Abstract
Background
Cholangiocarcinoma (CCA) is an invasive malignancy arising from biliary epithelial cells; it is the most common primary tumour of the bile tract and has a poor prognosis. The aim of this study was to screen prognostic biomarkers for CCA by integrated multiomics analysis.
Methods
The GSE32225 dataset was derived from the Gene Expression Omnibus (GEO) database and comprehensively analysed by using R software and The Cancer Genome Atlas (TCGA) database to obtain the differentially expressed RNAs (DERNAs) associated with CCA prognosis. Quantitative isobaric tags for relative and absolute quantification (iTRAQ) proteomics was used to screen differentially expressed proteins (DEPs) between CCA and nontumour tissues. Through integrated analysis of DERNA and DEP data, we obtained candidate proteins APOF, ITGAV and CASK, and immunohistochemistry was used to detect the expression of these proteins in CCA. The relationship between CASK expression and CCA prognosis was further analysed.
Results
Through bioinformatics analysis, 875 DERNAs were identified, of which 10 were associated with the prognosis of the CCA patients. A total of 487 DEPs were obtained by using the iTRAQ technique. Comprehensive analysis of multiomics data showed that CASK, ITGAV and APOF expression at both the mRNA and protein levels were different in CCA compared with nontumour tissues. CASK was found to be expressed in the cytoplasm and nucleus of CCA cells in 38 (45%) of 84 patients with CCA. Our results suggested that patients with positive CASK expression had significantly better overall survival (OS) and recurrence-free survival (RFS) than those with negative CASK expression. Univariate and multivariate analyses demonstrated that negative expression of CASK was a significantly independent risk factor for OS and RFS in CCA patients.
Conclusions
CASK may be a tumour suppressor; its low expression is an independent risk factor for a poor prognosis in CCA patients, and so it could be used as a clinically valuable prognostic marker.
Keywords: Cholangiocarcinoma, Bioinformatics, iTRAQ, CASK, Prognosis, Multiomics
Background
Cholangiocarcinoma is an invasive malignancy originating from biliary epithelial cells and is the most common primary tumour of the bile tract [1]. Over the past few decades, the incidence of CCA has steadily increased [2]. This trend brings more serious challenges to CCA research. The prognosis of CCA is very poor, and the disease not sensitive to chemoradiotherapy. Surgical treatment is the primary intervention at present [3]. However, patients with CCA often do not manifest obvious symptoms in the early stage, and the vast majority of clinically diagnosed CCA patients have often missed the opportunity for radical surgery [4]. In addition, although the survival rate of patients with radical resection is significantly higher than that of patients without resection, recurrence is still the leading cause of death in such patients, and the 5-year survival rate after surgery is still a depressing figure [5, 6]. Therefore, it is necessary to screen effective prognostic biomarkers to identify high-risk CCA patients and develop appropriate treatment strategies to improve their prognoses.
In recent decades, many biomarkers of CCA have been found through continuous CCA research [7, 8]. Carbohydrate antigen 19-9 (CA19-9) is the most commonly used diagnostic marker in clinical practice, but due to concomitant biliary inflammation and Lewis-gene-negative cases [9], its diagnostic efficacy is limited. Some studies have reported that increased preoperative CA19-9 levels are associated with poor postoperative prognosis in CCA patients [10, 11]. Gardini et al. found that low expression of connective tissue growth factor (CTGF) was an independent risk factor for a poor prognosis in CCA patients [12]. However, these biomarkers are still insufficient to fully understand the molecular mechanisms of CCA and to access CCA prognosis to optimize treatment strategies. We need to use more efficient methods to screen out new, more valuable biomarkers to promote CCA research [13] and develop and improve clinical, precise, individualized treatment schemes.
Isobaric tags for relative and absolute quantification (iTRAQ) technology is one of the most popular methods for differential protein identification; it allows the high-throughput, simultaneous comparison of protein expression among up to 8 samples with high labelling efficiency [14]. However, iTRAQ may yield up to hundreds or thousands of DEPs, and it is difficult to identify which proteins can serve as prognostic biomarkers for CCA. Fortunately, as advances in bioinformatics have continued to be developed, we can make bioinformatics predictions by analysing public CCA data [13]. Integrating the data obtained from iTRAQ and conducting a multiomics analysis can yield proteins that may serve as prognostic markers of CCA for further research. In addition, these prognostic biomarkers may help to further reveal the mechanism of tumourigenesis and development of cancer from a systematic perspective, providing an important basis for the individualized and precise treatments for CCA.
