Summary
Borrelia burgdorferi is a highly motile spirochete due to its periplasmic flagella. Unlike flagella of other bacteria, spirochetes’ periplasmic flagella possess a complex structure called the collar, about which little is known in terms of function and composition. Using various approaches, we have identified a novel protein, BB0326, as a key component of the collar. We show that a peripheral portion of the collar is diminished in the Δbb0326 mutant and restored in the complemented bb0326+ cells, leading us to rename BB0326 as periplasmic flagellar collar protein A or FlcA. The ΔflcA mutant cells produced fewer, abnormally tilted, and shorter flagella, as well as diminished stators, suggesting that FlcA is crucial for flagellar and stator assemblies. We provide further evidence that FlcA interacts with the stator and that this collar-stator interaction is essential for the high torque needed to power the spirochete’s periplasmic flagellar motors. These observations suggest that the collar provides various important functions to the spirochete’s periplasmic flagellar assembly and rotation.
Introduction
The Lyme disease spirochete Borrelia burgdorferi is a highly motile and invasive organism (Wolgemuth, 2015). The motility of the spirochete is generated by the rotation of the periplasmic flagellar motor, which is powered by the proton gradient across the cytoplasmic membrane (Chang et al., 2019, Islam et al., 2015). These flagella reside in the periplasmic space between spirochetes’ peptidoglycan layer and outer membrane. Motility is important for every stage of the Ixodes tick-vertebrate enzootic life cycle of B. burgdorferi (Motaleb et al., 2015, Sultan et al., 2013, Sultan et al., 2015), whose periplasmic flagella are not only responsible for motility, but also determine the spirochete’s characteristic flat-wave morphology (Motaleb et al., 2000, Sultan et al., 2013). Unlike other externally flagellated bacteria, B. burgdorferi possesses 7 to 11 periplasmic flagella at each pole of the cell that wrap inward as a flat ribbon along the cell body and overlap in the middle of the cell (Charon et al., 2009, Hovind-Hougen, 1984, Johnson et al., 1984, Sultan et al., 2015, Sze et al., 2011).
B. burgdorferi possesses more than 50 genes that encode proteins for chemotaxis and motility (Charon et al., 2012, Fraser et al., 1997). In contrast to the transcriptional hierarchy regulation of flagellar systems in well-established bacterial models (Chevance & Hughes, 2017, Chilcott & Hughes, 2000, Niehus et al., 2004, Syed et al., 2009, Wilhelms et al., 2013, Wilhelms et al., 2011), B. burgdorferi appears to possess only the housekeeping σ70 subunit of RNA polymerase to transcribe the chemotaxis and motility genes (Ge et al., 1997, Li et al., 2002, Motaleb et al., 2011b, Yang & Li, 2009). Periplasmic flagella share the highly conserved structures of the external flagella, such as the rod, P-, MS-, and C-rings, and the stator (Chen et al., 2011, Kudryashev et al., 2010, Liu et al., 2009, Murphy et al., 2006, Zhao et al., 2014, Qin et al., 2018). The stator is the torque generating unit consisting of MotA and MotB proteins, while the rotor is composed, minimally, of the MS ring (FliF), the rod, and the C-ring switch complex – FliG, FliM, and FliN (Chang et al., 2019, Sultan et al., 2015). Recently, the cytosolic ATPase complex of the export apparatus in B. burgdorferi was observed to form a novel spoke-hub structure attached to the C-ring, presumably facilitating flagellar assembly (Qin et al., 2018). The P-ring protein FlgI of B. burgdorferi is reported to be important for flagellar hook or filament assembly, however, the P-ring in the spirochete appears to be masked by the spirochete-specific collar structure (Liu et al., 2009).
Periplasmic flagella possess a unique collar structure, which is evidently important for periplasmic flagellar orientation, morphology, and motility of B. burgdorferi (Moon et al., 2016b, Moon et al., 2018). A similar structure in another spirochete has been postulated to limit the tilting of the flagellar hook and further stabilize the stators (Murphy et al., 2006). Yet, little is known about the composition or function of this collar structure. Using various comprehensive analyses, including characterizing almost all flagellar-related genes currently annotated in B. burgdorferi genome, we identified FlbB (BB0286) as the first collar protein (Moon et al., 2016b). FlbB appears to serve as the collar structure’s base and to be crucial for the motility, morphology, and orientation of periplasmic flagella of B. burgdorferi (Moon et al., 2016b). The collar, ~71 nm in diameter and ~24 nm in height, is a large structural component of the periplasmic flagellar basal body and, therefore, presumably composed of multiple proteins (Moon et al., 2016b). Subsequently, we used a protein-protein interaction map, developed in the syphilis-causing spirochete Treponema pallidum, to identify a hypothetical protein, BB0236, as the second collar protein (Moon et al., 2018, Rajagopala et al., 2007). However, given that the collar is a large structure, additional proteins are likely involved in its assembly and function.
In this study, we identified a third collar protein, BB0326, that is annotated as a hypothetical protein with unknown function in B. burgdorferi (Fraser et al., 1997). Using various comprehensive analyses, we found BB0326 to be a key component of the collar structure; therefore, we renamed it as periplasmic flagellar collar protein A (FlcA). More importantly, we found BB0326 or FlcA to directly interact with FlbB, providing further evidence that the collar is a multi-protein complex. Observing that collar-stator structures are adjacent in the basal body, and that all our collar mutants had diminished stator structures, we hypothesize that the collar is crucial for stator assembly (Moon et al., 2016b, Moon et al., 2018). Furthermore, another model already proposes that the stator binds to the peptidoglycan layer and stabilizes the stator complex within the cytoplasmic membrane during torque generation (Kojima et al., 2009, Kojima et al., 2018, Roujeinikova, 2008). Based on our FlcA and MotB interactions data, we propose that in addition to binding to the peptidoglycan layer for its immobilization, the stator interacts with the collar protein FlcA to provide additional support for the flagella to rotate in the periplasmic space.
Results
Identification of BB0326 as a potential flagellar collar protein in B. burgdorferi
Considering the large size of the periplasmic flagellar collar structure, we hypothesize that multiple proteins are involved in the assembly of the collar complex, in addition to established components FlbB and BB0236 (Moon et al., 2016b, Moon et al., 2018).
