Short abstract
Background
The average 5-year survival rate of lung adenocarcinoma patients is only 15% to 17%, which is primarily due to late-stage diagnosis and a lack of specific prognostic evaluations that can recommend effective therapies. Additionally, there is no clinically recognized biomarker that is effective for early-stage diagnosis.
Methods
Tissue samples from 10 lung adenocarcinoma patients (both tumor and non-tumor tissues) and 10 benign lung tumor samples were collected. The significantly differentially represented metabolites from the three groups were analyzed by liquid chromatography and tandem mass spectrometry.
Results
Pathway analysis indicated that central carbon metabolism was the top altered pathway in lung adenocarcinoma, while protein digestion and absorption, and central carbon metabolism were the top altered pathways in benign lung tumors. Receiver operating characteristic curve analysis revealed that adenosine 3′-monophosphate, creatine, glycerol, and 14 other differential metabolites were potential sensitive and specific biomarkers for the diagnosis and prognosis of lung adenocarcinoma.
Conclusion
Our findings suggest that the metabolomics approach may be a useful method to detect potential biomarkers in lung adenocarcinoma patients.
Keywords: Lung cancer, biomarker, metabolite, metabolomics, LC-MS/MS, diagnosis
Introduction
Lung cancer is a type of malignant tumor that seriously endangers human health and life because it has high morbidity and mortality worldwide.1 Lung adenocarcinoma, a subtype of lung cancer, is the leading cause of cancer-related deaths in the United States. Lung adenocarcinoma has an average 5-year survival rate of 15% to 17%, which is primarily due to late-stage diagnosis and no available clinical tests that can provide therapy recommendations.2 Additionally, the etiology of lung adenocarcinoma is complicated, and it is difficult to achieve early-stage diagnoses with existing imaging, histopathology, and bronchoscopy methods. Thus, most patients are diagnosed with advanced-stage lung adenocarcinoma on admission.3 Therefore, there is an urgent need to find early diagnostic markers for lung adenocarcinoma that are conducive to the early detection and treatment of this malignancy, as these would improve patient survival rates.
Proteomics is a cross-discipline that has emerged in the post-genomics era and is used to identify all proteins in a given sample. The goal of proteomics is to analyze the interactions and connections between proteins from a holistic perspective, revealing the rules of protein function and cellular activities.4 Among these methods, non-targeted metabolomics can quantify metabolites in biological systems, maximizing the information from metabolites.5 Because of the large number of small molecule metabolites in biological samples and the large dynamic range of their concentrations, chromatography-mass spectrometry is the most important tool for metabolomics research. Liquid chromatography and tandem mass spectrometry (LC-MS/MS) is a series analysis platform with high performance liquid chromatography as the separation system and high-resolution mass spectrometry as the detection system. Compared with other chromatographic-mass spectrometry techniques, LC-MS/MS is more suitable for the analysis of metabolites with low volatility or poor thermal stability. Ultra-high performance liquid chromatography columns packed with 1.7-μm ultrafine particles are at least 10× faster than conventional HPLC, with several times higher sensitivity and resolution.6 Currently, ultra-performance liquid chromatography and quadrupole-time-of-flight (Q-TOF) mass spectrometry have been widely used in metabolomics research. Therefore, protein metabolomics technologies have become an indispensable tool for studying tumor biology, and this field has shown rapid development.7
In the process of searching for lung cancer biomarkers, blood,7,8 urine,9,10 saliva,11 and lung tissue12 have been used as research samples. Protein metabolomics techniques are used to identify differences in metabolite expression between cancerous and normal lung tissues, thereby screening for biomarkers for the early diagnosis of lung cancer. So far, many lung cancer biomarkers have been identified. As previously reported, volatile organic compounds (VOCs)13 can be used as biomarkers for detecting lung cancer during breathing. Previous studies have clarified that metabolites such as cyclophilin (CYP-A), macrophage migration inhibitory factor (MIF),14 polymeric immunoglobulin receptor (PIGR), 14-3-3η,15 thymosin β4 (TMSB4), ubiquitin, acyl-CoA-binding protein (ACBP), cysteine protease inhibitor A (CSTA), Cytochrome C,16 thioredoxin (TXN), human S100 calcium binding protein A6 (S100A6), thymopoietin (TMPO), ribosomal proteins L39 and S30, peroxidase (peroxidase, PRDX) 1 and 3 (PRDX1, PRDX3), enolase-1 (ENO1), histone H2A.2,17 haptoglobin (HP),17 and SAA1 and SAA218 are overexpressed in cancer tissues, suggesting that they can be used as specific diagnostic biomarkers for lung cancer. Furthermore, Li et al.19 showed that leucine-rich alpha-2-glycoprotein (LRG1) is highly expressed in urine samples from lung cancer patients compared with healthy subjects. However, these studies are far from enough with regard to the complex associations between metabolomics and lung adenocarcinoma. A systemic analysis of metabolites of lung adenocarcinoma tissues is urgently needed to offer more candidates for the diagnosis and mechanism of early-stage lung adenocarcinoma.
In this study, a comprehensive metabolomics analysis of lung adenocarcinoma tissues was performed by LC-MS/MS. The selected differentially expressed metabolites could be used as clinical biomarkers for incipient diagnosis and prognosis of lung adenocarcinoma.
Materials and methods
Sample collection and preparation
In this study, tissue samples from 10 lung adenocarcinoma patients, including tumor and non-tumor tissues, and tissue samples from 10 benign lung tumor patients were collected. The clinical characteristics of these 20 patients are shown in Table 1. All patients provided written informed consent, and ethics approval was obtained from the Ethics Committees of the First Affiliated Hospital of University of South China.
Table 1.