In this study, we obtained 10 mRNAs related to the prognosis of CCA by obtaining CCA data from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) database and conducting a comprehensive analysis. Through integrated analysis with the proteome data obtained by iTRAQ, we screened three candidate proteins APOF, ITGAV and CASK, and verified their expression with immunohistochemistry. Finally, we identified CASK as a valuable biomarker in CCA patients for predicting the recurrence of CCA and a poor clinical outcome.
Materials and methods
Data obtaining and preprocessing
Matrix files from the GSE32225 dataset were obtained from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database. The expression profile of the GSE32225 dataset was recorded in GPL8432 (Illumina HumanRef-8 WG-DASL v3.0) and contained the gene expression data from 149 cancer samples and 6 normal tissues [15]. The gene probes were matched with the gene symbols in the platform annotation file of GPL8432 one by one. When the gene symbols were matched with multiple probes, the gene expressions were averaged.
Identification of differentially expressed RNAs
R software (version 3.6.0, https://www.r-project.org) was used to process the data. We used the “limma” package to identify DERNAs between CCA tissues and nontumour tissues [16]. Then, the expression values were log2 converted. Adjusted P values (FDR) < 0.05 and | log2FoldChange (FC) | > 1 were established as screening threshold values. Finally, we obtained the DERNAs, which will be used for further analyses.
Function and pathway analysis
To understand the underlying functions of these DERNAs, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov) web tool to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses [17, 18]. The results of enrichment analysis with P values < 0.05 were considered to be statistically significant.
Survival analysis and validation of expression
To determine whether these DERNAs could be used as biomarkers for the prognosis of CCA, we used Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cm), an analysis tool that provides customized gene expression data based on the TCGA databases, to perform a Kaplan–Meier survival analysis to assess the prognostic value of these DERNAs [19]. The statistically significant threshold was set as P < 0.05. CCA sequencing data from the TCGA database were analysed using GEPIA to verify the differential expression of previously obtained survival-related RNAs.
Selection of patients and collection of clinical samples
A total of 6 matched primary CCA tissues and paracancerous tissues were collected from patients undergoing surgery in The Department of Special Minimally Invasive Surgery, The First Hospital of Lanzhou University. All of the tissue specimens were diagnosed with primary CCA without radiotherapy or chemotherapy before surgery by two pathologists. These collected samples were used for iTRAQ followed by LC–MS. The basic clinical information of these patients is summarized in Table 1.
Table 1.
Summary of clinic parameters of CCA patients collected for iTRAQ
| Code | Pathology diagnosis | Age | Gender | Grade | Lymph node metastasis |
|---|---|---|---|---|---|
| 1 | Cholangiocarcinoma | 51 | Female | Well | Positive |
| 2 | Cholangiocarcinoma | 55 | Female | Poorly | Negative |
| 3 | Cholangiocarcinoma | 51 | Male | Poorly | Positive |
| 4 | Cholangiocarcinoma | 58 | Male | Moderately | Positive |
| 5 | Cholangiocarcinoma | 72 | Male | Moderately | Negative |
| 6 | Cholangiocarcinoma | 75 | Male | Moderately | Negative |
In addition, 7 cases of paraffin-embedded choledochal cysts and 86 cases of paraffin-embedded CCAs were obtained from patients receiving surgical treatment at The First Hospital of Lanzhou University from 2011 to 2016, and were used for immunohistochemical detection and tissue microarray (TMA) construction, respectively. This experimental scheme was approved by the Ethics Committee for Human Research, Lanzhou University. All tissue samples were obtained with written consent from the participants, and all diagnoses were confirmed by postoperative pathology. The inclusion criteria for CCA patients in this study were as follows: (1) the postoperative pathological diagnosis was primary cholangiocarcinoma; (2) no distant metastasis had occurred; (3) the patient had not undergone chemoradiotherapy before surgery; (4) the patient had undergone radical surgical resection; (5) no life-threatening preoperative and postoperative complications had occurred; and (6) adequate clinical and follow-up data were available. The clinical features of the CCA patients are summarized in Table 2.
