This review presents a molecular framework for how phytohormones regulate normal root hair development and how this is affected by changes in the rhizosphere, to enable greater understanding of the specialized functions of root hairs and their developmental and environmental plasticity.
Keywords: Arabidopsis, environment, hormone signalling, molecular pathway, root hairs, soil composition
Abstract
The main functions of plant roots are water and nutrient uptake, soil anchorage, and interaction with soil-living biota. Root hairs, single cell tubular extensions of root epidermal cells, facilitate or enhance these functions by drastically enlarging the absorptive surface. Root hair development is constantly adapted to changes in the root’s surroundings, allowing for optimization of root functionality in heterogeneous soil environments. The underlying molecular pathway is the result of a complex interplay between position-dependent signalling and feedback loops. Phytohormone signalling interconnects this root hair signalling cascade with biotic and abiotic changes in the rhizosphere, enabling dynamic hormone-driven changes in root hair growth, density, length, and morphology. This review critically discusses the influence of the major plant hormones on root hair development, and how changes in rhizosphere properties impact on the latter.
Introduction
Roots provide structural anchorage and access to water and nutrients, which is vital to plant survival. In addition, they form the site of symbiotic interactions with soil-living microorganisms (Haling et al., 2013; Grierson et al., 2014). As roots encounter heterogeneous soils in which nutrients and water are not equally distributed, their root system architecture is constantly modified. Dynamic modification of root hair growth, length, density, and morphology allows the plant to meet its nutrient demand in a variety of soil conditions, with the aim of creating an optimal soil-sampling root volume (Forde and Lorenzo, 2001; López-Bucio et al., 2003; Morris et al., 2017). The importance of root hairs is illustrated by the fact that overall plant fitness is negatively affected in several loss-of-function root hair mutants when grown in challenging soil conditions (Wu et al., 2007; Miguel et al., 2015). Consequently, dynamic root hair morphogenesis is a key trait for improving the acquisition of essential nutrients in heterogeneous soils. Therefore, a better understanding of the root hair developmental pathway is crucial. Various studies, mostly based on the model organism Arabidopsis, have led to a better understanding of the genetic and molecular cascade that lies at the base of root hair development (Balcerowicz et al., 2015; Schoenaers et al., 2017).
Nevertheless, it is far from clear how different plant hormones feed into this genetic framework. In addition, many environmental conditions influence different stages of root hair morphogenesis, in particular nutrient availability (phosphorus, nitrogen, iron, etc.), water deficiency, and microbe interaction. Currently, it is not fully understood how these different signals are integrated by the plant root, and how they fine-tune root hair development. Here, we review the current state-of-the-art regarding hormonal regulation and environmental impact on root hair development.
General hormonal regulation of root hair development
Root hair development starts with the determination of whether an epidermal cell becomes a root hair (H; trichoblast) or non-root hair (N; atrichoblast) cell, giving rise to distinct hair and non-hair cell files in the Arabidopsis root (Fig. 1; Dolan et al., 1994). Epidermal cell fate determination depends on a position-dependent signal originating from the underlying cortical cells. Signal perception in epidermal cells that overlie two cortical cells triggers a transcription factor cascade that ultimately inhibits the expression of GLABRA2 (GL2), a core non-root hair-determining transcription factor. As a result, ROOT HAIR DEFECTIVE 6 (RHD6) expression is initiated, leading to the initiation of a root hair bulge (Fig. 2). This bulge then begins tip growth before ultimately maturing. Root hair initiation and tip growth are each characterized by distinct yet interconnected molecular pathways (extensively reviewed in Balcerowicz et al., 2015; Schoenaers et al., 2017).
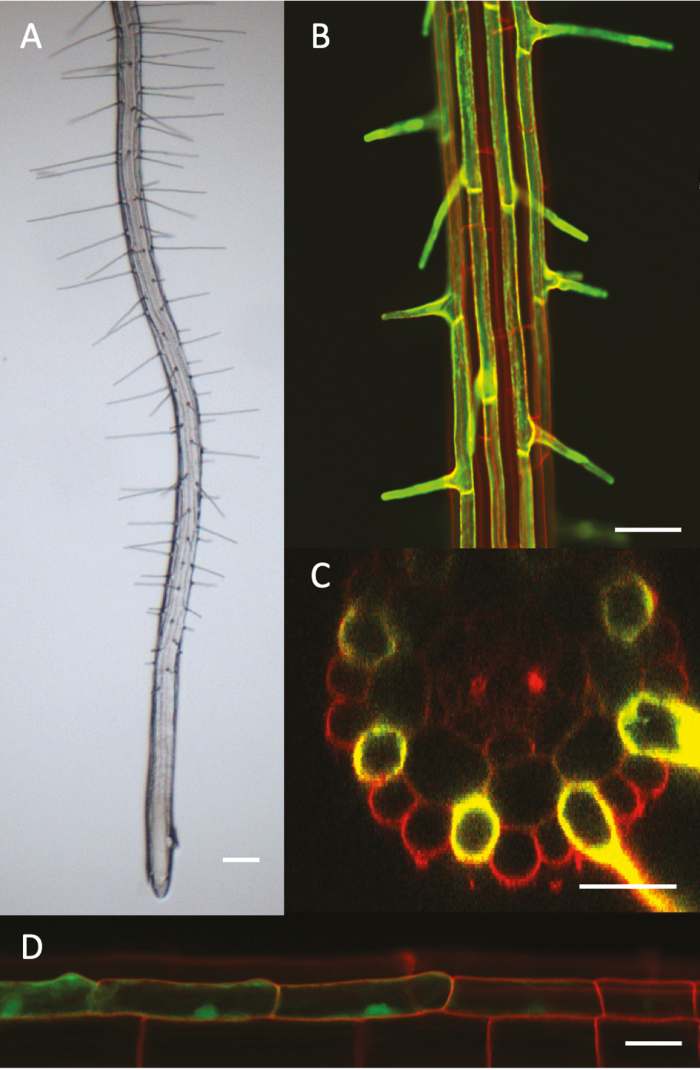
Fig. 1.
Arabidopsis root and root hairs. (A) Brightfield image of a root with root hairs. (B–D) Confocal images of a root hair-specific promoter::green fluorescent protein (GFP) line. (B) longitudinal section showing GFP in distinct root hair cell files; (C) cross-section showing root hair cell locations marked by GFP; (D) longitudinal section through an epidermal cell file showing GFP expression in two consecutive developmental stages: at the end of the determination phase (at the right) and in the root hair initiation phase (from the middle to the left). Scale bar: 200 μm (A), 50 μm (B), 20 μm (C, D).
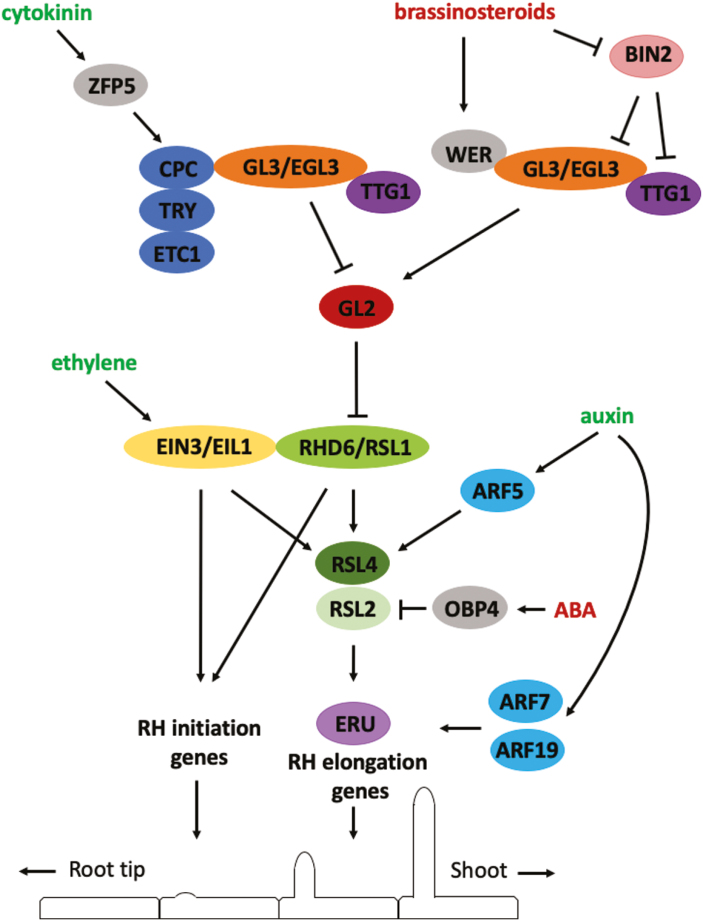
Fig. 2.
Model of the molecular and hormonal pathway for root hair development. Hormones stimulating root hair growth are shown in green, those repressing are shown in red. Root tip is at the left side in the schematic longitudinal section through a single epidermal cell layer with consecutive root hair developmental phases. ARF5/7/19, AUXIN RESPONSE FACTOR 5/7/19; BIN2, BRASSINOSTEROID-INSENSITIVE 2; CPC, CAPRICE; EGL3, ENHANCER OF GLABRA 3; EIN3, ETHYLENE INSENSITIVE 3; EIL1, ETHYLENE INSENSITIVE 3-LIKE 1; ETC1, ENHANCER OF TRY AND CPC1; GL2/3, GLABRA2/3; OBP4, OBF BINDING PROTEIN 4; RHD6, ROOT HAIR DEFECTIVE 6; RSL1–4, ROOT HAIR DEFECTIVE 6-LIKE 1–4; TRY, TRYPTICHON; TTG1, TRANSPARENT TESTA GLABRA 1; WER, WEREWOLF; ZFP5, ZINC FINGER PROTEIN 5.
This genetic framework, which guides root hair development (Fig. 2), is heavily controlled by hormonal cues. In particular the influence of auxin and ethylene is well documented. These hormonal signals are key to the plant’s ability to dynamically regulate root hair form and function in response to the changing properties of the surrounding soil environment. In addition, crosstalk exists between several hormone classes (Wang and Irving, 2011; Zhang et al., 2016; Liu et al., 2017; Zhang et al., 2018).