Clinical characteristics of the lung cancer and benign lung tumor patients.
| Lung cancer patients (Cancer 1–10) | Benign lung tumors patients (Lump 1–10) | |
|---|---|---|
| N (male/female) | 10 (6/4) | 10 (8/2) |
| Age (median/range) | 61/50–74 | 54/48–62 |
| Smoker/non-smoker | 4/6 | 5/5 |
| c or p stages (I–II/III–IV) | 8/2 | |
| Tumor metastasis (yes/no) | 3/7 | 0/10 |
c stage (clinical stage) and p stage (pathological stage) were based on the TNM classification.
Each sample weighed 60 mg and was sequentially added to 200 μL of water for homogenization, and then 800 μL of a pre-cooled methanol/acetonitrile solution (1:1, v/v). The mixture was vortexed and sonicated twice for 30 minutes, incubated at −20°C for 1 hour, centrifuged at 14,000 ×g for 4 minutes at 4°C, and then the supernatant was vacuum dried. The material obtained from vacuum drying was reconstituted in 100 μL of an aqueous acetonitrile solution (acetonitrile: water = 1:1, v/v), followed by vortexing and centrifugation at 14,000 ×g for 5 minutes at 4°C. Quality control (QC) samples, a mixture of the three samples in equal amounts, were used to determine the instrument state prior to injection, to balance the chromatography-mass spectrometry system, and to evaluate system stability throughout the experiment. The supernatant of the above samples were taken for LC-MS/MS analysis.
Chromatography and mass spectrometry
Samples were separated on an Agilent 1290 Infinity LC Ultra Performance Liquid Chromatography System (Agilent Technologies Inc., Santa Clara, CA, USA) with HILIC columns at 25°C, a flow rate of 0.3 mL/minute, and an injection volume of 2 μL. Solutions A (water, 25 mM ammonium acetate, and 25 mM ammonia) and B (acetonitrile) were used as the mobile phases. The gradient started at 95% B, reached 65% B from 1 to 14 minutes, 40% B in the next 2 minutes, and then reached 95% B from 18 to 18.1 minutes, and was maintained at 95% B from 18.1 to 23 minutes. Samples were placed in an autosampler at 4°C throughout the analysis. The separated samples were subjected to mass spectrometry using a Triple TOF 5600 mass spectrometer (AB SCIEX). Mass spectrometry was performed using electrospray ionization (ESI), with positive and negative ion modes, respectively.
Data processing and statistical analyses
Principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed to maximize the separation between groups using SIMCA-P+ 14.1 software (Umetrics, Umeå, Sweden). Statistical significance was analyzed using the Student’s t-test, and statistical significance was defined as p < 0.05.
Pathway analysis combined with expression data has recently been emphasized to reveal potential functional interactions between multiple candidate metabolites. Functional interactions between the different groups of differentially expressed metabolites were examined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (http://www.kegg.jp/).20
Receiver operating characteristic (ROC) curve analysis
We performed ROC curve analysis using IBM SPSS Statistics for Windows, version 19.0 (IBM Corp., Armonk, NY, USA) to analyze each candidate biomarker and inspect its utility for predicting lung adenocarcinoma or benign lung tumors. Sensitivity and specificity trade-offs were summarized for each variable using the area under the ROC curve (AUC). The AUCs of the selected metabolites were compared to judge the performance of the candidate metabolites in lung adenocarcinoma. An AUC value of 1.0 corresponded to a prediction model with 100% sensitivity and 100% specificity, whereas an AUC value of 0.5 corresponded to a poor predictive model. The level of significance was set at p < 0.05.
Results
Quality control of the experiments
The system stability of this project was analyzed and evaluated by QC sample spectrum comparison and PCA analysis. The results of comparing the total ion flow charts (TIC) of QC samples showed that the response intensity and retention time of each chromatographic peak basically overlapped, which indicated that the variation caused by instrument errors was small during the experiment. Additionally, the PCA of the total sample showed that the QC samples were closely clustered in the positive and negative ion modes, demonstrating that the experiment had good repeatability. Moreover, Hotelling’s T2 analysis of population samples showed that all samples were within 99% confidence interval, without outlier samples. Therefore, these findings clarified that the system of this study was stable and could be used for subsequent analysis.
Identification of differently expressed metabolites
The data produced by LC-MS/MS were analyzed to identify significantly differentially expressed metabolites. We compared the levels of metabolites between the lung adenocarcinoma and control groups. As shown in Supplemental Table 1 and Supplemental Figure 1, 119 metabolites in tumor tissues showed significant differences compared with non-tumor normal tissue (p < 0.05). Meanwhile, 105 metabolites were detected in benign lung tumors tissue, indicating significant differences compared with control groups (p < 0.05) (Supplemental Table 2 and Supplemental Figure 2). Additionally, 32 metabolites in lung adenocarcinoma tissue were significantly altered compared with benign lung tumors tissue (p < 0.05) (Supplemental Table 3 and Supplemental Figure 3). Therefore, these remarkably different metabolites were selected for subsequent bioinformatics analysis.