Table 2.
Summary of clinical characteristic of CCA patients collected for TMAs
| Characteristic | No. patients (%) |
|---|---|
| Age | |
| < 55 | 34 (39.5) |
| ≥ 55 | 52 (60.5) |
| Gender | |
| Female | 43 (50.0) |
| Male | 43 (50.0) |
| Histologic grade | |
| Poorly | 17 (19.8) |
| Moderate | 64 (74.4) |
| Well | 5 (5.8) |
| T classification | |
| T1 | 6 (7.0) |
| T2 | 59 (68.6) |
| T3 | 18 (20.9) |
| T4 | 3 (3.5) |
| N classification | |
| N0 | 58 (67.4) |
| N1 | 28 (32.6) |
| Vascular invasion | |
| No | 57 (66.3) |
| Yes | 29 (33.7) |
| Recurrence | |
| No | 31 (36.0) |
| Yes | 55 (64.0) |
Sample preparation and protein quantification
Frozen tissue samples were sectioned after thawing on ice. After adding lysis buffer (containing 0.1 mmol/L phenylmethylsulfonyl fluoride, 2 mol/L thiourea, 4% 3-(3-cholamidopropyl) dimethylammonium propane sulfonate (CHAPS), 7 mol/L urea and 65 mmol/L dithiothreitol) and 4% phenylmethylsulfonyl fluoride (PMSF), the tissue cubes were ground in liquid nitrogen and fully extracted. Then the obtained samples were placed on ice and sonicated for 3 min (0.8-s sonication, followed by a 0.8-s interval) with the power set at 80 W. The samples were thoroughly mixed with prechilled acetone at a prechilled acetone to sample ratio of 5:1 (v/v), and the resulting mixture was precipitated overnight at − 20 °C. Then, the mixture was centrifuged at 12,000×g and 4 °C for 10 min. After removing the supernatant, 2 mL prechilled acetone was added to the collected precipitate to shake and mix and the resulting mixture was centrifuged again with the above parameters for 15 min. Then we collected the precipitate and repeated the above steps. After removing the supernatant, the mixture was collected and dried at room temperature. The precipitate was added to 0.5 ml of 1 M triethylammonium bicarbonate buffer (TEAB; Sigma-Aldrich, Australia), and the mixture was thoroughly mixed and dissolved and then centrifuged at room temperature and 12,000×g for 15 min. After fully removing the insoluble impurities, the obtained supernatant was transferred to a new 1 mL test tube, and the protein concentration was measured by using the Bradford Protein Assay (Tiangen, Beijing, China).
Protein digestion and iTRAQ labelling
After protein quantification, prechilled acetone 5 times the volume of each sample was added to 100 µg of protein per group for precipitation, which was then fully precipitated at − 20 °C for 1 h. The resulting mixture was then centrifuged for 10 min at 4 °C and 12,000×g, and the precipitate was collected and dried by using a vacuum centrifuge. Then, 50 µL dissolution buffer was added to the precipitate to fully dissolve the protein, reacted with 4 µl reducing reagent for 1 h at 60 °C, and then alkylated with 2 µl cysteine blocking for 10 min at room temperature. Then, 50 µl trypsin (50 ng/µL) was added to the protein sample to fully dissolve it for 12 h at 37 °C. After the tryptic peptide mixtures were dried by vacuum centrifugation, iTRAQ reagent was added to the mixture, which was then labelled for 2 h at room temperature. Then, 100 ml distilled water was added to the samples to stop the reaction, and all the labelled samples were mixed in equal quantities. After the samples were completely dried by vacuum centrifugation, the samples were kept for further isolation and identification. The above iTRAQ labelling steps were performed in accordance with the reagent manufacturer’s instructions (AB SCIEX, Shanghai, China).