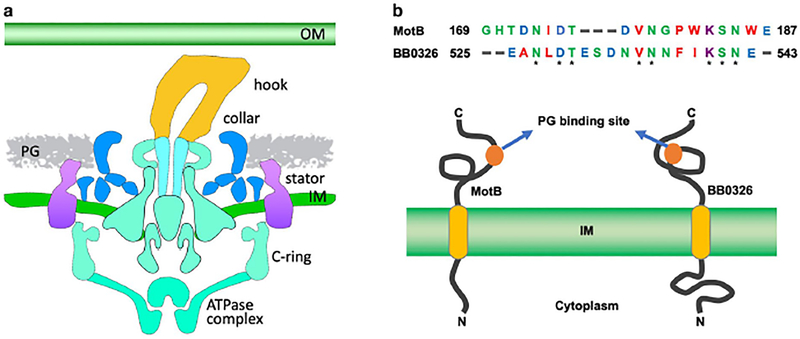
To identify additional collar proteins, we first observed that the stator and collar structures are adjacent in the basal body and surrounded by the peptidoglycan (PG) layer (Fig. 1A) (Murphy et al., 2006, Zhao et al., 2014). Since stator protein MotB binds to the PG, and the collar structure is adjacent to the stator, we predicted that the collar structure also binds to the PG (Fig. 1A). We also hypothesized that the collar protein that binds to the PG also conserves the PG-binding domain of MotB. To test these hypotheses, we first used a PG-binding domain of MotB as the query sequence to search B. burgdorferi protein banks using BLASTP, which yielded no proteins with significant amino acid sequence homology (Altschul et al., 1990, Altschul et al., 1997, Izard et al., 2009). Subsequently, we used a PG-binding loop of MotB (GHTDNIDTDVNGPWKSNWE) as the query sequence for the search. The BLAST search yielded a protein, BB0326, which is annotated as a hypothetical protein with unknown function in B. burgdorferi (Fig. 1B) (Fraser et al., 1997). BB0326 contains a transmembrane domain (aa 329–351) and three TPR motifs at the C-terminus (aa 736–820) (Fig. S1; 1B). Importantly, we found that one domain of BB0326 (aa 525–543) shares 42% amino acid sequence identity with the PG-binding loop of MotB (E-value, 0.019; Fig. 1B). This observation leads us to hypothesize that BB0326 is a flagellar protein involved in the formation of the collar. bb0326 is located within a 3-gene operon bb0325-bb0327 (Fig. S2A). None of the genes in this operon are predicted to be involved in motility or chemotaxis, as the gene product of bb0325 is annotated as a hypothetical protein with unknown function, and that of bb0327 is annotated as glycerol-3-phosphate O-acyltransferase (Fraser et al., 1997). Additionally, our BLAST searches suggest that BB0325 lacks any identifiable domain (not shown).
Figure 1. Identification of BB0326 as a potential collar protein in B. burgdorferi.
(A) An illustration of a periplasmic flagellar motor structure from wild type cells. Each structural component of the motor is labeled. The 2D cartoon is produced from our previously generated B. burgdorferi flagellar motors showing that the PG layer is in direct contact with the collar and stator. (B) Alignment of PG-binding loops of B. burgdorferi MotB (169–187 aa) sequence and BB0326 (525–543 aa) using Clustal Omega program: https://www.ebi.ac.uk/Tools/msa/clustalo/ (Top panel). Asterisks indicate identical amino acid residues between the two sequences. Schematic diagram of secondary structure prediction of MotB and BB0326 proteins showing the PG- and membrane-binding domains (Bottom). Diagram is not in scale.
BB0326 is essential for motility and morphology of B. burgdorferi
To determine the role of bb0326, we inactivated the gene using a kanamycin resistance cassette, PflgB-Kan (Fig. S2A) (Bono et al., 2000b, Motaleb et al., 2000, Motaleb et al., 2007). PCR analysis confirmed the successful construction of the bb0326 mutant (Δbb0326; Fig. S2B). Furthermore, to demonstrate that the Δbb0326 mutant’s phenotype (described below) was due solely to the mutation of bb0326 and not to other alterations elsewhere in the genome, we complemented the mutant in trans with a shuttle vector, pBSV2G, that contained an intact copy of bb0326, fused to a nonnative flagellar promoter flgB (Fig. S2A) (Elias et al., 2003). PCR analysis confirmed the construction of the complemented bb0326 clone (bb0326+; Fig. S2B). Furthermore, we performed qRT-PCR to determine if the Δbb0326 mutant exhibited any polar effect on downstream gene expression. The expression of the downstream (bb0327) or upstream gene (bb0325) was not altered in either Δbb0326 or bb0326+ cells compared to the wild-type B. burgdorferi, indicating that the insertion of PflgB-Kan within the bb0326 did not cause any polar effect (Fig. S3).
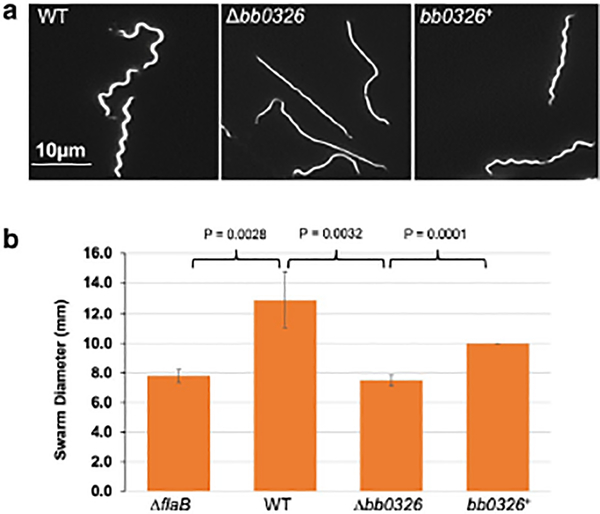
Using Dark-field microscopy and swarm plate assays, we analyzed the Δbb0326 mutant’s morphology and motility phenotypes (Motaleb et al., 2011a). The wild-type cells were motile and exhibited the typical flat-wave morphology, whereas the Δbb0326 cells were completely non-motile and rod-shaped (Fig. 2A). The Δbb0326 cells failed to swarm in the soft-agarose plate which is similar to the previously characterized flagellin-less ΔflaB mutants (Fig. 2B) (Motaleb et al., 2000, Sultan et al., 2013). Complemented bb0326+ cells restored the morphology and motility, though their swarming ability was less than that of wild type cells. This swarming deficiency was likely due to the loss of the multicopy shuttle vector from the bb0326+ cells during growth in the swarm plates that lacked the selective pressure for the complementing vector or overexpression of the bb0326 by the multicopy shuttle vector used for the complementation (Fig. 2B) (Samuels et al., 2018, Tilly et al., 2006).
Figure 2. Morphology and motility phenotypes of the mutant cells.
(A) Dark-field microscopic images showing the distinct rod-shaped morphology of Δbb0326 spirochetes whereas the WT and complemented bb0326+ cells exhibit the characteristic flat-wave morphology. Exponentially growing B. burgdorferi cells were visualized using a dark-field microscope (400x magnification) and images were captured using a digital camera. (B) Average swarm diameters from four independent swarm plates are shown in millimeter scale. A non-motile flagellar mutant ΔflaB was used as a control (Motaleb et al., 2000). Bars represent mean ± standard deviation of the mean from four plates. Statistical analysis was performed by using Student’s t-test. P-values between samples are shown at the top. A P-value of <0.05 was considered as significantly different.