Auxin
Auxin is a well-characterized hormone that influences many plant developmental processes and acts as a positive regulator of root hair development (Paque and Weijers, 2016). Most auxin responses occur as a result of transcriptional and translational changes. This signalling pathway is known as the canonical pathway. To the contrary, other responses occur too fast to be dependent on nuclear transcription. These non-canonical pathways (Kubeš and Napier, 2019) will not be discussed in this review. The TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX (TIR1/AFB) receptor complex that binds to auxin is the substrate-recognition subunit of the SUPPRESSOR OF KINETOCHORE PROTEIN 1 (SKP1)/CULLIN1/F-Box (SCF) E3 ubiquitin ligase complex. Auxin–TIR1 binding promotes the interaction between TIR1/AFB and AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) co-repressor proteins, which triggers the polyubiquitination and degradation of these Aux/IAAs. The latter causes dissociation of Aux/IAAs from auxin response factors (ARFs), allowing these transcription factors to regulate gene transcription (Fig. 3A; Teale et al., 2006; Weijers and Wagner, 2016; Ma et al., 2018).
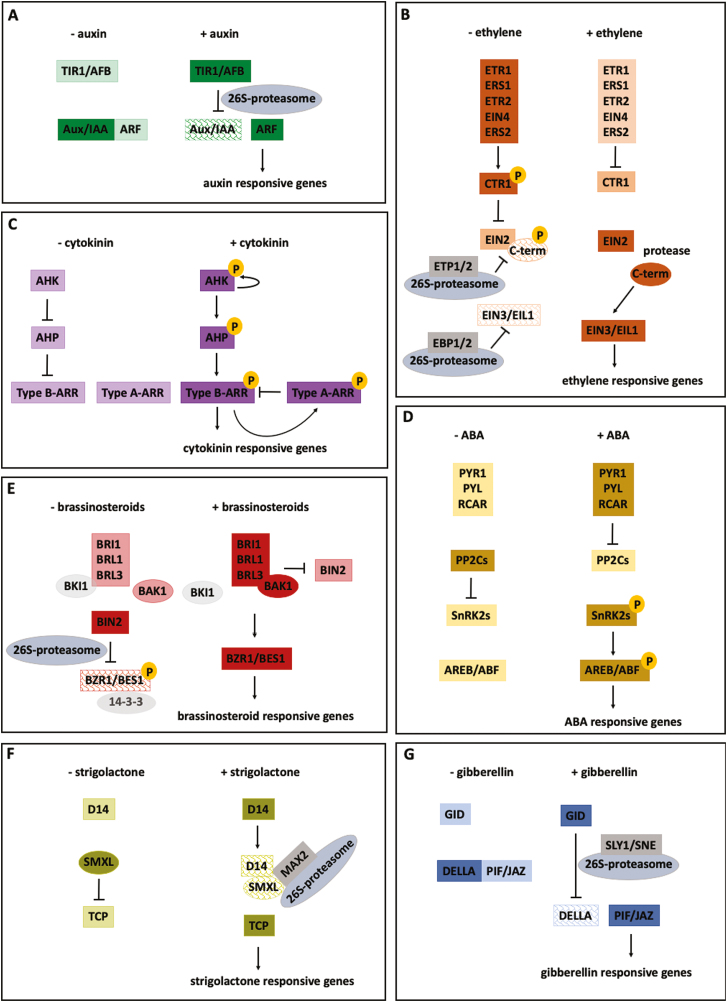
Fig. 3.
Models for hormone-responsive transcription. Simplified models describing how auxin (A), ethylene (B), cytokinin (C), abscisic acid (D), brassinosteroid (E), strigolactones (F), and gibberellin (G) regulate transcription of their responsive genes. Left: absence of hormone, and right: presence of hormone. Active proteins are represented by dark colour, inactive proteins by pale colour, and shingled colour indicates the proteins are targeted for ubiquitination and subsequent degradation by the 26S-proteasome.
Overexpression of ARF5, 7, and 8 enhances root hair growth (Mangano et al., 2017), while root hair-specific overexpression of ARF1–4, 9–11, and 16 inhibits root hair growth, indicating that root hair development depends on the relative transcriptional activity of ARFs, which function as activators (aARFs) or repressors (rARFs) of gene transcription (Choi et al., 2018). Several auxin-signalling mutants, including aux1, axr1, axr2/iaa7, arf5, arf7/arf19, axr3/iaa17, slr/iaa14, and iaa28 have altered root hair development (Masucci and Schiefelbein, 1996; Rahman et al., 2002; Okushima et al., 2005; Li et al., 2016), and supplementation or overproduction of auxin [e.g. gain-of-function mutant SHORT HYPOCOTYL 2 (SHY2/IAA3] leads to longer root hairs (Pitts et al., 1998; Knox et al., 2003). In addition, loss- and gain-of-function mutants of DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1), a protein that plays a major role in auxin oxidation and thus in regulating the cellular homeostasis of auxin, display altered root hair length (Porco et al., 2016). Together, these findings clearly illustrate the pivotal role of auxin in controlling root hair morphogenesis.
Masucci and Schiefelbein (1996) have shown that auxin is capable of restoring root hair growth even in the hairless rhd6 mutant, which lacks the core root hair-initiating basic helix–loop–helix (bHLH) transcription factor RHD6 (Fig. 2). According to Yi et al. (2010), this occurs through the induction of the bHLH transcription factor ROOT HAIR DEFECTIVE 6-LIKE4 (RSL4), which acts downstream of RHD6. As such, RSL4 is considered to be the first auxin-responsive gene in the timeline of root hair morphogenesis. During root hair initiation, a 4 h-long RSL4 expression pulse determines final root hair length. As shown in Fig. 2, auxin induces RSL4 expression through a direct interaction of ARF5 with the RSL4 promoter (Mangano et al., 2017). Surprisingly, to date, none of the genes functioning upstream of RSL4 has been shown to be auxin regulated, suggesting that auxin is not involved in early root hair development. Conversely, it has been shown that auxin is able to directly regulate the expression of genes downstream of RSL4. For instance, ARF7 and ARF19 directly interact with the promoter sequence of the RSL4-target gene ERULUS, a trichoblast-specific receptor-like kinase and core regulator of the root hair tip growth process (Schoenaers et al., 2018). The relative importance of direct (through ARFs) and indirect (through RSL4) auxin-controlled gene expression remains to be investigated.
Ethylene
The gaseous hormone ethylene is a positive regulator of root hair development, and frequently found to functionally interact with auxin signalling (Dubois et al., 2018). A family of five receptors perceives ethylene, ETHYLENE RESPONSE 1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR 1 (ERS1), ERS2, and ETHYLENE INSENSITIVE 4 (EIN4), which are functionally redundant (Dong et al., 2010; Ju and Chang, 2015). In the absence of ethylene, the ethylene receptors phosphorylate and activate the Ser/Thr kinase CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1). Subsequent phosphorylation of ETHYLENE INSENSITIVE 2 (EIN2) by CTR1 leads to the activation of EIN2 TARGETTING PROTEIN 1 and 2 (ETP1/2), which mediates ubiquitination-dependent degradation of the C-terminal domain of EIN2. As C-terminal EIN2 is required for stabilization of ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE 1 (EIL1) transcription factors in the nucleus, EIN3 and EIL1 remain inactive and are targeted for proteasome-dependent degradation, which requires EIN3 BINDING F-BOX 1 and 2 (EBF1/2) when ethylene is absent (Binder et al., 2007). The binding of ethylene inactivates the receptor signalling. As a consequence, EIN2 is not phosphorylated by CTR1, and a cytoplasmic C-end part is proteolytically cleaved off. The latter then moves to the nucleus, stabilizes EIN3 and EIL1, and facilitates expression of ethylene-responsive (ERF) genes (Fig. 3B; Ju and Chang, 2015; Moniuszko, 2015; Shakeel et al., 2015).
Loss of ETR1 and EIN2 function (leading to ethylene insensitivity) or blocking of ethylene biosynthesis inhibits root hair growth (Masucci and Schiefelbein, 1996; Rahman et al., 2002). Supplementation of the ethylene precursor aminocyclopropane carboxylic acid (ACC), on the other hand, stimulates root hair growth. Importantly, ACC supplementation also induces the formation of ectopic root hairs (Pitts et al., 1998). Together, these data show that ethylene is a positive regulator of root hair development that, unlike auxin, also regulates the epidermal cell fate determination pathway.
Ethylene stimulates the expression of different key root hair genes such as RHD6, RSL4, and RSL2, which regulate the later stages of root hair initiation and tip growth (Masucci and Schiefelbein, 1994; Yi et al., 2010, Zhang et al., 2016). Feng et al. (2017) showed that EIN3 physically interacts with RHD6, and that both transcription factors coactivate the expression of RSL4 in the presence of ethylene (Fig. 2). RHD6 forms a transcriptional complex with RSL1, another bHLH transcription factor of the RSL family that seems to be an upstream regulator of RSL2 (Pires et al., 2013). Together with the complex EIN3–EIL1, they are currently considered to be key regulators of root hair initiation and elongation (Feng et al., 2017). Whether EIN3 also interacts with RSL1 and whether EIL1 acts similarly is currently unknown. Furthermore, ethylene signalling seems to go specifically through RHD6 and RSL1 because root hair initiation cannot be rescued by ethylene in the rhd6 rsl1 double mutant. Transcriptional analysis revealed the genes that are regulated by EIN3–EIL1 and RHD6–RSL1, which potentially control root hair initiation and/or elongation (Feng et al., 2017). Further research is needed to better understand ethylene-controlled root hair morphogenesis and where and how auxin and ethylene intersect in the root hair formation pathway.
Cytokinin
Cytokinins control the cell cycle and several plant developmental processes (Werner and Schmülling, 2009; Bishopp et al., 2011; Kushwah et al., 2011; Muraro et al., 2011; Su et al., 2011).
There are few reports regarding the role of cytokinins in regulating root hair development. Recent findings have shed new light on the molecular basis of the cytokinin signalling pathway. Cytokinin binds to His kinase-like proteins such as ARABIDOPSIS HISTIDINE KINASE 2/3/4 (AHK2/AHK3/AHK4) triggering their autophosphorylation. Subsequent phosphate-group transfer towards an ARABIDOPSIS histidine phosphotransfer protein (AHP) leads to AHP nuclear translocation and the activation of type-B Arabidopsis response regulators (ARRs), which in turn induce cytokinin-mediated gene transcription. Finally, cytokinin signalling is subject to negative feedback regulation by expression of type-A ARRs, which are negative regulators of type-B ARRs (Fig. 3C; Hutchison and Kieber, 2002; Hwang et al., 2012; Kieber and Schaller, 2014; Huang et al., 2018b).