LC–MS/MS analysis
LC–MS/MS was carried out by Sangon Biotech (Shanghai) Co., Ltd. The freeze-dried samples were dissolved in 110 µl Nano-RPLC buffer A (0.1% formic acid and 2% ACN) and peptide separation was conducted by an Agilent 1200 HPLC system on a secondary RP analytical column (Analytical Guard Column 4.6 × 12.5 mm 5-μm; Narrow-Bore 2.1 × 150 mm 5 μm). The sample was then loaded with a flow rate of 0.3 mL/min for 1 h by using a linear binary gradient of 0–80% buffer B (350 mmol/L KCl in solvent A, pH = 2.8). The absorbance of the 6 collected SCX fractions was measured at 210 nm and 280 nm. The peptide mixture was redissolved in Nano-RPLC buffer A, and online Nano-RPLC liquid chromatography was performed using LC-20AD nano-RPLC (AB SCIEX). The analytical column was a C18 reversed-phase chromatographic column (75 μm × 15 cm C18 - 3 μm 120 A, ChromXP Eksigent). Subsequently, the gradient condition with phase B at a flow rate of 300 μL/min (5% phase B for 8 min, 25% phase B for 38 min, 40% phase B for 50 min, 90% phase B for 60 min, 2% phase B for 65 min, and 0 phase B for 70 min) were used to eluted the peptides. Mass spectrometry was performed on a Triple TOF 5600 (AB SCIEX) with 2.5 kV electrospray, and the information independent analysis (IDA) mode was used as the mass spectrometry scanning mode. In IDA mode, a hybrid quadrupole time-of-flight mass spectrometer (QStar hybrid LC/MS/MSQ-TOF, AB SCIEX, China) was operated to switch automatically between MS and MS/MS acquisition. The scanning time of the first level single MS was set as 250 ms, and a maximum of 35 s-level MS with charges of 2 + to 5 + and a single second count of more than 150 were collected under each IDA cycle. Dynamic exclusion was set to 18 s, approximately equal to the chromatographic half-peak width.
Protein identification and integrated analysis
The LC-MS/MS data were analysed by Sangon Biotech (Shanghai) Co., Ltd using the SEQUEST algorithm. The results were compared with the data in the NCBI Human RefSeq database. A cutoff value > 1.3 and peptides≥ 1 were used as thresholds for protein identification. If the FC value > 1.2 or < 0.87 in the cancer group compared with the nontumour group, the protein was regarded as a differentially expressed protein. A P value < 0.05 FC value for at least one dataset indicated significant differential expression. The validated prognosis-related RNA and the DEP data obtained by iTRAQ were integrated and analysed to obtain the intersection between the two sets of data. The candidate proteins in this intersection were reserved for further analysis.
Tissue microarray (TMA)
TMAs were constructed by the Pathology Department of the First Hospital of Lanzhou University, for which Shanghai Outdo Biotech Company (Shanghai, China) provided technical support. The TMA contained a total of 122 points, of which 31 included noncarcinoma foci. The paraffin-embedded tissue in each core was obtained from the non-necrotic area of the carcinoma and/or noncarcinoma foci.
Immunohistochemistry (IHC)
Integrin subunit alpha V (ITGAV) was detected by ITGAV rabbit polyclonal antibody (27,096-1-AP). Apolipoprotein F (APOF) was detected by APOF rabbit polyclonal antibody (16608-1-AP). Both antibodies were obtained from Proteintech (Wuhan, China). Peripheral plasma membrane protein CASK (CASK) was detected by rabbit polyclonal anti-CASK antibody (ab244393), which was purchased from Abcam (Shanghai, China). Goat serum and 3,3’-diaminobenzidine (DAB) substrate were purchased from Fuzhou Maixin Biotechnology Co. (Fuzhou, China). The tissue was embedded and fixed, and then the sections were dewaxed with xylene and diluted and hydrated with ethanol and distilled water, respectively. The sections and TMA were treated with antigenic thermal repair. Sections were incubated at room temperature with 3% hydrogen peroxidase for 10 min to eliminate endogenous peroxidase activity. The section and TMA were sealed with goat serum to prevent nonspecific staining. The sections and TMA were dripped with primary antibodies (ITGAV, 1:200; APOF, 1:50; CASK, 1:20) at 4 °C overnight. The sections and TMA were then dripped with secondary antibody and incubated at room temperature for 30 min. The sections were dripped with fresh DAB substrate and reacted for 5 min, then thoroughly rinsed with tap water, followed by counterstaining with haematoxylin. After dehydration with ethanol and xylene, the tablets were sealed with neutral gum.