Statistical analysis of differentially expressed metabolites
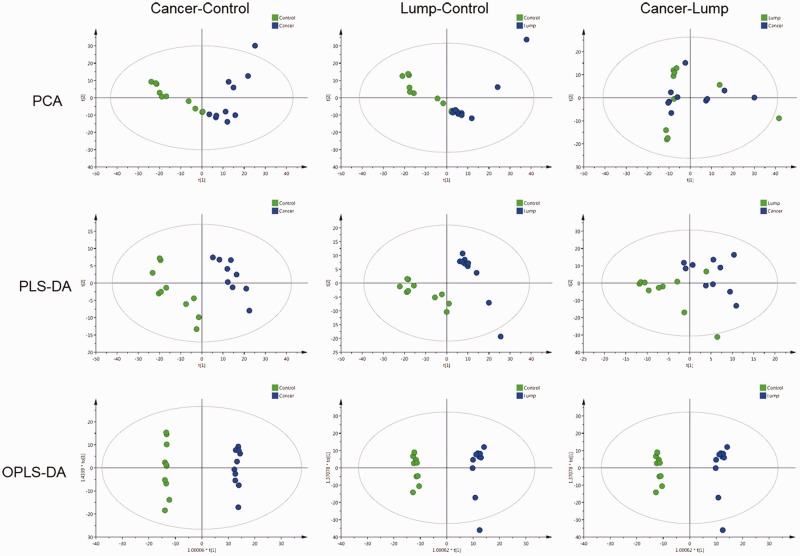
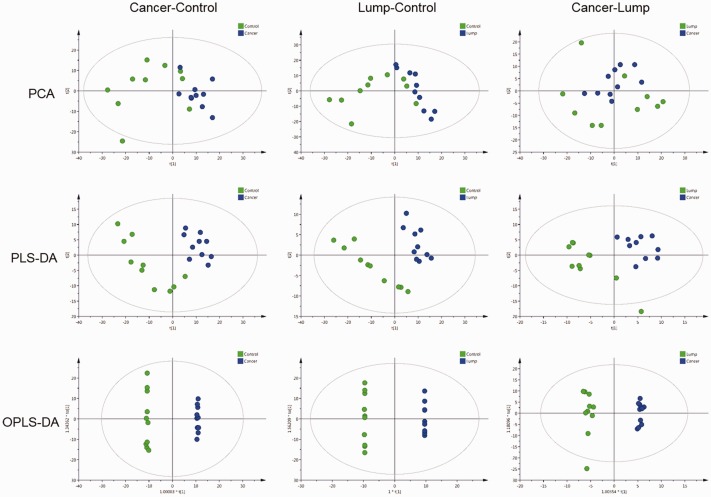
PCA, PLS-DA, and OPLS-DA were used to evaluate differences in the expression of tissue metabolites between lung adenocarcinoma patients (both tumor and non-tumor tissues) and the benign lung tumor patients. PCA analysis showed that there was apparent distinct clustering between the control and lung adenocarcinoma tissues of patients with lung adenocarcinoma. Meanwhile, the comparison between benign lung tumor tissue and control tissue was consistent with those of lung adenocarcinoma tissue and benign lung tumor tissue. To further distinguish the differences of benign lung tissue, lung adenocarcinoma tissue, and para-cancerous tissue, PLS-DA and OPLS-DA were used to supervise analyses of pattern recognition. In PLS-DA and OPLS-DA, the score plots showed good visual separation among benign lung tissue, lung adenocarcinoma, and para-cancerous tissue (Figure 1–2 and Table 2).
Figure 1.
PCA, PLS-DA, and OPLS-DA score maps from the positive ion mode. PCA, principle component analysis; PLS-DA, partial least squares discriminant analysis; OPLS-DA, orthogonal partial least squares discriminant analysis.
Figure 2.
PCA, PLS-DA, and OPLS-DA score maps from the negative ion mode. PCA, principle component analysis; PLS-DA, partial least squares discriminant analysis; OPLS-DA, orthogonal partial least squares discriminant analysis.
Table 2.
Model parameters for PCA, PLS-DA, and OPLS-DA.
| Models | Model parameter | Cancer-Control |
Lump-Control |
Cancer-Lump |
|||
|---|---|---|---|---|---|---|---|
| Positive ions | Negative ion | Positive ions | Negative ion | Positive ions | Negative ion | ||
| PCA | RX2 (cum) | 0.693 | 0.552 | 0.676 | 0.57 | 0.601 | 0.625 |
| PLS-DA | RY2 (cum) | 0.983 | 0.991 | 0.973 | 0.997 | 0.682 | 0.888 |
| Q2 (cum) | 0.839 | 0.892 | 0.892 | 0.878 | 0.0307 | 0.00537 | |
| OPLS-DA | R Y2 (cum) | 0.998 | 0.999 | 0.992 | 1 | 0.682 | 0.991 |
| Q2 (cum) | 0.876 | 0.783 | 0.892 | 0.848 | −0.00178 | 0.359 | |
PCA, principle component analysis; PLS-DA, partial least squares discriminant analysis; OPLS-DA, orthogonal partial least squares discriminant analysis.