The stator and the periphery of the collar are absent in the Δbb0326 mutant cells
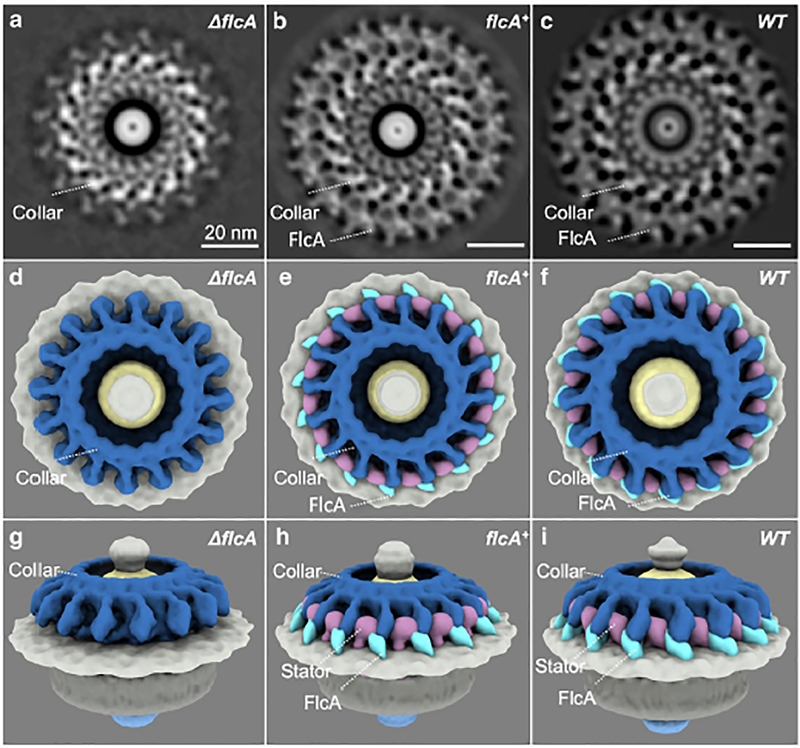
To determine the specific structure and function of BB0326, we determined the flagellar motor structures of Δbb0326 and BB0326+ cells using cryo-electron tomography (cryo-ET) and subtomogram averaging, and compared them with the WT motor structure (Liu et al., 2009, Zhao et al., 2014). We found that the peripheral portion of the collar is diminished in the Δbb0326 mutant and restored in the complemented bb0326+ spirochete motors, indicating that BB0326 is crucial for formation of the peripheral portion of the collar (compare Fig. 3A, D, G with B, E, H or C, F, I). Consequently, we renamed BB0326 as periplasmic flagellar collar protein A (FlcA). Additionally, we observed a diminished stator structure in the Δbb0326 or ΔflcA mutant cells, despite one of the stator proteins, MotB synthesis, remaining intact (compare Fig. 3G with H or I; Fig. S2C). Further comparing the motor structures of a ΔmotB-deletion mutant with those of the ΔflcA mutant, we were able to define the 3D structures and locations of the stator complexes, the periphery, and core parts of the collar (Fig. 3B, E, H; Video 1) (Chang et al., 2019, Sultan et al., 2015). Importantly, complementation of ΔflcA restored the stator complexes and the collar structure, indicating that FlcA is responsible for the formation of the periphery portion of the collar and the assembly of the stator complexes (Fig. 3).
Figure 3. Structures of the flagellar motor from ΔflcA, flcA+ and WT cells.
(A—C) A section of the subtomogram average of the Δbb0326 or ΔflcA, bb0326+ or flcA+ and WT motors, respectively. (D—F) A top view of the 3D rendering of the ΔflcA, flcA+, and WT motors, respectively. (G—I) Side views of the ΔflcA, flcA+, and WT motors, respectively. The core port of the collar is colored in steel blue. The gene products of flcA are colored in cyan. The stator complex is colored in plum. Note that the in flcA+ cells, the collar and other structures appear to be slightly different from the wild-type cells (B, C) likely due to the overexpression of flcA from the multicopy shuttle vector used to complement the mutant.
ΔflcA mutant cells show defects on the orientation and length of periplasmic flagella
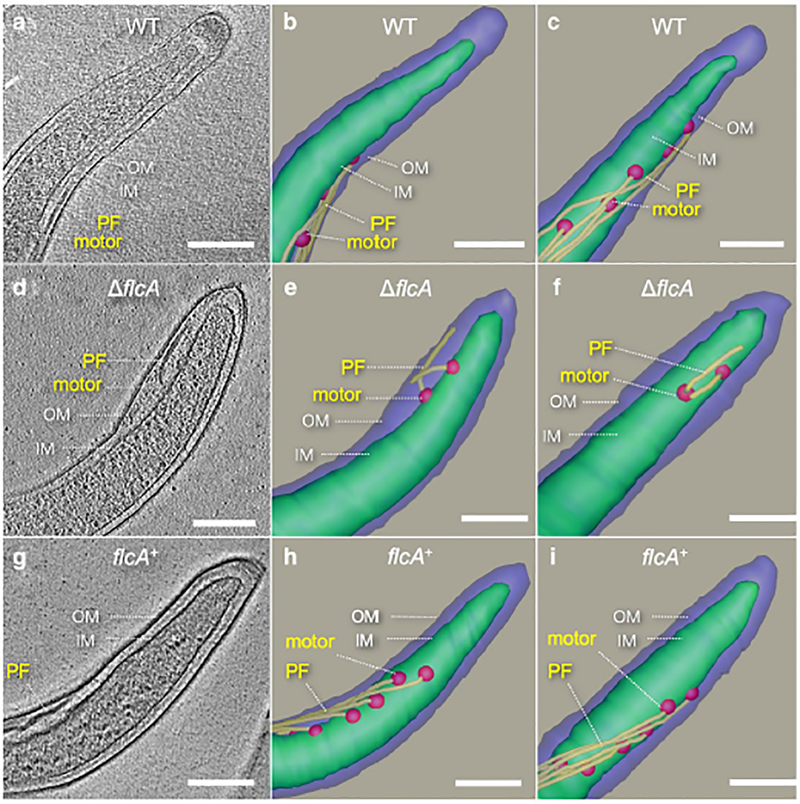
Previous studies have shown that periplasmic flagella are crucial for both motility and morphology of B. burgdorferi (Motaleb et al., 2000, Sultan et al., 2013). Since the ΔflcA mutant cells are rod-shaped and non-motile, like the flagellin-less ΔflaB cells, we predicted that the mutant cells may have defective periplasmic flagella. To confirm this, we used cryo-ET to visualize the mutant cells (Fig. 4). Cryo-ET reconstructions of the cell tips indicate that ΔflcA cells assembled relatively fewer flagellar motors, and thus fewer flagella per cell, compared to the wild-type or complemented cells (Table 1 and Fig. 4). On average, the ΔflcA cells possessed approximately 26% and 35% fewer motors than the WT and flcA+ cells, respectively (Table 1). Additionally, fewer flagellar filaments in the mutant cells resulted in fewer FlaB proteins than those in the wild-type or the complemented cells (see 41 kDa FlaB in SDS-PAGE/western blotting in Fig. S2C). Unlike the wild-type periplasmic flagella with normal orientation, in which the filaments of one pole extend toward the other pole of the cell as they wrap the cell cylinder, the mutant flagella appear to show abnormal orientations, with filaments extending toward their pole of origin. This abnormal orientation phenotype was also observed in our previously reported mutants in genes encoding collar proteins FlbB, BB0236 and FliL (Moon et al., 2016b, Moon et al., 2018, Motaleb et al., 2011a). Furthermore, the periplasmic flagella of the ΔflcA cells are also shorter in length (Fig. 4D—F). Specifically, 43% of periplasmic flagella are found to be shorter, with 34% oriented abnormally in the ΔflcA cells (Table 1). Complementation of ΔflcA restored the phenotypes to the wild-type level (Table 1; Fig. 4G—I). These results indicate that FlcA plays an important role in the assembly and orientation of the periplasmic flagella.