Evaluation of the results by scoring
Immunoreactivity images and their corresponding scores were assessed independently by two pathologists who had no knowledge of the experimental design, and any inconsistent scores were reassessed by both pathologists until a consistent score was obtained. The score for immunoreactivity included staining intensity and positive cell proportions. At 200× magnification, 10 fields were randomly selected for scoring. The staining intensity score was divided into 4 grades from 0 to 3, with 0 indicating invisible positive staining, 1 indicating weak staining, 2 indicating moderate staining and 3 indicating strong staining. The percentage score, which evaluates the estimated percentage of tumour cells with positive staining, was also classified into 4 levels from 0 to 3 (0: non-positive staining; 1: 0–20%; 2: 21–50%; 3: > 51%). The final immunoreactivity score was obtained by multiplying the intensity score by the percentage score. For the statistical analysis, we defined an immunostaining score of 0–2 as negative (0: completely negative; 1–2: weakly positive), while 3–9 was defined as positive (3–5: moderately positive; 6–9: strongly positive).
Statistical analysis
The Chi squared test, Fisher’s exact test, or t test were used to identify the association between CASK expression and the clinicopathological characteristics of patients with CCA. Kaplan–Meier analysis was applied to compare OS and RFS between positive and negative CASK-expression groups, and P values were calculated by the log-rank test to verify the difference in survival curves. Univariate and multivariate Cox regression analyses were applied to explore the potential prognostic risk factors in CCA patients. In the univariate analysis, we used 0.05 as the cutoff P value to select the analysis factors to be included in the multivariate analysis. P < 0.05 was considered statistically significant. All statistical analyses were performed by using SPSS 21.0 (SPSS Inc., Chicago, IL) and the R software package (version 3.6.0, https://www.r-project.org).
Results
Identification of DERNAs in CCA
By setting the thresholds for the adjusted P value (FDR) < 0.05 and | log2FoldChange (FC) | > 1 for differential expression analysis, we identified a total of 875 significantly DERNAs (372 upregulated and 503 downregulated) in GSE32225 (Additional file 1). The volcano plots (Additional file 2) and heatmap (Additional file 3) of identified DERNAs were up loaded in additional files.
Functional enrichment analysis
To further understand the potential biological functions of the identified DERNAs, we conducted GO and KEGG analyses using DAVID. The GO and KEGG analysis results with P < 0.05 were considered to be significant, and these results are displayed in Fig. 1. The significantly enriched GO terms in the biological process (BP) domain were extracellular matrix organization, acute-phase response, platelet degranulation, blood coagulation and collagen fibril organization (Fig. 1a). In the cellular component (CC) domain, the significantly enriched terms in which the DERNAs were involved were extracellular exosome, extracellular region, blood microparticle and platelet alpha granule lumen (Fig. 1b). In the molecular function (MF) domain, the DERNAs were mainly enriched in oxygen binding, arachidonic acid epoxygenase activity, lipase inhibitor activity, heme binding and iron ion binding (Fig. 1c). The KEGG pathway analysis revealed that the genes were mainly associated with Complement and coagulation cascades, ECM-receptor interaction, Metabolism of xenobiotics by cytochrome P450, Drug metabolism—cytochrome P450, Retinol metabolism and Chemical carcinogenesis (Fig. 1d).
Fig. 1.