Metabolic pathway and functional analysis
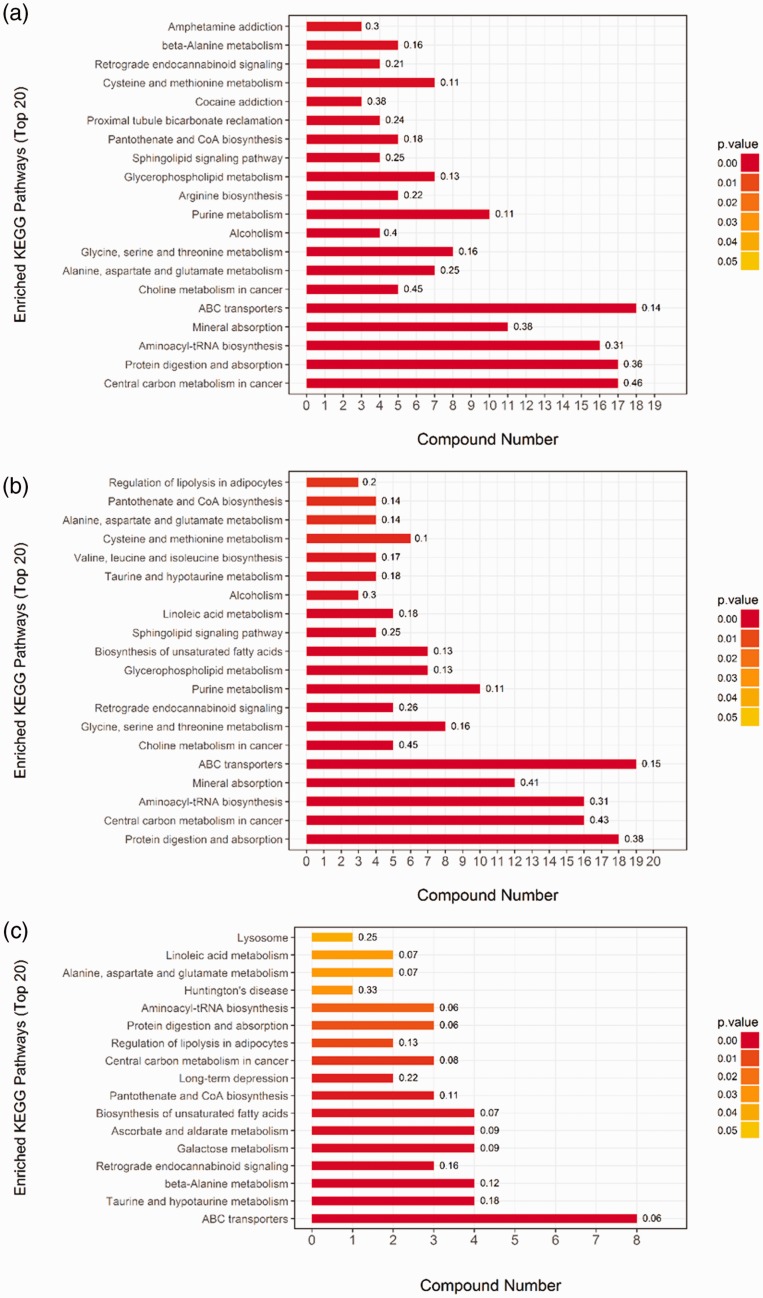
KEGG pathway analysis of the differential expression data was used to reveal potential functional interactions between multiple candidate metabolites by Student t-test. The results showed a significant enrichment of 43 pathways out of 112 pathways in lung adenocarcinoma tissue (p < 0.05). Among them, the most significantly enriched 10 signaling pathways were central carbon metabolism in cancer (p = 0), protein digestion and absorption (p = 8.88E-16), aminoacyl-tRNA biosynthesis (p = 1.26E-13), mineral absorption (p = 8.29E-11), ABC transporters (p = 4.85E-09), choline metabolism in cancer (p = 5.62E-06), alanine, aspartate, and glutamate metabolism (p = 6.84E-06), glycine, serine, and threonine metabolism (p = 4.89E-05), alcoholism (p = 9.78E-05), and purine metabolism (p = 1.65E-04) (Figure 3a).
Figure 3.
KEGG pathway analysis of differential expression data. (a) Cancer vs. control; (b) lump vs. control; and (c) cancer vs. lump. KEGG, Kyoto Encyclopedia of Genes and Genomes
A significant enrichment (p < 0.05) of 39 pathways out of 116 pathways was found by the KEGG pathway analysis in benign lung tumor tissue. The most significantly enriched 10 signaling pathways included protein digestion and absorption (p = 0), central carbon metabolism in cancer (p = 0), aminoacyl-tRNA biosynthesis (p = 4.17E-14), mineral absorption (p = 1.35E-12), ABC transporters (p = 1.92E-10), choline metabolism in cancer (p = 4.04E-06), glycine, serine, and threonine metabolism (p = 3.00E-05), retrograde endocannabinoid signaling (p = 8.64E-05), purine metabolism (p = 9.37E-05), and glycerophospholipid metabolism (p = 2.99E-04) (Figure 3b).
By comparing lung adenocarcinoma tissue with benign lung tumor tissue, we found that 17 of the 85 pathways were significantly enriched (p < 0.05). The most significantly enriched 10 signaling pathways included ABC transporters (p = 3.28E-05), taurine and hypotaurine metabolism (p = 6.08E-05), beta-alanine metabolism (p = 0.000278), retrograde endocannabinoid signaling (p = 0.00087), galactose metabolism (p = 0.001138), ascorbate and aldarate metabolism (p = 0.001235), unsaturated fatty acid biosynthesis (p = 0.00208), pantothenate and CoA biosynthesis (p = 0.002759), long-term depression (p = 0.003555), and central carbon metabolism in cancer (p = 0.006142) (Figure 3c).
Variation in the levels of metabolites involved in central carbon metabolism
Among the metabolites involved in central carbon metabolism, 17 were observed in lung adenocarcinoma tissue and para-cancerous tissue, and the levels of the following metabolites were significantly changed: L-alanine, L-arginine, L-asparagine, L-aspartate, L-glutamate, L-glutamine, L-histidine, L-leucine, L-malic acid, L-methionine, L-tryptophan, L-tyrosine, L-valine, D-glucose 6-phosphate, glycine, L-isoleucine, and L-serine. In the benign lung tumor tissue, 16 metabolites were significantly changed, including, L-alanine, L-arginine, L-asparagine, L-glutamate, L-glutamine, L-histidine, L-leucine, L-methionine, L-phenylalanine, L-tryptophan, L-tyrosine, L-valine, D-glucose 6-phosphate, glycine, L-isoleucine, and L-serine.
Variation in the levels of metabolites involved in protein digestion and absorption
Seventeen metabolites that were significantly altered in lung adenocarcinoma tissue and para-cancerous tissue were related to protein digestion and absorption, including indole, L-alanine, L-arginine, L-asparagine, L-aspartate, L-glutamate, L-glutamine, L-histidine, L-leucine, L-methionine, L-tryptophan, L-tyrosine, L-valine, glycine, L-isoleucine, L-serine, and L-threonine. In benign lung tumor tissue, levels of the following 18 metabolites were significantly altered compared with the control group: L-alanine, L-arginine, L-asparagine, L-glutamate, L-glutamine, L-histidine, L-leucine, L-methionine, L-phenylalanine, L-tryptophan, L-tyrosine, L-valine, glycine, L-cystine, L-isoleucine, L-serine, L-threonine, and tyramine.