Figure 4. Periplasmic flagella of WT, ΔflcA and flcA+ cells.
(A) A tomographic slice of a WT cell shows normal regular periplasmic flagella. (B, C) Two surface views of the WT cell in two different orientations, respectively. (D) A representative tomographic slice of a ΔflcA cell showing short periplasmic flagella with irregular orientation. (E, F) Two surface views of the ΔflcA cell. (G) A representative tomographic slice of a flcA+ cell. (H, I) Two surface views of the flcA+ cell. The motors are colored in red, the filaments in yellow. The scale bar is 200 nm.
Table 1.
Phenotype of ΔflcA mutant’s periplasmic flagella detected by cryo-ET.
| Strain | No. of cells analyzed | Total no. of flagella detected* | No. of irregular flagella (%) | No. of short flagella (%) |
|---|---|---|---|---|
| WT | 26 | 180 | 0 (0%) | 4 (2.2%) |
| ΔflcA | 26 | 134 | 46 (34.3) | 58 (43.3%) |
| flcA+ | 46 | 366 | 0 (0%) | 10 (2.7%) |
Total number of periplasmic flagella (normal and abnormal) were detected from the cells analyzed shown in the left column. Percentages were determined by dividing the number of irregular periplasmic flagella by the total number of periplasmic flagella. A normal flagellum is defined as being oriented toward the other pole of the cell or cell body. An abnormal or irregular periplasmic flagellum is defined as being tilted toward the cell pole from where they were originated.
FlcA directly interacts with other collar proteins
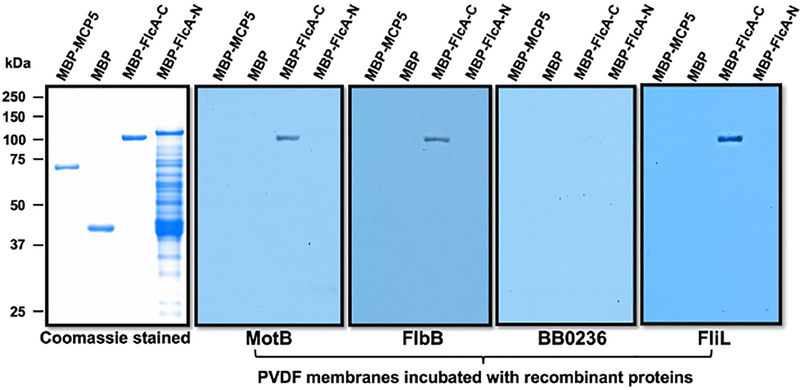
Based on the comparative flagellar motor analyses, we propose that the multiprotein collar complex is assembled sequentially like other periplasmic flagellar structures (Zhao et al., 2013, Moon et al., 2018). If FlcA is a component of the multiprotein collar complex, then one would imagine that FlcA interacts with the other collar proteins, such as FlbB, BB0236 or FliL. To confirm this or to determine which collar proteins FlcA interacts with, we performed affinity blotting or far-western blotting using recombinant proteins. Since FlcA possesses a membrane-binding domain (aa 329–351), we truncated it into two proteins, FlcA-N (aa 2–328) and FlcA-C (aa 360–931). As shown in Fig. 5, FlcA-C specifically interacts with FlbB and FliL, but not BB0236, validating our finding that this new collar protein localizes in the periphery of the collar structure where FliL and FlbB were also found to be (Moon et al., 2016b, Moon et al., 2018, Motaleb et al., 2011a). Identifying FlbB, BB0236, and FlcA as components of the collar further supports the idea that the structure is comprised of multiple proteins.
Figure 5. Specific interactions between FlcA and other flagellar proteins by affinity blotting.
Approximately 1 μg of MBP-tagged proteins shown on top of each panel were subjected to SDS-PAGE and then stained with Coomassie blue or transferred to a PVDF membrane. The membranes were incubated with 1xFLAG tagged MotB, FlbB, BB0236 or FliL, and then immunoblotted with anti-FLAG monoclonal antibodies.
FlcA interacts with the stator protein MotB
The current model for bacterial motility presume that interactions between peptidoglycan and MotB stabilize the stator complex within the cytoplasmic membrane, providing a platform against which the rotor and rod rotate to generate torque (Murphy et al., 2006, Murphy et al., 2008). Furthermore, using the spirochete periplasmic flagellar high-resolution structures as guides, it has been shown that the peptidoglycan contacts the stator (Liu et al., 2009, Murphy et al., 2006). These observations are supported by the report that stator protein MotB interacts with the peptidoglycan (Kojima et al., 2018). Interestingly, the flagellar structural models also show that the stator structures are in direct contact with other flagellar proteins likely to further immobilize or stabilize the stator (Gao et al., 2014, Hizukuri et al., 2010, Okabe et al., 2005, Terashima et al., 2006). For example, in spirochetes, stator structures are found to be in direct physical contact with collar structures (Kudryashev et al., 2010, Murphy et al., 2006, Zhao et al., 2014). These stator structures are found to be diminished when the genes encoding the collar structure are deleted in the FlbB, BB0236 and FlcA, even though a stator protein MotB is synthesized in those mutant cells (Fig. S2C) (Moon et al., 2016b, Moon et al., 2018). This leads us to hypothesize that the collar structure assembly occurs before the stator and that the assembly of the stator structure is stabilized by the collar. However, such a collar protein that interacts and stabilizes the stator has not been previously identified. Since FlcA is a flagellar collar protein, and the stator structure is missing in the ΔflcA mutant (Fig. 3), we tested if FlcA interacts with MotB using biochemical interactions of recombinant proteins. The affinity blotting results shown in Fig. 5 indicate that FlcA specifically interacts with MotB but not the control proteins MBP-MCP5 or MBP.
Discussion
The periplasmic flagellar collar is a unique spirochete-specific feature that is comprised of multiple proteins. Here, we characterized the third collar protein BB0326 or FlcA, which possesses some unique characteristics. We provided evidence for the first time that FlcA is important for the motility and morphology of B. burgdorferi (Fig. 2). These motility and morphology phenotypes are similar to those exhibited by the ΔmotB, ΔflbB, and Δbb0236 mutants (Moon et al., 2016b, Moon et al., 2018, Sultan et al., 2015). In all these mutants, including the ΔflcA, the stator complex is not assembled, and its absence is the cause for the observed motility-deficient phenotype (Fig. 2, 3).