Functional enrichment analysis. a The top 10 enriched terms in biological process. b The top 10 enriched terms in cellular component. c The top 10 enriched terms in molecular function. d The top 10 enriched pathway in KEGG analysis
Survival analysis and Validation of DERNAs
By using TCGA data for survival analysis, we evaluated the prognostic value of these DERNAs. The GEPIA analysis tool is based on clinical data from the TCGA database, and clinical data from 36 patients were incorporated in the survival analysis. The results indicate that a total of 18 out of 875 DERNAs were observably associated with OS. Then, with the help of GEPIA, the expression of survival related DERNAs was verified in the TCGA data. We found that the CASK, APOF, NOL3, CD2BP2, CHD7, FAM98C, STXBP2, TRIM59, YIPF6 and ITGAV expression levels were consistent with our analysis results (Fig. 2). We then removed the inconsistent DERNAs in further analyses. The validated survival-related DERNAs are shown in Fig. 3.
Fig. 2.
Validation of the DERNAs. A, B Scatter plots of expression of DERNAs associated with survival in the GSE32225 dataset. a, b The expression levels of DERNAs associated with survival in TCGA dataset. Red block is CCA tissues, * stands for P < 0.05, and the difference is statistically significant
Fig. 3.
Survival analysis of the DERNAs. The relationship between DERNAs and survival of CCA patients was evaluated by performing Kaplan–Meier survival curves
Differentially expressed proteins (DEPs) and integrated analysis
By using the iTRAQ technique, we identified a total of 2886 confidential proteins in 6 matched primary and nontumour tissues. Among these, 487 proteins showed statistically significant in differential expression between CCA and paracancerous tissues, among which 235 was upregulated and 252 downregulated. We integrated the DEP data with the prognosis-related DERNA data validated by the TCGA dataset. By intersecting the two sets of data, we obtained 3 candidate proteins for further study (Fig. 4). Figure 4 was created by using the VENNY 2.1 tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html). These candidate proteins are summarized in Table 3.
Fig. 4.

Integrated data analysis. Venn diagram present the intersection of DEPs with survival related DERNAs
Table 3.
The Basic Properties of candidate protein
ITGAV or APOF expression in choledochal cysts and CCA
APOF was highly expressed in the cytoplasm of 7 choledochal cysts (Fig. 5a), and ITGAV was poorly expressed in the cell cytoplasm of 2 choledochal cysts (Fig. 5b). APOF was expressed at low levels in the cytoplasm in 3 of 14 CCA cases (Fig. 5A). ITGAV was expressed in the cytoplasm in 6 of 14 CCA tissues (Fig. 5B).
Fig. 5.

Immunohistochemical assay. a APOF is highly expressed in choledochal cyst. A APOF is lowly expressed in CCA. b ITGAV is poorly expressed in choledochal cyst. B ITGAV is moderately expressed in CCA. c, C CASK was strongly positive expressed in CCA. d, D CASK was moderately positive expressed in CCA. e, E CASK was weakly positive expressed in CCA. f, F CASK was completely negative expressed in interlobular bile duct. The red arrow shows the interlobular bile duct
CASK expression in TMA
CASK expression was evaluated using a TMA comprising 31 interlobular bile ducts and 86 CCA tissues (Fig. 5). CASK was detected in the nuclei and cytoplasm of carcinoma cells in 38 of 86 CCA tissues in TMA. In the following analysis, 1 case was excluded due to incomplete follow-up data and 1 patient was excluded due to tissue loss.
Relationship between CASK expression and clinicopathological features of CCA patients
The association between CASK expression and the clinicopathological characteristics of CCA patients is summarized in Table 4. CASK expression was found to be significantly correlated with vascular invasion (P = 0.009) and T classification (P = 0.017).
Table 4.
