Abstract
A plethora of previous studies have been focused on the role of indoleamine 2,3-dioxygenase 1 (IDO1) in cancer immunity; however, the alternative way of targeting tryptophan 2,3-dioxygenase (TDO2) in cancer immunotherapy has been largely ignored. In particular, the specific role of TDO2 in breast cancer remains unclear. In the present study, we systematically explored and validated the expression and prognostic value of TDO2 in breast cancer using large-scale transcriptome data. We observed overexpression of TDO2 in many types of cancer tissues compared with adjacent normal tissues. TDO2 overexpression was revealed to be positively correlated with malignancy and tumor grade in breast cancer. TDO2 expression was higher in estrogen-negative breast cancer and triple-negative breast cancer, and it was correlated with worse outcome in breast cancer patients. TDO2 expression was correlated with immune infiltrates and tryptophan metabolism-related genes (IDO1 and kynureninase [KYNU]). Therefore, our results indicated that TDO2 plays a pivotal role in regulating the immune microenvironment and tryptophan metabolism in breast cancer, and it predicts poor prognosis in breast cancer, which suggests that TDO2 might be a promising novel immunotherapy target for breast cancer. Additionally, we established the concept that tryptophan-catabolizing enzymes (IDO1, IDO2, TDO2, and KYNU) may function through co-regulating the immunological microenvironment, and thus immunotherapy targeting IDO1 alone might be insufficient.
Keywords: breast cancer, cancer immunotherapy, TDO2, IDO1, IDO2, KYNU, immune infiltrates
Graphical Abstract
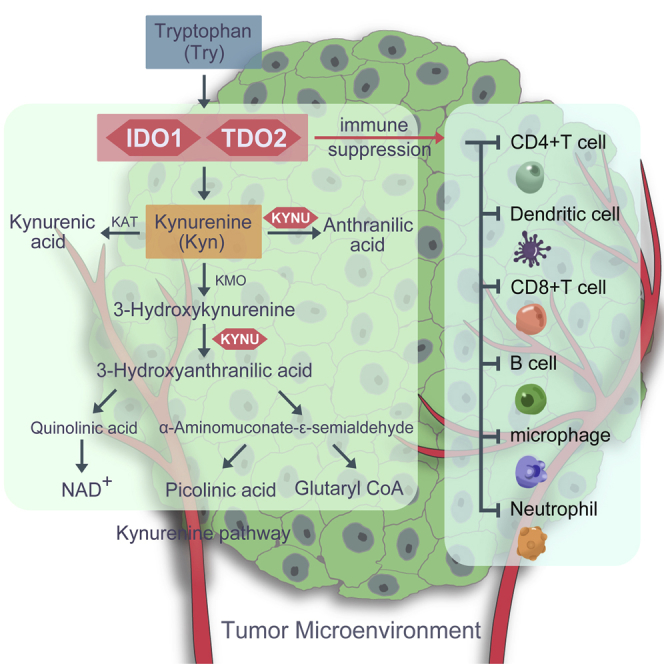
Indoleamine 2,3-dioxygenase 1 (IDO1) and tryptophan 2,3-dioxygenase (TDO2) can both catalyze the first step of the kynurenine pathway, but the specific role of TDO2 in breast cancer is unclear. Liu et al. comprehensively investigated the molecular and clinical characteristics of TDO2 in breast cancer, implicating the critical role of TDO2 in manipulating the breast cancer immune microenvironment.
Introduction
Breast cancer (BC) is the most common type of cancer in women in the United States, accounting for 30% of all new cancer diagnoses in women.1,2 Triple-negative BC (TNBC) is defined by the negative expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2/neu),3 which makes it more difficult to treat. TNBC patients who have failed to respond to chemotherapy are more likely to have a poor prognosis for tumor recurrences or metastasis.4 Therefore, novel therapeutic strategies are urgently needed to improve the treatment efficacy of TNBC patients. Along with increasing knowledge of cancer immunity, cancer immunotherapy has shed a light on TNBC treatment. Recently, numerous studies have elucidated the key role of immune checkpoints in modulating the immune response in malignancies. Programmed cell death 1 (PD1) is a prominent immune checkpoint reported to induce an immunosuppressive effect in tumors by interacting with programmed death ligand 1 (PD-L1).5,6 Indeed, PD-L1 was overexpressed in TNBC compared with other BC subtypes,5,6 and numerous active clinical trials are evaluating the efficacy of immunotherapy targeting PD1, but the anti-PD1 agent pembrolizumab treatment in TNBC patients showed limited efficacy.7,8 Thus, it is vital to undertake more endeavors to find novel potential targets for TNBC immunotherapy.
The kynurenine pathway (KP) has been demonstrated to be involved in regulating the immunosuppressive microenvironment in human tumors.9, 10, 11, 12 Tryptophan degradation is thought to produce immunosuppressive effects partly from the direct effects of tryptophan catabolites and partly from tryptophan depletion,13,14 which stimulated interest in therapeutically targeting the main rate-limiting enzymes, including indoleamine 2,3-dioxygenase 1 (IDO1), IDO2, and tryptophan 2,3-dioxygenase (TDO2) as well as kynurenine monooxygenase (KMO).15 Importantly, both IDO1 and TDO2 catalyze the first and rate-limiting step of tryptophan degradation along the KP and thereby regulate systemic tryptophan and kynurenine levels.10 A plethora of studies have been focused on investigating the role of IDO1 in inducing immunosuppressive effects in human cancers and supporting the key role of IDO1 in immune regulation,16 but the specific role of TDO2 in cancer immunotherapy remains unclear. However, although small-molecule inhibitors targeting IDO1 alone showed promising results in early-stage cancer clinical trials, a phase III trial result was negative,17 suggesting that targeting IDO1 alone might not be sufficient to reverse tumor immune invasion.
A previous study reported that TDO2 overexpression is correlated with cancer stem cells and poor prognosis in esophageal squamous cell carcinoma.18 In BC, a recent study reported that TDO2 was involved in facilitating anoikis resistance and metastasis in TNBC19 and inhibiting CD8 T cell viability.20 More recently, evidence indicated that TDO2-mediated production of the immunosuppressive metabolite kynurenine can be modulated by the miR-200c family.21 Although TDO2 was a promising immunotherapy target for BC, the specific role of TDO2 in BC and its association with immune infiltrates are still unclear. In the present study, we systematically investigated the expression status and prognostic value of TDO2 in BC, using large-scale transcriptome data. In addition, we also analyzed the association between TDO2 expression and immune infiltrates, which might provide novel implications for future studies.
Results
mRNA Expression Levels of TDO2 in Different Types of Human Cancers
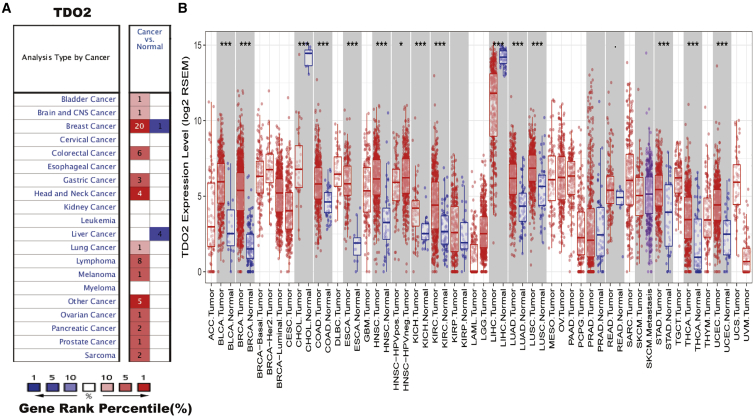
To determine mRNA expression of TDO2 in tumor and normal tissues, we compared the mRNA levels of TDO2 between tumors and normal tissues of multiple cancer types using the Oncomine database.22 It was revealed that TDO2 expression was higher in most of the tumor tissues when compared with normal tissues, especially in BC (the detailed expression profile of TDO2 in different BC datasets is listed in Table 1), and lower expression of TDO2 was observed in only one type of cancer, that is, liver cancer (Figure 1A). To validate the mRNA levels of TDO2 in multiple human cancers, we analyzed expression of TDO2 using RNA-sequencing (RNA-seq) data derived from The Cancer Genome Atlas (TCGA) database. The expression of TDO2 in tumor and adjacent normal tissues across all tumors in TCGA is shown in Figure 1B. TDO2 expression was significantly higher in BLCA (bladder urothelial carcinoma), BRCA (breast invasive carcinoma), COAD (colon adenocarcinoma), ESCA (esophageal carcinoma), HNSC (head and neck cancer), KICH (kidney chromophobe), KIRC (kidney renal clear cell carcinoma), LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma), STAD (stomach adenocarcinoma), THCA (thyroid carcinoma), and UCEC (uterine corpus endometrial carcinoma), when compared with adjacent normal tissues. However, TDO2 expression was significantly lower in only two types of cancers, that is, CHOL (cholangiocarcinoma) and LIHC (liver hepatocellular carcinoma), compared with adjacent normal tissues.
