [DOI] [PubMed] [Google Scholar]
- 75.Hakimi A.A., Voss M.H., Kuo F., Sanchez A., Liu M., Nixon B.G., Vuong L., Ostrovnaya I., Chen Y.B., Reuter V., et al. Transcriptomic profiling of the tumor microenvironment reveals distinct subgroups of clear cell renal cell cancer—data from a randomized phase iii trial. Cancer Discov. 2019;9:510–525. doi: 10.1158/2159-8290.CD-18-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDermott D.F., Huseni M.A., Atkins M.B., Motzer R.J., Rini B.I., Escudier B., Fong L., Joseph R.W., Pal S.K., Reeves J.A., et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018;24:749–757. doi: 10.1038/s41591-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joseph R.W., Millis S.Z., Carballido E.M., Bryant D., Gatalica Z., Reddy S., Bryce A.H., Vogelzang N.J., Stanton M.L., Castle E.P., et al. Pd-1 and pd-l1 expression in renal cell carcinoma with sarcomatoid differentiation. Cancer Immunol. Res. 2015;3:1303–1307. doi: 10.1158/2326-6066.CIR-15-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamoun A., de Reynies A., Allory Y., Sjodahl G., Robertson A.G., Seiler R., Hoadley K.A., Groeneveld C.S., Al-Ahmadie H., Choi W., et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur. Urol. 2019;77:420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R., Schrock A., Campbell B., Shlien A., Chmielecki J., et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Velasco G., Miao D., Voss M.H., Hakimi A.A., Hsieh J.J., Tannir N.M., Tamboli P., Appleman L.J., Rathmell W.K., Van Allen E.M., et al. Tumor mutational load and immune parameters across metastatic renal cell carcinoma risk groups. Cancer Immunol. Res. 2016;4:820–822. doi: 10.1158/2326-6066.CIR-16-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miao D., Margolis C.A., Gao W., Voss M.H., Li W., Martini D.J., Norton C., Bosse D., Wankowicz S.M., Cullen D., et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dudley J.C., Lin M.T., Le D.T., Eshleman J.R. Microsatellite instability as a biomarker for pd-1 blockade. Clin. Cancer Res. 2016;22:813–820. doi: 10.1158/1078-0432.CCR-15-1678. [DOI] [PubMed] [Google Scholar]
- 83.Derosa L., Hellmann M.D., Spaziano M., Halpenny D., Fidelle M., Rizvi H., Long N., Plodkowski A.J., Arbour K.C., Chaft J.E., et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018;29:1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautes-Fridman C., Fridman W.H., et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gerlinger M., Catto J.W., Orntoft T.F., Real F.X., Zwarthoff E.C., Swanton C. Intratumour heterogeneity in urologic cancers: From molecular evidence to clinical implications. Eur. Urol. 2015;67:729–737. doi: 10.1016/j.eururo.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 86.Suva M.L., Tirosh I. Single-cell rna sequencing in cancer: Lessons learned and emerging challenges. Mol. Cell. 2019;75:7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 87.Kim K.T., Lee H.W., Lee H.O., Song H.J., Jeong da E., Shin S., Kim H., Shin Y., Nam D.H., Jeong B.C., et al. Application of single-cell rna sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016;17:80. doi: 10.1186/s13059-016-0945-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roelants C., Pillet C., Franquet Q., Sarrazin C., Peilleron N., Giacosa S., Guyon L., Fontanell A., Fiard G., Long J.A., et al. Ex-vivo treatment of tumor tissue slices as a predictive preclinical method to evaluate targeted therapies for patients with renal carcinoma. Cancers (Basel) 2020;12:232. doi: 10.3390/cancers12010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meijer T.G., Naipal K.A., Jager A., van Gent D.C. Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Sci. OA. 2017;3:FSO190. doi: 10.4155/fsoa-2017-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seo Y.D., Jiang X., Sullivan K.M., Jalikis F.G., Smythe K.S., Abbasi A., Vignali M., Park J.O., Daniel S.K., Pollack S.M., et al. Mobilization of cd8( + ) t cells via cxcr4 blockade facilitates pd-1 checkpoint therapy in human pancreatic cancer. Clin. Cancer Res. 2019;25:3934–3945. doi: 10.1158/1078-0432.CCR-19-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dijkstra K.K., Cattaneo C.M., Weeber F., Chalabi M., van de Haar J., Fanchi L.F., Slagter M., van der Velden D.L., Kaing S., Kelderman S., et al. Generation of tumor-reactive t cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–1598. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bensch F., van der Veen E.L., Lub-de Hooge M.N., Jorritsma-Smit A., Boellaard R., Kok I.C., Oosting S.F., Schroder C.P., Hiltermann T.J.N., van der Wekken A.J., et al. (89)zr-atezolizumab imaging as a non-invasive approach to assess clinical response to pd-l1 blockade in cancer. Nat. Med. 2018;24:1852–1858. doi: 10.1038/s41591-018-0255-8. [DOI] [PubMed] [Google Scholar]
- 93.Sun R., Limkin E.J., Vakalopoulou M., Dercle L., Champiat S., Han S.R., Verlingue L., Brandao D., Lancia A., Ammari S., et al. A radiomics approach to assess tumour-infiltrating cd8 cells and response to anti-pd-1 or anti-pd-l1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 94.Gradilone A., Iacovelli R., Cortesi E., Raimondi C., Gianni W., Nicolazzo C., Petracca A., Palazzo A., Longo F., Frati L., et al. Circulating tumor cells and “suspicious objects” evaluated through cellsearch(r) in metastatic renal cell carcinoma. Anticancer. Res. 2011;31:4219–4221. [PubMed] [Google Scholar]
- 95.He H., Magi-Galluzzi C. Epithelial-to-mesenchymal transition in renal neoplasms. Adv. Anat. Pathol. 2014;21:174–180. doi: 10.1097/PAP.0000000000000018. [DOI] [PubMed] [Google Scholar]
- 96.Liu S., Tian Z., Zhang L., Hou S., Hu S., Wu J., Jing Y., Sun H., Yu F., Zhao L., et al. Combined cell surface carbonic anhydrase 9 and cd147 antigens enable high-efficiency capture of circulating tumor cells in clear cell renal cell carcinoma patients. Oncotarget. 2016;7:59877–59891. doi: 10.18632/oncotarget.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santoni M., Cimadamore A., Cheng L., Lopez-Beltran A., Battelli N., Massari F., Scarpelli M., Galosi A.B., Bracarda S., Montironi R. Circulating tumor cells in renal cell carcinoma: Recent findings and future challenges. Front. Oncol. 2019;9:228. doi: 10.3389/fonc.2019.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]