Variation in the levels of metabolites involved in fatty acid metabolism
Twenty-five significantly altered fatty acids and their derivatives were detected in tissues from lung adenocarcinoma patients and the control groups: 1-Aminocyclopropanecarboxylic acid, all cis-(6,9,12)-linolenic acid, cis-9-ialmitoleic acid, DL-Indole-3-lactic acid, D-pipecolinic acid, palmitic acid, L-pipecolic acid, L-pyroglutamic acid, trans-2-hydroxycinnamic acid, argininosuccinic acid, phenyllactic acid, 2-hydroxy-3-methylbutyric acid, caproic acid, caprylic acid, DL-3-phenyllactic acid, DL-mandelic acid, erucic acid, hydroxyphenyllactic acid, N-acetylneuraminic acid, stearic acid, 2-oxoadipic acid, alpha-linolenic acid, dodecanoic acid, L-malic acid, and N-acetyl-L-aspartic acid. In the benign lung tumor tissues, levels of the following 21 metabolites were notably changed compared with the control groups: 16-hydroxypalmitic acid, 1-aminocyclopropanecarboxylic acid, 20-hydroxyarachidonic acid, all cis-(6,9,12)-linolenic acid, arachidonic Acid (peroxide free), DL-indole-3-lactic acid, eicosapentaenoic acid, linoleic acid, L-pyroglutamic acid, N-acetylneuraminic acid, cis-9-palmitoleic acid, D-pipecolinic acid, phenyllactic acid, trans-2-hydroxycinnamic acid, 2-oxoadipic acid, erucic acid, hydroxyphenyllactic acid, alpha-linolenic acid, caproic acid, caprylic acid, and palmitic acid.
Variation in the levels choline-associated metabolites
In the lung adenocarcinoma patients, levels of 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine, 1-palmitoyl-sn-glycero-3-phosphocholine, 1-stearoyl-2-hydroxy-sn-glycero-3-phosphocholine, 1-stearoyl-sn-glycerol 3-phosphocholine, choline, glycerophosphocholine, phosphorylcholine, 1-oleoyl-sn-glycero-3-phosphocholine, and 1-stearoyl-2-oleoyl-sn-glycerol 3-phosphocholine (SOPC) were significantly changed compared with the control group. In tissues from benign lung tumor patients, levels of 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine, 1-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-sn-glycero-3-phosphocholine, 1-stearoyl-2-hydroxy-sn-glycero-3-phosphocholine, 1-stearoyl-sn-glycerol 3-phosphocholine, choline, glycerophosphocholine, phosphorylcholine, and SOPC were significantly changed compared with the control group.
Differential metabolites involved in enrichment pathways between lung adenocarcinoma and benign lung tumor tissues
By comparing lung adenocarcinoma tissue with benign lung tumor tissue, we found 17 differential metabolites involved in ABC transporters, taurine and hypotaurine metabolism, and beta-alanine metabolism. The specific metabolites that were significantly differentially expressed in lung adenocarcinoma and benign lung tumor tissues were acetyl phosphate, D-galactarate, eicosapentaenoic acid, glycerol, L-alanine, L-glutamate (both positive and negative ions), L-gulonic gamma-lactone, linoleic acid, L-threonate, oleic acid, arachidonic acid (peroxide free), myo-inositol, dihydrouracil, L-histidine, stachyose, and uracil.
Validation of differential metabolites as potential biomarkers for lung adenocarcinoma or benign lung tumors
All metabolites involved in the 10 most significantly enrichment pathways from each group were compared with the differential metabolites from the mass spectrometry results, and finally metabolites involved in both were selected. The relationship between sensitivity and 1−specificity was plotted to construct a ROC curve, and the AUC was calculated; the ROC and AUC values of the selected candidate biomarkers were calculated using binary logistic regression.
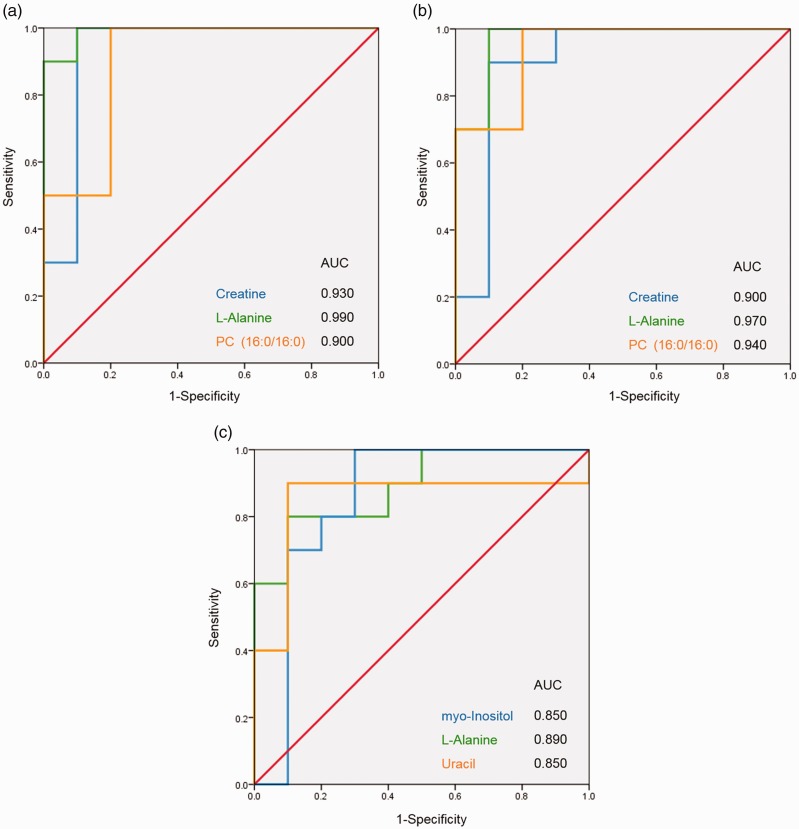
The ROC curves that distinguished cancer from the control group showed that the AUC values of the following 15 metabolites were all greater than 0.850: adenosine 3′-monophosphate, creatine, glycerol, guanosine 5′-monophosphate (GMP), indole, L-alanine, L-glutamate, phosphorylcholine, taurine, xanthine, xanthosine, glycine, L-serine, N-acetyl-D-glucosamine, and phosphatidylcholine (PC) (16:0/16:0) (p < 0.05) (Table 3). Similarly, in the lump group vs. control group, the ROC curve showed the AUC values of 16 metabolites were greater than 0.850 (p < 0.05): adenosine 3′-monophosphate, creatine, GMP, inosine, L-alanine, L-asparagine, L-methionine, O-phosphoethanolamine, phosphorylcholine, sn-glycerol 3-phosphoethanolamine, arachidonic acid (peroxide free), glycerophosphocholine, L-cystine, L-threonine, N-acetyl-D-glucosamine, and PC (16:0/16:0) (Table 4). Moreover, compared with the lump group, only four cancer-associated metabolites had AUC values greater than 0.850: D-galactarate (0.890 ± 0.074), L-alanine (0.890 ± 0.073), myo-inositol (0.850 ± 0.100), and uracil (0.850 ± 0.104) (all, p < 0.05) (Table 5). To make the results more intuitive, three different metabolites were selected for display as ROC curves (Figure 4).
