Numerous genes have been identified that regulate leaf growth, which can be grouped into regulatory modules. Here, we review six important gene modules that affect cell proliferation during leaf development.
Keywords: Arabidopsis thaliana, cell cycle, cell proliferation, leaf development, leaf size, organ growth
Abstract
Leaves are the primary organs for photosynthesis, and as such have a pivotal role for plant growth and development. Leaf development is a multifactorial and dynamic process involving many genes that regulate size, shape, and differentiation. The processes that mainly drive leaf development are cell proliferation and cell expansion, and numerous genes have been identified that, when ectopically expressed or down-regulated, increase cell number and/or cell size during leaf growth. Many of the genes regulating cell proliferation are functionally interconnected and can be grouped into regulatory modules. Here, we review our current understanding of six important gene regulatory modules affecting cell proliferation during Arabidopsis leaf growth: ubiquitin receptor DA1–ENHANCER OF DA1 (EOD1), GROWTH REGULATING FACTOR (GRF)–GRF-INTERACTING FACTOR (GIF), SWITCH/SUCROSE NON-FERMENTING (SWI/SNF), gibberellin (GA)–DELLA, KLU, and PEAPOD (PPD). Furthermore, we discuss how post-mitotic cell expansion and these six modules regulating cell proliferation make up the final leaf size.
Introduction
Plants develop and grow mainly post-embryonically, forming two types of organs: organs such as roots with an indeterminate growth and therefore a theoretical unlimited growth potential; and organs such as leaves and flowers with a determinate growth and a fixed final size (Tsukaya, 2003; Rodriguez et al., 2014). Leaves are the organs in which photosynthesis predominantly occurs. Leaves also contribute significantly to plant biomass, since the energy and carbohydrates produced during photosynthesis are used by the rest of the plant to sustain its growth and complete its life cycle (Demura and Ye, 2010). These features render leaf size control a highly interesting field of study.
In Arabidopsis thaliana (Arabidopsis), leaves grow through cell proliferation and cell expansion, two highly interconnected developmental processes, which are partially overlapping during leaf development (Asl et al., 2011; Gonzalez et al., 2012). Leaves are initiated by a group of founder cells emerging at the flanks of the shoot apical meristem (Reinhardt et al., 2000; Efroni et al., 2010; Kalve et al., 2014). These leaf primordium founder cells undergo extensive cell division, resulting in an increased cell number that contributes to final leaf size (Gonzalez et al., 2012). After a predefined developmental time frame, cells at the tip of the leaf exit the mitotic cell cycle and start to expand, marking the beginning of the cell expansion phase. In Arabidopsis, a cell cycle arrest front then moves through the leaf in a tip to base manner (Andriankaja et al., 2012). However, some cells dispersed throughout the leaf epidermis retain their meristematic activity. These stem cell-like cells, called meristemoids, continue to divide asymmetrically for several rounds before giving rise to stomata, pores located in the epidermis that allow gas and water vapor exchange with the environment (Bergmann and Sack, 2007). In Arabidopsis, the increase in number of stomatal cells takes place in a tip to base direction as well, suggesting the occurrence of a secondary cell cycle arrest front corresponding to the arrest of meristemoid asymmetric divisions (White, 2006; Andriankaja et al., 2012). Altogether, at least six major cellular processes contribute to final leaf size and shape: the number of cells recruited to the leaf primordium from the shoot apical meristem; the rate and duration of cell proliferation; the rate and duration of cell expansion; and the extent of meristemoid division (Gonzalez et al., 2012; Hepworth and Lenhard, 2014). Impinging on one of these processes often results in an alteration in cell number or cell size, affecting final leaf size (Gonzalez et al., 2012). Therefore, the correct regulation of cell proliferation and cell expansion mechanisms is fundamental to determine final leaf size.
In this review, we describe current advances in Arabidopsis leaf growth regulation, mainly focusing on six gene regulatory modules involved in cell proliferation: DA1–ehhancer of DA1 (EOD1), GROWTH REGULATORY FACTOR (GRF)–GRF-INTERACTING FACOR (GIF), SWITCH/SUCROSE NON-FERMENTING (SWI/SNF), gibberellin (GA)–DELLA, KLU, and PEAPOD. We describe not only the components within each regulatory module, but also the connections between the modules, and how they are connected with the cell cycle, and, to a lesser extent, with the post-mitotic cell expansion machinery (Fig. 1; Table 1). The majority of the genes discussed throughout this review affect cell proliferation, demonstrating how the cell cycle and its machinery are central in mediating leaf growth. There are also many other aspects of leaf development including mechanisms controlling cell growth of dividing cells, such as leaf initiation (Ichihashi and Tsukaya, 2015; Sluis and Hake, 2015), leaf shape (Nikolov et al., 2019; Sapala et al., 2019), and the effect of environmental stress (Dubois et al., 2018). Many of the processes are governed by plant hormones (Du et al., 2018). However, to keep this review concise, emphasis is given to the regulation of cell division and to a lesser extent the contribution of post-mitotic cell expansion to leaf size.
Fig. 1.
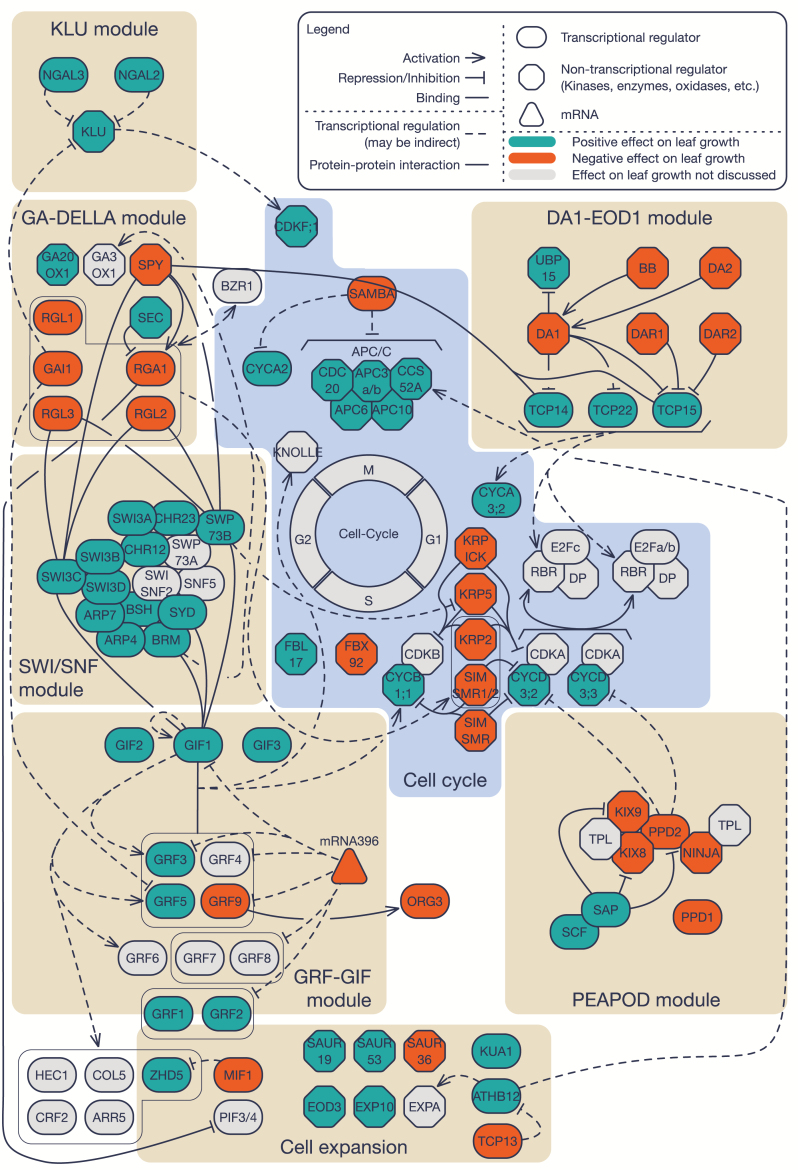
Overview of six of the gene regulatory modules known to be involved in cell proliferation and/or cell expansion: DA1–EOD1, GRF–GIF, SWI/SNF, GA–DELLA, KLU, and PEAPOD. The cell cycle is shown in the center and is surrounded by the core cell cycle proteins whose expression/activity is affected by one or more of the six regulatory modules. Proteins involved in cell expansion and their interaction with some of the modules are also indicated. Transcriptional (pill shapes) or non-transcriptional (octagonal shapes) regulators with a positive (teal blue) or negative (orange) effect on leaf growth are indicated. Gray proteins/transcriptional regulators are proteins whose effect on leaf growth is unknown or not presented in this review. The type of arrowhead indicates an activating (arrow) or repressing (T-junction) action, while absence of an arrowhead represents binding. These three actions are at either a transcriptional (dotted lines) or protein (solid lines) level. Abbreviations: APC/C (ANAPHASE PROMOTING COMPLEX/CYCLOSOME), ARP (ACTIN RELATED PROTEINS), ARR (ARABIDOPSIS THALIANA RESPONSE REGULATOR), ATHB (ARABIDOPSIS THALIANA HOMEOBOX), BB (BIG BROTHER), BRM (BRAHMA), BSH (BUSHY), BZR (BRASSINAZOLE RESISTANT), CCS52A (CELL-CYCLE SWITCH PROTEIN), CDC20 (CELL DIVISION CYCLE 20), CDK (CYCLIN DEPENDANT KINASE), COL5 (CONSTANS-LIKE 5), CRF2 (CYTOKININ RESPONSE FACTOR 2), CYC (CYCLIN), DAR (DA1-RELATED), DP (DIMERISATION PROTEIN), EOD (ENHANCER OF DA1), EXP (EXPANSIN), GA20OX1 (GIBBERELLIN 20-OXIDASE 1), GA3OX1 (GIBBERELLIN 3-OXIDASE 1), GAI1 (GA INSENSITIVE), GIF (GRF-INTERACTING FACTOR), GRF (GROWTH REGULATING FACTOR), HEC1 (HECATE 1), KIX (KINASE-INDUCIBLE DOMAIN INTERACTING), KRP/ICK (KIP-RELATED PROTEIN/INTERACTOR OF CDKs), KUA1 (KUODA1), MIF1 (MINI ZINC-FINGER 1), CHR (CHROMATIN REMODELING), NGAL (NGATHA-LIKE PROTEIN), NINJA (NOVEL INTERACTOR OF JAZ), ORG3 (OBP3-RESPONSIVE GENE 3), PIF (PHYTOCHROME INTERACTING FACTOR), PPD (PEAPOD), RBR (RETINOBLASTOMA-RELATED), RGA1 (REPRESSOR OF ga1-3), RGL (RGA-LIKE), SAP (STERILE APETALA), SAUR (SMALL AUXIN UP RNA), SCF (SKP1/CULLIN1/F-BOX PROTEIN), SEC (SECRET AGENT), SIM (SIAMESE), SMR (SIAMESE-RELATED), SNF5 (SUCROSE NON-FERMENTING 5), SPY (SPINDLY), SWI/SNF (SWITCH/SUCROSE NON-FERMENTING), SWI3 (SWITCH), SWP73 (SWI/SNF ASSOCIATED PROTEIN 73), SYD (SPLAYED), TCP (TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR), TPL (TOPLESS), UBP15 (UBIQUITIN SPECIFIC PROTEASE 15), ZHD5 (ZINC-FINGER HOMEODOMAIN 5).