The role of cytokinin in root hair development remained unclear until the transcription factor ZINC FINGER PROTEIN5 (ZFP5) was discovered. Loss-of-function zfp5 mutants exhibit a short root hair phenotype, caused by slower growth, which cannot be rescued by cytokinin addition. In wild-type plants, however, cytokinin supplementation induces an increase in final root hair length, while lowering endogenous cytokinin levels results in a short root hair phenotype. ZFP5, which is cytokinin-inducible, seems to lie at the base of cytokinin-regulated root hair morphogenesis and directly promotes the expression of CPC, a positive root hair regulator (Fig. 2; An et al., 2012; Zhang et al., 2016). At the moment, it needs to be investigated whether other genes in the root hair developmental pathway are (direct) targets of cytokinin signalling and whether cytokinin acts through ZFP5 only.
Abscisic acid
Abscisic acid (ABA) controls seed dormancy, germination (Kermode, 2005), inhibition of root growth, and the response to abiotic stresses such as drought and salinity (Wang et al., 2007; Luo et al., 2014; Lombardo and Lamattina, 2018). ABA is perceived by cytosolic receptors, designated as PYRABACTIN RESISTANCE1 (PYR1), PYR1-LIKE (PYL), or REGULATORY COMPONENTS OF ABA RECEPTOR (RCAR). Under the absence or low levels of ABA, protein phosphatases of type 2C (PP2Cs) dephosphorylate SNF1-related protein kinases (SnRKs), thereby reducing SnRK2 kinase activity and inhibiting ABA-induced gene transcription. Under higher ABA concentrations, the RCAR receptors mediate the inactivation of the PP2Cs. As a consequence, SnRK2s activate several transcription factors from the ABA RESPONSIVE ELEMENT (ABRE) BINDING proteins (AREBs)/ABRE BINDING FACTORs (ABFs), initiating ABA-induced gene expression (Fig. 3D; Ma et al., 2009; Fujii et al., 2009; Melcher et al., 2009; Li et al., 2018).
Abscisic acid is a negative regulator of root hair growth since supplementation results in reduced root hair growth, a response that is absent in ABA-insensitive mutants. Recent work shows that ABA controls root hair growth by inhibiting the expression of the key root hair transcription factor RSL2 through the up-regulation of OBF BINDING PROTEIN4 (OBP4; Fig. 2). This transcription factor in turn binds to the RSL2 promoter sequence and negatively regulates its transcription. In line with the latter, root hairs of rsl2 were found to be irresponsive to ABA treatment (Rymen et al., 2017).
Brassinosteroids
Brassinosteroids (BR) are endogenous plant hormones that are involved in multiple developmental processes (Müssig et al., 2003; Kuppusamy et al., 2009; Clouse, 2011; Wei and Li, 2016). They are perceived by the receptor kinases BRASSINOSTEROID-INSENSITIVE 1 (BRI1) and its paralogs BRI1-LIKE 1 (BRL1) and BRI1-LIKE 3 (BRL3). BRI1 is expressed throughout the root, while BRL1 and BRL3 are mainly found in the stem cell niche (Planas-Riverola et al., 2019). BRI1–BR association leads to BRI1 binding to BRI1-ASSOCIATED RECEPTOR KINASE (BAK1). This BRI1–BAKI heterodimer complex then initiates a phosphorylation relay cascade involving BRI1 KINASE INHIBITOR (BKI1) dissociation, which finally ends with dephosphorylation and activation of the transcription factors BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR (BES1), allowing them to regulate BR-induced gene transcription. In the absence of BR, the kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2) phosphorylates BZR1/BES1, which inactivates them and promotes binding to 14-3-3 proteins. This interaction leads to their cytoplasmic retention and degradation (Fig. 3E; Zhu et al., 2013; Nolan et al., 2017; Yu et al., 2018; Planas-Riverola et al., 2019).
BRs seem essential for position-dependent epidermal cell fate determination in roots (Wei and Li, 2016). BR-deficient mutants display ectopic root hair formation, whereas BR-signal-enhanced mutants produce fewer root hairs. In line with this, treatment with brassinolide or bikinin produced fewer root hairs, while brassinazole, a BR-synthesis inhibitor, led to a change in epidermal cell fate determination resulting in the formation of more trichoblasts and thus more root hairs (Cheng et al., 2014). Kuppusamy et al. (2009) found that BRs positively regulate the expression of WER and GL2, both negative regulators of early root hair development (Fig. 2). Root hair formation is determined by transcription factor complex formation and lateral communication between H- and N-cells (Bernhardt et al., 2005; Savage et al., 2008). Cheng et al. (2014) have demonstrated that BRs interfere with this process, leading to aberrant cell-specific GL2 and WER activity. At low BR concentrations, the formation and activity of the transcription factor complex (WER–GL3/EGL3–TTG1) is inhibited in both H- and N-cells since WER expression is lower and the kinase BIN2 phosphorylates EGL3 and TTG1, which reduces the functionality of the transcription factor complex. This suppresses GL2 expression and thus results in formation of more root hairs as hair cell fate is promoted. At high BR concentrations, WER expression is increased in both N- and H-cells, the WER–GL3/EGL3–TTG1 transcription factor complex is fully functional (since BIN2 is not active), promoting GL2 expression and determination of the non-hair cell fate in all epidermal cells (Cheng et al., 2014; Wei and Li, 2016).
Strigolactones
Strigolactones (SLs) were recently classified as a class of phytohormones (Brewer et al., 2013). DWARF 14 (D14) receptor proteins perceive strigolactones, leading to a conformational change and subsequent signal transduction. Similar to other hormone-perception mechanisms, SL signal transduction depends on the initial ubiquitin-dependent degradation of SL-suppressor proteins (e.g. SUPPRESSOR OF MORE AXILLARY GROWTH 2 (MAX2)-LIKE family proteins; SMXLs). SMXL degradation allows downstream signalling through TCP-domain transcription factors (Fig. 3F; Machin et al., 2020).
Kapulnik et al. (2011a,b) have described that supplementation with synthetic SL (GR24) enhances root hair growth. This positive effect of SLs depends, at least in part, on MAX2 F-box proteins that function in ubiquitin-dependent protein degradation. MAX2/3/4 loss-of-function plants display a short root hair phenotype. However, contrary to max3 and max4 mutants, the max2 root hair length can only be restored at high GR24 concentrations. These data highlight an emerging role for SLs in controlling root hair morphogenesis.
Gibberellin
Among other things, gibberellins (GA) control cell elongation, root apical meristem (RAM) growth, and the formation of lateral roots (Ogawa et al., 2003; Tyler et al., 2004; Davière and Achard, 2013, 2016; Hu et al., 2018), yet little is currently known on GA involvement in root hair development. GA signal perception works through three orthologs of GIBBERELLIN INSENSITIVE DWARF 1 (GID1A–C), soluble nuclear receptors, which consist of a GA-binding pocket and a flexible N-terminal extension. GID1–GA binding leads to a GID1 conformational change that enables subsequent interaction with DELLA repressor proteins, leading to the formation of a GA–GID1–DELLA complex. Upon GA–GID1–DELLA complex formation, the DELLA proteins undergo small conformational changes that enhance recognition with the F-box proteins SLEEPY 1 (SLY1) and SNEEZY (SNE/SLY2). This promotes DELLA ubiquitination and subsequent destruction of DELLAs by the 26S proteasome. As a result, the transcription factors (PIFs, JAZs) that were inactivated by DELLAs become de-repressed and GA-induced signal transduction is initiated (Fig. 3G; Davière and Achard, 2013).
There are no reports on direct root hair growth responses towards GA in Arabidopsis. Nevertheless, Devlin and Brown (1969) have shown that GA has a positive effect on the elongation rate of Agrostis alba root hairs. A similar effect was seen in Datura innoxia, where exogenous GA resulted in more hairy roots and increased root hair elongation (Ohkawa et al., 1989). How GA feeds into the genetic root hair growth framework needs further investigation.
The complex crosstalk among phytohormones during root hair development
Hormonal crosstalk is instrumental in plant development. The root hair developmental pathway is fundamentally controlled by input from several hormonal classes. Hence, understanding how hormone signalling pathways are interconnected is a crucial step towards building an integrated view of root hair morphogenesis.
Auxin and ethylene
A large effort has gone into understanding the heavily interconnected auxin and ethylene signalling pathways during root hair growth (Vanstraelen and Benková, 2012; Muday et al., 2012; Robles et al., 2013; Lee and Cho, 2013; Qin and Huang, 2018). A functional interaction is illustrated by the finding that auxin biosynthesis is up-regulated by ethylene in the root tip and during polar root hair initiation (Stepanova et al., 2005, 2007; Růzicka et al., 2007; Swarup et al., 2007; Ikeda et al., 2009). Vice versa, Strader et al. (2010) demonstrated that the aux1 mutation suppresses the long hair phenotype of the eto1 mutant, suggesting that ethylene needs auxin transport during the regulation of some phases of root hair development. In addition, the rhd6 root hair-less phenotype can be rescued by both auxin and ACC (Masucci and Schiefelbein, 1994), showing that both hormones can influence root hair development downstream of RHD6-controlled gene transcription.
The importance of both auxin and ethylene signalling is further illustrated by the fact that root hairs are defective in the aux1 ein2 double mutant (Rahman et al., 2002) and that transcriptome analysis identified a large group of common root hair genes that are both up-regulated by auxin and ethylene supplementation (Bruex et al., 2012). The nature of auxin–ethylene crosstalk is, however, complex. Hence, auxin and ethylene signalling are not per definition mutually interdependent. For instance, the phenotype of aux1 etr1 double mutants can be suppressed by administration of IAA, but not by ACC addition (Masucci and Schiefelbein, 1996). To the contrary, ein3 eil1 rhd6 rsl1 quadruple mutant roots remain hairless after auxin application, suggesting that the link between ethylene and auxin signalling is not that straightforward after all during root hair morphogenesis (Feng et al., 2017). Nonetheless, auxin–ethylene crosstalk is embedded throughout a variety of plant developmental processes (Muday et al., 2012). In general, it is now believed that functional auxin signalling is required for optimal ethylene responses. In line with the latter, the aux1 and tir1 auxin mutants show reduced sensitivity to ethylene treatment. Vice versa, it was found that the expression of auxin homeostasis and polar transport genes is increased as a result of increased ethylene levels (Zemlyanskaya et al., 2018).