Periplasmic flagella of the mutant cells are shorter than the wild-type cells and they are abnormally oriented (Fig. 4 and Table 1). Shorter and abnormally tilted periplasmic flagella were also observed in other collar mutants, such as ΔflbB, Δbb0236, and ΔfliL, highlighting that collar and FliL proteins are crucial for the normal orientation of periplasmic flagella (Moon et al., 2016b, Moon et al., 2018, Motaleb et al., 2011a). We have also found that a small fraction of ΔmotB mutant’s periplasmic flagella are shorter than the those of wild type cells (Sultan et al., 2015). The mutant cells also exhibit rod-shaped morphology, whereas the wild-type cells are flat-wave (Fig. 2). Previously, we have reported that motor rotation or active periplasmic flagella wrapping around the cell cylinder is required for the spirochete’s wave-like morphology (Sultan et al., 2015). However, in the ΔflcA mutant, shorter flagella are not wrapped around the cell cylinder leading to the rod-shaped morphology. Furthermore, ΔflcA and other collar mutants produced fewer flagella per cell than wild type cells (Moon et al., 2016b, Moon et al., 2018). While we do not completely understand why a collar mutant would produce fewer, shorter, and abnormally tilted flagella, we propose that these effects resulted from a combinatorial effect of collar, stator, and FliL structures.
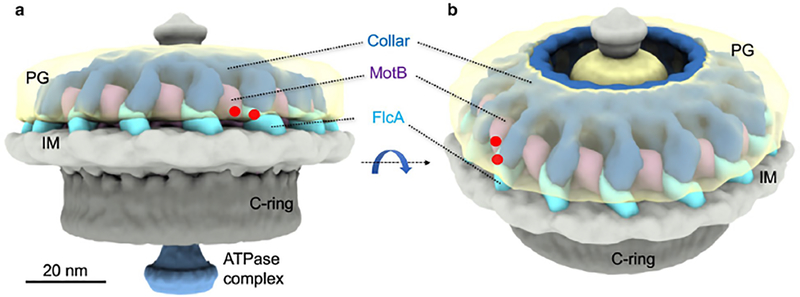
B. burgdorferi stator and collar structures appear to be attached to the peptidoglycan layer. Amino acid sequence analysis and cryo-ET data suggest that FlcA localizes at the periphery of the collar structure and interacts with the peptidoglycan using its peptidoglycan-binding domain/loop. Similar to the stator MotB proteins of E. coli and Helicobacter pylori (Hizukuri et al., 2009, Roujeinikova, 2008), the conserved peptidoglycan-binding domain of B. burgdorferi MotB may hold the stator in place during the motor rotation by binding to the cross-linked peptidoglycan layer. However, it appears that the interaction between the peptidoglycan and the stator is not sufficient for motor rotation in spirochetes and other bacteria without a collar-like structure. The interaction between MotB and FlcA suggests that the spirochete-specific collar structure adds another layer of immobilization to the stator (Fig. 5, 6). The stator’s binding to a flagellar protein, in addition to the peptidoglycan, has also been suggested for bacteria that lack a collar-like structure, such as Salmonella typhimurium, Campylobacter jejuni, and Vibrio cholerae (Gao et al., 2014, Hizukuri et al., 2010, Okabe et al., 2005, Terashima et al., 2006). The FlgI proteins that construct the flagellar P-rings in E. coli or Salmonella generally bind the stator, however, in B. burgdorferi, the P-ring is masked by the collar structure leading us to propose that the binding of the stator with the collar protein FlcA may substitute the FlgI-MotB interactions reported in other bacteria (Hizukuri et al., 2010, Liu et al., 2009). Our data and the fact that collar protein FlcA contains a peptidoglycan-binding domain/loop (Fig. 1B, S1) strongly supports our model – that the collar structure not only provides direct contact to stabilize or immobilize the stator and itself by binding to the peptidoglycan layer (Fig. 6), but also functions as the prop for stator’s assembly.
Figure 6. A schematic of FlcA and its interactions with PG and the stator complex.
(A) A side view and (B) a tilted view of a motor from the wild type cells show that FlcA is located at the periphery of the collar. Both FlcA and the stator complex appear to interact with the PG layer. Their interaction sites to PG are highlighted by red dots.
High-resolution cryo-ET and western blotting data show that neither the FliL structure nor protein synthesis was affected in the ΔflcA cells, suggesting that the FliL structure assembles before FlcA (Fig. S2C, S4). The other collar protein, FlbB synthesis, was not altered in the ΔflcA mutant cells compared to the WT or flcA+ complemented bacteria (Fig. S2C). Unchanged FlbB levels suggest that FlcA assembles after the FlbB structure, which further supports our previous notion that the assembled FlbB serves as the base of the collar structure (Moon et al., 2016b).
FlcA possesses three TPR motifs at its C-terminus (aa 736–820; Fig. S1). TPR motifs were first identified and named in yeast (Hirano et al., 1990, Sikorski et al., 1990), which was later found ubiquitously among all kingdoms of life with the conserved amino acid preference and sequence variations (Cerveny et al., 2013, D’Andrea & Regan, 2003). TPR domain-containing proteins participate in a variety of cellular processes, such as transcriptional regulation, protein translocation, mitosis, chaperone activities, etc. (Cerveny et al., 2013, D’Andrea & Regan, 2003). Genome sequence analysis suggests that B. burgdorferi encodes many TPR domain-containing proteins, among which only three proteins – BB0324 (BamD), BB0238, and BB0236 – have thus far been characterized, though all were shown to be involved in protein-protein interactions and/or chaperon for multiprotein complexes (Dunn et al., 2015, Groshong et al., 2014, Lenhart et al., 2012, Moon et al., 2018, Thakur et al., 2017). Since FlcA interacts with FlbB, FliL, and MotB, and flcA mutation resulted in diminishing a portion of the collar, we suggest that the TPR containing C-terminal domain of FlcA is involved in protein-protein interactions, whereas the N-terminal domain may be involved in the assembly of the collar structure’s periphery or incorporation into the inner membrane (Fig. 1B, 5).
In summary, our findings support the notion that the periplasmic collar of B. burgdorferi is a multi-protein complex and its assembly is well organized. However, as the major subunits of the collar are still intact in ΔflcA mutant cells, the corresponding subunit genes are yet to be determined.
Experimental procedures
Bacterial strains and growth conditions
The high-passage B. burgdorferi strain B31-A was used as the wild-type (WT) clone throughout the study (Bono et al., 2000a, Elias et al., 2002). Constructions of the Δbb0326 or ΔflcA mutant and complemented flcA+ strains are described below. B. burgdorferi cells were cultured in liquid Barbour-Stoenner-Kelly (BSK-II) broth or plated using plating BSK (P-BSK) containing 0.5% agarose, and grown at 35°C in a 2.5% CO2 incubator as described previously (Motaleb et al., 2007). Antibiotics, when required, were supplemented properly in B. burgdorferi mediums at the following concentrations: 200 μg/ml kanamycin and/or 40 μg/ml gentamicin. E. coli strains were cultivated in Luria-Bertani (LB) broth (1% tryptone, 1% NaCl, 0.5% yeast extract) or plated on LB agar (Bertani, 1951, Bertani, 2004). When required, 100 μg/ml ampicillin, 50 μg/ml kanamycin or 4 μg/ml gentamicin was supplemented into LB medium.