Table 1.
AT code, gene name, and description of genes discussed or mentioned in this review as well as the module to which they belong
| Module | AT code | Gene name | Gene description |
|---|---|---|---|
| – | AT1G75080 | BZR1 | BRASSINAZOLE RESISTANT 1 |
| – | AT1G75950 | SKP1/ASK1/UIP1 | S PHASE KINASE-ASSOCIATED PROTEIN 1/ARABIDOPSIS SKP1 HOMOLOGUE 1/UFO INTERACTING PROTEIN 1 |
| – | AT3G48100 | ARR5/IBC6 | ARABIDOPSIS THALIANA RESPONSE REGULATOR 5/INDUCED BY CYTOKININ 6 |
| – | AT3G56980 | ORG3/BHLH039 | OBF-BINDING PROTEIN 3 (OBP3)-RESPONSIVE GENE 3/BASIC HELIX-LOOP-HELIX 39 |
| – | AT4G02570 | CUL1/ICU13 | CULLIN 1/INCURVATA 13 |
| – | AT4G18710 | BIN2 | BRASSINOSTEROID-INSENSITIVE 2 |
| – | AT4G23750 | CRF2/TMO3 | CYTOKININ RESPONSE FACTOR 2/TARGET OF MONOPTEROS 3 |
| – | AT5G57660 | COL5/BBX6 | CONSTANS-LIKE 5/B-BOX DOMAIN PROTEIN 6 |
| – | AT5G67060 | HEC1 | HECATE 1 |
| Cell expansion | AT1G09530 | PIF3 | PHYTOCHROME INTERACTING FACTOR 3 |
| Cell expansion | AT1G19840 | SAUR53 | SMALL AUXIN UPREGULATED RNA 53 |
| Cell expansion | AT1G26770 | EXP10 | EXPANSIN 10 |
| Cell expansion | AT1G74660 | MIF1 | MINI ZINC-FINGER 1 |
| Cell expansion | AT1G75240 | ZHD5/HB33 | ZINC-FINGER HOMEODOMAIN 5/HOMEOBOX PROTEIN 33 |
| Cell expansion | AT2G43010 | PIF4 | PHYTOCHROME INTERACTING FACTOR 4 |
| Cell expansion | AT2G45210 | SAUR36/SAG201 | SMALL AUXIN UPREGULATED 36/SENESCENCE-ASSOCIATED GENE 201 |
| Cell expansion | AT2G46660 | EOD3/CYP78A6 | ENHANCER OF DA-1 3/CYTOCHROME P450, FAMILY 78, SUBFAMILY A, POLYPEPTIDE 6 |
| Cell expansion | AT3G02150 | TCP13/PTF1 | TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR 13/PLASTID TRANSCRIPTION FACTOR 1 |
| Cell expansion | AT3G61890 | ATHB12 | ARABIDOPSIS THALIANA HOMEOBOX 12 |
| Cell expansion | AT5G18010 | SAUR19 | SMALL AUXIN UP RNA 19 |
| Cell expansion | AT5G47390 | KUA1/MYBH | KUODA1/MYB HYPOCOTYL ELONGATION-RELATED |
| Cell expansion | Gene group | PP2C | 2C PROTEIN PHOSPHATASE |
| Cell expansion | Gene group | EXPA | EXPANSIN A |
| Cell cycle machinery | AT1G08560 | KNOLLE/SYP111 | SYNTAXIN OF PLANTS 111 |
| Cell cycle machinery | AT1G32310 | SAMBA | SAMBA |
| Cell cycle machinery | AT3G07870 | FBX92 | F-BOX PROTEIN 92 |
| Cell cycle machinery | AT3G54650 | FBL17 | F-BOX LIKE 17 |
| Cell cycle machinery | Gene group | APC/C | ANAPHASE PROMOTING COMPLEX/CYCLOSOME |
| Cell cycle machinery | Gene group | CCS52A | CELL CYCLE SWITCH PROTEIN 52 A |
| Cell cycle machinery | Gene group | CDC20 | CELL DIVISION CYCLE 20 |
| Cell cycle machinery | Gene group | CDK | CYCLIN DEPENDANT KINASE |
| Cell cycle machinery | Gene group | CYC | CYCLIN |
| Cell cycle machinery | Gene group | DP | DIMERISATION PROTEIN |
| Cell cycle machinery | Gene group | KRP/ICK | KIP-RELATED PROTEIN/INTERACTOR OF CDKs |
| Cell cycle machinery | Gene group | RBR | RETINOBLASTOMA-RELATED |
| Cell cycle machinery | Gene group | SIM | SIAMESE |
| Cell cycle machinery | Gene group | SMR | SIAMESE-RELATED |
| DA1–EOD1 | AT1G14920 | GAI/RGA2 | GIBBERELLIC ACID INSENSITIVE/RESTORATION ON GROWTH ON AMMONIA 2 |
| DA1–EOD1 | AT1G15550 | GA3OX1 | GIBBERELLIN 3-OXIDASE 1 |
| DA1–EOD1 | AT1G17110 | UBP15/SOD2 | UBIQUITIN-SPECIFIC PROTEASE 15/SUPPRESSOR OF DA1 2 |
| DA1–EOD1 | AT1G19270 | DA1 | DA1 |
| DA1–EOD1 | AT1G69690 | TCP15 | TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR 15 |
| DA1–EOD1 | AT1G72010 | TCP22 | TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR 22 |
| DA1–EOD1 | AT1G78420 | DA2 | DA2 |
| DA1–EOD1 | AT2G39830 | DAR2 | DA1-RELATED PROTEIN 2 |
| DA1–EOD1 | AT3G47620 | TCP14 | TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR 14 |
| DA1–EOD1 | AT3G63530 | BB/EOD1 | BIG BROTHER/ENHANCER1 OF DA1 |
| DA1–EOD1 | AT4G25420 | GA20OX1 | GIBBERELLIN 20-OXIDASE 1 |
| DA1–EOD1 | AT4G36860 | DAR1 | DA1-RELATED PROTEIN 1 |
| GA–DELLA | AT1G66350 | RGL1 | RGA-LIKE 1 |
| GA–DELLA | AT2G01570 | RGA1 | REPRESSOR OF GA1-3 |
| GA–DELLA | AT3G03450 | RGL2 | RGA-LIKE 2 |
| GA–DELLA | AT3G04240 | SEC | SECRET AGENT |
| GA–DELLA | AT3G11540 | SPY | SPINDLY |
| GA–DELLA | AT4G24210 | SLY | SLEEPY 1 |
| GA–DELLA | AT5G17490 | RGL3 | RGA-LIKE PROTEIN 3 |
| GA–DELLA | GID2 | GIBBERELLIN INSENSITIVE DWARF 2 | |
| GRF–GIF | AT1G01160 | GIF2 | GRF1-INTERACTING FACTOR 2 |
| GRF–GIF | AT2G06200 | GRF6 | GROWTH-REGULATING FACTOR 6 |
| GRF–GIF | AT2G22840 | GRF1 | GROWTH-REGULATING FACTOR 1 |
| GRF–GIF | AT2G36400 | GRF3 | GROWTH-REGULATING FACTOR 3 |
| GRF–GIF | AT2G45480 | GRF9 | GROWTH-REGULATING FACTOR 9 |
| GRF–GIF | AT3G13960 | GRF5 | GROWTH-REGULATING FACTOR 5 |
| GRF–GIF | AT3G52910 | GRF4 | GROWTH-REGULATING FACTOR 4 |
| GRF–GIF | AT4G00850 | GIF3 | GRF1-INTERACTING FACTOR 3 |
| GRF–GIF | AT4G24150 | GRF8 | GROWTH-REGULATING FACTOR 8 |
| GRF–GIF | AT4G37740 | GRF2 | GROWTH-REGULATING FACTOR 2 |
| GRF–GIF | AT5G28640 | GIF1/AN3 | GRF1-INTERACTING FACTOR 1/ANGUSTIFOLIA 3 |
| GRF–GIF | AT5G53660 | GRF7 | GROWTH-REGULATING FACTOR 7 |
| KLU | AT1G13710 | KLU/CYP78A5 | KLUH/CYTOCHROME P450, FAMILY 78, SUBFAMILY A, POLYPEPTIDE 5 |
| KLU | AT3G11580 | NGAL2/SOD7 | NGATHA-LIKE PROTEIN 2/SUPRESSOR OF DA1 7 |
| KLU | AT5G06250 | NGAL3/DPA4 | NGATHA-LIKE PROTEIN 3/DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
| PPD | AT1G15750 | TPL/WSIP1 | TOPLESS/WUS-INTERACTING PROTEIN 1 |
| PPD | AT3G24150 | KIX8 | KINASE-INDUCIBLE DOMAIN INTERACTING 8 |
| PPD | AT4G14713 | PPD1 | PEAPOD 1 |
| PPD | AT4G14720 | PPD2 | PEAPOD 2 |
| PPD | AT4G28910 | NINJA | NOVEL INTERACTOR OF JAZ |
| PPD | AT4G32295 | KIX9 | KINASE-INDUCIBLE DOMAIN INTERACTING 9 |
| PPD | AT5G35770 | SAP/SOD3 | STERILE APETALA/SUPRESSOR OF DA1 3 |
| SWI/SNF | AT1G18450 | ARP4 | ACTIN-RELATED PROTEIN 4 |
| SWI/SNF | AT2G28290 | SYD/CHR3 | SPLAYED/CHROMATIN REMODELING COMPLEX SUBUNIT R 3 |
| SWI/SNF | AT2G46020 | BRM | BRAHMA |
| SWI/SNF | AT3G01890 | SWP73A/CHC2 | SWI/SNF ASSOCIATED PROTEIN 73 A |
| SWI/SNF | AT3G06010 | CHR12 | CHROMATIN REMODELING 12 |
| SWI/SNF | AT3G17590 | BSH | BUSHY GROWTH |
| SWI/SNF | AT3G17590 | SNF5 | SUCROSE NON-FERMENTING 5 |
| SWI/SNF | AT3G60830 | ARP7 | ACTIN-RELATED PROTEIN 7 |
| SWI/SNF | AT5G14170 | SWP73B/CHC1 | SWI/SNF ASSOCIATED PROTEIN 73 B |
| SWI/SNF | AT5G19310 | CHR23 | CHROMATIN REMODELING 23 |
| SWI/SNF | Gene group | SWI/SNF | SWITCH/SUCROSE NON-FERMENTING |
| SWI/SNF | Gene group | SWI3 | SWITCH |
Protein groups or families represent multiple genes and therefore have no AT code.