Table 3.
The AUC, specificity, and sensitivity of the diagnostic efficacy of potential lung cancer biomarkers in a comparison of the cancer and control groups (AUC > 0.850).
| Potential biomarkers | p value | AUC ± Sem | Sensitivity | Specificity |
|---|---|---|---|---|
| Adenosine 3'-monophosphate | 0.000924 | 0.860 ± 0.083 | 0.800 | 0.800 |
| Creatine | 0.00061 | 0.930 ± 0.069 | 1.000 | 0.900 |
| Glycerol | 0.000229 | 0.960 ± 0.044 | 1.000 | 0.900 |
| Guanosine 5'-monophosphate (GMP) | 0.001083 | 0.930 ± 0.056 | 1.000 | 0.800 |
| Indole | 0.00019 | 0.980 ± 0.026 | 0.900 | 1.000 |
| L-Alanine | 8.13E-05 | 0.990 ± 0.016 | 0.900 | 1.000 |
| L-Glutamate | 0.002521 | 0.880 ± 0.080 | 0.800 | 0.900 |
| Phosphorylcholine | 0.015352 | 0.970 ± 0.035 | 0.900 | 1.000 |
| Taurine | 0.00155 | 0.880 ± 0.078 | 0.800 | 0.900 |
| Xanthine | 0.000752 | 0.860 ± 0.087 | 1.000 | 0.700 |
| Xanthosine | 0.003153 | 0.900 ± 0.073 | 1.000 | 0.800 |
| Glycine | 0.002043 | 0.900 ± 0.076 | 1.000 | 0.800 |
| L-Serine | 0.005319 | 0.870 ± 0.085 | 0.900 | 0.800 |
| N-Acetyl-D-glucosamine | 0.000394 | 0.950 ± 0.045 | 0.800 | 1.000 |
| PC (16:0/16:0) | 0.000512 | 0.900 ± 0.073 | 1.000 | 0.800 |
AUC, area under the ROC curve.
Table 4.
The AUC, specificity, and sensitivity of the diagnostic efficacy of potential lung cancer biomarkers in a comparison of the lump and control groups (AUC > 0.850).
| Potential biomarkers | p value | AUC ± Sem | Sensitivity | Specificity |
|---|---|---|---|---|
| Adenosine 3'-monophosphate | 0.000271 | 0.910 ± 0.065 | 0.800 | 0.900 |
| Creatine | 0.001514 | 0.900 ± 0.080 | 0.900 | 0.900 |
| Guanosine 5'-monophosphate (GMP) | 0.000218 | 0.970 ± 0.032 | 0.800 | 1.000 |
| Inosine | 0.024261 | 0.850 ± 0.085 | 0.600 | 1.000 |
| L-Alanine | 0.000193 | 0.970 ± 0.035 | 1.000 | 0.900 |
| L-Asparagine | 0.009937 | 0.860 ± 0.099 | 0.900 | 0.800 |
| L-Methionine | 0.012289 | 0.860 ± 0.083 | 0.600 | 1.000 |
| O-Phosphoethanolamine | 0.004335 | 0.880 ± 0.080 | 0.700 | 1.000 |
| Phosphorylcholine | 0.02622 | 0.880 ± 0.075 | 0.600 | 1.000 |
| sn-Glycerol 3-phosphoethanolamine | 0.010908 | 0.870 ± 0.087 | 0.900 | 0.800 |
| Arachidonic Acid (peroxide free) | 0.002591 | 0.880 ± 0.075 | 0.900 | 0.700 |
| Glycerophosphocholine | 0.004279 | 0.900 ± 0.073 | 1.000 | 0.800 |
| L-Cystine | 0.037211 | 0.890 ± 0.073 | 0.800 | 0.900 |
| L-Threonine | 0.005467 | 0.880 ± 0.078 | 0.900 | 0.800 |
| N-Acetyl-D-glucosamine | 0.002906 | 0.920 ± 0.060 | 0.900 | 0.800 |
| PC (16:0/16:0) | 0.000388 | 0.940 ± 0.051 | 1.000 | 0.800 |
AUC, area under the ROC curve.
Table 5.