Mechanistically, the interconnection of auxin and ethylene signalling can be explained by the presence of an EIN3 binding region in the AUXIN RESISTANT 1 (AUX1) promoter (Zažímalová et al., 2007). Similarly, ACC transport is promoted by LYSINE HISTIDINE TRANSPORTER 1 (LHT1), which in turn is under the control of auxin (Lewis et al, 2013). Moreover, the transcription of 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE (ACS) genes is up-regulated by auxin in an ARF-dependent manner (Stepanova et al., 2007). Auxin and ethylene seem to impinge on the same core root hair pathway, through RSL4 and RHD6, respectively, suggesting that both hormonal signals need to be considered in order to understand root hair growth.
Cytokinin, auxin and ethylene
A functional auxin–cytokinin interaction during plant development is emerging. Surprisingly, cytokinin and auxin were found to interact in a synergistic or antagonistic manner during plant development. For instance, both hormones were found to positively regulate root/stem growth and flowering (Srivastava, 2002; Moubayidin et al., 2013; El-Showk et al., 2013; Schaller et al., 2015; Liu et al., 2017). Contrastingly, auxin and cytokinin have an antagonistic role in lateral root formation. On one hand, type-B ARR1 stimulates the expression of SHY2, which encodes IAA3 (Tian and Reed, 1999) and negatively influences auxin signalling. This indirectly causes degradation of PIN1 and thereby suppresses auxin transport (Dello Ioio et al., 2008; Marhavý et al., 2014), which leads to an antagonistic relationship between auxin and cytokinin. On the other hand, auxin can inhibit SHY2/IAA3 through degradation via the SCF complex, resulting in PIN expression and subsequent auxin transport. In addition, Schaller et al. (2015) have shown that auxin directly activates the transcription of ARR7 and ARR15, both type-A ARRs known to be negative regulators of the cytokinin response. During early lateral root development auxin promotes the cytokinin inhibitor AHP6, which represses cytokinin signalling by modulating PIN1 and, as such, auxin distribution (Moreira et al., 2013).
Cytokinin, together with ethylene, stimulates the abscission of different plant organs. In fact, the stability of ACC synthase can be increased by cytokinin, resulting in higher ethylene production (Chae et al., 2003; Hansen et al., 2009). For root hair development, however, the effect of cytokinin might be independent of auxin and ethylene, especially since cytokinin can rescue root hair elongation when auxin (axr1) and ethylene signalling (etr1-1) are disturbed (Zhang et al., 2016). Furthermore, auxin and ethylene can stimulate root hair elongation when cytokinin is defective in an overexpression line of CKX2, a cytokinin oxidase (Zhang et al., 2016). It is therefore plausible that several common genes in the root hair-forming network can be targets for auxin, ethylene, and cytokinin. This idea is strengthened by the observation that the phenotype of the rhd6 mutant can be rescued by exogenous auxin and ethylene, but also by cytokinin (Masucci and Schiefelbein, 1994; Zhang et al., 2016). Candidates downstream of RHD6 are RSL1/2/4 (Fig. 2). In addition, auxin regulates cytokinin biosynthesis (Nordström et al., 2004) as the auxin response factors ARF5 and ARF7 control cytokinin biosynthesis by up-regulating CYTOKININ RESPONSE FACTOR (CRF) genes and cytokinin biosynthetic enzymes Liu et al. (2017). Furthermore, it has been demonstrated that cytokinin can regulate ethylene biosynthesis (El-Showk et al., 2013), showing a complex interplay of cytokinin with other hormones and as such influencing root hair development.
Strigolactones, auxin, and ethylene
Strigolactones have emerged as major players in regulating plant growth and development (Brewer et al., 2013). Several studies have shown that SLs functionally interact with auxin transport through effects on PIN and TIR1 protein abundance (Kapulnik et al., 2011a,b; Ruyter-Spira et al., 2011; Mayzlish-Gati et al., 2012; Machin et al., 2020). These changes in auxin transport lead to changes in auxin levels, which can impact on root hair development. However, how exactly SLs regulate PIN abundances remains unclear.
Treatment of mutants showed that SL signalling is not required for ethylene-responses in root hairs and that SL responses need ethylene synthesis and signalling. In line with this, SL treatment induced ACC synthase genes, encoding key rate-determining enzymes in ethylene synthesis (Yamagami et al., 2003). Besides, SL signalling is not required for auxin to elicit its root hair growth stimulating effect, while auxin perception is needed to some extent for the SL effect on root hairs. In addition, auxin and SL have an additive effect on root hair development. Treatment of aux1-7 ein2-1 mutants, which are impaired in sensitivity towards auxin and ethylene, showed reduced sensitivity to SLs. These observations, together with the finding that SLs can regulate PIN abundances in an unknown manner, strongly suggest that there is intimate crosstalk between SLs, ethylene, and auxin in the regulation of root hair elongation. Evidence indicates that SL affects root hair elongation via up-regulation of the ethylene pathway, which in turn acts on root hair development and interacts with the auxin pathway. The latter is supported by the fact that SLs affect PIN abundances, auxin transport capacities, and thus cellular auxin (Kapulnik et al., 2011a,b; Ruyter-Spira et al., 2011; Mayzlish-Gati et al., 2012; Machin et al., 2020).
These descriptions clearly show that hormonal regulation of root/root hair development cannot be described by a linear signalling cascade. Instead, the mode of action of different hormones is integrated in a complex, interconnected network of which the developmental output is the result of crosstalk between the latter and direct and indirect effects. It is crucial to emphasize that there are still many outstanding questions regarding hormone interactions during root hair development.
Influence of environmental stressors on root hair development and their interaction with hormones
Rhizosphere composition dramatically impacts root hair development. Nutrients and abiotic stressors are unevenly distributed throughout the soil. Hence, roots encounter heterogeneous conditions along their growth axis, forcing them to dynamically regulate root system architecture and, for example, root hair morphogenesis (Morris et al., 2017; Shahzad and Amtmann, 2017).
Environmental stressors affecting root hair development
Phosphorus (Pi) is a crucial soil-immobile macronutrient that is taken up and recycled by the plant root. It is compartmentalized specifically to the top-soil, as its main source is plant detritus. Plants have evolved specialized mechanisms for Pi sensing, uptake and transport. Mechanistically, Pi status is sensed by SPX-domain-containing proteins. They act as cellular receptors for inositol pyrophosphate molecules (PP-InsP), a proxy for cellular P-content, and regulate Pi homeostasis (Puga et al., 2017). SPX proteins negatively regulate PHOSPHATE STARVATION RESPONSE (PHR) transcription factor and its homologs PHR1-LIKE 1 (PHLs) (Puga et al., 2014; Qi et al., 2017). The SPX–PHR1 complex dissociates in the absence of PP-InsPs, releasing PHR1 to interact with the promoters of Pi starvation induced genes (PSI) (Bustos et al., 2010; Wild et al., 2016). Intriguingly, Ried et al. (2019, Preprint) recently described that InsP8/SPX-induced PHR1 oligomerization is required for PHR-mediated transcriptional regulation during Pi homeostasis. Choi et al. (2017) showed that sustained exposure to Pi-deficient conditions results in a decrease of cellular ATP levels. The latter triggers PP-InsP generation and degradation by VIH1/2, two bifunctional inositol pyrophosphate kinases/phosphatases (Zhu et al., 2019), allowing the plant to maintain its Pi/ATP homeostasis and regulate the Pi starvation response.
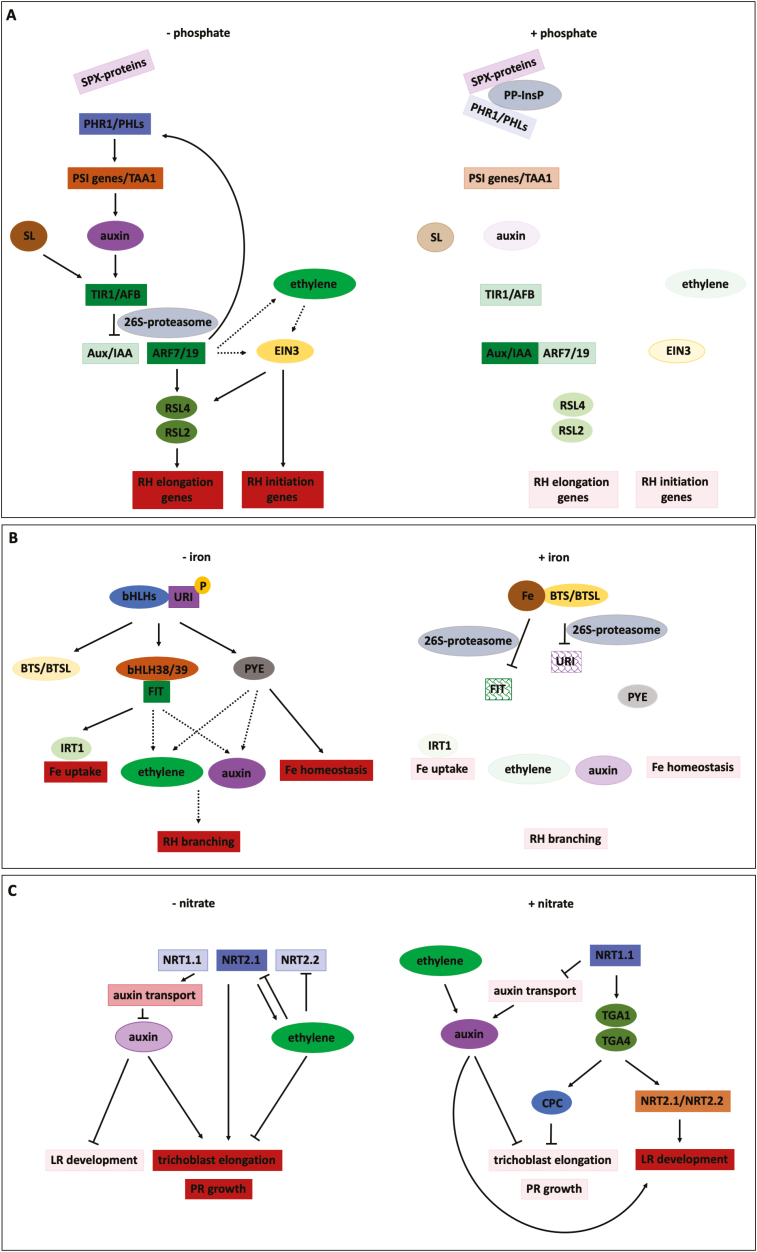
In low phosphate conditions the root is shorter, more lateral roots are formed, and root hairs grow much longer and denser (Fig. 4), facilitating top-soil foraging and increased nutrient uptake (Ma et al., 2001; Koevoets et al., 2016; Zhang et al., 2018). These adaptations occur through local up-regulation of auxin biosynthesis (Bhosale et al., 2018) and ARF7/19-induced PHR1 expression, which promotes root hair growth (Huang et al., 2018a). Recently Bhosale et al. (2018) found that Pi deficiency is first sensed at the root tip, causing up-regulation of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1), a crucial factor in the auxin biosynthetic pathway. AUX1-mediated auxin influx in the lateral root cap and adjacent epidermal cells is then required for increased auxin transport towards the epidermal root hair zone. Subsequently, an auxin-induced genetic cascade is initiated, which involves ARF19, RSL4, and RSL2, enhancing root hair elongation (Fig. 5A). Conversely, high phosphate conditions are able to strongly repress RSL4 expression and root hair growth. Mangano et al. (2018) revealed that when root hairs encounter two conflicting growth signals, i.e. high phosphate levels (inhibits root hair growth) and high auxin levels (enhances root hair growth), RSL2 plays an important role in restoring reactive oxygen species homeostasis and concomitantly in root hair growth recovery. The importance of RSL2 rather than RSL4 in the integration of conflicting growth signals is further illustrated by the finding that the rsl4 mutant displays auxin-enhanced root hair length in either low or high Pi conditions, whereas in the presence of high phosphate, auxin supplementation of rsl2 roots has no phenotypic effect.