The summary of relationship between CASK expression and clinicopathological features of CCA patients
| Category | No. patients (%) | CASK | P value | |
|---|---|---|---|---|
| Negative | Positive | |||
| Age | 0.974 | |||
| < 55 | 33 (39.3) | 18 | 15 | |
| ≥ 55 | 51 (60.7) | 28 | 23 | |
| Gender | 0.130 | |||
| Female | 43 (51.2) | 27 | 16 | |
| Male | 41 (48.8) | 19 | 22 | |
| Histologic grade | 0.300 | |||
| Poorly | 17 (20.2) | 12 | 5 | |
| Moderate | 62 (73.8) | 32 | 30 | |
| Well | 5 (6.0) | 2 | 3 | |
| Stage | 0.491 | |||
| I | 8 (9.5) | 3 | 5 | |
| II | 38 (45.2) | 19 | 19 | |
| III | 25 (29.8) | 16 | 9 | |
| IV | 13 (15.5) | 8 | 5 | |
| T classification | 0.017 | |||
| T1 | 6 (7.1) | 2 | 4 | |
| T2 | 58 (69.0) | 30 | 28 | |
| T3 | 17 (20.2) | 14 | 3 | |
| T4 | 3 (3.7) | 0 | 3 | |
| N classification | 0.757 | |||
| N0 | 56 (66.7) | 30 | 26 | |
| N1 | 28 (33.3) | 16 | 12 | |
| Vascular invasion | 0.009 | |||
| No | 56 (66.7) | 25 | 31 | |
| Yes | 28 (33.3) | 21 | 7 | |
| Recurrence | 0.117 | |||
| No | 30 (35.7) | 13 | 17 | |
| Yes | 54 (64.3) | 33 | 21 | |
Italics values indicate P < 0.05
Relationship between positive CASK expression and overall survival rate
Kaplan–Meier curves showed that the survival rate of patients with positive CASK expression was significantly better than that of patients with negative CASK expression (P = 0.01; Fig. 6a). Univariate analysis of prognostic factors of CCA showed that pathological grade (P = 0.019), stage (P = 0.002), vascular invasion (P = 0.001) and negative CASK expression (P = 0.012) were significant risk factors for the outcome of CCA patients. Multivariate analysis showed that negative CASK expression (P = 0.027) was an independent risk factor for OS among CCA patients. The results of the univariate and multivariate analyses are summarized in Table 5.
Fig. 6.
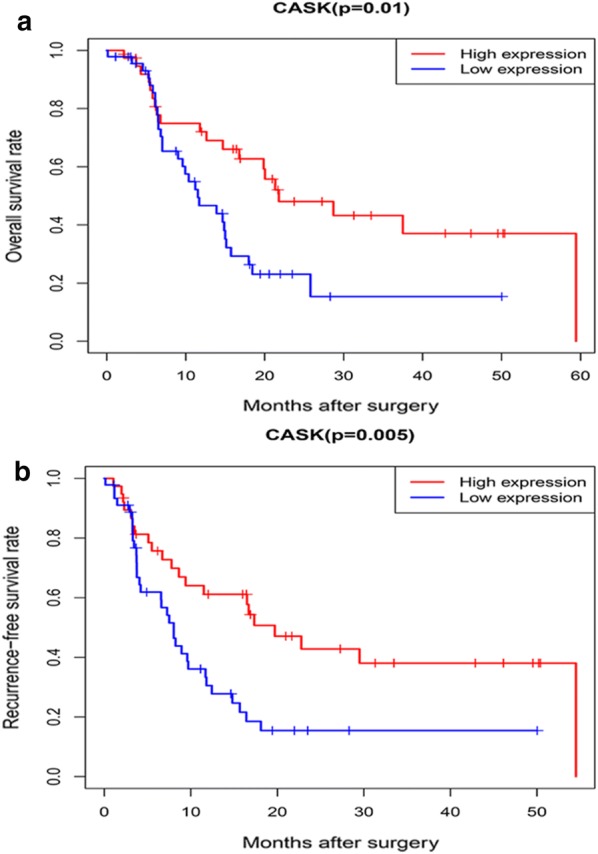
Kaplan–Meier survival analysis. a Kaplan–Meier survival analysis was used to analyse the relationship between CASK expression and OS of CCA patients. b Kaplan–Meier survival analysis was used to analyse the relationship between CASK expression and RFS of CCA patients
Table 5.