Table 1.
TDO2 Expression in Different Microarray Datasets
| Dataset | Comparisons | Fold Change | p Value |
|---|---|---|---|
| Curtis breast | invasive ductal breast carcinoma versus normal | 2.094 | 1.4E−162 |
| ductal breast carcinoma in situ versus normal | 3.434 | 8.27E−05 | |
| medullary breast carcinoma versus normal | 3.658 | 3.66E−12 | |
| invasive breast carcinoma versus normal | 2.263 | 9.79E−07 | |
| breast carcinoma versus normal | 2.173 | 0.000258 | |
| TCGA breast | invasive breast carcinoma versus normal | 9.789 | 1.41E−35 |
| mucinous breast carcinoma versus normal | 12.152 | 3.48E−08 | |
| mixed lobular and ductal breast carcinoma versus normal | 5.977 | 2.04E−06 | |
| invasive ductal breast carcinoma versus normal | 7.576 | 8.45E−38 | |
| invasive lobular breast carcinoma versus normal | 3.738 | 2E−12 | |
| male breast carcinoma versus normal | 4.212 | 0.001 | |
| Perou breast | lobular breast carcinoma versus normal | 6.968 | 0.004 |
| ductal breast carcinoma versus normal | 5.071 | 0.002 | |
| Richardson breast 2 | ductal breast carcinoma versus normal | 5.009 | 5.61E−11 |
| Sorlie breast 2 | ductal breast carcinoma versus normal | 4.398 | 0.001 |
| Sorlie breast | ductal breast carcinoma versus normal | 4.053 | 0.002 |
| Radvanyi breast | invasive ductal breast carcinoma versus normal | 2.347 | 0.005 |
| Ma breast 4 | invasive ductal breast carcinoma stroma versus normal | 2.322 | 0.005 |
| ductal breast carcinoma in situ stroma versus normal | 4.825 | 0.003 |
Figure 1.
TDO2 Expression Levels in Multiple Types of Human Cancers
(A) Higher or lower TDO2 expression in multiple tumors when compared with normal tissues in the Oncomine database. (B) TDO2 expression levels in all tumors and adjacent normal tissues across TCGA (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
TDO2 Expression Status in BC
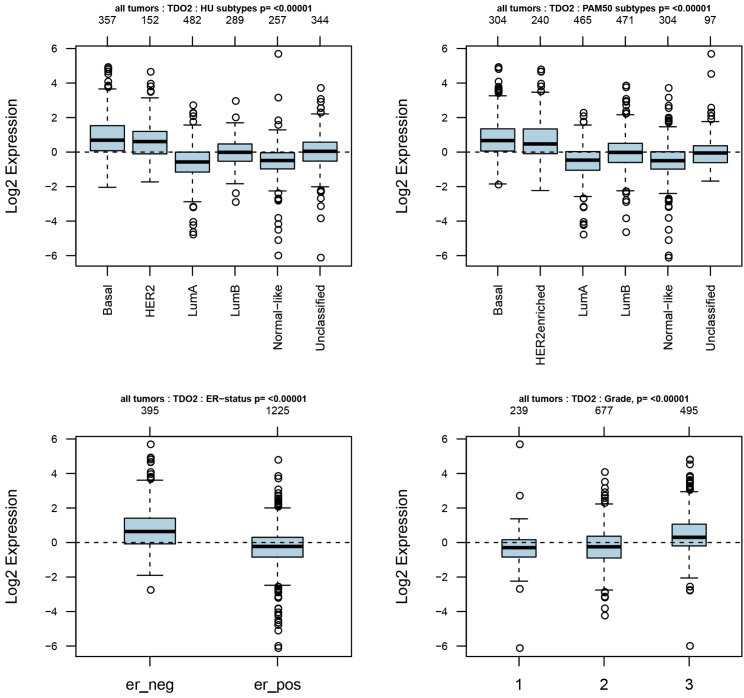
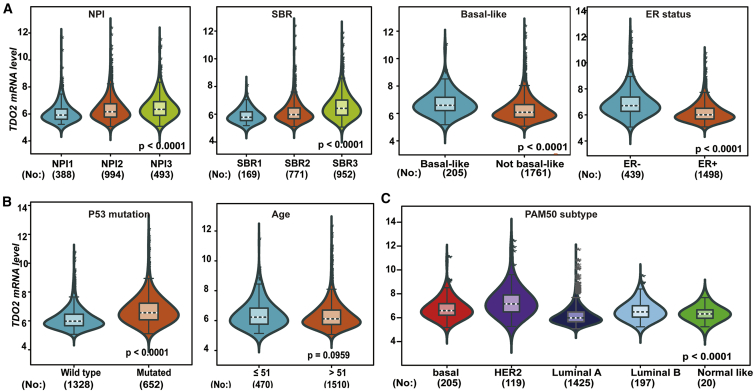
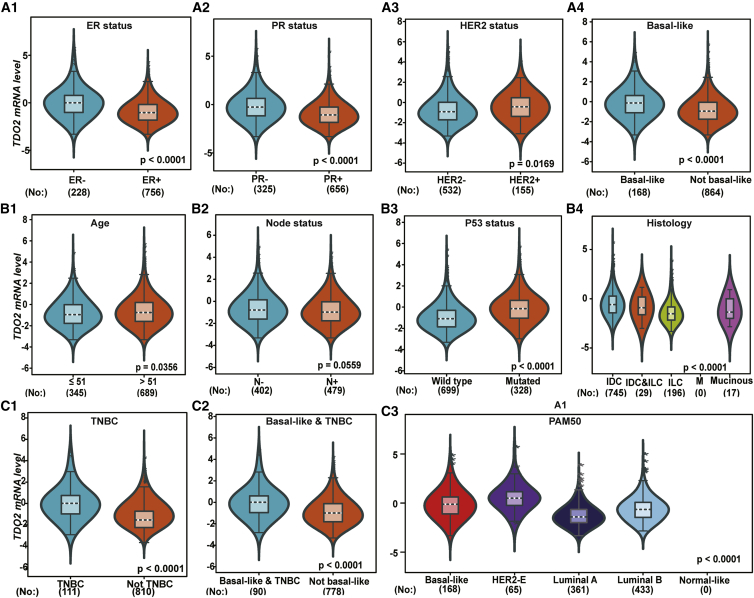
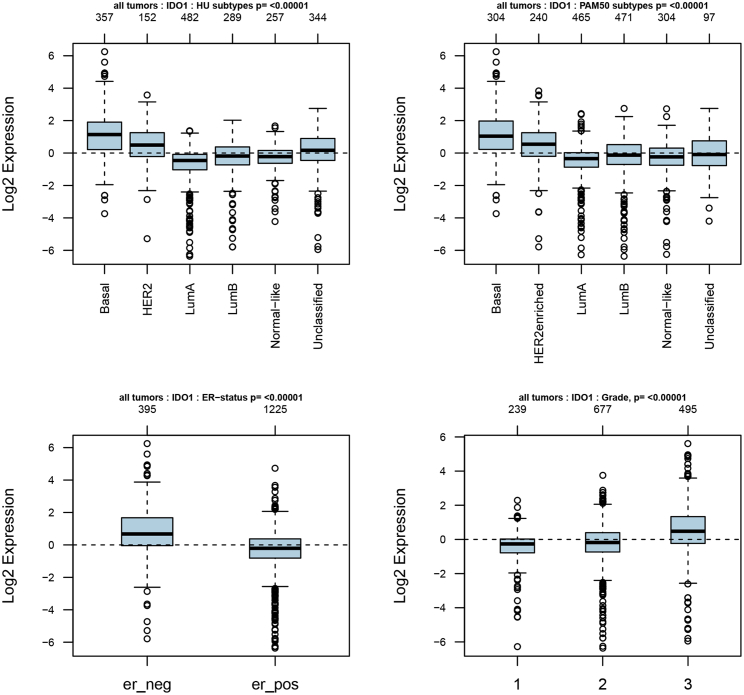
As TDO2 showed significantly higher expression in BC, we focused on exploring the expression profile of TDO2 in different clinical subtypes and stages. First, we evaluated the expression of TDO2 in an 1,881-sample breast tumor dataset generated on Affymetrix U133A microarrays, using the GOBO database.23 GOBO analysis showed that TDO2 expression was significantly higher in basal-like and HER2-enriched clinical subtypes, and significantly higher expression of TDO2 was found in ER- BC when compared with ER+ BC; surprisingly, higher TDO2 expression was found in a higher grade (Figure 2), which suggests that TDO2 might be correlated with progression and development of BC. Second, to further validate the expression status of TDO2 in different clinical subtypes and associations with characteristics, we examined TDO2 expression in the largest BC transcriptomic data using the Breast Cancer Gene-Expression Miner v4.4 database (bc-GenExMiner v4.4),24 which contains a total of 10,001 microarray samples and 4,712 RNA-seq samples. We systematically analyzed TDO2 expression in the METABRIC dataset,25 which