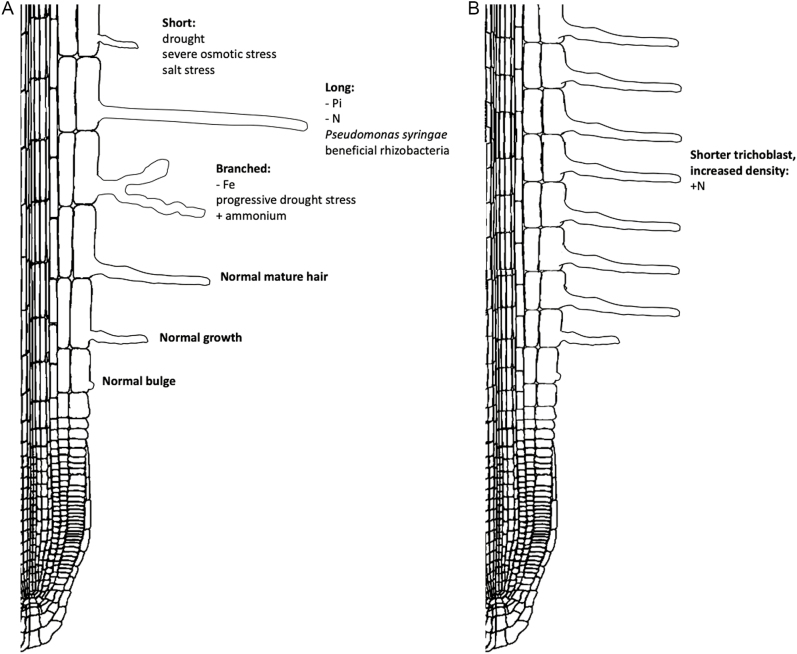
Fig. 4.
Effects of environmental conditions on Arabidopsis root hair development. Schematic representation of a longitundal section through an Arabidopsis root showing root hair development under normal conditions (A) and in altered root environments (B).
Fig. 5.
Models for signalling pathways downstream of phosphate, iron, and nitrate. Simplified models describing how phosphate (A), iron (B) and nitrate (C) deficiency or presence (left and right, respectively) regulate root hair development. Active proteins are represented in dark colour, inactive proteins are in pale colour, and shingled colour indicates the proteins are targeted for ubiquitination and subsequent degradation by the 26S-proteasome. For hormones, full colour and faded colour represent high and low concentrations. Dotted lines represent unknown relationships/interactions. ARF7/19, AUXIN RESPONSE FACTOR 7/19; Aux/IAA, AUXIN/INDOLE-3-ACETIC ACID; bHLHs, basic helix–loop–helix proteins; BTS, BRUTUS; BTSL, BRUTUS-LIKE; CPC, CAPRICE; EIN3, ETHYLENE INSENSITIVE 3; FIT, FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR; IRT1, IRON-REGULATED TRANSPORTER 1; LR, lateral root; NRT1.1/NRT2.1/NRT2.2, NITRATE TRANSPORTER 1.1/2.1/2.2; PHL, PHOSPHATE STARVATION RESPONSE 1-LIKE; PHR1, PHOSPHATE STARVATION RESPONSE 1; PR, primary root; PSI, phosphate starvation induced; PYE, POPEYE; RH, root hair; RSL2/4, ROOT HAIR DEFECTIVE 6-LIKE 2/4; SL, strigolactone; SPX-proteins, SPX-domain containing proteins; TAA1, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1; TGA1/4, TGACG SEQUENCE-SPECIFIC BINDING PROTEIN 1/4; TIR1/AFB, TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-BOX; URI, UPSTREAM REGULATOR OF IRT 1.
According to Song et al. (2016), Pi starvation indirectly leads to ectopic root hair formation by elevation of EIN3 protein levels, in turn up-regulating root hair initiation and elongation genes (Fig. 5A). This was found in a dominant negative mutation of an ethylene receptor (ERS1). It is currently unknown whether the EIN3 level increases due to an increase in ethylene synthesis or ethylene-dependent auxin signalling. Besides auxin, also SLs play a role in Pi-regulated root hair growth. Hence, SL concentrations increase under low Pi and nitrogen levels (Pérez-Torres et al., 2008). In the presence of MAX2, the transcription of the auxin receptor TIR1 is stimulated. This promotes auxin signalling and results in an increase in root hair density and induction of PSI genes (Ma et al., 2001; Chiou and Lin, 2011). Mayzlish-Gati et al. (2012) have shown that the supply of GR24 restores this response in SL-synthesis mutants, pointing to the importance of SL (Fig. 5A).
Iron (Fe) is a crucial micronutrient for plants as it is a key cofactor for various enzymatic reactions. However, Fe solubility is low in aerobic and neutral pH environments, leading to limited bioavailability and consequent Fe deficiency (Ivanov et al., 2012). Arabidopsis roots alter Fe solubility by acidifying the rhizosphere through the action of the H+-ATPase AHA2. Iron is then reduced from ferric (Fe3+) to ferrous (Fe2+) iron by FERRIC REDUCTASE-OXIDASE 2 (FRO2), and the bivalent ion is then imported into root cells by IRON-REGULATED TRANSPORTER 1 (IRT1). In Arabidopsis, UPSTREAM REGULATOR OF IRT1 (URI) becomes phosphorylated under Fe deficiency and forms heterodimers with specific bHLH transcription factors to induce expression of many Fe-deficiency-induced genes, including BRUTUS (BTS), BRUTUS-LIKE 1 (BTSL1), POPEYE (PYE), and other class bHLHs. The latter induce the expression of the transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT), which forms heterodimers with bHLH38/39/100/101 (Wang et al., 2013) and regulates various genes involved in Fe uptake, such as IRT1 and FRO2 (Lan et al., 2012; Kobayashi and Nishizawa, 2015). In parallel, PYE regulates the expression of genes that are involved in Fe homeostasis (Kim et al., 2019). Under low Fe, BTS/BTSLs are inactive, but when Fe availability increases or is restored, Fe binding to the proteins activates their ubiquitin ligase activity (Kobayashi and Nishizawa, 2015) and they interact with the different transcription factor complexes, leading to their degradation (Kim et al., 2019) (Fig. 5B).
Under low Fe conditions, rather than forming ectopic root hairs, plant roots increase their absorptive surface by inducing root hair branching (Müller and Schmidt, 2004; Lan et al., 2012). Several lines of evidence suggest that both auxin and ethylene are involved in this response. The auxin-insensitive mutant axr2-1 (Wilson et al., 1990), mutated in IAA7 (Nagpal et al., 2000), also displays branched root hairs, as under Fe-deficient conditions. Transcriptome analysis showed that Fe deficiency causes differential expression of several genes involved in ethylene biosynthesis and auxin/ethylene signalling (Thimm et al., 2001). It is plausible that these ethylene- and auxin-related genes are targets of the FIT and PYE transcription factors. The latter would explain the altered root hair morphology (due to changes in auxin responsiveness) and (low) percentage of ectopic root hair formation (by ethylene) (Fig. 5B). Importantly, Lan et al. (2011) reported that post-transcriptional and post-translational processes are also involved in the low Fe response.
Nitrogen is a major limiting mineral nutrient that is available in the soil as ammonium (NH4+) and nitrate (NO3−) (Forde, 2002). Besides being an essential nutrient, nitrate also functions as a signalling molecule regulating many different aspects of development, including root development (Walch-Liu et al., 2006). This mobile nutrient is distributed by vertical water flow, known as leaching, and accumulates in deeper soils (Jobbágy and Jackson, 2001). Expectedly, low nitrate induces primary root growth yet inhibits lateral root development, enabling root systems to reach deeper soil layers (Bouguyon et al., 2015; Araya et al., 2016). In contrast, high nitrate conditions lead to slower primary root growth and an increase in root branching, which aids in the foraging for nutrients in that particular nitrate-rich soil patch or layer. When external nitrate concentrations are even higher, lateral root development is arrested in the early stages after emergence. Because of its high concentration in the soil, nitrate accumulates in the shoot and limits basipetal auxin fluxes, which results in lower root auxin and negative regulation of lateral root development (Walch-Liu et al., 2006). Root hairs seem to play an important role in nitrate uptake, as root hair defective mutants accumulate significantly less nitrate than wild-types, while mutants with ectopic root hairs show an increase in nitrate content. Furthermore, root hairs seem important in detecting or sensing changes in nitrogen (Shin et al., 2005). In addition, nitrate stimulates an increase in root hair density (Vatter et al., 2015) by decreasing trichoblast cell lengths and not by inducing ectopic root hair formation, nor by stimulating root hair elongation (Canales et al., 2017).