The AUC, specificity, and sensitivity of the diagnostic efficacy of potential lung cancer biomarkers in a comparison of the cancer and lump groups (AUC > 0.850).
| Potential biomarkers | p value | AUC ± Sem | Sensitivity | Specificity |
|---|---|---|---|---|
| D-Galactarate | 0.044737 | 0.890 ± 0.074 | 1.000 | 0.700 |
| L-Alanine | 0.015104 | 0.890 ± 0.073 | 0.800 | 0.900 |
| myo-Inositol | 0.011372 | 0.850 ± 0.100 | 1.000 | 0.700 |
| Uracil | 0.008811 | 0.850 ± 0.104 | 0.900 | 0.900 |
AUC, area under the ROC curve.
Figure 4.
ROC curve analysis was used to examine the diagnostic efficacy of the metabolite candidates. (a) Cancer vs. control; (b) lump vs. control; and (c) cancer vs. lump. AUC, area under the ROC curve; ROC, receiver operating characteristic.
Discussion
In this study, adenocarcinoma and para-cancerous tissues from 10 patients with lung cancer, as well as benign lung tumor tissue from 10 patients with benign tumors were studied using LC-MS/MS-based metabolomics to discover potential lung adenocarcinoma biomarkers for the diagnosis and prognosis of early-stage lung adenocarcinoma. The results showed that compared with para-cancerous tissue, 119 and 105 significant differential metabolites were identified from lung adenocarcinoma and benign tumor tissues, respectively. Moreover, the comparison between the cancer group and lump group screened 32 significant differential metabolites. Based on the KEGG pathway analysis, 43 and 39 significantly enriched metabolic pathways were determined from lung adenocarcinoma and benign lung tumor samples, respectively. Moreover, when the lung adenocarcinoma group was compared with the lump group, 17 enriched pathways were detected.
Previous studies have demonstrated that lung cancer may alter levels of the metabolites involved in the TCA cycle and its related signaling pathways.21,22 In this study, levels of metabolites involved with central carbon metabolism in cancer; i.e., L-alanine, L-arginine, L-glutamate, and L-asparagine were found. Most cancer cells depend on aerobic glycolysis rather than oxidative phosphorylation for energy production, and the presence of glutamine has a significant effect on the production of ATP in cancer cells. Additionally, glutamine is also a major component of cancer cells,23 which is in line with our results. Therefore, central carbon metabolism for energy production in cancer cells is different from that in normal cells. Thus, differences in metabolic pathways may result in changes in the levels of certain metabolites in lung adenocarcinoma tissues.
Several studies have revealed that aminoacyl-tRNAs function to transfer amino acids to ribosomes during protein synthesis; therefore, the increased protein synthesis rate of cancer cells indicates that the level of aminoacyl-tRNA in cancer tissues is significantly higher than in normal tissue.23–25 In this study, levels of following metabolites related to the aminoacyl-tRNA biosynthesis in cancer were altered: L-alanine, L-arginine, and L-asparagine. These differential metabolites in lung adenocarcinoma tissue affect the synthesis of aminoacyl-tRNA biosynthesis, which affect protein synthesis and further regulate tumor cell proliferation. Levels of most amino acids in lung adenocarcinoma tissues were higher than in normal tissues. Therefore, the levels of metabolites related to the protein digestion and absorption were significantly altered in cancer and normal tissues.
Choline is essential for the synthesis of the major membrane phospholipid phosphatidylcholine (PC), the methyl donor betaine, and the neurotransmitter acetylcholine (ACh). It has been reported that abnormal choline metabolism is a metabolic hallmark of oncogenesis and tumor progression.26 Previous studies have demonstrated abnormalities in choline uptake and choline phospholipid metabolism in cancer cells by imaging tumors with positron emission tomography (PET) and magnetic resonance spectroscopy (MRS).27 Higher levels of choline and up-regulated choline kinase activity have been detected in various cancers.26,28 Consistently, metabolites in this study, such as 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine, 1-palmitoyl-sn-glycero-3-phosphocholine, and 1-stearoyl-2-hydroxy-sn-glycero-3-phosphocholine, increased choline levels in lung adenocarcinoma and benign lung tumor tissues compared with non-cancer tissues.
Furthermore, it was worth noting that seven metabolites, namely adenosine 3′-monophosphate, creatine, GMP, L-alanine, phosphorylcholine, N-acetyl-D-glucosamine, and PC (16:0/16:0), were involved in the enriched pathways and were significantly differentially expressed in either the cancer group or the lump group compared with the control group. Early studies have reported the expression and application of L-alanine, phosphorylcholine, and N-acetyl-D-glucosamine in lung malignancies.29–31 In a recent study of non-small cell carcinoma, Ye et al.32 combined N-acetyl-D-glucosamine with TRAIL, and their results uncovered the molecular mechanism through which GlcNAc sensitized cancer cells to TRAIL-induced apoptosis. Thus, with more verification in the future, these metabolites could be used for lung tumor screening.
In summary, this study presents preliminary comparative proteomics data from the discovery of serum biomarkers in lung adenocarcinoma, and generates a robust set of candidate proteins for lung adenocarcinoma diagnosis. However, the sample size of this study was relatively small, and a larger sample size is required for future systematic studies. To the best of our knowledge, this is the first time that differences between metabolites and metabolic pathways have been detected by LC-MS/MS among different lung tissues. The clinical utility of these candidate lung adenocarcinoma serum biomarker proteins needs to be validated with additional analytical platforms as well as in independent case/control sample sets in the future.
Acknowledgements
Not applicable.
Availability of data and materials
Not applicable.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Funding
This study was supported by the scientific research project education department of Hunan (17C1389).
Patient consent for publication
Not applicable.
ORCID iD
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Karri K, Bastola DR. Immune based prognostic biomarkers for multiple anticancer therapies in lung adenocarcinoma. In: 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2016.