is an independent microarray dataset containing 1,980 samples; higher expression of TDO2 was found in higher Nottingham Prognostic Index (NPI)26 and Scarff-Bloom-Richardson (SBR)27 grade (Figure 3A); TDO2 expression was also higher in basal-like, HER2-enriched molecular subtypes and in ER- BC (Figures 3A–3C), which is consistent with results from the GOBO database. In addition, TDO2 expression was higher in the P53-mutated group when compared with the P53-wild group, which implicates that TDO2 expression might be correlated with P53 mutation; no association was found between TDO2 expression and patient age (Figure 3B). Third, these results were further validated in the merged microarray datasets containing a total of 10,001 samples (Figure 4). TDO2 expression was also higher in PR- BC (Figure 4B1) and invasive ductal carcinoma (IDC) (Figure 4B3), and no association was found between TDO2 expression and lymph node status; however, significantly increased TDO2 expression was found in patients over 51 years old, which is inconsistent with the results in the independent METABRIC dataset.
Figure 2.
Expression of TDO2 in 1,881-Sample Microarray Dataset
er_neg, ER- breast cancer; er_pos, ER+ breast cancer.
Figure 3.
TDO2 Expression in METABRIC Dataset (n = 1,980)
TDO2 expression pattern by NPI, SBR, basal-like status, and ER status (A); Association of TDO2 expression and P53 mutation status and age group (B); TDO2 expression pattern by different subtypes of breast cancer.
NPI, Nottingham Prognostic Index (used to determine prognosis following surgery for breast cancer); SBR, Scarff-Bloom-Richardson grade (an important prognostic factor in breast cancer).
Figure 4.
TDO2 Expression in 10,001-Sample Merged Microarray Dataset
TDO2 expression pattern by NPI, SBR, basal-like status and ER status (A1-A4); Association of TDO2 expression and PR status, HER2 status, histology group, and lymphnode status (B1-B4); TDO2 expression pattern by age group, P53 mutation status and different subtypes of breast cancer (C1-C3).
IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; M, micropapillary breast cancer.
Validation of TDO2 Expression in Independent RNA-Seq Datasets
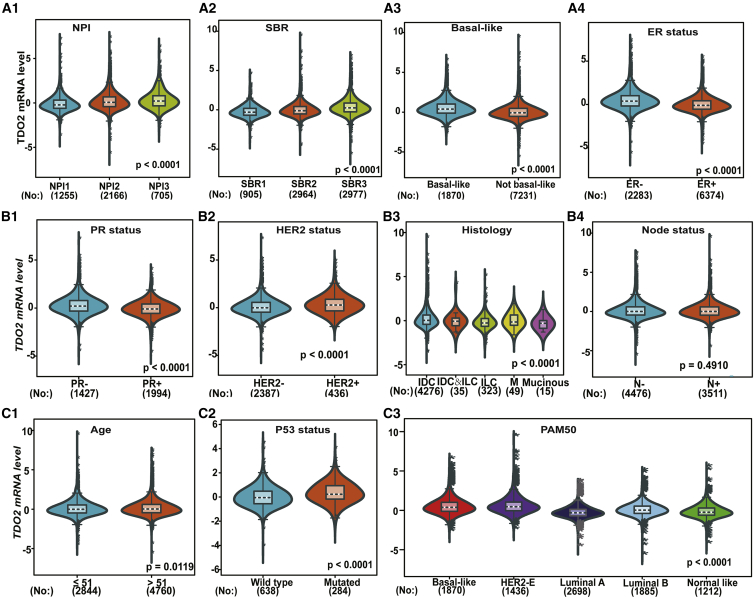
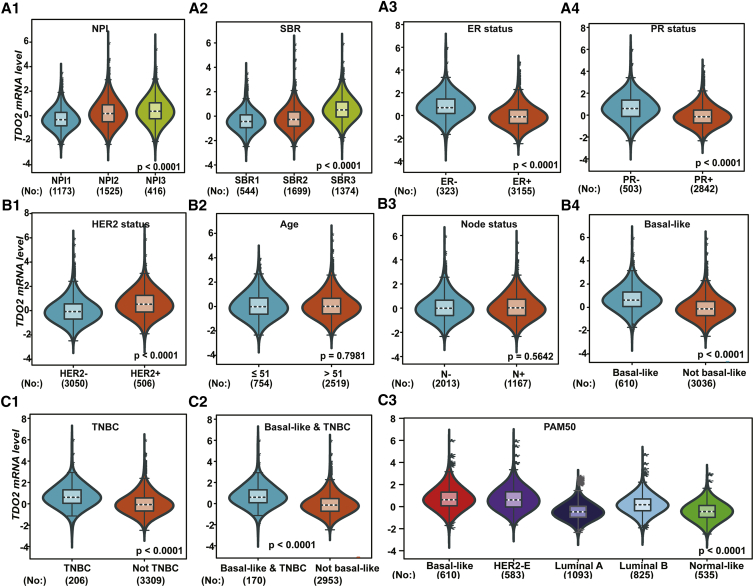
To further validate the expression of TDO2 in BC subtypes and associations with clinical characteristics, we investigated the expression of TDO2 using two independent RNA-seq datasets. First, TDO2 expression was examined in TCGA RNA-seq dataset containing a total of 984 tumor samples, and all analysis results were consistent with results in the 10,001-sample microarray dataset. In particular, TDO2 is also overexpressed in TNBC tissues compared with non-TNBC tissues (Figure 5). Second, we also analyzed TDO2 expression in another independent dataset (GEO: GSE81540)28 containing a total of 3,678 samples, the results of which were consistent with microarray data analysis (Figure 6); however, no association was found between TDO2 expression and patient age (Figure 6B2), which was inconsistent with results in 10,001-sample microarray data.
Figure 5.