Two families of transporters participate in the uptake of nitrate in the root, NITRATE TRANSPORTER 1 and 2 (NRT1/2), where NRT1.1 and NRT2.1 are thought to be the most important (Okamoto et al., 2003). To be in the active form as a nitrate transporter, NRT2.1 has to interact with NAR2.1 (also called NRT3.1) and form an NRT2.1–NAR2.1 hetero-oligomer (Yong et al., 2010), whose activity is under post-translational control (Jacquot et al., 2019, Preprint). Currently, NRT1.1 is the only known transporter for nitrate in Arabidopsis (Ho et al., 2009; Giehl and von Wirén, 2015) and functions both as a transporter and as a receptor, called a transceptor. In line with the latter, NRT1-1 loss-of-function mutants failed to show an increase in nitrate-induced root hair density (Vatter et al., 2015). Downstream of NRT1.1, two transcription factors, TGACG SEQUENCE-SPECIFIC BINDING PROTEIN 1 and 4 (TGA1/TGA4), play a redundant role in increasing the expression of CPC, which seems to be required for the reduction of trichoblast cell length and the subsequent increase in root hair density (Canales et al., 2017). In addition, both transcription factors regulate the expression of NRT2.1 and NRT2.2, which are required for the induction of lateral root development but not for the reduction of cell elongation in the primary root (and resulting increase in root hair density). Furthermore, the nitrate transport function of NRT1.1 is needed for the increase in expression of the auxin receptor AFB3 in pericycle cells and its downstream target, NAC4 (Álvarez et al., 2014; Vidal et al., 2014). The involvement of auxin is further evidenced by the fact that NRT1.1 can facilitate the uptake of both nitrate and auxin. Consequently, in low nitrate conditions, auxin is taken up, while in higher nitrate conditions, nitrate outcompetes auxin leading to decreased auxin uptake through NRT1.1 and, consequently, auxin accumulation. This increased auxin level then affects lateral root development (Krouk et al., 2010). Tian et al. (2009) have described a rapid rise in ethylene production upon exposure to high nitrate levels. It is known that increased ethylene concentrations lead to reduced root epidermal cell elongation and thus an increase in root hair density (Swarup et al., 2007; Markakis et al., 2013). The observation that both ethylene and auxin impact epidermal cell length could also explain the involvement of NRT1.1 in the shorter trichoblast cell length in high nitrate conditions (Fig. 5C).
In nitrate-deficient conditions, simultaneous NRT1.1, NRT2.1, and NRT2.2 expression is required for maintaining root growth (Ye et al., 2019). Zheng et al. (2013) have demonstrated that NTR2.1 is strongly up-regulated in low nitrate conditions in an attempt to enhance nitrate uptake. In parallel, ethylene biosynthesis is up-regulated and dependent on NRT2.1. Surprisingly, ethylene reduced the expression of NRT1.1 and NRT2.1 and concomitantly the uptake of nitrate, which acts as a negative feedback loop (Fig. 5C; Tian et al., 2009). No data on root hair responses are available.
Furthermore, Arabidopsis possesses six AMMONIUM TRANSPORTERS (AMT1.1–5 and AMT2; Yuan et al., 2007). The expression of AMT1.1 and AMT2 is up-regulated in N-deprived roots (Sohlenkamp et al., 2002; Lanquar et al., 2009), and root hair elongation is increased in this condition (Bloch et al., 2011). In contrast, ammonium was found to stimulate root hair branching, yet inhibit their elongation (Yang et al., 2011).
During drought, plants are confronted with osmotic stress, causing cell dehydration and membrane/macromolecule damage (Lisar et al., 2012). Primary root growth, lateral root formation, and root hair development are reduced upon osmotic stress, a process that is heavily controlled by ABA in a complex interacting network with cytokinin and other phytohormones (De Smet et al., 2003; Nakashima et al., 2014; Singh and Laxmi, 2015; Rowe et al., 2016; Tiwari et al., 2017). ABA stimulates the expression of the gene for ABSCISIC ACID INSENSITIVE4 (ABI4) transcription factor, among many other genes, which in turn inhibits PIN1 expression, leading to reduced polar auxin transport (Shkolnik-Inbar and Bar-Zvi, 2010) and thus lower auxin levels in the root. Besides having a direct inhibitory effect on root hair development, ABA could thus also act indirectly through changing the levels of other hormones. Under progressive drought stress, root hairs seemed to branch more often, which was also mediated by ABA and auxin, probably in a similar (concentration-dependent) manner (Bobrownyzky, 2016).
Increased salinity inhibits plant growth by creating osmotic stress and inhibiting photosynthesis in the shoot (Koevoets et al., 2016). In addition, primary and lateral root growth and root hair development are suppressed during salt stress (Wang et al., 2008; Mabrouk and Heikal, 2008). Salt exposure causes down-regulation of PIN genes and stabilization of AUXIN RESISTANT3 (AXR3)/IAA17, reducing polar auxin transport and inhibiting auxin signalling, respectively. The resulting reduction in auxin levels and signalling then negatively affects root hair development (Liu et al., 2015).
Biotic factors, such as fungi, bacteria, insects, and pathogens can influence plant development. The main actors in the defence against such biotic factors are salicylic acid (SA), which offers biotroph resistance, and jasmonic acid (JA), which mediates necrotroph resistance (Takatsuji and Jiang, 2014). From the six major hormones discussed in this review, only ethylene and cytokinin positively interact with SA and JA and provoke the synthesis of pathogenesis-related proteins (Yang et al., 2015). Pathogenic strains of Pseudomonas syringae inhibit primary root elongation and stimulate lateral root and root hair formation/elongation, which requires functional ethylene signalling and does not seem to be associated with changed auxin levels (Pecenková et al., 2017). The same effect is noticeable in environments containing beneficial rhizobacteria (Pecenková et al., 2017) and mycorrhizal, saprotrophic, and pathogenic fungi. In their presence, a peculiar clumped root phenotype is observed, marked by a short primary root and more or longer lateral roots (Casarrubia et al., 2016). Unfortunately, the effect on root hair development has not been reported.
Environmental stressors with no reported effect on root hair development
Temperature changes greatly impact on root functioning. For instance, seedling growth at 26–29 °C stimulates primary root elongation (Hanzawa et al., 2013; Wang et al., 2016; Martins et al., 2017; Feraru et al., 2019). Although both auxin and BR were reported to be involved in this response, the underlying hormonal mechanism is currently under debate. Yet, it was recently shown that enhanced primary root growth at higher temperatures coincided with increased auxin signalling. The latter is linked to the inhibition of PIN-LIKES6 (PILS6) proteins that limit nuclear auxin by sequestering auxin to the ER (Feraru et al., 2019). The effect of high temperature exposure on root hair development has not been studied/reported to date.
Shibasaki et al. (2009) have shown that acute cold stress (4 °C) leads to inhibition of root elongation due to blocked basipetal auxin transport. The latter was caused by reduced intracellular trafficking of PIN2 and PIN3. After low temperature acclimation, the elongation rate recovers. It is plausible to think that cold-affected auxin concentrations regulate root hair development, but the exact effect of low temperatures on root hair development is still largely unexplored.
Conclusion and future remarks
A large effort has gone into understanding the regulatory mechanisms controlling root hair development, with the aim of harnessing this knowledge to improve root functioning in future environmental conditions (e.g. nutrient depletion, water deprivation). The combination of state-of-the-art tools to study the molecular-genetic background of plant growth has led to a drastically improved understanding of how different plant hormones impinge on the root hair growth pathway.
In this review we have highlighted the general root hair developmental scheme and how hormones control this. Combined, it is clear that highly complex hormonal crosstalk and sensing of rhizosphere conditions lie at the base of root hair morphogenesis. At this stage, constructing an integrated view of the root hair signalling cascade, including hormonal and environmental control, is too optimistic. Yet, with the scientific community increasingly embracing the complexity and interconnectivity of the involved pathways, and harnessing the power of -omic datasets and hormone marker and sensor lines, we are closing in on deciphering the role of novel genes in the different phases of root hair development. Integration of all these data allows us to link the environmental changes to changes in hormone levels and finally to their effect on the (sub)cellular processes that actually guide root hair growth.
Acknowledgements
The authors acknowledge the financial support by the University of Antwerp (BOF-DOCPRO4) and the Research Foundation Flanders (FWO Post-doc fellowship).