The pivotal role of the cell cycle machinery during leaf growth
During division, cells separate their duplicated genetic information into two daughter cells. This process, referred to as the cell cycle, can be subdivided into four phases: the S-phase during which the nuclear DNA is duplicated, the M-phase or mitosis during which the chromosomes are separated and distributed to the daughter cells, and two gap phases (G1 and G2) to prepare the cells for DNA replication or mitosis, respectively (Inzé and De Veylder, 2006). To ensure correct transmission of the genetic information, progression through these different phases is tightly controlled by different groups of core cell cycle proteins; the CYCLINS (CYCs) complexed with CYCLIN-DEPENDENT KINASES (CDKs), the E2F/DIMERISATION PROTEIN (DP) transcriptional regulatory proteins, KIP-RELATED PROTEIN/INTERACTOR OF CDKs (KRP/ICK), and SIAMESE/SIAMESE-RELATED (SIM/SMR) proteins (Inzé and De Veylder, 2006; Harashima et al., 2013).
In Arabidopsis, CYCs include A-type CYCs (CYCA), B-type CYCs (CYCB), and D-type CYCs (CYCD), while CDKs include A-type CDKs (CDKA) and B-type CDKs (CDKB), the latter being plant specific (Vandepoele et al., 2002; Inzé and De Veylder, 2006). The composition and activity of the CDK–CYC complexes are highly cell cycle phase specific, with CYCAs and CYCDs mainly involved in G1 progression and G1 to S transition, and CYCBs mainly regulating the G2 to M transition and progression through mitosis (Inzé and De Veylder, 2006; Van Leene et al., 2011; Zhao et al., 2012). In parallel, CDKAs are essential at both G1 to S and G2 to M phases, whereas CDKBs mainly control the G2 to M phase, progression through mitosis, and cell cycle exit (Inzé and De Veylder, 2006; Harashima et al., 2013). The expression of genes required for G1 to S transition and S-phase progression is predominantly controlled by three E2F proteins (E2Fa, E2Fb, and E2Fc) that form heterodimeric complexes with DP proteins (DPa and DPb) (Magyar et al., 2000; Kosugi and Ohashi, 2002; Desvoyes et al., 2006; Yao et al., 2018). While the E2Fc/DP complex is a transcriptional inhibitor, E2Fa/DP and E2Fb/DP complexes are transcriptional activators, whose activity is inhibited by binding to RETINOBLASTOMA-RELATED (RBR) proteins (Desvoyes et al., 2006; Inzé and De Veylder, 2006). During the G1 to S transition, CYCD proteins are predominantly complexed with CDKA;1 (Boruc et al., 2010; Van Leene et al., 2011) that binds and phosphorylates RBR proteins associated with the E2Fa-b/DP complex, causing RBR degradation (Huntley et al., 1998; Nakagami et al., 1999; del Pozo et al., 2006). The activated E2Fa-b/DP transcription factor complex triggers the expression of numerous target genes involved in cell cycle progression, transcription, chromatin dynamics, and DNA replication (Vandepoele et al., 2005; Yao et al., 2018). During transition between the G2- and M-phase, CDKA–CYCB complexes activate MYB3R proteins that in their turn activate several M-phase-related genes such as KNOLLE and CYCB1;1 itself, guiding cell cycle exit into mitosis (De Veylder et al., 2007). Alternatively, however, cells can continue to duplicate their genomic content (S-phase) for several rounds without subsequent division, called endoreduplication (Inzé and De Veylder, 2006; Breuer et al., 2014).
The activity of the CDK–CYC complexes is tightly regulated by multiple mechanisms, acting at a transcriptional and a mainly post-translational level (Inzé and De Veylder, 2006; De Veylder et al., 2007; Breuer et al., 2014; Edgar et al., 2014). These regulatory mechanisms include phosphorylation, interaction with cell cycle inhibitor proteins of the KRP/ICK and SIM/SMR family, and proteolysis mediated by the anaphase-promoting complex/cyclosome (APC/C) and the SKP1/CULLIN1/F-BOX PROTEIN (SCF) complexes (Inzé and De Veylder, 2006; Heyman and De Veylder, 2012). KRP/ICK proteins predominantly inhibit CDKA–CYCD complexes (Van Leene et al., 2010). In lines overexpressing KRP proteins, mitosis is hampered, leading to a drastic decrease in cell number that is partially compensated by an increase in cell size (De Veylder et al., 2001, 2011). While single krp mutants do not show drastic effects, triple (krp4/6/7), quadruple (krp1/2/6/7), and quintuple (krp1/2/5/6/7) krp mutants have longer and enlarged leaves, which are narrow and curled downwards as a result of an increased cell number (Cheng et al., 2013). A septuple krp mutant, in which all seven KRP/ICK genes are inactivated, produces leaves with an increased leaf size, similar to that in the quintuple krp mutant (Cao et al., 2018). The SIM/SMR proteins inhibit CDKA–CYCD and CDKB–CYCB complexes, blocking progression through the cell cycle and promoting endoreduplication (Walker et al., 2000; Churchman et al., 2006; Van Leene et al., 2010). Although sim mutants do not have an altered leaf phenotype, they have multicellular and clustered trichomes, and SIM-overexpressing plants are dramatically reduced in size (Walker et al., 2000; Churchman et al., 2006; Kumar et al., 2015). The APC/C complex is a multiple subunit E3 ligase that controls cell cycle progression and endocycle entry, and altered expression levels of APC/C complex members, their activators, or their inhibitors impair plant morphology. APC10 is an essential component of the APC/C complex and, upon APC10 overexpression, epidermal cells divide more quickly owing to a faster degradation of the mitotic cyclin CYCB1;1, resulting in the formation of enlarged leaves (Eloy et al., 2011). Down-regulation of APC10 or APC6, encoding another APC/C subunit, results in the production of smaller and curled leaves that show a reduced cell area (Marrocco et al., 2009). In Arabidopsis, two isoforms exist for the APC/C subunit APC3: APC3a/CDC27a and APC3b/HOBBIT (Heyman and De Veylder, 2012). These proteins act with APC10 as receptors for the APC/C activators CELL CYCLE SWITCH PROTIEN 52 A/B (CCS52A/B) and CELL DIVISION CYCLE 20 (CDC20) (Fülöp et al., 2005; Eloy et al., 2011; Kevei et al., 2011; Breuer et al., 2012). Plants highly overexpressing CCS52A have a reduced leaf area as a result of a decreased cell number, slightly compensated by an increased cell area. Milder overexpression of CCS52A, however, results in larger plants because of increased cell divisions during the early stages of leaf development (Baloban et al., 2013). Overexpression of APC3a/CDC27a increases leaf size owing to an increased cell number, whereas plants in which the expression of APC3b/HOBBIT is down-regulated are extremely dwarfed (Willemsen et al., 1998; Heyman and De Veylder, 2012). APC/C is negatively regulated by SAMBA. Loss-of-function mutation in SAMBA (samba) results in plants that produce a larger shoot apical meristem, larger leaf primordia, and enlarged mature leaves, proposed to result, at least partially, from an increase in leaf primordium founder cells (Eloy et al., 2012). SAMBA targets mitotic cyclins such as CYCLIN A2 (CYCA2) for APC/C-mediated degradation and eventually cell cycle exit (Eloy et al., 2012). Accordingly, CYCA2s are stabilized in samba mutants throughout early leaf development, stimulating cell division (Eloy et al., 2012).
F-box proteins are a major type of E3 ligases some of which are involved in cell cycle control, marking proteins for ubiquitin-mediated proteasomal degradation (Skaar et al., 2013). Recently, it was described that overexpression of F-BOX PROTEIN 92 (AtFBX92) results in the formation of smaller leaves as a result of a decreased cell number, though slightly compensated by an increased cell size (Baute et al., 2017). Conversely, plants with a decreased expression of AtFBX92 (amiFBX92) exhibited larger leaves, resulting from an increased cell division rate (Baute et al., 2017). In addition, the F-box protein F-BOX LIKE 17 (FBL17) was characterized as a positive growth regulator, because fbl17 mutants display a drastic reduction in leaf area due to a decrease in cell number compared with wild-type plants (Noir et al., 2015).
The DA1–EOD1 module
The DA1–EOD1 module has an important role in controlling leaf growth by regulating several key growth regulatory proteins in a post-translational manner. Plants with a dominant-negative point mutation in the gene encoding peptidase DA1 (da1-1) display enlarged leaves that contain more cells owing to a prolonged cell proliferation phase (Li et al., 2008; Dong et al., 2017; Vanhaeren et al., 2017). In these plants, not only is the leaf area increased, but also the size of flowers, fruits, and seeds. In contrast, a decreased leaf size is observed upon overexpression of GFP-DA1, probably because DA1 is stabilized by the fluorescent tag, demonstrating that DA1 is a negative regulator of leaf growth (Vanhaeren et al., 2017).