Bioinformatics
Basic local alignment search tool (BLAST) (Altschul et al., 1990, Altschul et al., 1997) was used to determine protein homologs from the sequence database. The lower the E-value, or the closer it is to zero, the more significant the match is. However, note that virtually identical short alignments have relatively high E values. This is because the calculation of the E value takes into account the length of the query sequence. These high E values make sense because shorter sequences have a higher probability of occurring in the database purely by chance.
Overexpression of recombinant proteins in E. coli
Multiple attempts to express full-length soluble B. burgdorferi BB0326 or FlcA protein in E. coli were unsuccessful. Both the His6 and Maltose binding protein (MBP) tagged FlcA recombinant full-length proteins were found to form inclusion bodies in the host E. coli cells. Since this protein possesses a membrane-binding domain in the middle (aa 329–351), we truncated FlcA into two proteins, FlcA-N (aa 2–328) and FlcA-C (aa 360–931). Even so, His6 tagged FlcA-N or FlcA-C proteins were found to be insoluble. However, MBP tagged FlcA-N and FlcA-C were able to be expressed and retained in the soluble fraction of the cell lysates. FlcA-N and FlcA-C coding regions were PCR amplified from B. burgdorferi genomic DNA, using primers PF_HisThromBB0326NdeI (GGCAGCCATATGCCTGATGTAGATAAGATAATACAG) and PR_HisThroBB326N328aaNotI (GCTCGAGTGCGGCCGCTTACCTTGAAACCCTGTAGCTCAA), and PF_HisThroBB326C572aaNdeI (GGCAGCCATATGGCCTCTGAGAGCAAGTATAAAGAG) and PR_HisThromBB0326NotI (GCTCGAGTGCGGCCGCTTAAAGTTTTTCGGATAAATTTTC), respectively (restriction sites are underlined). The PCR products were subsequently cloned into pMAL c5x (NEB Inc.) using NdeI and NotI restriction sites. MBP-tagged FlbB and FliL were similarly constructed using primers PF_FlbB-TM35aa-NdeI (GATTTCACATATGAATACTAAAAGATATTTCCCCG) and PR_FlbB-TM35aa-NotI (GACGATATCGCGGCCGCtTACTCCAATGAACTAACAGAC), and BamHI_Trun_FliL_F (ggatccgtgtctaaaatggtggtaagcc) and FliL_R_PstI (ctgcagttacatatcaaaaatatcaatttggg) respectively. 1xFLAG (DYKDDDDK) tagged MotB, BB0236, FlbB, FliL were also constructed for affinity blotting. Briefly, 1xFLAG tag coding sequence (GACTACAAAGACGATGACGACAAG) was fused to the coding regions of MotB (without the transmembrane domain, aa 1–44), BB0236 (without the predicted signal peptide, aa 1–20), FlbB (without the transmembrane domain, aa 1–42), and FliL (without the transmembrane domain, aa 1–50) either at C-terminus or N-terminus through PCR amplification, which were then cloned into pET28a(+) (Novagen Inc.) or pMAL c5x (NEB Inc.). Expression of 6xHis tagged FliL and FlbB, MBP-BB0236 were described elsewhere (Moon et al., 2018, Motaleb et al., 2011a).
All E. coli strains were induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at room temperature and purifications of recombinant proteins were performed with proper affinity resins (amylose resin for MBP and HisPure Ni-NTA resin for His6).
SDS-PAGE, immunoblot and affinity blotting
Sodium dodecyl sulfate polyacrylamide gel electrophoresis or SDS-PAGE and immunoblotting with an enhanced chemiluminescent detection were performed as described (GE Health Inc.) (Motaleb et al., 2000, Sultan et al., 2013). Protein concentrations were determined using a Bio-Rad protein assay kit with bovine serum albumin as the standard. Unless specified, 1 μg of protein were subjected to SDS-PAGE. Biochemical interactions of recombinant proteins such as Far-western or affinity blot assays were described previously (Toker & Macnab, 1997, Kariu et al., 2015, Moon et al., 2016a). Briefly, 1 μg purified recombinant proteins was subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked in the blocking solution (5% skim milk, 10mM Tris, 150mM NaCl and 0.3% Tween 20, pH 7.4) with gentle shaking for 4 to 6 hours at room temperature, and then incubated with purified 1xFLAG tagged proteins at concentration 2 μg/ml in blocking solution overnight. The membranes were washed 3 times with the washing buffer (10mM Tris, 150mM NaCl and 0.3% Tween 20, pH 7.4), and then probed with monoclonal anti-FLAG® M2 antibody (Sigma-Aldrich Co. LLC).
Construction of the bb0326 or flcA mutant and complemented strains
Construction of the bb0326 or flcA inactivation plasmids, electroporation, and plating conditions were described previously (Motaleb et al., 2000, Sultan et al., 2013). Briefly, the bb0326 gene region was first amplified by PCR from the chromosomal DNA of B. burgdorferi strain B31-A using primers BB0326_F (ttaaagtttttcggataaattttcataaaa) and BB0326_R (cctgatgtagataagataatacag), and the product obtained was cloned into plasmid pGEM-T Easy (Promega Inc.). The bb0326 gene was inactivated using a kanamycin resistance cassette (PflgB-Kan) (Bono et al., 2000b). Plasmid containing bb0326-PflgB-Kan was linearized by NotI restriction digestion to remove the ampicillin marker of the vector and electroporated into competent B31-A cells. Kanamycin-resistant transformants were screened by PCR.
The Δbb0326 or ΔflcA mutant was complemented in trans using the B. burgdorferi shuttle vector pBSV2G that was prepared to contain the flgB promoter (PflgB) fused to an intact copy of bb0326 or flcA gene, as described (Elias et al., 2003, Sultan et al., 2015). The vector pBSV2G::PflgB-bb0326 was electroporated into the Δbb0326 mutant cells, followed by selection with both kanamycin and gentamicin. The resistant clones were analyzed by PCR using appropriate primers.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Exponentially growing B. burgdorferi wild type, ΔflcA and flcA+ cells were treated with RNAprotect® Bacteria Reagent (Qiagen Inc.) followed by the total RNA isolation using the RNeasy mini kit (Qiagen Inc.). Contaminating DNA in the RNA samples was digested by RNase-free Turbo™ DNase I (Invitrogen by Thermo Fisher Scientific Inc.) for 1 hour at 37°C and the removal of DNA was confirmed by PCR amplification of the B. burgdorferi bb0327 gene. For RT-PCR, cDNA was prepared from 200 ng RNA using the AffinityScript QPCR cDNA synthesis kit according to the manufacturer’s protocol (Agilent Technologies Inc.). The Power SYBR® Green detection system (Applied Biosystems by Thermo Fisher Scientific Inc.) was used to measure bb0327 and bb0325 genes transcript levels according to the manufacturer’s instructions. qRT-PCR of B. burgdorferi bb0327 and bb0325 genes was done with primers PF bb0327RT (TTCCAATAGCTGGAGTTAAG) and PR bb0327RT (TTTGCCTAGCATGTTCTAAACC), and primers PF bb0325RT (GTTTATGTTGAGGCTCATGAAG) and PR bb0325RT (CCGAATTTCATCTCCATTAACC) respectively. B. burgdorferi enolase gene was used as reference gene using primers RT-enolase-F (TGGAGCGTACAAAGCCAACATT) and RT-enolase-R (TGAAAAACCTCTGCTGCCATTC) (Novak et al., 2016, Pitzer et al., 2011, Sultan et al., 2011, Sultan et al., 2010, Xu et al., 2017). The result of relative quantification of the copies of bb0327 or bb0325 was normalized to 105 copies of enolase gene in each strain.