TDO2 Expression in TCGA RNA-Seq Dataset
Association of TDO2 expression and ER status, PR status, HER2 status, and basal-like status (A1-A4); Association of TDO2 expression and age group, lymphnode status, P53 mutation status as well as histology group (B1-B4); TDO2 expression pattern by TNBC status and different breast cancer subtypes (C1-C3).
Figure 6.
TDO2 Expression in an Independent RNA-Seq Dataset (GEO: GSE81540; n = 3,678)
TDO2 expression pattern by NPI, SBR, ER status and PR status (A1-A4); Association of TDO2 expression and HER2 status, age group, lymphnode status as well as basal-like status (B1-B4); TDO2 expression pattern by TNBC group and different subtypes of breast cancer (C1-C3).
TDO2-Related Biological Process
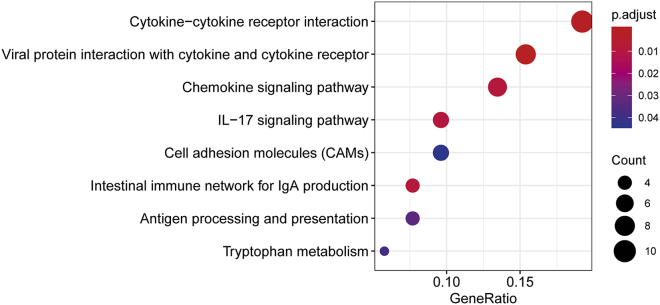
To explore the potential functional role of TDO2, genes correlated with TDO2 expression (Pearson |R| ≥ 0.4) were screened out (n = 77) (Table S1); these genes were further used to do functional enrichment analysis in R using the clusterProfiler package.29 Gene Ontology (GO) analysis revealed that these genes were mainly involved in the immune-related biological response; that is, more than half of the biological processes (BPs) were significantly enriched in immune-related biological processes (Table S2). Interestingly, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis also revealed that these genes were significantly enriched in immune-related pathways, including antigen processing and presentation and the intestinal immune network for immunoglobulin A (IgA) production, and these genes were also significantly enriched in tryptophan metabolism (IDO1 and kynureninase [KYNU]) (Figure 7); additionally, IDO1 has been reported to play a pivotal role in BC progression and cancer immunotherapy.16
Figure 7.
KEGG Pathway Enrichment Results of TDO2-Correlated Genes
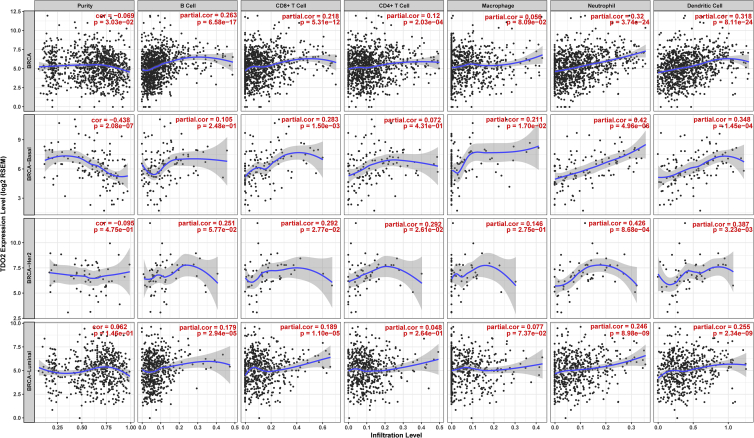
TDO2 Expression Is Correlated with Immune Infiltrates in BC
We estimated the correlations between TDO2 expression and immune cell populations from transcriptomic data in different molecular subtypes and the entire TCGA-BRCA cohort using the TIMER database.30 TDO2 expression was significantly correlated with purity and six types of immune cell infiltrates, including B cells, CD8+ T cells, CD4+ T cells, macrophages, and neutrophils as wells as dendritic cells. Interestingly, significant moderate correlations were found between TDO2 and neutrophils as well as dendritic cells in both basal-like and HER2-enriched BC subtypes (Figure 8). TDO2 expression was also found to be negatively correlated with purity in basal-like BC (correlation = −0.438).
Figure 8.
Correlations between TDO2 Expression and Immune Infiltrates in Breast Cancer
Correlation of TDO2 Expression and Immune Marker Gene Sets
To further investigate the role of TDO2 in immune-related responses, we estimated its association with a total of 57 immune marker genes of different immune cells,31, 32, 33 including CD8+ T cells, T cells (general), B cells, monocytes, tumor-associated macrophages, M1 and M2 macrophages, neutrophils, NK cells, and dendritic cells, and also different functional T cells, including T helper 1 (Th1) cells, Th2 cells, T follicular helper (Tfh) cells, Th17 cells, regulatory T cells (Tregs), and exhausted T cells. Surprisingly, we found that the TDO2 expression level was significantly correlated with 52 of 57 immune marker genes after the correlation adjustment by purity (Table 2).
Table 2.
Correlation Analysis between TDO2 Markers of Immune Cells in TIMER
| Descriptions | Gene Markers | None |
Purity Adjusted |
||
|---|---|---|---|---|---|
| Correlation | p | Correlation | p | ||
| CD8+ T cell | CD8A | 0.245174 | ∗∗∗ | 0.247093 | ∗∗∗ |
| CD8B | 0.208377 | ∗∗∗ | 0.207519 | ∗∗∗ | |
| T cell (general) | CD3D | 0.295458 | ∗∗∗ | 0.290558 | ∗∗∗ |
| CD3E | 0.284067 | ∗∗∗ | 0.278572 | ∗∗∗ | |
| CD2 | 0.334967 | ∗∗∗ | 0.331989 | ∗∗∗ | |
| B cell | CD19 | 0.231611 | ∗∗∗ | 0.223418 | ∗∗∗ |