- 3.Latimer KM. Lung cancer: clinical presentation and diagnosis. FP Essent 2018; 464: 23–26. [PubMed] [Google Scholar]
- 4.Forester CM, Zhao Q, Phillips NJ, et al. Revealing nascent proteomics in signaling pathways and cell differentiation. Proc Natl Acad Sci U S A 2018; 115: 201707514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JH, Jo JH, Seo KA, et al. Non-targeted metabolomics-guided sildenafil metabolism study in human liver microsomes. J Chromatogr B Analyt Technol Biomed Life Sci 2018; 1072: 86–93. [DOI] [PubMed] [Google Scholar]
- 6.Yang CL, Chang HC, Chen CY, et al. Determination of isosteroidal alkaloids in Fritillaria spp. by LC-MS/MS. Journal of Chinese Mass Spectrometry Society 2017; 1: 11–18. [Google Scholar]
- 7.Ivanisevic J, Zhu ZJ, Plate L, et al. Toward ‘omic scale metabolite profiling: a dual separation–mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal Chem 2013; 85: 6876–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng X, Hood BL, Zhao T, et al. Lung cancer serum biomarker discovery using label free LC-MS/MS. J Thorac Oncol 2011; 70: 4564–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia X, Lu JJ, Zhang SS, et al. Midkine is a serum and urinary biomarker for the detection and prognosis of non-small cell lung cancer. Oncotarget 2016; 7: 87462–87472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolen BM, Lomakin A, Marrangoni A, et al. Urinary protein biomarkers in the early detection of lung cancer. Cancer Prev Res (Phila) 2015; 8: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Xiao H, Zhou H, et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 2012; 69: 3341–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Zhang X, Wang W, et al. Quantitative proteomics analysis of mitochondrial proteins in lung adenocarcinomas and normal lung tissue using iTRAQ and tandem mass spectrometry. Am J Transl Res 2017; 9: 3918. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong X, Li D, Du W, et al. Rapid recognition of volatile organic compounds with colorimetric sensor arrays for lung cancer screening. Anal Bioanal Chem 2018; 410: 3671–3681. [DOI] [PubMed] [Google Scholar]
- 14.Lechien JR, Nassri A, Kindt N, et al. Role of macrophage migration inhibitory factor in head and neck cancer and novel therapeutic targets: a systematic review. Head Neck 2017; 39: 2573–2584. [DOI] [PubMed] [Google Scholar]
- 15.Yue X, Ai J, Xu Y, et al. Polymeric immunoglobulin receptor promotes tumor growth in hepatocellular carcinoma. Hepatology 2017; 65: 1948–1962. [DOI] [PubMed] [Google Scholar]
- 16.Rahman SMJ, Gonzalez AL, Ming L, et al. Lung cancer diagnosis from proteomic analysis of preinvasive lesions. Cancer Res 2011; 71: 3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiyoshi Y, Shuta T, Yukako S, et al. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst 2007; 99: 858–867. [DOI] [PubMed] [Google Scholar]
- 18.Hye-Jin S, Jung-Mo A, Yeon-Hee Y, et al. Identification and validation of SAA as a potential lung cancer biomarker and its involvement in metastatic pathogenesis of lung cancer. J Proteome Res 2011; 10: 1383–1395. [DOI] [PubMed] [Google Scholar]
- 19.Yanyan L, Yuan Z, Feng Q, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 2011; 32: 1976–1983. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012; 40: D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deberardinis RJ, Sayed N, Ditsworth D, et al. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 2008; 18: 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori S, Nishiumi S, Kobayashi K, et al. A metabolomic approach to lung cancer. Lung Cancer 2011; 74: 284–292. [DOI] [PubMed] [Google Scholar]
- 23.Vulimiri SV, Manoj M, Hamm JT, et al. Effects of mainstream cigarette smoke on the global metabolome of human lung epithelial cells. Chem Res Toxicol 2009; 22: 492–503. [DOI] [PubMed] [Google Scholar]
- 24.Motzik A, Amir E, Erlich T, et al. Post-translational modification of HINT1 mediates activation of MITF transcriptional activity in human melanoma cells. Oncogene 2017; 36: 4732–4738. [DOI] [PubMed] [Google Scholar]
- 25.Seung-Hun S, Ho-Shik K, Seung-Hyun J, et al. Implication of leucyl-tRNA synthetase 1 (LARS1) over-expression in growth and migration of lung cancer cells detected by siRNA targeted knock-down analysis. Exp Mol Med 2008; 40: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaezi AE, Gerold B, Bhagwat NR, et al. Choline phosphate cytidylyltransferase-α is a novel antigen detected by the anti-ERCC1 antibody 8F1 with biomarker value in patients with lung and head and neck squamous cell carcinomas. Cancer 2014; 120: 1898–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inazu M. . Choline transporter-like proteins CTLs/SLC44 family as a novel molecular target for cancer therapy. Biopharm Drug Dispos 2014; 35: 431–449. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Ma Z, Zhong J, et al. Simultaneous quantification of serum monounsaturated and polyunsaturated phosphatidylcholines as potential biomarkers for diagnosing non-small cell lung cancer. Sci Rep 2018; 8: 7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang LI, Miller DP, Sai Y, et al. Manganese superoxide dismutase alanine-to-valine polymorphism at codon 16 and lung cancer risk. J Natl Cancer Inst 2001; 93: 1818–1821. [DOI] [PubMed] [Google Scholar]
- 30.Chen CH, Statt S, Chiu CL, et al. Targeting myristoylated alanine-rich C kinase substrate phosphorylation site domain in lung cancer. Mechanisms and therapeutic implications. Am J Respir Crit Care Med 2014; 190: 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camussi G, Pawlowski I, Tetta C, et al. Acute lung inflammation induced in the rabbit by local instillation of 1-0-octadecyl-2-acetyl-sn-glyceryl-3-phosphorylcholine or of native platelet-activating factor. Am J Pathol 1983; 112: 78–88. [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Y, Xu W, Liu S, et al. N-acetyl-glucosamine sensitizes non-small cell lung cancer cells to TRAIL-induced apoptosis by activating death receptor 5. Cell Physiol Biochem 2018; 45: 2054–2070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.