Dark-field microscopy and swarm plate assays
Exponentially growing B. burgdorferi cells were observed using a Zeiss Axio Imager M1 dark-field microscope connected to an AxioCam digital camera to determine the morphology and motility as described previously (Motaleb et al., 2007, Motaleb et al., 2011a). Swarm plate assay was performed to determine spirochetes’ motility using our established protocol (Motaleb et al., 2007, Motaleb et al., 2011a).
Cryo-electron tomography
Frozen-hydrated specimens were prepared as described previously (Liu et al. 2009; Zhao et al. 2013). Briefly, various clones of exponentially growing B. burgdorferi cells were centrifuged individually at 5,000 x g for 5 min and the resulting pellets were suspended in PBS to achieve a cell concentration approximately 1×108/ml. After adding 10 nm gold marker solution, 5 μl of the cell suspension was placed on freshly glow-discharged holey carbon grid (Quantifoil Cu R2/2, 200 mesh) for 25 s. The grids were blotted with filter paper for 3 to 5 s and rapidly frozen in liquid ethane, using a home-made plunger apparatus as described previously (Liu et al., 2009, Zhao et al., 2013).
Spirochete specimens were imaged using a 300-kV electron microscope (Titan Krios) equipped with a field emission gun, a Volta Phase Plate (VPP) and a Direct Electron Detector (Gatan K2 Summit). SerialEM was used to collect tilt series at focus. A total dose of 55 e−/Å2 is distributed among 35 tilt images covering angles from −51° to +51° at tilt steps of 3°. For every single tilt series collection, dose-fractionated mode was used to generate 11 frames per projection image. Collected dose-fractionated data were first subjected to the Motioncorr2 (Li et al., 2013) to generate drift-corrected files. IMOD software was used to align the tilt series and to generate tomograms (Kremer et al., 1996, Zhao et al., 2013). In total, 38 of tomographic reconstructions of Δbb0326 or ΔflcA cells and 42 of tomographic reconstructions of bb0326+ or flcA+ cells were generated.
Subtomogram averaging
206 and 356 flagellar motor subtomograms were picked from tomograms of ΔflcA and flcA+ cells, respectively. The initial orientation of each particle was manually estimated by the C-ring and the hook. To accelerate image analysis, 4×4×4 binned sub-tomograms were used for initial alignment. The original data was then used for the refinement and averaging as described previously (Liu et al., 2009, Zhao et al., 2013).
Three-Dimensional visualization
The software package UCSF Chimera X was mainly used for 3D visualization and surface rendering of sub-tomogram averages (Goddard et al., 2018). Segmentations of representative reconstructions from WT, ΔflcA and flcA+ cells were constructed using IMOD (Kremer et al., 1996). The filaments, the outer and inner membranes were manually segmented.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases grants 2R01AI087946 (J.L.) and 1R01AI132818 (M.A.M). We thank Dr. Shiwei Zhu for cryo-EM sample preparation, Jonathan Sigworth for careful reading of the manuscript and Zhou Yu for technical assistance.
References
- Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ, (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W & Lipman DJ, (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G, (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G, (2004) Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol 186: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko Iii JJ, Stevenson B, Tilly K & Rosa P, (2000a) Efficient targeted mutagenesis in Borrelia burgdorferi.. Journal of Bacteriology 182: 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JJ 3rd, Stevenson B, Tilly K & Rosa P, (2000b) Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol 182: 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny L, Straskova A, Dankova V, Hartlova A, Ceckova M, Staud F & Stulik J, (2013) Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun 81: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Moon KH, Zhao X, Norris SJ, Motaleb MA & Liu J, (2019) Structural insights into flagellar stator-rotor interactions. Elife 8: e48979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA & Wolgemuth CW, (2012) The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66: 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ & Rowe N, (2009) The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol 191: 600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ & Jensen GJ, (2011) Structural diversity of bacterial flagellar motors. EMBO J 30: 2972–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF & Hughes KT, (2017) Coupling of Flagellar Gene Expression with Assembly in Salmonella enterica. Methods Mol Biol 1593: 47–71. [DOI] [PubMed] [Google Scholar]
- Chilcott GS & Hughes KT, (2000) Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev 64: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD & Regan L, (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Dunn JP, Kenedy MR, Iqbal H & Akins DR, (2015) Characterization of the beta-barrel assembly machine accessory lipoproteins from Borrelia burgdorferi. BMC Microbiol 15: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ 3rd, Stewart PE, Krum JG & Rosa PA, (2003) New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol 6: 29–40. [DOI] [PubMed] [Google Scholar]
- Elias AF, Schmutzhard J, Stewart PE, Schwan TG & Rosa P, (2002) Population dynamics of a heterogeneous Borrelia burgdorferi B31 strain in an experimental mouse-tick infectious cycle. Wien Klin Wochenschr 114: 557–561. [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO & Venter JC, (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586. [DOI] [PubMed] [Google Scholar]
- Gao B, Lara-Tejero M, Lefebre M, Goodman AL & Galan JE, (2014) Novel components of the flagellar system in epsilonproteobacteria. MBio 5: e01349–01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Old IG, Saint Girons I & Charon NW, (1997) Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J Bacteriol 179: 2289–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH & Ferrin TE, (2018) UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groshong AM, Fortune DE, Moore BP, Spencer HJ, Skinner RA, Bellamy WT & Blevins JS, (2014) BB0238, a presumed tetratricopeptide repeat-containing protein, is required during Borrelia burgdorferi mammalian infection. Infect Immun 82: 4292–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K & Yanagida M, (1990) Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+. Cell 60: 319–328. [DOI] [PubMed] [Google Scholar]
- Hizukuri Y, Kojima S & Homma M, (2010) Disulphide cross-linking between the stator and the bearing components in the bacterial flagellar motor. J Biochem 148: 309–318. [DOI] [PubMed] [Google Scholar]
- Hizukuri Y, Morton JF, Yakushi T, Kojima S & Homma M, (2009) The peptidoglycan-binding (PGB) domain of the Escherichia coli pal protein can also function as the PGB domain in E. coli flagellar motor protein MotB. J Biochem 146: 219–229. [DOI] [PubMed] [Google Scholar]
- Hovind-Hougen K, (1984) Ultrastructure of spirochetes isolated from Ixodes ricinus and Ixodes dammini. Yale J Biol Med 57: 543–548. [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Morimoto YV, Kudo S & Nakamura S, (2015) H(+) and Na(+) are involved in flagellar rotation of the spirochete Leptospira. Biochem Biophys Res Commun 466: 196–200. [DOI] [PubMed] [Google Scholar]
- Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M & Radolf JD, (2009) Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol 191: 7566–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CR, Schmid GP, Hyde FW, Steigerwalt AG & Brenner DJ, (1984) Borrelia burgdorferi sp. nov.: Etiologic Agent of Lyme Disease International Journal of Systematic Bacteriology 34: 496–497. [Google Scholar]
- Kariu T, Sharma K, Singh P, Smith AA, Backstedt B, Buyuktanir O & Pal U, (2015) BB0323 and novel virulence determinant BB0238: Borrelia burgdorferi proteins that interact with and stabilize each other and are critical for infectivity. J Infect Dis 211: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, Homma M & Namba K, (2009) Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol Microbiol 73: 710–718. [DOI] [PubMed] [Google Scholar]