| CD79A | 0.215303 | ∗∗∗ | 0.206306 | ∗∗∗ | |
| Monocyte | CD86 | 0.394471 | ∗∗∗ | 0.399875 | ∗∗∗ |
| CD115 (CSF1R) | 0.19243 | ∗∗∗ | 0.194018 | ∗∗∗ | |
| TAM | CCL2 | 0.377172 | ∗∗∗ | 0.376796 | ∗∗∗ |
| CD68 | 0.37994 | ∗∗∗ | 0.391855 | ∗∗∗ | |
| IL10 | 0.414858 | ∗∗∗ | 0.431181 | ∗∗∗ | |
| M1 macrophage | INOS (NOS2) | 0.075771 | ∗ | 0.081674 | ∗ |
| IRF5 | 0.253776 | ∗∗∗ | 0.255338 | ∗∗∗ | |
| COX2(PTGS2) | 0.049728 | 0.09926 | 0.047829 | 0.131641 | |
| M2 macrophage | CD163 | 0.384785 | ∗∗∗ | 0.396327 | ∗∗∗ |
| VSIG4 | 0.230386 | ∗∗∗ | 0.241997 | ∗∗∗ | |
| MS4A4A | 0.337152 | ∗∗∗ | 0.348611 | ∗∗∗ | |
| Neutrophils | CD66b (CEACAM8) | 0.037856 | 0.209634 | 0.037391 | 0.238641 |
| CD11b (ITGAM) | 0.179053 | ∗∗∗ | 0.179193 | ∗∗∗ | |
| CCR7 | 0.263431 | ∗∗∗ | 0.255275 | ∗∗∗ | |
| Natural killer cell | KIR2DL1 | 0.248415 | ∗∗∗ | 0.262995 | ∗∗∗ |
| KIR2DL3 | 0.270892 | ∗∗∗ | 0.270489 | ∗∗∗ | |
| KIR2DL4 | 0.377769 | ∗∗∗ | 0.381431 | ∗∗∗ | |
| KIR3DL1 | 0.261523 | ∗∗∗ | 0.25955 | ∗∗∗ | |
| KIR3DL2 | 0.276474 | ∗∗∗ | 0.269903 | ∗∗∗ | |
| KIR3DL3 | 0.147658 | ∗∗∗ | 0.154768 | ∗∗∗ | |
| KIR2DS4 | 0.210854 | ∗∗∗ | 0.210864 | ∗∗∗ | |
| Dendritic cell | HLA-DPB1 | 0.177509 | ∗∗∗ | 0.177217 | ∗∗∗ |
| HLA-DQB1 | 0.203416 | ∗∗∗ | 0.204322 | ∗∗∗ | |
| HLA-DRA | 0.295315 | ∗∗∗ | 0.293571 | ∗∗∗ | |
| HLA-DPA1 | 0.232387 | ∗∗∗ | 0.233481 | ∗∗∗ | |
| BDCA-1(CD1C) | 0.057982 | 0.054546 | 0.044691 | 0.158939 | |
| BDCA-4(NRP1) | 0.082773 | ∗∗ | 0.099865 | ∗∗ | |
| CD11c (ITGAX) | 0.278226 | ∗∗∗ | 0.277131 | ∗∗∗ | |
| Th1 | T-bet (TBX21) | 0.302566 | ∗∗∗ | 0.298116 | ∗∗∗ |
| STAT4 | 0.251384 | ∗∗∗ | 0.247076 | ∗∗∗ | |
| STAT1 | 0.415731 | ∗∗∗ | 0.421584 | ∗∗∗ | |
| IFN-γ (IFNG) | 0.357511 | ∗∗∗ | 0.358795 | ∗∗∗ | |
| TNF-α (TNF) | 0.217439 | ∗∗∗ | 0.208063 | ∗∗∗ | |
| Th2 | GATA3 | −0.31958 | ∗∗∗ | −0.30945 | ∗∗∗ |
| STAT6 | −0.20079 | ∗∗∗ | −0.20855 | ∗∗∗ | |
| STAT5A | −0.03753 | 0.213554 | −0.04649 | 0.142772 | |
| IL13 | 0.184803 | ∗∗∗ | 0.171661 | ∗∗∗ | |
| Tfh | BCL6 | −0.12198 | ∗∗∗ | −0.13259 | ∗∗∗ |
| IL21 | 0.286359 | ∗∗∗ | 0.291807 | ∗∗∗ | |
| Th17 | STAT3 | −0.06799 | ∗ | −0.06881 | ∗ |
| IL17A | 0.214299 | ∗∗∗ | 0.224482 | ∗∗∗ | |
| Treg | FOXP3 | 0.405434 | ∗∗∗ | 0.396407 | ∗∗∗ |
| CCR8 | 0.438307 | ∗∗∗ | 0.437575 | ∗∗∗ | |
| STAT5B | −0.1762 | ∗∗∗ | −0.17439 | ∗∗∗ | |
| TGFβ (TGFB1) | 0.031626 | 0.294651 | 0.041407 | 0.191877 | |
| T cell exhaustion | PD-1 (PDCD1) | 0.279633 | 3.29E−21 | 0.272028 | ∗∗∗ |
| CTLA4 | 0.421275 | 1.49E−48 | 0.41456 | ∗∗∗ | |
| LAG3 | 0.39448 | 2.85E−42 | 0.395075 | ∗∗∗ | |
| TIM-3 (HAVCR2) | 0.340601 | 2.79E−31 | 0.350868 | ∗∗∗ | |
| GZMB | 0.435824 | 3.28E−52 | 0.429831 | ∗∗∗ | |
TAM, tumor associated macrophage. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Correlation of TDO2 Expression and Tryptophan Metabolism-Related Molecules
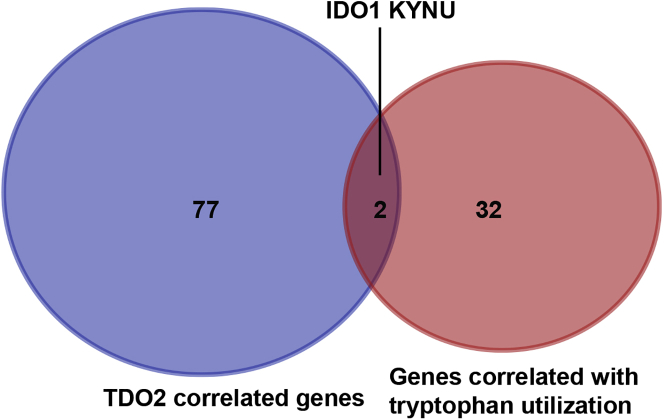
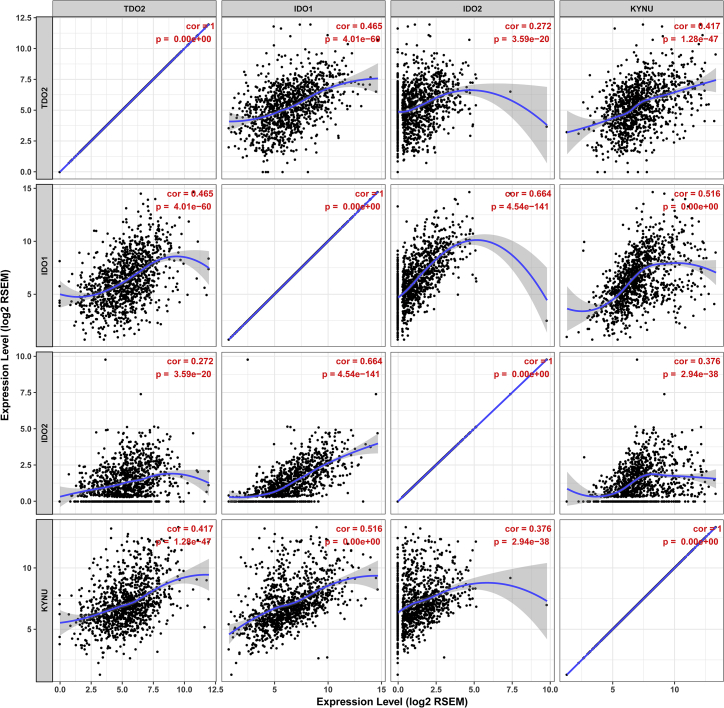
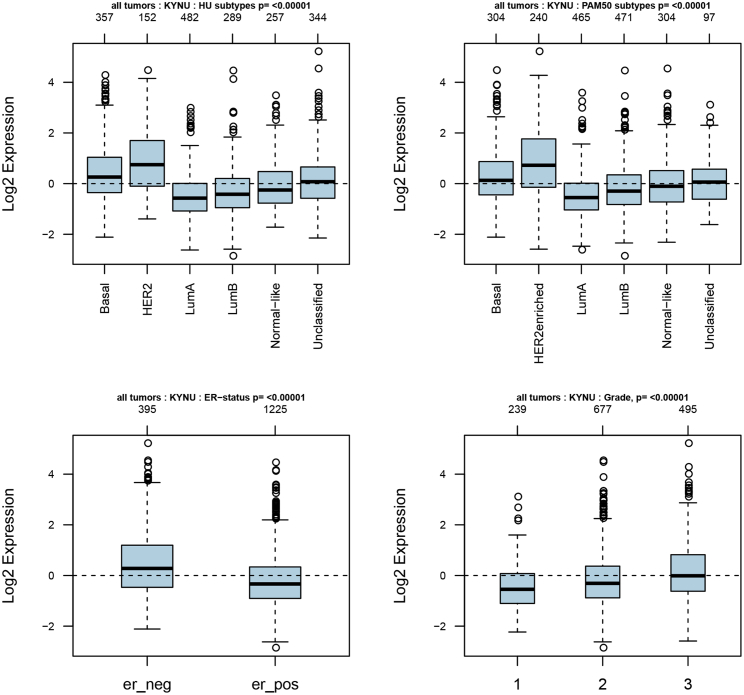
32 genes in the superpathway of tryptophan collected from the PathCards database (https://pathcards.genecards.org) were defined as tryptophan metabolism-related gene sets (Table S3). Interestingly, we found two genes (IDO1 and KYNU) that were found in both the tryptophan metabolism-related gene sets and TDO2-correlated genes (Figure 9). In particular, IDO1 and KYNU were also the genes enriched in the KEGG pathway (tryptophan metabolism). Correlations between TDO2, IDO1, IDO2, and KYNU were analyzed using the TIMER database; interestingly, we found that these genes were significantly correlated with each other (Figure 10). Therefore, we further investigated the expression of IDO1 and KYNU in a 1,881-sample microarray dataset. As expected, the expression patterns of both IDO1 and KYNU were similar to TDO2; that is, higher expression was found in basal-like and HER2-enriched subtypes; increased expression levels of IDO1 and KYNU were also found in ER- BC, as well as a higher grade (Figures 11 and 12).