The peptidase activity of DA1 is activated upon multiple mono-ubiquitination by the E3 ligases BIG BROTHER/ENHANCER OF DA1 (BB/EOD1; referred to from hereon as BB) and DA2 (Xia et al., 2013; Dong et al., 2017). BB mutants (bb-1) exhibit smaller but shorter leaves, leaving total leaf area unchanged, and larger floral organs (Disch et al., 2006). Overexpression of BB in the bb-1 mutant background decreases leaf size drastically by restricting cell proliferation duration (Disch et al., 2006). Plants in which DA2 is mutated (da2-1) display larger leaves and have an increased biomass compared with the wild type, whereas overexpressing lines form smaller plants with a decreased leaf area (Xia et al., 2013). While overexpression of BB or DA2 dramatically decreases leaf size (Disch et al., 2006; Xia et al., 2013), bb and da2 mutations in the da1-1 mutant background synergistically enhance the da1-1 phenotype (Li et al., 2008; Xia et al., 2013; Dong et al., 2017; Vanhaeren et al., 2017).
Several targets of DA1 have so far been described. Among others, DA1 negatively regulates the stability of the deubiquitinating enzyme SUPPRESSOR OF DA1 2/UBIQUITIN SPECIFIC PROTEASE 15 (SOD2/UBP15, referred to from hereon as UBP15) (Liu et al., 2008; Du et al., 2014; Dong et al., 2017). Overexpression of UBP15 leads to the formation of larger leaves, roots, flowers, and seeds as a result of increased cell divisions, mimicking the da1-1 mutant phenotype (Liu et al., 2008; Du et al., 2014). In agreement with this, ubp15-1 mutants have smaller organs compared with the wild type (Liu et al., 2008; Du et al., 2014), and the da1-1 enlarged seed phenotype is repressed in da1-1/ubp15 double mutants (Du et al., 2014). In addition to UBP15, DA1 also inactivates TEOSINTE BRANCHED 1/CYCLOIDEA/PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR 14 (TCP14), TCP15, and TCP22, transcription factors that positively regulate cell division duration (Dong et al., 2017). More specifically, TCP14 and TCP15 repress the transition from mitosis to endoreduplication by inducing the expression of RBR and CYCA3;2 (Li et al., 2012; Peng et al., 2015). The stability of TCP14 and TCP15 is modulated not only by DA1, but also by its close family members DA1-RELATED 1 (DAR1) and DAR2 (Peng et al., 2015). Nonetheless, whereas the da1-ko/dar1-1/dar2-1 triple mutant produces enlarged flowers and seeds, leaf size is decreased compared with wild-type plants, suggesting that DA1, DAR1, and DAR2 may regulate plant growth and development in an organ-specific manner (Peng et al., 2015).
The GRF–GIF module
The GRF–GIF module plays an important role in cell number determination in leaves. It consists of several interacting proteins of which ANGUSTIFOLIA3/GRF-INTERACTING FACTOR 1 (AN3/GIF1, referred to from hereon as GIF1) and members of the GROWTH REGULATING FACTOR (GRF) are transcriptional regulators (Kim and Kende, 2004; Debernardi et al., 2014). The three GIF family members, GIF1, GIF2, and GIF3, are transcriptional co-activators that act, at least partially, redundantly to activate cell proliferation in the leaf primordia (Kim and Kende, 2004; Horiguchi et al., 2005). Overexpression of GIF1 results in plants that form enlarged organs resulting from an increased cell proliferation, reflected by an increased expression of CYCB1;1 and other cell cycle-related genes (Lee et al., 2009). In contrast, gif1 mutants display smaller and narrower leaves that contain fewer cells compared with the wild type (Kim and Kende, 2004; Horiguchi et al., 2005; Lee et al., 2009). Accordingly, overexpression of GIF2 and GIF3 also increases leaf size by an increased cell number, demonstrating that GIF proteins act as positive regulators of cell proliferation (Lee et al., 2009). Recently, it was shown that GIF1 might act as a mobile growth factor that diffuses through the leaf using plasmodesmata and, as such, establishes a long-range gradient along the leaf proximal–distal axis to determine the cell proliferation domain (Kawade et al., 2017).
GIF1 was shown to interact with six out of the nine members comprising the GRF protein family in Arabidopsis: GRF1, GRF2, GRF3, GRF4, GRF5, and GRF9 (Kim and Kende, 2004; Horiguchi et al., 2005; Debernardi et al., 2014; Vercruyssen et al., 2015). Overexpression of GRF5 results in larger organs owing to an increased cell number, whereas down-regulation of GRF5 results in the formation of narrower leaves that contain fewer cells (Horiguchi et al., 2005; Kim and Tsukaya, 2015; Vercruyssen et al., 2015). Several other members of the GRF family are also positive regulators of growth, such as GRF1 and GRF2, of which overexpression results in the formation of larger leaves (Kim and Tsukaya, 2015; Omidbakhshfard et al., 2015). In contrast, however, GRF9 negatively regulates leaf growth, since overexpression of GRF9 decreases organ size and the grf9 mutant produces bigger leaf primordia, rosette leaves, and petals, resulting from an increased cell proliferation compared with wild-type plants (Omidbakhshfard et al., 2018). GRF9 acts as a growth repressor by activating the expression of OBF-BINDING PROTEIN 3 (OBP3)-RESPONSIVE GENE 3 (ORG3/bHLH039, referred to from hereon as ORG3), which encodes a basic LEUCINE-ZIPPER (bZIP) transcription factor (Omidbakhshfard et al., 2018). Whereas org3 loss-of-function mutants produce leaves with an increased area as a result of an increased cell number compared with wild-type plants, the opposite phenotype is observed in plants overexpressing ORG3 (Omidbakhshfard et al., 2018). Consistent with the genetic interaction between GRF9 and ORG3, the decreased leaf area in plants overexpressing GRF9 (GRF9ox) is completely restored in GRF9ox/org3 double mutants (Omidbakhshfard et al., 2018). Several downstream target genes of GIF1 have been identified so far, including GIF1 itself, GRF3, GRF5, GRF6, TARGET OF MONOPTEROS 3/CYTOKININ RESPONSE FACTOR 2 (TMO3/CRF2), B-BOX DOMAIN PROTEIN 6/CONSTANS-LIKE 5 (BBX6/COL5), HECATE (HEC1), ZINC-FINGER HOMEODOMAIN 5/HOMEOBOX PROTEIN 33 (ZHD5/HB33, referred to from hereon as ZHD5), and ARABIDOSPIS THALIANA RESPONSE REGULATOR 5 (ARR5) (Vercruyssen et al., 2014).
Except for GRF5 and GRF6, GRF family members are regulated at the transcript level by miR396-mediated RNA cleavage (Liu et al., 2009; Rodriguez et al., 2010; Debernardi et al., 2014). miR396 expression increases throughout leaf development in a basipetal direction, following the cell cycle arrest front, restricting GRF expression to the basal part of the leaf (Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). Since the balance between GRF genes and miR396 regulates cell number in a quantitative manner, miR396-overexpressing plants produce small and narrow leaves containing fewer cells owing to a shorter cell proliferation phase (Liu et al., 2009; Rodriguez et al., 2010; Wang et al., 2011). In contrast, overexpression of a miR396-resistant version of GRF3 (rGRF3) prolongs cell proliferation, resulting in the formation of larger leaves that contain more cells (Debernardi et al., 2014).
The SWI/SNF chromatin remodeling module
The SWITCH/SUCROSE NON-FERMENTING (SWI/SNF) chromatin remodeling complex is closely linked with the GRF–GIF module and can activate and/or repress transcription by disrupting DNA–histone interactions, thereby altering chromatin accessibility (Han et al., 2015; Archacki et al., 2017). The SWI/SNF complex comprises a functional core including a SWI2/SNF2 ATPase family member, BRAHMA (BRM), SPLAYED (SYD), CHROMATIN REMODELING 12 (CHR12) or CHR23 (Han et al., 2015), an SNF5 subunit, BUSHY (BSH), two SWI/SNF ASSOCIATED PROTEINS 73 (SWP73A/CHC2 and SWP73B), two ACTIN RELATED PROTEINS (ARP4 and ARP7), and a pair of SWI3 subunits: SWI3A, SWI3B, SWI3C, or SWI3D (Vercruyssen et al., 2014). SWI/SNF subunits are important for transcriptional regulation of key developmental processes (Wagner and Meyerowitz, 2002; Farrona et al., 2004; Hurtado et al., 2006; Kwon et al., 2006). Loss of function in the double knockout CHR12/CHR23 (minu1/minu2), SWI3A, SWI3B, or ARP7 causes embryonic lethality. Whereas plants with a single mutation in BRM, SYD, SWI3C, or SWI3D or showing silencing of BSH, SWP73B, or ARP4 do manage to develop, they display severe embryonal defects with limited leaf and flower development, often resulting in sterility (Kandasamy et al., 2005a, b; Sarnowski et al., 2005; Sang et al., 2012; Sacharowski et al., 2015). The brm mutant exhibits pleiotropic phenotypic alterations, resulting in an overall reduced plant size accompanied by a downward curling of the leaves (Farrona et al., 2004; Hurtado et al., 2006; Tang et al., 2008). Furthermore, overexpression of SWI3C enhances leaf growth by increasing the number of cells (Vercruyssen et al., 2014), whereas swi3c mutants display small rosettes constituted of curled leaves (Sacharowski et al., 2015). GIF1 associates with the SWI/SNF complex through several subunits, including BRM, SYD, and SWP73B, to induce transcription of several downstream cell cycle-related genes (Vercruyssen et al., 2014).
The GA–DELLA module
Gibberellins (GAs) play an important role in both cell proliferation and cell expansion, and mutations in genes involved in GA signaling or homeostasis can drastically affect plant organ size (Achard et al., 2009). Overexpression of GIBBERELLIN 20-OXIDASE 1 (GA20OX1), encoding a rate-limiting enzyme essential for GA biosynthesis, results in increased levels of active GA, leading to the formation of enlarged leaves that contain more and larger cells (Coles et al., 1999; Gonzalez et al., 2010). In contrast, plants with reduced GA levels or a reduced GA sensitivity display a dwarfed phenotype (Olszewski et al., 2002). In Arabidopsis, there are five DELLA proteins; GA INSENSITIVE (GAI), REPRESSOR OF ga1-3 (RGA), RGA-LIKE 1 (RGL1), RGL2, and RGL3. All five DELLA proteins function as key repressors of GA-responsive growth, inhibiting GA-regulated gene expression (Sun and Gubler, 2004; de Lucas et al., 2008). GA binds to the GIBBERELLIN INSENSITIVE DWARF 2 (GID2) receptor, which causes ubiquitination of the DELLA proteins, marking them for protein degradation with the help of F-box protein SLEEPY 1 (SLY1) and the SCFSLY1/GID2 E3 ligase complex (McGinnis et al., 2003; Dill et al., 2004; Ueguchi-Tanaka et al., 2007). Plants in which DELLA proteins are stabilized (sleepy1), which are GA deficient (ga1-3), or in which SLY is mutated (sly1-10), show a dwarfed phenotype (Olszewski et al., 2002; Dill et al., 2004; Fu et al., 2004). In contrast, the quadruple DELLA mutant (gai-t6/rga-t2/rgl1-1/rgl2-1), mimicking constitutive GA signaling, displays increased cell division rates, and consequently larger leaves (Achard et al., 2009).