References
- Álvarez JM, Riveras E, Vidal EA, et al. . 2014. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- An L, Zhou Z, Sun L, et al. . 2012. A zinc finger protein gene ZFP5 integrates phytohormone signaling to control root hair development in Arabidopsis. The Plant Journal 72, 474–490. [DOI] [PubMed] [Google Scholar]
- Araya T, Kubo T, von Wirén N, Takahashi H. 2016. Statistical modeling of nitrogen-dependent modulation of root system architecture in Arabidopsis thaliana. Journal of Integrative Plant Biology 58, 254–265. [DOI] [PubMed] [Google Scholar]
- Balcerowicz D, Schoenaers S, Vissenberg K. 2015. Cell fate determination and the switch from diffuse growth to planar polarity in Arabidopsis root epidermal cells. Frontiers in Plant Science 6, 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. 2005. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, 291–298. [DOI] [PubMed] [Google Scholar]
- Bhosale R, Giri J, Pandey BK, et al. . 2018. Author Correction: A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nature Communications 9, 1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. 2007. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. The Plant Cell 19, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A, Lehesranta S, Vatén A, Help H, El-Showk S, Scheres B, Helariutta K, Mähönen AP, Sakakibara H, Helariutta Y. 2011. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Current Biology 21, 927–932. [DOI] [PubMed] [Google Scholar]
- Bloch D, Monshausen G, Singer M, Gilroy S, Yalovsky S. 2011. Nitrogen source interacts with ROP signaling in root hair tip-growth. Plant Cell and Environment 34, 76–88. [DOI] [PubMed] [Google Scholar]
- Bobrownyzky J. 2016. Production of branched root hairs under progressive drought stress in Arabidopsis thaliana. Cytology and Genetics 50, 324–329. [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, et al. . 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1, 15015. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA. 2013. Diverse roles of strigolactones in plant development. Molecular Plant 6, 18–28. [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, et al. . 2012. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genetics 8, e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. 2010. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genetics 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales J, Contreras-López O, Álvarez JM, Gutiérrez RA. 2017. Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. The Plant Journal 92, 305–316. [DOI] [PubMed] [Google Scholar]
- Casarrubia S, Sapienza S, Fritz H, Daghino S, Rosenkranz M, Schnitzler JP, Martin F, Perotto S, Martino E. 2016. Ecologically different fungi affect Arabidopsis development: contribution of soluble and volatile compounds. PLoS One 11, e0168236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. 2003. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. The Plant Cell 15, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhu W, Chen Y, Ito S, Asami T, Wang X. 2014. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 3, e02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Choi HS, Seo M, Cho HT. 2018. Two TPL-binding motifs of ARF2 are involved in repression of auxin responses. Frontiers in Plant Science 9, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Rajagopal A, Xu YF, Rabinowitz JD, O’Shea EK. 2017. A systematic genetic screen for genes involved in sensing inorganic phosphate availability in Saccharomyces cerevisiae. PLoS One 12, e0176085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. 2011. Brassinosteroids. The Arabidopsis Book 9, e0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Achard P. 2013. Gibberellin signaling in plants. Development 140, 1147–1151. [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P. 2016. A pivotal role of DELLAs in regulating multiple hormone signals. Molecular Plant 9, 10–20. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Devlin RM, Brown DP. 1969. Effect of gibberellic acid on the elongation rate of Agrostis alba root hairs. Plant Physiology 22, 759–763. [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H. 2003. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. The Plant Journal 33, 543–555. [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. 1994. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120, 2465–2474. [Google Scholar]
- Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C. 2010. Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signalling, RTE1. The Journal of Biological Chemistry 285, 40706–40713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Van den Broeck L, Inzé D. 2018. The pivotal role of ethylene in plant growth. Trends in Plant Science 23, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S, Ruonala R, Helariutta Y. 2013. Crossing paths: cytokinin signalling and crosstalk. Development 140, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Feng Y, Xu P, Li B, et al. . 2017. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proceedings of the National Academy of Sciences, USA 114, 13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Barbez E, Waidmann S, Sun L, Gaidora A, Kleine-Vehn J. 2019. PILS6 is a temperature-sensitive regulator of nuclear auxin input and organ growth in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 116, 3893–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. 2002. Local and long-range signaling pathways regulating plant responses to nitrate. Annual Review of Plant Biology 53, 203–224. [DOI] [PubMed] [Google Scholar]
- Forde BG, Lorenzo H. 2001. The nutritional control of root development. Plant Soil 232, 51–68. [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl RF, von Wirén N. 2015. Nitrate signalling: functions of a nitrate transceptor. Nature Plants 1, 15021. [DOI] [PubMed] [Google Scholar]
- Grierson C, Nielsen E, Ketelaarc T, Schiefelbein J. 2014. Root hairs. The Arabidopsis Book 12, e0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haling RE, Brown LK, Bengough AG, Young IM, Hallett PD, White PJ, George TS. 2013. Root hairs improve root penetration, root-soil contact, and phosphorus acquisition in soils of different strength. Journal of Experimental Botany 64, 3711–3721. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chae HS, Kieber JJ. 2009. Regulation of ACS protein stability by cytokinin and brassinosteroid. The Plant Journal 57, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa T, Shibasaki K, Numata T, Kawamura Y, Gaude T, Rahman A. 2013. Cellular auxin homeostasis under high temperature is regulated through a SORTING NEXIN1-dependent endosomal trafficking pathway. The Plant Cell 25, 3424–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhou L, Huang M, He X, Yang Y, Liu X, Li Y, Hou X. 2018. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nature Plants 4, 289–298. [DOI] [PubMed] [Google Scholar]
- Huang KL, Ma GJ, Zhang ML, et al. . 2018. a The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiology 178, 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hou L, Meng J, You H, Li Z, Gong Z, Yang S, Shi Y. 2018b The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Molecular Plant 11, 970–982. [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Kieber JJ. 2002. Cytokinin signaling in Arabidopsis. The Plant Cell 14 Suppl, S47–S59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. 2012. Cytokinin signaling networks. Annual Review of Plant Biology 63, 353–380. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M. 2009. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nature Cell Biology 11, 731–738. [DOI] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Bauer P. 2012. Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Molecular Plant 5, 27–42. [DOI] [PubMed] [Google Scholar]
- Jacquot A, Chaput V, Mauries A, et al. . 2019. NRT2.1 phosphorylation prevents root high affinity nitrate uptake activity in Arabidopsis thaliana. bioRxiv, 583542. Preprint. [DOI] [PubMed] [Google Scholar]
- Jobbágy EG, Jackson RB. 2001. The distribution of soil nutrients with depth: global patterns and the imprint of plants. Biogeochemistry 53, 51–77. [Google Scholar]
- Ju C, Chang C. 2015. Mechanistic insights in ethylene perception and signal transduction. Plant Physiology 169, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux PM, Resnick N, et al. . 2011. a Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216. [DOI] [PubMed] [Google Scholar]
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H. 2011b Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. Journal of Experimental Botany 62, 2915–2924. [DOI] [PubMed] [Google Scholar]
- Kermode AR. 2005. Role of abscisic acid in seed dormancy. Journal of Plant Growth Regulation 24, 319–344. [Google Scholar]
- Kieber JJ, Schaller GE. 2014. Cytokinins. The Arabidopsis Book 12, e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, LaCroix IS, Gerber SA, Guerinot ML. 2019. The iron deficiency response in Arabidopsis thaliana requires the f-phosphorylated transcription factor URI. Proceedings of the National Academy of Sciences, USA 116, 24933–24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. 2003. AXR3 and SHY2 interact to regulate root hair development. Development 130, 5769–5777. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2015. Intracellular iron sensing by the direct binding of iron to regulators. Frontiers in Plant Science 6, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koevoets IT, Venema JH, Elzenga JT, Testerink C. 2016. Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Frontiers in Plant Science 7, 1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, et al. . 2010. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell 18, 927–937. [DOI] [PubMed] [Google Scholar]
- Kubeš M, Napier R. 2019. Non-canonical auxin signalling: fast and curious. Journal of Experimental Botany 70, 2609–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Chen AY, Nemhauser JL. 2009. Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proceedings of the National Academy of Sciences, USA 106, 8073–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwah S, Jones AM, Laxmi A. 2011. Cytokinin-induced root growth involves actin filament reorganization. Plant Signaling & Behavior 6, 1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Schmidt W. 2012. Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiology 159, 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W. 2011. iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiology 155, 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Loqué D, Hörmann F, Yuan L, Bohner A, Engelsberger WR, Lalonde S, Schulze WX, von Wirén N, Frommer WB. 2009. Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. The Plant Cell 21, 3610–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RD, Cho HT. 2013. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Frontiers in Plant Science 4, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK. 2013. A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. The Plant Cell 25, 3329–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SB, Xie ZZ, Hu CG, Zhang JZ. 2016. A review of auxin response factors (ARFs) in plants. Frontiers in Plant Science 7, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li G, Li Y, Kong X, Zhang L, Wang J, Li X, Yang Y. 2018. ABA receptor subfamily III enhances abscisic acid sensitivity and improves the drought tolerance of Arabidopsis. International Journal of Molecular Sciences 19, E1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisar SYS, Motafakkerazad R, Hossain MM, Rahman IMM. 2012. Water stress in plants: causes, effects and responses. In: Rahman IMM, Hasegawa H, eds. Water stress. London: InTech, 1–14. [Google Scholar]
- Liu J, Moore S, Chen C, Lindsey K. 2017. Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Molecular Plant 10, 1480–1496. [DOI] [PubMed] [Google Scholar]
- Liu W, Li RJ, Han TT, Cai W, Fu ZW, Lu YT. 2015. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiology 168, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MC, Lamattina L. 2018. Abscisic acid and nitric oxide modulate cytoskeleton organization, root hair growth and ectopic hair formation in Arabidopsis. Nitric Oxide 80, 89–97. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. 2003. The role of nutrient availability in regulating root architecture. Current Opinion in Plant Biology 6, 280–287. [DOI] [PubMed] [Google Scholar]
- Luo X, Chen Z, Gao J, Gong Z. 2014. Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. The Plant Journal 79, 44–55. [DOI] [PubMed] [Google Scholar]
- Ma Q, Grones P, Robert S. 2018. Auxin signaling: a big question to be addressed by small molecules. Journal of Experimental Botany 69, 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Ma Z, Bielenberg DG, Brown KM, Lynch JP. 2001. Regulation of root hair density by phosphorus availability in Arabidopsis thaliana. Plant, Cell and Environment 24, 459–467. [Google Scholar]
- Mabrouk YM, Heikal H. 2008. Characterization of a T-DNA insertion mutant for NaCl-hypersensitivity in Arabidopsis thaliana. Australian Journal of Basic and Applied Sciences 2, 82–89. [Google Scholar]
- Machin DC, Hamon-Josse M, Bennett T. 2020. Fellowship of the rings: a saga of strigolactones and other small signals. New Phytologist 225, 621–636. [DOI] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Choi HS, et al. . 2017. Molecular link between auxin and ROS-mediated polar growth. Proceedings of the National Academy of Sciences, USA 114, 5289–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Marzol E, Borassi C, Estevez JM. 2018. High auxin and high phosphate impact on RSL2 expression and ROS-homeostasis linked to root hair growth in Arabidopsis thaliana. Frontiers in Plant Science 9, 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benková E. 2014. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Current Biology 24, 1031–1037. [DOI] [PubMed] [Google Scholar]
- Markakis MN, Boron AK, Van Loock B, Saini K, Cirera S, Verbelen JP, Vissenberg K. 2013. Characterization of a small auxin-up RNA (SAUR)-like gene involved in Arabidopsis thaliana development. PLoS One 8, e82596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S, Montiel-Jorda A, Cayrel A, Huguet S, Roux CP, Ljung K, Vert G. 2017. Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nature Communications 8, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1994. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiology 106, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. 1996. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. The Plant Cell 8, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, et al. . 2012. Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiology 160, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. . 2009. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel MA, Postma JA, Lynch JP. 2015. Phene synergism between root hair length and basal root growth angle for phosphorus acquisition. Plant Physiology 167, 1430–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniuszko G. 2015. Ethylene signaling pathway is not linear, however its lateral part is responsible for sensing and signaling of sulfur status in plants. Plant Signaling & Behavior 10, e1067742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S, Bishopp A, Carvalho H, Campilho A. 2013. AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PLoS One 8, e56370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, et al. . 2017. Shaping 3D root architecture. Current Biology 27, R919–R930. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sozzani R, et al. . 2013. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Developmental Cell 26, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM. 2012. Auxin and ethylene: collaborators or competitors? Trends in Plant Science 17, 181–195. [DOI] [PubMed] [Google Scholar]