Figure 9.
Venn Diagram Depicts Intersection of TDO2 Correlated Genes and Genes Correlated with Tryptophan Utilization
Figure 10.
Correlations between TDO2, IDO1, and IDO2 as well as KYNU
Figure 11.
Expression of IDO1 in 1,881-Sample Microarray Dataset
er_neg, ER- breast cancer; er_pos, ER+ breast cancer.
Figure 12.
Expression of KYNU in 1,881-Sample Microarray Dataset
er_neg, ER- breast cancer; er_pos, ER+ breast cancer.
TDO2 Expression Is Correlated with Survival of BC Patients
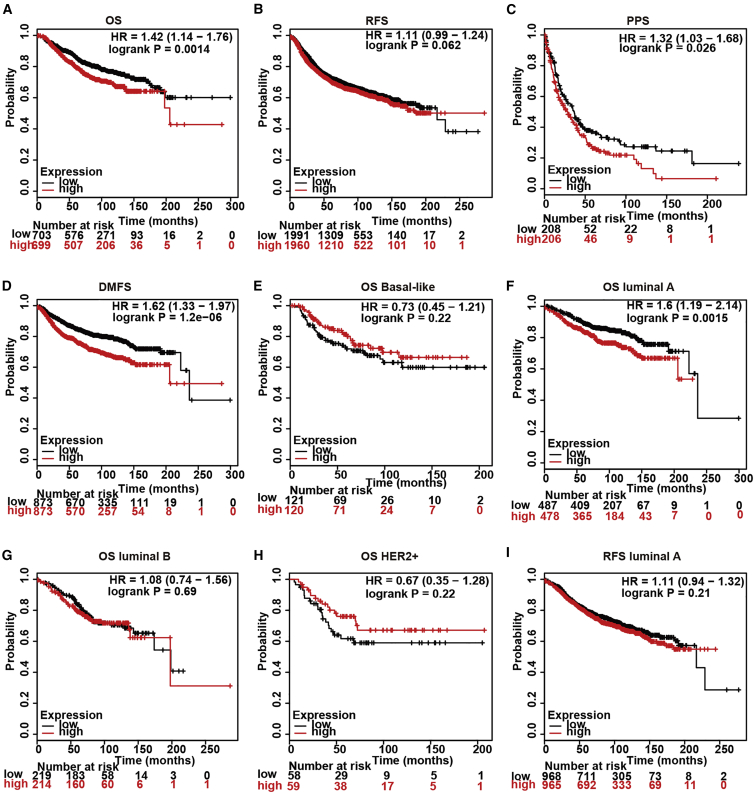
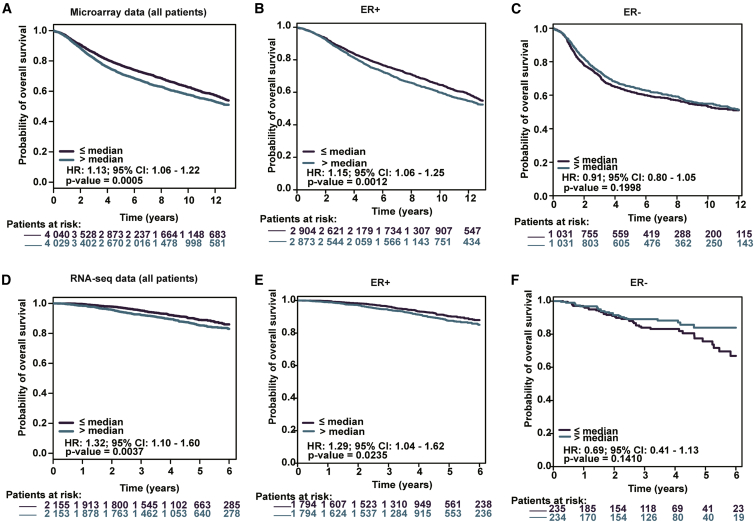
AS TDO2 expression was correlated with BC subtypes and stages, we then explored the the prognostic value of TDO2 using the Kaplan-Meier plotter database containing a total of 6,243 BC samples. Kaplan-Meier analysis demonstrated that higher TDO2 expression was associated with both worse overall survival (OS), relapse-free survival (RFS), post-progression survival (PPS), and distant metastasis-free (DMFS) survival (Figures 13A–13D). We further analyzed the prognostic value of TDO2 in subtype levels and found that higher TDO2 expression predicts worse OS in the luminal A subtype, but not in basal-like, luminal B, and HER2+ subtypes (Figures 13E–13I). To validate the prognostic value of TDO2 in BC, we further performed prognostic analysis of TDO2 in an 8,069-sample microarray dataset and an independent 4,258-sample RNA-seq dataset using the bc-GenExMiner v4.4 database.24 Higher TDO2 expression level significantly predicts worse OS in the entire BC cohort and ER- BC, but not ER+ BC (Figures 14A–14F), which is consistent with results from the Kaplan-Meier plotter database.
Figure 13.
Kaplan-Meier Survival Curves Comparing the High and Low Expression of TDO2 in Breast Cancer Using the Kaplan-Meier Plotter Database
Kaplan-Meier survival curve of TDO2 expression by OS, RFS, PPS, DMFS in all breast cancer patients of Kaplan-Meier Plotter Database (A-D); Prognostic value of TDO2 in different molecular subtypes of breast cancer (E-I).
OS, overall survival; RFS, relapse-free survival; PPS, post-progression survival; DMFS, distant metastasis-free; HR, hazard ratio.
Figure 14.
Validation of Kaplan-Meier Survival Curves Comparing the High and Low Expression of TDO2 in Microarray Dataset (n = 8,049) and RNA-seq Dataset (n = 4,308)
Prognostic value of TDO2 in microarray data set (A-C); Prognostic value of TDO2 in RNA-seq data set (D-F).
HR, hazard ratio; 95% CI, 95% confidence interval.
Discussion
To our knowledge, this is the largest and most comprehensive study involving more than 10,000 samples that systematically investigated the expression pattern of TDO2 together with its prognostic value and related biological processes in BC. Our data revealed that TDO2 expression tends to be upregulated in BC with a higher grade and degree of malignancy. No correlation was found between TDO2 expression and lymph node status, and it is still unclear whether TDO2 expression was correlated with patient age due to varying results in different datasets. Genes correlated with TDO2 were mainly involved in biological processes and pathways related to immune regulation. Higher expression of TDO2 was associated with worse prognosis in BC patients. We also observed that TDO2 expression was significantly correlated with immune cell infiltrates and purity, and it was also correlated with most of the immune marker genes (52/57). These results implicate the critical role of TDO2 in manipulating the BC immune microenvironment.