To regulate transcription, DELLA proteins exert their inhibiting function through protein–protein interactions with other transcriptional regulators (de Lucas et al., 2008). Among others, RGA is known to interact with and inhibit the transcriptional activity of PHYTOCRHOME INTERACTING FACTOR 3 (PIF3) and PIF4, basic HELIX-LOOP-HELIX (bHLH) factors involved in light signaling and mediators of cell elongation (de Lucas et al., 2008). Further downstream, DELLA proteins promote the expression of the cell cycle inhibitor-encoding genes KRP2, SIM, SMR1, and SMR2, because the expression of these cell cycle genes is elevated in GA-deficient plants, suggesting that the resulting dwarfed phenotype is caused by inhibition of the cell cycle (Achard et al., 2009). In addition to their involvement in the GA pathway, DELLA proteins are linked to the brassinosteroid pathway, since they regulate and are regulated by BRASSINAZOLE RESISTANT 1 (BZR1), which in its turn is inhibited by BRASSINOSTEROID INSENSITIVE 2 (BIN2), known to positively affect cell proliferation. Furthermore, DELLA proteins are regulated through protein modification by SECRET AGENT (SEC) and SPINDLY (SPY) (Zentella et al., 2017). SEC acts as a positive growth regulator by inducing a closed conformation of the DELLA protein RGA1 through the addition of O-β-N-acetylglucosamine, inhibiting the repressor activity of RGA1 (Zentella et al., 2017). The loss-of-function mutant sec-2 displays a reduction in leaf length compared with wild-type plants (Hartweck et al., 2006). In contrast, SPY acts as a negative regulator of growth by enhancing the capacity of RGA1 to bind to PIF3, PIF4, and BZR1 (Zentella et al., 2017). Reduced SPY activity partially suppresses the dwarfed phenotype caused by ga1 that lacks an early GA biosynthesis enzyme (Filardo and Swain, 2003). In contrast, mutations in SEC do not reverse the dwarfed phenotypes in a ga1 background, demonstrating that its role might be GA signaling specific (Hartweck et al., 2006).
The KLU module
KLU/KLUH/CYP78A5 (referred to from hereon as KLU) is a plant-specific cytochrome P450 protein belonging to the CYP78A subfamily. The CYP78A subfamily consists of six members in Arabidopsis termed CYP78A5–CYP78A10, and stimulates cell proliferation during leaf, flower, seed, and fruit development (Anastasiou et al., 2007; Adamski et al., 2009; Eriksson et al., 2010). It is proposed that KLU stimulates cell proliferation in a non-cell-autonomous manner, either by producing a mobile growth-promoting molecule or by degrading a, so far unknown, growth-inhibiting signal (Anastasiou et al., 2007; Eriksson et al., 2010). Loss of KLU function also shortens the time between successive leaf initiation events, referred to as the plastochron, leading to an increased final leaf number (Wang et al., 2008). Accordingly, KLU is expressed at the boundary between the shoot apical meristem and developing organ primordia, further strengthening its putative role in leaf initiation (Zondlo and Irish, 1999). KLU is proposed to stimulate cell proliferation, at least to some extent, redundantly with the closely related protein CYP78A7, because the loss-of-function cyp78a7 mutant does not show a clear phenotype, whereas seedlings of cyp78a5/cyp78a7 double mutants are smaller compared with wild-type plants (Wang et al., 2008).
The expression of KLU is repressed by SUPPRESSOR OF DA1-1 7/NGATHA-LIKE PROTEIN 2 (SOD7/NGAL2), a B3 transcription factor that binds directly to the KLU promoter (Zhang et al., 2015). Accordingly, the smaller leaf phenotype in the dominant sod7-1D mutant may directly result from an increased expression of KLU, though largely unexplored so far. The closest homolog of NGAL2, DEVELOPMENT-RELATED PcG TARGET IN THE APEX4 (DPA4)/NGAL3, also seems to regulate plant size since, in the absence of DPA4/NGAL3, leaves appear smaller as a result of a decreased cell number compared with the wild type (Zhang et al., 2015). Additionally, KLU is regulated by the DELLA protein GAI1, which may link the KLU module with the GA–DELLA module, although this is largely underexplored so far (Claeys et al., 2014).
The PEAPOD module
In the epidermis of Arabidopsis leaves, 48% of the pavement cells are estimated to originate from the repeating asymmetric divisions of meristemoids, stem cell-like precursor cells of the stomatal lineage (Larkin et al., 1997; Geisler et al., 2000). Consequently, the extent of meristemoid division also contributes significantly to final leaf size (White, 2006; Gonzalez et al., 2015). Meristemoid asymmetric division is negatively regulated by PEAPOD 1 (PPD1) and PPD2, putative DNA-binding proteins that belong to the TIFY protein family, a plant-specific group of proteins with a broad range of functions (White, 2006; Zhang et al., 2012; Gonzalez et al., 2015). Landsberg erecta (Ler) plants in which the PPD locus is deleted (∆ppd) and Col-0 plants expressing an artificial miRNA targeting the PPD transcripts (ami-ppd) both form enlarged rosettes with enlarged dome-shaped leaves that contain more cells owing to an increased meristemoid division compared with wild-type leaves (White, 2006; Gonzalez et al., 2015). In contrast, overexpression of the PPD genes results in the formation of leaves that are smaller and flatter, containing fewer cells compared with wild-type leaves (White, 2006).
PPD proteins interact with KINASE-INDUCIBLE DOMAIN INTERACTING 8 (KIX8), KIX9 and NOVEL INTERACTOR OF JAZ (NINJA), acting as adaptor proteins for the co-repressor TOPLESS (TPL) (Gonzalez et al., 2015; Baekelandt et al., 2018). The kix8/kix9 double mutant phenocopies both the ami-ppd leaf size and shape, suggesting that KIX8 and KIX9 act in a redundant manner and are pivotal for PPD functionality (Gonzalez et al., 2015). Whereas ninja mutants also show dome-shaped leaves, they lack the leaf size increase observed in ami-ppd and kix8/kix9 plants (Baekelandt et al., 2018). PPD2 is known to bind to the promoters of two out of the three D3-type CYCLIN genes, CYCD3;2 and CYCD3;3, repressing their transcription, and, accordingly, the expression of CYCD3;2 and CYCD3;3 is increased in ami-ppd, kix8/kix9, and ninja leaves compared with the wild type (Gonzalez et al., 2015; Baekelandt et al., 2018). Interestingly, meristemoid initiation and activity are reduced in the cycd3;1/cycd3;2/cycd3;3 triple mutant compared with the wild type (Dewitte et al., 2007; Elsner et al., 2012; Lau et al., 2014). More recently, it has been shown that plants overexpressing CYCD3;2 display propeller-like rosettes with narrow dome-shaped leaves, though lacking the leaf size increase observed in ppd and kix8/kix9 mutants (Baekelandt et al., 2018). Down-regulation of the expression of two out of the three CYCD3 genes, CYCD3;1 and CYCD3;2, can partially complement the ami-ppd leaf curvature phenotype, suggesting that CYCD3 genes are direct PPD2 target genes involved in controlling leaf shape (Baekelandt et al., 2018). In contrast, overexpression of CYCD3;3 does not affect leaf shape, but results in an overall reduced growth, indicative that, though considered to act redundantly, CYCD3 proteins may have specific functions during leaf shape control (Baekelandt et al., 2018).
In Arabidopsis, the activity of the PPD/KIX complex is regulated by the SCF complex containing the F-box protein STERILE APETALA/SUPPRESSOR OF DA1 3 (SAP/SOD3, referred to from hereon as SAP) (Wang et al., 2016; Li et al., 2018). Polyubiquitination of the PPD/KIX complex by SCFSAP results in proteasome-dependent degradation of the protein complex (Wang et al., 2016; Li et al., 2018). Consistently, Arabidopsis plants overexpressing SAP produce enlarged leaves with uneven lamina growth and have an increased expression of the PPD/KIX downstream target genes CYCD3;2 and CYCD3;3 compared with wild-type plants (Wang et al., 2016; Li et al., 2018).
Connecting the growth regulatory modules with the cell cycle
During recent years, more and more studies demonstrate that the six growth regulatory modules discussed here do not operate independently, and several links between the different modules and with the core cell cycle machinery have already been discussed (Fig. 1). DA1-mediated proteolysis of TCP14/15/22 results in the induction of CYCA3;2 and RBR expression, whereas the PPD module regulates CYCD3;2 and CYCD3;3 expression (Baekelandt et al., 2018), demonstrating that both modules regulate the G1/S transition of the cell cycle. Furthermore, the SWP73B subunit of the SWI/SNF complex is known to bind to the promoter of KRP5, encoding a cell cycle inhibitor that regulates endoreduplication and interacts with D-type CYCs, thereby also regulating the G1/S transition (Jégu et al., 2013). Also the downstream target genes of the GRF transcription factors include many cell cycle-related genes, such as KNOLLE, which is active during the M-phase, when cell plate formation occurs (Lauber et al., 1997; Touihri et al., 2011), and CYCB1;1, pivotal for the G2/M transition (Debernardi et al., 2014; Vercruyssen et al., 2014). Additionally, inducible KLU overexpression in the klu-2 mutant background causes up-regulation of CDKF;1, a CDK-ACTIVATING KINASE (CAK) affecting the activity of the CDK/CYC complexes throughout the cell cycle by phosphorylation (Umeda et al., 2005; Takatsuka et al., 2009). Plants lacking functional CDKF;1 exhibit a dwarfed phenotype because of a decreased cell number and cell size (Takatsuka et al., 2009). Finally, DELLA proteins activate the expression of several genes encoding cell cycle inhibitors, such as KRP2, SIM, SMR1, and SMR2, that are responsible for the onset of endoreduplication and as such contribute to the balance between cell proliferation and endoreduplication during leaf development (Achard et al., 2009; Kumar et al., 2015).