- Kojima S, Takao M, Almira G, Kawahara I, Sakuma M, Homma M, Kojima C & Imada K, (2018) The Helix Rearrangement in the Periplasmic Domain of the Flagellar Stator B Subunit Activates Peptidoglycan Binding and Ion Influx. Structure 26: 590–598 e595. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN & McIntosh JR, (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76. [DOI] [PubMed] [Google Scholar]
- Kudryashev M, Cyrklaff M, Wallich R, Baumeister W & Frischknecht F, (2010) Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169: 54–61. [DOI] [PubMed] [Google Scholar]
- Lenhart TR, Kenedy MR, Yang X, Pal U & Akins DR, (2012) BB0324 and BB0028 are constituents of the Borrelia burgdorferi beta-barrel assembly machine (BAM) complex. BMC Microbiol 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC & Charon NW, (2002) Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc Natl Acad Sci U S A 99: 6169–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA & Cheng Y, (2013) Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10: 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H & Norris SJ, (2009) Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191: 5026–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Hobbs G & Motaleb MA, (2016a) Borrelia burgdorferi CheD Promotes Various Functions in Chemotaxis and the Pathogenic Life Cycle of the Spirochete. Infect Immun 84: 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Zhao X, Manne A, Wang J, Yu Z, Liu J & Motaleb MA, (2016b) Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol 102: 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Zhao X, Xu H, Liu J & Motaleb MA, (2018) A tetratricopeptide repeat domain protein has profound effects on assembly of periplasmic flagella, morphology and motility of the lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 110: 634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS & Charon NW, (2000) Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A 97: 10899–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Liu J & Wooten RM, (2015) Spirochetal motility and chemotaxis in the natural enzootic cycle and development of Lyme disease. Curr Opin Microbiol 28: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Bakker RG, Li C & Charon NW, (2007) Isolation and characterization of chemotaxis mutants of the Lyme disease Spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol 422: 421–437. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Pitzer JE, Sultan SZ & Liu J, (2011a) A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol 193: 3324–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Sultan SZ, Miller MR, Li C & Charon NW, (2011b) CheY3 of Borrelia burgdorferi is the key response regulator essential for chemotaxis and forms a long-lived phosphorylated intermediate. J Bacteriol 193: 3332–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GE, Leadbetter JR & Jensen GJ, (2006) In situ structure of the complete Treponema primitia flagellar motor. Nature 442: 1062–1064. [DOI] [PubMed] [Google Scholar]
- Murphy GE, Matson EG, Leadbetter JR, Berg HC & Jensen GJ, (2008) Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol 67: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus E, Gressmann H, Ye F, Schlapbach R, Dehio M, Dehio C, Stack A, Meyer TF, Suerbaum S & Josenhans C, (2004) Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol Microbiol 52: 947–961. [DOI] [PubMed] [Google Scholar]
- Novak EA, Sekar P, Xu H, Moon KH, Manne A, Wooten RM & Motaleb MA, (2016) The Borrelia burgdorferi CheY3 response regulator is essential for chemotaxis and completion of its natural infection cycle. Cell Microbiol 18: 1782–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Yakushi T & Homma M, (2005) Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J Biol Chem 280: 25659–25664. [DOI] [PubMed] [Google Scholar]
- Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR & Motaleb MA, (2011) Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect Immun 79: 1815–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Tu J, Lin T, Norris SJ, Li C, Motaleb MA & Liu J, (2018) Cryo-electron tomography of periplasmic flagella in Borrelia burgdorferi reveals a distinct cytoplasmic ATPase complex. PLoS Biol 16: e3000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala SV, Titz B, Goll J, Parrish JR, Wohlbold K, McKevitt MT, Palzkill T, Mori H, Finley RL Jr. & Uetz P, (2007) The protein network of bacterial motility. Mol Syst Biol 3: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roujeinikova A, (2008) Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc Natl Acad Sci U S A 105: 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, Drecktrah D & Hall LS, (2018) Genetic Transformation and Complementation. Methods Mol Biol 1690: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Goebl M & Hieter P, (1990) A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60: 307–317. [DOI] [PubMed] [Google Scholar]
- Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA, Charon NW & Motaleb MA, (2013) Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infect Immun 81: 2012–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR & Motaleb MA, (2011) Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect Immun 79: 3273–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Miller MR & Motaleb MA, (2010) Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol 77: 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Sekar P, Zhao X, Manne A, Liu J, Wooten RM & Motaleb MA, (2015) Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infect Immun 83: 1765–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJ, Yildiz F & Klose KE, (2009) The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol 191: 6555–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Morado DR, Liu J, Charon NW, Xu H & Li C, (2011) Carbon storage regulator A (CsrA(Bb)) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol 82: 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Fukuoka H, Yakushi T, Kojima S & Homma M, (2006) The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol Microbiol 62: 1170–1180. [DOI] [PubMed] [Google Scholar]
- Thakur M, Sharma K, Chao K, Smith AA, Herzberg O & Pal U, (2017) A protein-protein interaction dictates Borrelial infectivity. Sci Rep 7: 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Vanraden MJ, Stewart P & Rosa P, (2006) Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect Immun 74: 3554–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker AS & Macnab RM, (1997) Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol 273: 623–634. [DOI] [PubMed] [Google Scholar]
- Wilhelms M, Gonzalez V, Tomas JM & Merino S, (2013) Aeromonas hydrophila lateral flagellar gene transcriptional hierarchy. J Bacteriol 195: 1436–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelms M, Molero R, Shaw JG, Tomas JM & Merino S, (2011) Transcriptional hierarchy of Aeromonas hydrophila polar-flagellum genes. J Bacteriol 193: 5179–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth CW, (2015) Flagellar motility of the pathogenic spirochetes. Semin Cell Dev Biol 46: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sultan S, Yerke A, Moon KH, Wooten RM & Motaleb MA, (2017) Borrelia burgdorferi CheY2 Is Dispensable for Chemotaxis or Motility but Crucial for the Infectious Life Cycle of the Spirochete. Infect Immun 85: e00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y & Li C, (2009) Transcription and genetic analyses of a putative N-acetylmuramyl-L-alanine amidase in Borrelia burgdorferi. FEMS Microbiol Lett 290: 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Norris SJ & Liu J, (2014) Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53: 4323–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Pitzer J, Motaleb MA, Norris S & Liu J, (2011) Molecular Architecture of Bacterial Flagellar Stator Revealed by Cryo-Electron Tomography. 17: 110–111. [Google Scholar]
- Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C & Liu J, (2013) Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A 110: 14390–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.