Intriguingly, we found that TDO2 expression was significantly correlated with other rate-limiting enzymes involved in catalyzing the conversion of tryptophan to kynurenine (Figure 10), including IDO1 and IDO2 as well as KYNU. More importantly, the expression patterns of IDO1 and KYNU in BC were similar to TDO2. A large body of evidence has demonstrated that IDO1 contributes to immunosuppressive effects by tryptophan depletion and direct effects of tryptophan catabolites.14,34,35 IDO1 is expressed in the placenta and many types of human tumors.35,36 On the basis of the tumor-promoting effects of IDO1, a large number of small-molecule inhibitors have been investigated for cancer therapy; however, the negative result of a recent phase III trial performed in patients with unresectable or metastatic melanoma clearly represents a setback in the development of IDO1 inhibitors in cancer immunotherapy.15
Therefore, it is important to learn more about the mechanism of IDO1 in inhibiting cancer and to exploit novel immunotherapy targets. One possible explanation for the failure of IDO1 inhibitors might be that targeting IDO1 alone is not sufficient, since it is still unknown whether the inhibition of IDO1 has impacts on regulating the expression of other rate-limiting enzymes. Notably, our results indicated the significant correlations between IDO1 and other enzymes, including IDO2 (correlation = 0.664) and TDO2 (correlation = 0.465) as well as KYNU (correlation = 0.516). Critically, IDO2 is another rate-limiting enzyme involved in tryptophan metabolism and was expressed in multiple human tumors; however, little is known about the specific role of IDO2 in modulating the tumor immune microenvironment. Most recently, evidence has supported that IDO2 is also an important immune modulator that is altering the tumor microenvironment, including regulating tryptophan accumulation and kynurenine reduction, thereby promoting immune cell invasion. IDO2 expression has a strong correlation with IDO1, which suggests that IDO2 might function as a co-regulator in the IDO1-mediated immunosuppressive microenvironment. Therefore, more endeavors are warranted to explore the specific relationship between IDO1 and IDO2 in the regulation of the tumor microenvironment.
KYNU is an enzyme downstream of TDO2 that is encoded by the protein-coding gene KYNU; however, little is known about the role of KYNU in cancer. A recent study examined expression of KYNU in 137 primary BC tissues using immunohistochemistry.37 The authors claimed that KYNU was positively correlated with the expressions of ER and negatively correlated with tumor grade, suggesting that overexpression of KYNU inhibited cell proliferation and might function as a tumor suppressor in BC. However, our results indicated that KYNU was overexpressed in ER- BC and a higher grade as well as basal-like BC. Notably, evidence provided in a paper published in Cancer Research indicated that KYNU was elevated in suspended TNBC cell lines, and also overexpressed in ER- BC cell lines;19 in addition, previous studies have demonstrated suspended TNBC cells upregulate proteins involved in anoikis resistance and motility,38,39 these results are in line with our observations. We also found that KYNU expression was significantly correlated with IDO1 (correlation = 0.516) and IDO2 (correlation = 0.376) as well as TDO2 (correlation = 0.417). These results suggest that KYNU may also function as a co-regulator of the tumor microenvironment in BC.
Critically, our results also reported significant correlations between TDO2 expression and immune cell infiltrates in BC. Currently, little is known about the specific role of TDO2 in modulating the immune response. A recent study has reported that TDO2 was involved in inhibiting CD8+ T cell viability in TNBC.20 However, the interactions between TDO2 and other immune cells remain unclear. Our results indicated moderate correlations between TDO2 expression and neutrophils as well as dendritic cells. Future studies focused on investigating the role of TDO2 in regulating the immune response might reveal novel insights for TDO2-mediated immunotherapy.
In summary, our observations support the critical role of TDO2 in mediating the immunosuppressive microenvironment for BC, and we highlight that tryptophan-catabolizing enzymes (i.e., IDO1, IDO2, TDO2, and KYNU) might function by co-modulating the tumor immune microenvironment, instead of functioning by one gene alone, which suggests that targeting one or two enzymes in cancer immunotherapy might not be sufficient to reverse tumor immune resistance. The concept of developing inhibitors targeting multiple enzymes should be established in cancer immunotherapy.
Materials and Methods
Oncomine Database Analysis
Oncomine22 is a comprehensive microarray database for cancer research containing a total of 715 datasets and 86,733 samples. The mRNA expression level of the TDO2 in multiple types of human cancers was identified in this database: https://www.oncomine.org/resource/login.html. The threshold for analysis was set as follows: p value of 0.05, fold change of 2, and gene ranking of the top 10%.
TIMER Database Analysis
The TIMER database: https://cistrome.shinyapps.io/timer/ is a comprehensive web platform containing 10,897 samples for systematic analyses of immune infiltrates across 32 cancer types from TCGA database.30 The “DiffExp” module was used to explore the differential expression of TDO2 between tumor and adjacent normal tissues, and the Wilcoxon test was applied to determine statistical significance of differential expression. The “gene” module was used to visualize the correlation of TDO2 expression with immune infiltration level (B cells, CD4 T cells, CD8 T cells, neutrophils, macrophages, and dendritic cells) in BC and its molecular subtypes. The “correlation” module was applied to draw the expression scatterplots between gene pairs, together with the Spearman’s correlation and estimated statistical significance. In addition, correlations between TDO2 expression and immune marker genes were calculated using the correlation module; immune marker gene sets are referenced in previous studies.31, 32, 33
GOBO Database Analysis
GOBO is a comprehensive online database: http://co.bmc.lu.se/gobo/gsa_cellines.pl that allows rapid assessment of gene expression levels in a 1,881-sample BC dataset.23 The tumor dataset consists of 1,225 ER+ tumors and 395 ER- tumors, 927 untreated tumors, and 326 tamoxifen-treated tumors. Expression analysis of TDO2, IDO1, and KYNU in a 1,881-sample dataset was performed using the GOBO database.
bc-GenExMiner v4.4 Database Analysis
bc-GenExMiner v4.4 database: http://bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1 is a comprehensive statistical mining tool of published annotated BC transcriptomic data,24 containing a total of 10,001 microarray samples and 4,712 RNA-seq samples. Expression of TDO2 in the METABRIC dataset (n = 1,980), a merged microarray dataset (n = 10,001), TCGA RNA-seq dataset (n = 1,034), and GEO: GSE81540 (n = 3,678) was determined using the “expression analysis” module; in this module, the global significant difference between groups was assessed by Welch’s test (the corresponding p value is indicated on the bottom right of the figure). When a global significant difference existed (p < 0.05) and there were more than two groups, a Dunnett-Tukey-Kramer’s test was calculated for each pairwise comparison. Prognostic analysis of TDO2 was validated using the “prognostic analysis” module.
cBioPortal Database Analysis
The cBioPortal for Cancer Genomics: http://cbioportal.org/ is a comprehensive resource for exploring, visualizing, and analyzing multidimensional cancer genomics data.40 Spearman’s correlations between TDO2 and all 20,171 genes were identified using the “co-expression” module in TCGA BC RNA-seq dataset.
Functional enrichment analysis
GO20 and KEGG pathway enrichment analysis21 was performed using the clusterProfiler package22 in statistical software R version 3.6.0. (http://www.r-project.org/). GO terms and KEGG pathways with an adjusted p value less than 0.05 were considered to be statistically significant. Dot plots of enriched KEGG pathways were plotted using the clusterProfiler package.
Kaplan-Meier Plotter Database Analysis
The Kaplan-Meier plotter41 is capable of assessing the effect of 54,000 genes on survival in 21 cancer types; BC is the largest dataset in the Kaplan-Meier plotter, containing a total of 6,234 samples. The effect of TDO2 expression on survival together with the hazard ratio (HR) with a 95% confidence interval and log rank p value in BC was estimated by the Kaplan-Meier plotter: http://kmplot.com/analysis.
Data Availability
The data generated or analyzed during this study are included in this article, or, if absent, are available from a corresponding author upon reasonable request.
Author Contributions
Q.L. and J.Z. carried out the primary literature search; Q.L. performed the data analysis, Q.L. drafted the manuscript; Q.L., X.K., Z.W., and Y.F. performed the literature search and revised the manuscript; Q.L., J.Z., X.K., Y.F., X.W., Z.W., and J.W. discussed, revised, and edited the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This article was partially sponsored by grants from the National Natural Science Foundation of China, no. 81872160 (J.W.), Beijing Municipal Natural Science Foundation (Key program, No. 7191009) (J.W.), Capital Public Health Education, Beijing Science and Technology Program (No. Z161100000116093) (J.W.) and CAMS Initiative for Innovative Medicine (No. 2016-12M-1-007) (J.W.). This article was partially supported by PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (No. 3332015157) (Y.F.), as well as Capital Public Health Education, Beijing Science and Technology Program (No. Z171100000417028) (Y.F.). The first author is particularly indebted to Miss Ping He for valuable help during the process of data analysis and drafting the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.03.013.