In addition to the direct connections with the cell cycle, several interactions between the members of different regulatory modules have been described. The SWI/SNF and the GA–DELLA modules are directly connected through SWI3C, a subunit of the SWI/SNF complex, that interacts with the DELLA proteins RGL2 and RGL3, and the DELLA regulatory protein SPY (Sarnowska et al., 2013). Furthermore, SPY is known to physically interact with TCP14 and TCP15, which are degraded in a DA1-dependent manner and repressed by DELLA proteins, connecting the SWI/SNF, GA–DELLA, and DA1–EOD1 modules (Steiner et al., 2012; Davière et al., 2014; Resentini et al., 2015). Additionally, the BRM subunit was found to bind to the promoters of GA3ox1 (Sarnowska et al., 2013; Archacki et al., 2016), affecting GA biosynthesis. The GRF–GIF and SWI/SNF modules are also closely connected, because GIF1 associates with the SWI/SNF complex through several subunits, including BRM, SYD, and SWP73B, to induce the expression of the downstream target genes (Vercruyssen et al., 2014). Finally, upon expression of an inducible non-degradable form of GAI in proliferating leaf cells, GRF5 and KLU transcripts are decreased, putatively linking the GA–DELLA, KLU, and GRF–GIF modules (Claeys et al., 2014).
Phenotypic effects observed upon misexpression of individual members of distinct modules may also be balanced at the leaf level. For instance, whereas the DA1–EOD1 module predominantly affects the primary arrest front, the PPD module is mainly involved in establishing the secondary arrest front (Gonzalez et al., 2012). Taken together, both are involved in determining cell proliferation, and therefore cell number and final leaf size. In agreement, at least two SOD mutants were identified in forward genetic screens that could so far not be directly linked with the DA1–EOD1 module: SAP that is part of the PPD module and NGAL2 that is part of the KLU module (Zhang et al., 2015; Wang et al., 2016). In both cases, it seems that the da1-1 phenotype can be complemented by affecting distinct core cell cycle genes or impinging on different processes of leaf development.
The importance of post-mitotic cell expansion for leaf growth
Besides cell proliferation, cell expansion contributes significantly to final leaf size, and a close coordination between cell proliferation and cell expansion is fundamental for proper organogenesis (Andriankaja et al., 2012). Cell expansion is proposed to be predominantly regulated by EXPANSINs (EXPs), XYLOGLUCAN ENDOTRANSGLUCOSEYLASE/HYDROLASEs (XTHs), PECTIN METHYLESTERASEs (PMEs), and reactive oxygen species (ROS) (Cosgrove, 2015; Schmidt et al., 2016). Auxin-induced acidification of the apoplast by ATPases importing H+ ions results in the activation of cell wall-associated EXPs that facilitate cell wall loosening (Cosgrove, 2000, 2005). Plants ectopically expressing EXP10 display larger leaves and longer petioles containing larger cells, whereas down-regulation of EXP10 has the inverse effect (Cosgrove, 2015). Also, SMALL AUXIN UP RNA (SAUR)-type proteins are proposed to promote ATPase activity by inhibiting 2C protein phosphatase (PP2C) proteins, resulting in the acidification of the apoplast and stimulating cell expansion (Chae et al., 2012; Hou et al., 2013). Plants ectopically expressing green fluorescent protein (GFP)-stabilized SAUR19 protein display an increased leaf area owing to the production of larger cells (Spartz et al., 2012, 2014). In contrast, saur36 mutants produce bigger leaves containing larger cells, demonstrating that SAUR36 acts as a negative regulator of cell expansion (Hou et al., 2013). Furthermore, SAUR53 has also been identified to positively regulate cell elongation, because ectopic expression of SAUR53 results in the elongation of cells and organs (Kathare et al., 2018). Another link between auxin and cell expansion was demonstrated by Katano et al. (2016). They showed that in fugu5 mutants, lacking the AVP1-encoded H+-pyrophosphatase, cell division is inhibited, thus triggering auxin-induced compensated cell expansion (Katano et al., 2016).
Besides EXP10 and several members of the SAUR family, only few other proteins have been described to impinge on the cell expansion phase, including GRF1, GRF2, EOD3/CYP78A6, ZHD5, KUODA 1 (KUA1), and ARABIDOPSIS THALIANA HOMEOBOX 12 (ATHB12) (Hong et al., 2011; Fang et al., 2012; Lu et al., 2014; Hur et al., 2015; Omidbakhshfard et al., 2015; Tsukaya, 2015). In contrast to the increased cell numbers in plants overexpressing GRF5 or GRF9, the increased leaf area in GRF1- and GRF2-overexpressing plants results from an increased cell area (Lee et al., 2009; Omidbakhshfard et al., 2015). Also in plants overexpressing EOD3, encoding a cytochrome P450 similar to KLU, seeds and leaves are bigger as a result of increased cell expansion, whereas EOD3 down-regulation leads to smaller leaves that consist of smaller cells (Fang et al., 2012). Also the transcriptional regulators ZHD5, KUA1, and ATHB12 positively regulate leaf growth, and their overexpression results in larger leaves and seeds owing to an increased cell area compared with the wild type (Hong et al., 2011; Fang et al., 2012; Lu et al., 2014; Hur et al., 2015). ZHD5 is part of the zinc-finger homeodomain (ZF-HD) class of transcription factors, which comprises 14 members in Arabidopsis that can homo- and heterodimerize (Tan and Irish, 2006; Hu et al., 2008). ZHD5 activity can be abolished by MINI ZINC-FINGER 1 (MIF1), which also contains a zinc-finger domain but lacks a DNA-binding domain (Hu and Ma, 2006; Hong et al., 2011). In this way, MIF1 acts as a competitive inhibitor peptide and, upon overexpression, blocks binding of ZHD5 to the DNA, resulting in dwarfed plants (Hu and Ma, 2006; Hong et al., 2011). KUA1 encodes a MYB-like transcription factor that positively regulates leaf growth by promoting cell wall relaxation (Lu et al., 2014; Schmidt et al., 2016). ATHB12 is involved in cell expansion as well as ploidy determination, since overexpression of ATHB12 induces the expression of CCS52A and CCS52B, encoding components of the APC/C complex, regulating endoreduplication onset, as well as the expression of EXPA, involved in cell expansion (Hur et al., 2015). Recently, TCP13 was found to repress ATHB12 expression, and overexpression of TCP13 results in a decreased leaf length and size owing to a reduction in cell size (Hur et al., 2019). Similarly, down-regulation of TCP13 and its paralogs, TCP5 and TCP17, results in enlarged leaf cells, suggesting that TCP13 regulates cell expansion through transcriptional control of ATHB12 (Hur et al., 2019).
The alterations in organ size in mutants with an impaired cell division are often not as pronounced as one would expect based on the reduction in cell numbers (Ferjani et al., 2007; Horiguchi and Tsukaya, 2011). This is because inhibition of cell division in organs with determinate growth, such as leaves, is often compensated by excessive post-mitotic cell expansion, a phenomenon called compensation (Hisanaga et al., 2015). Interestingly, such compensatory mechanisms often occur in mutants of core cell cycle genes (Blomme et al., 2014). For instance, the decreased cell number in the triple cycd3 mutant is compensated by an increased cell area (Dewitte et al., 2007). Also, gif1 mutants and plants overexpressing KRP2 show only a slight decrease in leaf area, because the decrease in cell number is partially restored by an increase in cell size (Mizukami and Fischer, 2000; De Veylder et al., 2001; Horiguchi et al., 2005; Kawade et al., 2010). Analogously, the increased cell number in plants that ectopically express E2Fa is partially restored by a decreased cell size, resulting in the formation of slightly enlarged cotyledons and leaves (De Veylder et al., 2002). Altogether, these findings strengthen the putative presence of complex interactions between cell division and cell expansion, coordinated by distinct mechanisms (Ferjani et al., 2007; Horiguchi and Tsukaya, 2011). In this way, inhibition of one process may, at least partially, be restored by an increased activity of another process to ensure that the genetically determined size is attained as well as possible (Horiguchi et al., 2006; Horiguchi and Tsukaya, 2011; Hisanaga et al., 2015). The underlying molecular mechanisms, however, are often still largely underexplored (Ferjani et al., 2007; Horiguchi and Tsukaya, 2011; Hisanaga et al., 2015).
Concluding remarks
In this review, we presented six modules that are important for Arabidopsis leaf size determination and showed that for most of them, direct links with the cell cycle machinery have been revealed. In addition, we demonstrate that connections between these different modules are revealed with an increasing pace. This demonstrates that the modules described throughout this review do not stand on their own, but that leaf growth is an intricate process that requires the cooperation of various interconnected key players that are part of complex regulatory networks. In the future, additional work will be required to further complete our view on these regulatory networks and the connections residing therein. There are also many genes affecting leaf size that were not presented in this review, largely because there are, to our knowledge, so far no links with any of the modules discussed here. In the future, more research will be required to also map these regulators in the bigger network of leaf growth regulation. Ultimately, mathematical modeling may enable us to fully grasp the complexity of the organ growth machinery.
Acknowledgements
The growth regulatory machinery is immensely complex and the authors apologize for not having cited all the relevant work in this field. The authors would like to thank Annick Bleys for proofreading and submitting this manuscript, as well as the present and former members of the System Biology of Yield group for fruitful discussion and contributions. This work was supported by Ghent University (BOF-Methusalem projects BOFMET2015000201 & B/09140/02) and by the Research Foundation Flanders (FWO research project 3G038719).