- Müller M, Schmidt W. 2004. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiology 134, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D, Byrne H, King J, Voss U, Kieber J, Bennett M. 2011. The influence of cytokinin-auxin cross-regulation on cell-fate determination in Arabidopsis thaliana root development. Journal of Theoretical Biology 283, 152–167. [DOI] [PubMed] [Google Scholar]
- Müssig C, Shin GH, Altmann T. 2003. Brassinosteroids promote root growth in Arabidopsis. Plant Physiology 133, 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. 2000. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiology 123, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. 2014. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Frontiers in Plant Science 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan T, Chen J, Yin Y. 2017. Crosstalk of brassinosteroid signalling in controlling growth and stress responses. The Biochemical Journal 474, 2641–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G. 2004. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proceedings of the National Academy of Sciences, USA 101, 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. 2003. Gibberellin biosynthesis and response during Arabidopsis seed germination. The Plant Cell 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H, Kamada H, Sudo H, Harada H. 1989. Effects of gibberellic acid on hairy root growth in Datura innoxia. Journal of Plant Physiology 134, 633–636. [Google Scholar]
- Okamoto M, Vidmar JJ, Glass AD. 2003. Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant & Cell Physiology 44, 304–317. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, et al. . 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. The Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Weijers D. 2016. Q&A: Auxin: the plant molecule that influences almost anything. BMC Biology 14, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecenková T, Janda M, Ortmannová J, Hajná V, Stehlíková Z, Žárský V. 2017. Early Arabidopsis root hair growth stimulation by pathogenic strains of Pseudomonas syringae. Annals of Botany 120, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. 2008. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. The Plant Cell 20, 3258–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L. 2013. Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proceedings of the National Academy of Sciences, USA 110, 9571–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. 1998. Auxin and ethylene promote root hair elongation in Arabidopsis. The Plant Journal 16, 553–560. [DOI] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI. 2019. Brassinosteroid signalling in plant development and adaptation to stress. Development 146, dev151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S, Pěnčík A, Rashed A, et al. . 2016. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, 11016–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, et al. . 2014. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proceedings of the National Academy of Sciences, USA 111, 14947–14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares J. 2017. Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Current Opinion in Plant Biology 39, 40–49. [DOI] [PubMed] [Google Scholar]
- Qi W, Manfield IW, Muench SP, Baker A. 2017. AtSPX1 affects the AtPHR1-DNA-binding equilibrium by binding monomeric AtPHR1 in solution. The Biochemical Journal 474, 3675–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Huang R. 2018. Auxin controlled by ethylene steers root development. International Journal of Molecular Sciences 19, 3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S. 2002. Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiology 130, 1908–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried MK, Wild R, Zhu J, Broger L, Harmel RK, Hothorn LA, Fiedler D, Hothorn M. 2019. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. bioRxiv, 875393. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles L, Stepanova A, Alonso J. 2013. Molecular mechanisms of ethylene-auxin interaction. Molecular Plant 6, 1734–1737. [DOI] [PubMed] [Google Scholar]
- Rowe JH, Topping JF, Liu J, Lindsey K. 2016. Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytologist 211, 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C, Kohlen W, Charnikhova T, et al. . 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiology 155, 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E. 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. The Plant Cell 19, 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Schäfer S, et al. . 2017. ABA suppresses root hair growth via the OBP4 transcriptional regulator. Plant Physiology 173, 1750–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage NS, Walker T, Wieckowski Y, Schiefelbein J, Dolan L, Monk NA. 2008. A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biology 6, e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. The Plant Cell 27, 44–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenaers S, Balcerowicz D, Breen G, et al. . 2018. The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Current Biology 28, 722–732.e6. [DOI] [PubMed] [Google Scholar]
- Schoenaers S, Balcerowicz D, Vissenberg K. 2017. Molecular mechanisms regulating root hair tip growth: a comparison with pollen tubes. In: Obermeyer G, Feijó JA, eds. Pollen tip growth. Berlin, Heidelberg: Springer, 167–243. [Google Scholar]
- Shakeel SN, Gao Z, Amir M, Chen YF, Rai MI, Haq NU, Schaller GE. 2015. Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana. The Journal of Biological Chemistry 290, 12415–12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad Z, Amtmann A. 2017. Food for thought: how nutrients regulate root system architecture. Current Opinion in Plant Biology 39, 80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki K, Uemura M, Tsurumi S, Rahman A. 2009. Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. The Plant Cell 21, 3823–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman D. 2005. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorous and potassium deficiency. Plant and Cell Physiology 46, 1350–1357. [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D. 2010. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. The Plant Cell 22, 3560–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Laxmi A. 2015. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Frontiers in Plant Science 6, 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK. 2002. Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiology 130, 1788–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yu H, Dong J, Che X, Jiao Y, Liu D. 2016. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genetics 12, e1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava LM. 2002. Plant growth and development: Hormones and environment. Oxford: Academic Press. [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. 2005. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. The Plant Cell 17, 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM. 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. The Plant Cell 19, 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Chen GL, Bartel B. 2010. Ethylene directs auxin to control root cell expansion. The Plant Journal 64, 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Liu YB, Zhang XS. 2011. Auxin-cytokinin interaction regulates meristem development. Molecular Plant 4, 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Perry P, Hagenbeek D, Van Der Straeten D, Beemster GT, Sandberg G, Bhalerao R, Ljung K, Bennett MJ. 2007. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. The Plant Cell 19, 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H, Jiang CJ. 2014. Plant hormone crosstalks under biotic stresses. In: Tran LS, Pal S, eds. Phytohormones: a window to metabolism, signalling and biotechnological applications. New York: Springer. [Google Scholar]
- Teale WD, Paponov IA, Palme K. 2006. Auxin in action: signalling, transport and the control of plant growth and development. Nature Reviews. Molecular Cell Biology 7, 847–859. [DOI] [PubMed] [Google Scholar]
- Thimm O, Essigmann B, Kloska S, Altmann T, Buckhout TJ. 2001. Response of Arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiology 127, 1030–1043. [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW. 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Tian QY, Sun P, Zhang WH. 2009. Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytologist 184, 918–931. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Lata C, Chauhan PS, Prasad V, Prasad M. 2017. A functional genomic perspective on drought signalling and its crosstalk with phytohormone-mediated signalling pathways in plants. Current Genomics 18, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. 2004. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiology 135, 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M, Benková E. 2012. Hormonal interactions in the regulation of plant development. Annual Review of Cell and Developmental Biology 28, 463–487. [DOI] [PubMed] [Google Scholar]
- Vatter T, Neuhäuser B, Stetter M, Ludewig U. 2015. Regulation of length and density of Arabidopsis root hairs by ammonium and nitrate. Journal of Plant Research 128, 839–848. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Álvarez JM, Gutiérrez RA. 2014. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signaling & Behavior 9, e28501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG. 2006. Nitrogen regulation of root branching. Annals of Botany 97, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ. 2013. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Molecular Plant 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M. 2016. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nature Communications 7, 10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu C, Li K, et al. . 2007. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Molecular Biology 64, 633–644. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Li K, Sun F, Han C, Wang Y, Li X. 2008. Salt-induced plasticity of root hair development is caused by ion disequilibrium in Arabidopsis thaliana. Journal of Plant Research 121, 87–96. [DOI] [PubMed] [Google Scholar]
- Wang YH, Irving HR. 2011. Developing a model of plant hormone interactions. Plant Signaling & Behavior 6, 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li J. 2016. Brassinosteroids regulate root growth, development, and symbiosis. Molecular Plant 9, 86–100. [DOI] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annual Review of Plant Biology 67, 539–574. [DOI] [PubMed] [Google Scholar]
- Werner T, Schmülling T. 2009. Cytokinin action in plant development. Current Opinion in Plant Biology 12, 527–538. [DOI] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, et al. . 2016. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. 1990. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Molecular & General Genetics 222, 377–383. [DOI] [PubMed] [Google Scholar]
- Wu J, Kurten EL, Monshausen G, Hummel GM, Gilroy S, Baldwin IT. 2007. NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. The Plant Journal 52, 877–890. [DOI] [PubMed] [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. 2003. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. The Journal of Biological Chemistry 278, 49102–49112. [DOI] [PubMed] [Google Scholar]
- Yang N, Zhu C, Gan L, Ng D, Xia K. 2011. Ammonium-stimulated root hair branching is enhanced by methyl jasmonate and suppressed by ethylene in Arabidopsis thaliana. Journal of Plant Biology 54, 92–100. [Google Scholar]
- Yang Y, Ahammed GJ, Wu C, Fan S-Y, Zhou Y-H. 2015. Crosstalk among jasmonate, salicylate and ethylene signalling pathways in plant disease and immune responses. Current Protein and Peptide Science 16, 450–461. [DOI] [PubMed] [Google Scholar]
- Ye JY, Tian WH, Jin CW. 2019. A reevaluation of the contribution of NRT1.1 to nitrate uptake in Arabidopsis under low-nitrate supply. FEBS Letters 593, 2051–2059. [DOI] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. 2010. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nature Genetics 42, 264–267. [DOI] [PubMed] [Google Scholar]
- Yong Z, Kotur Z, Glass AD. 2010. Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. The Plant Journal 63, 739–748. [DOI] [PubMed] [Google Scholar]
- Yu MH, Zhao ZZ, He JX. 2018. Brassinosteroid signalling in plant-microbe interactions. International Journal of Molecular Sciences 19, 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Loqué D, Kojima S, Rauch S, Ishiyama K, Inoue E, Takahashi H, von Wirén N. 2007. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. The Plant Cell 19, 2636–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zažímalová E, Křeček P, Skůpa P, Hoyerová K, Petrášek J. 2007. Polar transport of the plant hormone auxin – the role of PIN-FORMED (PIN) proteins. Cellular and Molecular Life Sciences 64, 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemlyanskaya EV, Omelyanchuk NA, Ubogoeva EV, Mironova VV. 2018. Deciphering auxin-ethylene crosstalk at a systems level. International Journal of Molecular Sciences 19, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-J, Yang Y-J, Liu C-Y, Zhang F, Wu Q-S. 2018. Root hair growth and development in response to nutrients and phytohormones. In: Giri B, Prasad R, Varma A, eds. Root biology. Soil biology, Vol. 52 Cham: Springer, 65–84. [Google Scholar]
- Zhang S, Huang L, Yan A, Liu Y, Liu B, Yu C, Zhang A, Schiefelbein J, Gan Y. 2016. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. Journal of Experimental Botany 67, 6363–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Han X, An YI, Guo H, Xia X, Yin W. 2013. The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant, cell & Environment 36, 1328–1337. [DOI] [PubMed] [Google Scholar]
- Zhu J, Lau K, Puschmann R, et al. . 2019. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 8, e43582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Sae-Seaw J, Wang ZY. 2013. Brassinosteroid signalling. Development 140, 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]