Contributor Information
Zhongzhao Wang, Email: wangzhongzhao206@sina.com.
Yi Fang, Email: fangyi@cicams.ac.cn.
Jing Wang, Email: wwwjjj1234@vip.sina.com.
Supplemental Information
References
- 1.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 4.Denkert C., von Minckwitz G., Brase J.C., Sinn B.V., Gade S., Kronenwett R., Pfitzner B.M., Salat C., Loi S., Schmitt W.D. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 2015;33:983–991. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 5.Zou W., Wolchok J.D., Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016;8:328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 7.Nanda R., Chow L.Q., Dees E.C., Berger R., Gupta S., Geva R., Pusztai L., Pathiraja K., Aktan G., Cheng J.D. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J. Clin. Oncol. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwa M.J., Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): where to go from here. Cancer. 2018;124:2086–2103. doi: 10.1002/cncr.31272. [DOI] [PubMed] [Google Scholar]
- 9.Opitz C.A., Litzenburger U.M., Sahm F., Ott M., Tritschler I., Trump S., Schumacher T., Jestaedt L., Schrenk D., Weller M. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 10.Pilotte L., Larrieu P., Stroobant V., Colau D., Dolusic E., Frédérick R., De Plaen E., Uyttenhove C., Wouters J., Masereel B., Van den Eynde B.J. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller A.J., DuHadaway J.B., Donover P.S., Sutanto-Ward E., Prendergast G.C. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 12.Uyttenhove C., Pilotte L., Théate I., Stroobant V., Colau D., Parmentier N., Boon T., Van den Eynde B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 13.Lee G.K., Park H.J., Macleod M., Chandler P., Munn D.H., Mellor A.L. Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology. 2002;107:452–460. doi: 10.1046/j.1365-2567.2002.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munn D.H., Shafizadeh E., Attwood J.T., Bondarev I., Pashine A., Mellor A.L. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J. Exp. Med. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Platten M., Nollen E.A.A., Röhrig U.F., Fallarino F., Opitz C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019;18:379–401. doi: 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 16.Cheong J.E., Sun L. Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy—challenges and opportunities. Trends Pharmacol. Sci. 2018;39:307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Garber K. A new cancer immunotherapy suffers a setback. Science. 2018 doi: 10.1126/science.360.6389.588. [DOI] [PubMed] [Google Scholar]
- 18.Pham Q.T., Oue N., Sekino Y., Yamamoto Y., Shigematsu Y., Sakamoto N., Sentani K., Uraoka N., Yasui W. TDO2 overexpression is associated with cancer stem cells and poor prognosis in esophageal squamous cell carcinoma. Oncology. 2018;95:297–308. doi: 10.1159/000490725. [DOI] [PubMed] [Google Scholar]
- 19.D’Amato N.C., Rogers T.J., Gordon M.A., Greene L.I., Cochrane D.R., Spoelstra N.S., Nemkov T.G., D’Alessandro A., Hansen K.C., Richer J.K. A TDO2-AhR signaling axis facilitates anoikis resistance and metastasis in triple-negative breast cancer. Cancer Res. 2015;75:4651–4664. doi: 10.1158/0008-5472.CAN-15-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greene L.I., Bruno T.C., Christenson J.L., D’Alessandro A., Culp-Hill R., Torkko K., Borges V.F., Slansky J.E., Richer J.K. A role for tryptophan-2,3-dioxygenase in CD8 T-cell suppression and evidence of tryptophan catabolism in breast cancer patient plasma. Mol. Cancer Res. 2019;17:131–139. doi: 10.1158/1541-7786.MCR-18-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers T.J., Christenson J.L., Greene L.I., O’Neill K.I., Williams M.M., Gordon M.A., Nemkov T., D’Alessandro A., Degala G.D., Shin J. Reversal of triple-negative breast cancer EMT by miR-200c decreases tryptophan catabolism and a program of immunosuppression. Mol. Cancer Res. 2019;17:30–41. doi: 10.1158/1541-7786.MCR-18-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes D.R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B.B., Barrette T.R., Anstet M.J., Kincead-Beal C., Kulkarni P. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringnér M., Fredlund E., Häkkinen J., Borg Å., Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS ONE. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jézéquel P., Campone M., Gouraud W., Guérin-Charbonnel C., Leux C., Ricolleau G., Campion L. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 25.Milioli H.H., Vimieiro R., Tishchenko I., Riveros C., Berretta R., Moscato P. Iteratively refining breast cancer intrinsic subtypes in the METABRIC dataset. BioData Min. 2016;9:2. doi: 10.1186/s13040-015-0078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galea M.H., Blamey R.W., Elston C.E., Ellis I.O. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res. Treat. 1992;22:207–219. doi: 10.1007/BF01840834. [DOI] [PubMed] [Google Scholar]
- 27.Le Doussal V., Tubiana-Hulin M., Friedman S., Hacene K., Spyratos F., Brunet M. Prognostic value of histologic grade nuclear components of Scarff-Bloom-Richardson (SBR). An improved score modification based on a multivariate analysis of 1262 invasive ductal breast carcinomas. Cancer. 1989;64:1914–1921. doi: 10.1002/1097-0142(19891101)64:9<1914::aid-cncr2820640926>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Brueffer C., Vallon-Christersson J., Grabau D., Ehinger A., Häkkinen J., Hegardt C., Malina J., Chen Y., Bendahl P.-O., Manjer J. Clinical value of RNA sequencing–based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden Cancerome analysis network—breast initiative. JCO Precis. Oncol. 2018;2:1–18. doi: 10.1200/PO.17.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siemers N.O., Holloway J.L., Chang H., Chasalow S.D., Ross-MacDonald P.B., Voliva C.F., Szustakowski J.D. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS ONE. 2017;12:e0179726. doi: 10.1371/journal.pone.0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danaher P., Warren S., Dennis L., D’Amico L., White A., Disis M.L., Geller M.A., Odunsi K., Beechem J., Fling S.P. Gene expression markers of tumor infiltrating leukocytes. J. Immunother. Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sousa S., Määttä J. The role of tumour-associated macrophages in bone metastasis. J. Bone Oncol. 2016;5:135–138. doi: 10.1016/j.jbo.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terness P., Bauer T.M., Röse L., Dufter C., Watzlik A., Simon H., Opelz G. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J. Exp. Med. 2002;196:447–457. doi: 10.1084/jem.20020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munn D.H., Zhou M., Attwood J.T., Bondarev I., Conway S.J., Marshall B., Brown C., Mellor A.L. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 36.Godin-Ethier J., Hanafi L.-A., Piccirillo C.A., Lapointe R. Indoleamine 2,3-dioxygenase expression in human cancers: clinical and immunologic perspectives. Clin. Cancer Res. 2011;17:6985–6991. doi: 10.1158/1078-0432.CCR-11-1331. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., Feng X., Lai J., Yi W., Yang J., Du T., Long X., Zhang Y., Xiao Y. A novel role of kynureninase in the growth control of breast cancer cells and its relationships with breast cancer. J. Cell. Mol. Med. 2019;23:6700–6707. doi: 10.1111/jcmm.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howe E.N., Cochrane D.R., Richer J.K. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13:R45. doi: 10.1186/bcr2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howe E.N., Cochrane D.R., Cittelly D.M., Richer J.K. miR-200c targets a NF-κB up-regulated TrkB/NTF3 autocrine signaling loop to enhance anoikis sensitivity in triple negative breast cancer. PLoS ONE. 2012;7:e49987. doi: 10.1371/journal.pone.0049987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy Á., Lánczky A., Menyhárt O., Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated or analyzed during this study are included in this article, or, if absent, are available from a corresponding author upon reasonable request.