Author contributions
JV and AB wrote the manuscript, and NG and DI supervised and complemented the writing.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. 2009. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Current Biology 19, 1188–1193. [DOI] [PubMed] [Google Scholar]
- Adamski NM, Anastasiou E, Eriksson S, O’Neill CM, Lenhard M. 2009. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proceedings of the National Academy of Sciences, USA 106, 20115–20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. 2007. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Developmental Cell 13, 843–856. [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, et al. 2012. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Developmental Cell 22, 64–78. [DOI] [PubMed] [Google Scholar]
- Archacki R, Yatusevich R, Buszewicz D, et al. 2017. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Research 45, 3116–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asl LK, Dhondt S, Boudolf V, Beemster GT, Beeckman T, Inzé D, Govaerts W, De Veylder L. 2011. Model-based analysis of Arabidopsis leaf epidermal cells reveals distinct division and expansion patterns for pavement and guard cells. Plant Physiology 156, 2172–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekelandt A, Pauwels L, Wang Z, et al. 2018. Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiology 178, 217–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloban M, Vanstraelen M, Tarayre S, Reuzeau C, Cultrone A, Mergaert P, Kondorosi E. 2013. Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytologist 198, 1049–1059. [DOI] [PubMed] [Google Scholar]
- Baute J, Polyn S, De Block J, Blomme J, Van Lijsebettens M, Inzé D. 2017. F-box protein FBX92 affects leaf size in Arabidopsis thaliana. Plant & Cell Physiology 58, 962–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. 2007. Stomatal development. Annual Review of Plant Biology 58, 163–181. [DOI] [PubMed] [Google Scholar]
- Blomme J, Inzé D, Gonzalez N. 2014. The cell-cycle interactome: a source of growth regulators? Journal of Experimental Botany 65, 2715–2730. [DOI] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E. 2010. Functional modules in the Arabidopsis core cell cycle binary protein–protein interaction network. The Plant Cell 22, 1264–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Braidwood L, Sugimoto K. 2014. Endocycling in the path of plant development. Current Opinion in Plant Biology 17, 78–85. [DOI] [PubMed] [Google Scholar]
- Breuer C, Morohashi K, Kawamura A, Takahashi N, Ishida T, Umeda M, Grotewold E, Sugimoto K. 2012. Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. The EMBO Journal 31, 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Wang S, Venglat P, Zhao L, Cheng Y, Ye S, Qin Y, Datla R, Zhou Y, Wang H. 2018. Arabidopsis ICK/KRP cyclin-dependent kinase inhibitors function to ensure the formation of one megaspore mother cell and one functional megaspore per ovule. PLoS Genetics 14, e1007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW. 2012. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. The Plant Journal 71, 684–697. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Cao L, Wang S, et al. 2013. Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. The Plant Journal 75, 642–655. [DOI] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, et al. 2006. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. The Plant Cell 18, 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, De Bodt S, Inzé D. 2014. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends in Plant Science 19, 231–239. [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P. 1999. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. The Plant Journal 17, 547–556. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2000. Loosening of plant cell walls by expansins. Nature 407, 321–326. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews. Molecular Cell Biology 6, 850–861. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2015. Plant expansins: diversity and interactions with plant cell walls. Current Opinion in Plant Biology 25, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P. 2014. Class I TCP–DELLA interactions in inflorescence shoot apex determine plant height. Current Biology: CB 24, 1923–1928. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, et al. 2002. Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa–DPa transcription factor. The EMBO Journal 21, 1360–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. 2001. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. The Plant Cell 13, 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D. 2007. The ins and outs of the plant cell cycle. Nature Reviews. Molecular Cell Biology 8, 655–665. [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. 2011. Molecular control and function of endoreplication in development and physiology. Trends in Plant Science 16, 624–634. [DOI] [PubMed] [Google Scholar]
- Debernardi JM, Mecchia MA, Vercruyssen L, Smaczniak C, Kaufmann K, Inze D, Rodriguez RE, Palatnik JF. 2014. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. The Plant Journal 79, 413–426. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. 2006. The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin–SCFSKP2A pathway in Arabidopsis. The Plant cell 18, 2224–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. [DOI] [PubMed] [Google Scholar]
- Demura T, Ye ZH. 2010. Regulation of plant biomass production. Current Opinion in Plant Biology 13, 299–304. [DOI] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. 2006. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiology 140, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, et al. 2007. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proceedings of the National Academy of Sciences, USA 104, 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. 2004. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. The Plant Cell 16, 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disch S, Anastasiou E, Sharma VK, Laux T, Fletcher JC, Lenhard M. 2006. The E3 ubiquitin ligase BIG BROTHER controls arabidopsis organ size in a dosage-dependent manner. Current Biology 16, 272–279. [DOI] [PubMed] [Google Scholar]
- Dong H, Dumenil J, Lu FH, et al. 2017. Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes & Development 31, 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Guan C, Jiao Y. 2018. Molecular mechanisms of leaf morphogenesis. Molecular Plant 11, 1117–1134. [DOI] [PubMed] [Google Scholar]
- Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y. 2014. The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. The Plant Cell 26, 665–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Van den Broeck L, Inzé D. 2018. The pivotal role of ethylene in plant growth. Trends in Plant Science 23, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. 2014. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nature Reviews. Molecular Cell Biology 15, 197–210. [DOI] [PubMed] [Google Scholar]
- Efroni I, Eshed Y, Lifschitz E. 2010. Morphogenesis of simple and compound leaves: a critical review. The Plant Cell 22, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy NB, de Freitas Lima M, Van Damme D, Vanhaeren H, Gonzalez N, De Milde L, Hemerly AS, Beemster GT, Inzé D, Ferreira PC. 2011. The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. The Plant Journal 68, 351–363. [DOI] [PubMed] [Google Scholar]
- Eloy NB, Gonzalez N, Van Leene J, et al. 2012. SAMBA, a plant-specific anaphase-promoting complex/cyclosome regulator is involved in early development and A-type cyclin stabilization. Proceedings of the National Academy of Sciences, USA 109, 13853–13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner J, Michalski M, Kwiatkowska D. 2012. Spatiotemporal variation of leaf epidermal cell growth: a quantitative analysis of Arabidopsis thaliana wild-type and triple cyclinD3 mutant plants. Annals of Botany 109, 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Stransfeld L, Adamski NM, Breuninger H, Lenhard M. 2010. KLUH/CYP78A5-dependent growth signaling coordinates floral organ growth in Arabidopsis. Current Biology 20, 527–532. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang Z, Cui R, Li J, Li Y. 2012. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. The Plant Journal 70, 929–939. [DOI] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, Bowman JL, Reyes JC. 2004. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965–4975. [DOI] [PubMed] [Google Scholar]
- Ferjani A, Horiguchi G, Yano S, Tsukaya H. 2007. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiology 144, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo FF, Swain SM. 2003. SPYing on GA signaling and plant development. Journal of Plant Growth Regulation 22, 163–175. [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. 2004. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. The Plant Cell 16, 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülöp K, Tarayre S, Kelemen Z, et al. 2005. Arabidopsis anaphase-promoting complexes: multiple activators and wide range of substrates might keep APC perpetually busy. Cell Cycle 4, 4084–4092. [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD. 2000. Oriented asymmetric divisions that generate the stomatal spacing pattern in arabidopsis are disrupted by the too many mouths mutation. The Plant Cell 12, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, et al. 2010. Increased leaf size: different means to an end. Plant Physiology 153, 1261–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Pauwels L, Baekelandt A, et al. 2015. A repressor protein complex regulates leaf growth in Arabidopsis. The Plant Cell 27, 2273–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D. 2012. Leaf size control: complex coordination of cell division and expansion. Trends in Plant Science 17, 332–340. [DOI] [PubMed] [Google Scholar]
- Han SK, Wu MF, Cui S, Wagner D. 2015. Roles and activities of chromatin remodeling ATPases in plants. The Plant Journal 83, 62–77. [DOI] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A. 2013. Cell cycle control across the eukaryotic kingdom. Trends in Cell Biology 23, 345–356. [DOI] [PubMed] [Google Scholar]
- Hartweck LM, Genger RK, Grey WM, Olszewski NE. 2006. SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. Journal of Experimental Botany 57, 865–875. [DOI] [PubMed] [Google Scholar]
- Hepworth J, Lenhard M. 2014. Regulation of plant lateral-organ growth by modulating cell number and size. Current Opinion in Plant Biology 17, 36–42. [DOI] [PubMed] [Google Scholar]
- Heyman J, De Veylder L. 2012. The anaphase-promoting complex/cyclosome in control of plant development. Molecular Plant 5, 1182–1194. [DOI] [PubMed] [Google Scholar]
- Hisanaga T, Kawade K, Tsukaya H. 2015. Compensation: a key to clarifying the organ-level regulation of lateral organ size in plants. Journal of Experimental Botany 66, 1055–1063. [DOI] [PubMed] [Google Scholar]
- Hong SY, Kim OK, Kim SG, Yang MS, Park CM. 2011. Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. Journal of Biological Chemistry 286, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. 2006. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. Journal of Plant Research 119, 37–42. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Kim GT, Tsukaya H. 2005. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. The Plant Journal 43, 68–78. [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Tsukaya H. 2011. Organ size regulation in plants: insights from compensation. Frontiers in Plant Science 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou K, Wu W, Gan SS. 2013. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiology 161, 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, dePamphilis CW, Ma H. 2008. Phylogenetic analysis of the plant-specific zinc finger-homeobox and mini zinc finger gene families. Journal of Integrative Plant Biology 50, 1031–1045. [DOI] [PubMed] [Google Scholar]
- Hu W, Ma H. 2006. Characterization of a novel putative zinc finger gene MIF1: involvement in multiple hormonal regulation of Arabidopsis development. The Plant Journal 45, 399–422. [DOI] [PubMed] [Google Scholar]
- Huntley R, Healy S, Freeman D, et al. 1998. The maize retinoblastoma protein homologue ZmRb-1 is regulated during leaf development and displays conserved interactions with G1/S regulators and plant cyclin D (CycD) proteins. Plant Molecular Biology 37, 155–169. [DOI] [PubMed] [Google Scholar]
- Hur Y-S, Kim J, Kim S, Son O, Kim W-Y, Kim G-T, Ohme-Takagi M, Cheon C-I. 2019. Identification of TCP13 as an upstream regulator of ATHB12 during leaf development. Genes 10, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur YS, Um JH, Kim S, et al.. 2015. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytologist 205, 316–328. [DOI] [PubMed] [Google Scholar]
- Hurtado L, Farrona S, Reyes JC. 2006. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Molecular Biology 62, 291–304. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Tsukaya H. 2015. Behavior of leaf meristems and their modification. Frontiers in Plant Science 6, 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D, De Veylder L. 2006. Cell cycle regulation in plant development. Annual Review of Genetics 40, 77–105. [DOI] [PubMed] [Google Scholar]
- Jégu T, Latrasse D, Delarue M, et al. 2013. Multiple functions of Kip-related protein5 connect endoreduplication and cell elongation. Plant Physiology 161, 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalve S, De Vos D, Beemster GT. 2014. Leaf development: a cellular perspective. Frontiers in Plant Science 5, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, Deal RB, McKinney EC, Meagher RB. 2005. Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. The Plant Journal 41, 845–858. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Deal RB, Meagher RB. 2005. Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiology 138, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katano M, Takahashi K, Hirano T, Kazama Y, Abe T, Tsukaya H, Ferjani A. 2016. Suppressor screen and phenotype analyses revealed an emerging role of the monofunctional peroxisomal enoyl-CoA hydratase 2 in compensated cell enlargement. Frontiers in Plant Science 7, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathare PK, Dharmasiri S, Dharmasiri N. 2018. SAUR53 regulates organ elongation and apical hook development in Arabidopsis. Plant Signaling & Behavior 13, e1514896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Tsukaya H. 2010. Non-cell-autonomously coordinated organ size regulation in leaf development. Development 137, 4221–4227. [DOI] [PubMed] [Google Scholar]
- Kawade K, Tanimoto H, Horiguchi G, Tsukaya H. 2017. Spatially different tissue-scale diffusivity shapes ANGUSTIFOLIA3 gradient in growing leaves. Biophysical Journal 113, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z, Baloban M, Da Ines O, Tiricz H, Kroll A, Regulski K, Mergaert P, Kondorosi E. 2011. Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS One 6, e20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kende H. 2004. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proceedings of the National Academy of Sciences, USA 101, 13374–13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Tsukaya H. 2015. Regulation of plant growth and development by the growth-regulating factor and grf-interacting factor duo. Journal of Experimental Botany 66, 6093–6107. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. 2002. E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. Journal of Biological Chemistry 277, 16553–16558. [DOI] [PubMed] [Google Scholar]
- Kumar N, Harashima H, Kalve S, et al. 2015. Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. The Plant Cell 27, 3065–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D. 2006. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133, 3223–3230. [DOI] [PubMed] [Google Scholar]
- Larkin JC, Marks MD, Nadeau J, Sack F. 1997. Epidermal cell fate and patterning in leaves. The Plant Cell 9, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC. 2014. Direct roles of speechless in the specification of stomatal self-renewing cells. Science 345, 1605–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. 1997. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. Journal of Cell Biology 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Ko JH, Lee S, Lee Y, Pak JH, Kim JH. 2009. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiology 151, 655–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Liu Z, Wang Z, et al. 2018. STERILE APETALA modulates the stability of a repressor protein complex to control organ size in Arabidopsis thaliana. PLoS Genetics 14, e1007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng L, Corke F, Smith C, Bevan MW. 2008. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes & Development 22, 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li B, Dong AW. 2012. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Molecular Plant 5, 270–280. [DOI] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D. 2009. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiologia Plantarum 136, 223–236. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang F, Zhang H, He H, Ma L, Deng XW. 2008. Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. The Plant Journal 55, 844–856. [DOI] [PubMed] [Google Scholar]
- Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JH. 2014. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nature Communications 5, 3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inzé D. 2000. Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Letters 486, 79–87. [DOI] [PubMed] [Google Scholar]
- Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC. 2009. The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development 136, 1475–1485. [DOI] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. 2003. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. The Plant Cell 15, 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. 2000. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences, USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Sekine M, Murakami H, Shinmyo A. 1999. Tobacco retinoblastoma-related protein phosphorylated by a distinct cyclin-dependent kinase complex with Cdc2/cyclin D in vitro. The Plant Journal 18, 243–252. [DOI] [PubMed] [Google Scholar]
- Nikolov LA, Runions A, Das Gupta M, Tsiantis M. 2019. Leaf development and evolution. Current Topics in Developmental Biology 131, 109–139. [DOI] [PubMed] [Google Scholar]
- Noir S, Marrocco K, Masoud K, Thomann A, Gusti A, Bitrian M, Schnittger A, Genschik P. 2015. The control of Arabidopsis thaliana growth by cell proliferation and endoreplication requires the F-box protein FBL17. The Plant Cell 27, 1461–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. 2002. Gibberellin signaling: biosynthesis, catabolism, and response pathways. The Plant Cell 14(Suppl), S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Fujikura U, Olas JJ, Xue GP, Balazadeh S, Mueller-Roeber B. 2018. GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PLoS Genetics 14, e1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B. 2015. Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Molecular Plant 8, 998–1010. [DOI] [PubMed] [Google Scholar]
- Peng Y, Chen L, Lu Y, Wu Y, Dumenil J, Zhu Z, Bevan MW, Li Y. 2015. The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. The Plant Cell 27, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. 2000. Auxin regulates the initiation and radial position of plant lateral organs. The Plant Cell 12, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resentini F, Felipo-Benavent A, Colombo L, Blázquez MA, Alabadí D, Masiero S. 2015. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Molecular Plant 8, 482–485. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Debernardi JM, Palatnik JF. 2014. Morphogenesis of simple leaves: regulation of leaf size and shape. Wiley Interdisciplinary Reviews. Developmental Biology 3, 41–57. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. 2010. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacharowski SP, Gratkowska DM, Sarnowska EA, et al. 2015. SWP73 subunits of Arabidopsis SWI/SNF chromatin remodeling complexes play distinct roles in leaf and flower development. The Plant Cell 27, 1889–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Silva-Ortega CO, Wu S, Yamaguchi N, Wu MF, Pfluger J, Gillmor CS, Gallagher KL, Wagner D. 2012. Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. The Plant Journal 72, 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapala A, Runions A, Smith RS. 2019. Mechanics, geometry and genetics of epidermal cell shape regulation: different pieces of the same puzzle. Current Opinion in Plant Biology 47, 1–8. [DOI] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, et al. 2013. DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiology 163, 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski TJ, Ríos G, Jásik J, et al. 2005. SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. The Plant cell 17, 2454–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Kunkowska AB, Schippers JH. 2016. Role of reactive oxygen species during cell expansion in leaves. Plant Physiology 172, 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. 2013. Mechanisms and function of substrate recruitment by F-box proteins. Nature Reviews. Molecular Cell Biology 14, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluis A, Hake S. 2015. Organogenesis in plants: initiation and elaboration of leaves. Trends in Genetics 31, 300–306. [DOI] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. 2012. The SAUR19 subfamily of small auxin up RNA genes promote cell expansion. The Plant Journal 70, 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. The Plant Cell 26, 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. 2012. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. The Plant Cell 24, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. 2004. Molecular mechanism of gibberellin signaling in plants. Annual Review of Plant Biology 55, 197–223. [DOI] [PubMed] [Google Scholar]
- Takatsuka H, Ohno R, Umeda M. 2009. The Arabidopsis cyclin-dependent kinase-activating kinase CDKF;1 is a major regulator of cell proliferation and cell expansion but is dispensable for CDKA activation. The Plant Journal 59, 475–487. [DOI] [PubMed] [Google Scholar]
- Tan QK, Irish VF. 2006. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiology 140, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Hou A, Babu M, et al.. 2008. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiology 147, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touihri S, Knöll C, Stierhof YD, Müller I, Mayer U, Jürgens G. 2011. Functional anatomy of the Arabidopsis cytokinesis-specific syntaxin KNOLLE. The Plant Journal 68, 755–764. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2003. Organ shape and size: a lesson from studies of leaf morphogenesis. Current Opinion in Plant Biology 6, 57–62. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. 2015. Yield increase: GRFs provide the key. Nature Plants 2, 15210. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. 2007. Gibberellin receptor and its role in gibberellin signaling in plants. Annual Review of Plant Biology 58, 183–198. [DOI] [PubMed] [Google Scholar]
- Umeda M, Shimotohno A, Yamaguchi M. 2005. Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant & Cell Physiology 46, 1437–1442. [DOI] [PubMed] [Google Scholar]
- Van Leene J, Boruc J, De Jaeger G, Russinova E, De Veylder L. 2011. A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends in Plant Science 16, 141–150. [DOI] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, et al. 2010. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Molecular Systems Biology 6, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D. 2002. Genome-wide analysis of core cell cycle genes in Arabidopsis. The Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GT, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. 2005. Genome-wide identification of potential plant E2F target genes. Plant Physiology 139, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaeren H, Nam YJ, De Milde L, Chae E, Storme V, Weigel D, Gonzalez N, Inzé D. 2017. Forever young: the role of ubiquitin receptor DA1 and E3 ligase big brother in controlling leaf growth and development. Plant Physiology 173, 1269–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Tognetti VB, Gonzalez N, Van Dingenen J, De Milde L, Bielach A, De Rycke R, Van Breusegem F, Inzé D. 2015. Growth regulating FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiology 167, 817–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruyssen L, Verkest A, Gonzalez N, et al. 2014. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. The Plant cell 26, 210–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Meyerowitz EM. 2002. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Current Biology 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. 2000. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127, 3931–3940. [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. 2008. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. The Plant Cell 20, 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X. 2011. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. Journal of Experimental Botany 62, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li N, Jiang S, Gonzalez N, Huang X, Wang Y, Inzé D, Li Y. 2016. SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nature Communications 7, 11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DW. 2006. PEAPOD regulates lamina size and curvature in Arabidopsis. Proceedings of the National Academy of Sciences, USA 103, 13238–13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B. 1998. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531. [DOI] [PubMed] [Google Scholar]
- Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y. 2013. The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis. The Plant Cell 25, 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Yang H, Zhu Y, Xue J, Wang T, Song T, Yang Z, Wang S. 2018. The canonical E2Fs are required for germline development in Arabidopsis. Frontiers in Plant Science 9, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Sui N, Barnhill B, et al. 2017. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nature Chemical Biology 13, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Du L, Xu R, Cui R, Hao J, Sun C, Li Y. 2015. Transcription factors SOD7/NGAL2 and DPA4/NGAL3 act redundantly to regulate seed size by directly repressing KLU expression in Arabidopsis thaliana. The Plant Cell 27, 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X. 2012. Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One 7, e44465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Harashima H, Dissmeyer N, et al. 2012. A general G1/S-phase cell-cycle control module in the flowering plant Arabidopsis thaliana. PLoS Genetics 8, e1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zondlo SC, Irish VF. 1999. CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. The Plant Journal 19, 259–268. [DOI] [PubMed] [Google Scholar]