Univariate analysis and multivariate analysis of the relationship of CASK expression with OS among CCA patients
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.018 | 0.560–1.854 | 0.952 | |||
| Gender | 0.746 | 0.422–1.318 | 0.313 | |||
| Histologic grade | 2.050 | 1.126–3.732 | 0.019 | 1.795 | 0.939–3.430 | 0.077 |
| Stage | 1.681 | 1.208–2.340 | 0.002 | 1.528 | 0.944–2.474 | 0.085 |
| T classification | 1.262 | 0.852–1.869 | 0.246 | |||
| N classification | 1.659 | 0.930–2.960 | 0.087 | |||
| Vascular invasion | 2.644 | 1.501–4.656 | 0.001 | 1.236 | 0.523–2.918 | 0.629 |
| CASK | 2.155 | 1.188–3.909 | 0.012 | 2.031 | 1.083–3.809 | 0.027 |
Italics values indicate P < 0.05
HR hazard ratio, CI confidence interval
Relationship between positive CASK expression and recurrence-free survival rate
As shown by the Kaplan–Meier curves for RFS in Fig. 6b, patients with CCA who expressed CASK positively were associated with a better RFS (P = 0.005). In univariate analysis, pathological grade (P = 0.017), stage (P = 0.001), vascular invasion (P = 0.00007) and negative CASK expression (P = 0.006) were independent risk factors for RFS in patients with CCA. The multivariate analysis showed that pathological grade (P = 0.036), stage (P = 0.035) and negative CASK expression (P = 0.014) were statistically significant independent risk factors among CCA patients. A summary of the univariate and multivariate analyses is shown in Table 6.
Table 6.
Univariate analysis and multivariate analysis of the relationship of CASK expression with RFS among CCA patients
| Parameter | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.157 | 0.655–2.045 | 0.616 | |||
| Gender | 0.835 | 0.485–1.437 | 0.515 | |||
| Histologic grade | 1.983 | 1.130–3.481 | 0.017 | 1.918 | 1.044–3.526 | 0.036 |
| Stage | 1.750 | 1.273–2.408 | 0.001 | 1.612 | 1.034–2.514 | 0.035 |
| T classification | 1.377 | 0.946–2.004 | 0.095 | |||
| N classification | 1.591 | 0.910–2.780 | 0.103 | |||
| Vascular invasion | 2.797 | 1.625–4.816 | 0.000 | 1.325 | 0.616–2.851 | 0.472 |
| CASK | 2.229 | 1.259–3.947 | 0.006 | 2.114 | 1.163–3.844 | 0.014 |
Italics values indicate P < 0.05
HR hazard ratio, CI confidence interval
Discussion
In most published studies, many single-molecule biomarkers have been identified and clinically applied for CCA; for example, CA19-9 can be used as a diagnostic marker for CCA [20], SMAD4 expression is associated with the prognosis of CCA patients [21], and PPP3CA expression is an independent prognostic risk factor for CCA patients [22]. With the continuous improvement of multiomics technology, we can now screen cancer biomarkers at different levels through the combined application of multiple omics data to avoid the randomness of single omics data, improve the accuracy of diagnostic and prognostic biomarkers, and search for possible therapeutic targets [23, 24]. However, the special clinical characteristics of CCA, combined with restrictions from limited, CCA-specific research funds, make it difficult to obtain CCA tissue samples and therefore to realize the combined application of multiple omics to analyse this data. Fortunately, as bioinformatics has continued developing and public databases such as GEO and TCGA have been established, the realization of combined multiomics applications has become more acceptable. By sharing transcriptome microarray data uploaded by experimenters around the world, we can apply combinations of multiple omics techniques to provide a comprehensive and systematic perspective to advance the understanding of the molecular mechanism of CCA [25–27].
In our study, we first obtained CCA transcriptome data from the GEO database by using a bioinformatics method and then obtained 875 DERNAs through differential analysis of the microarray data by R software language. We further performed GO and KEGG functional analysis on these DERNAs. These DERNAs were basically enriched in the pathways of Complement and coagulation cascades, ECM-receptor interaction, Metabolism of xenobiotics by cytochrome P450, Drug metabolism—cytochrome P450, Retinol metabolism and Chemical carcinogenesis, which are closely related to tumour occurrence and development. Then, we used GEPIA to analyse CCA transcriptome data and the corresponding clinical information in the TCGA database. We explored the relationship between these DERNAs and the prognosis of patients and verified the differential expression of these prognostic related DERNAs in the TCGA data. We found that CASK, APOF, NOL3, CD2BP2, CHD7, FAM98C, STXBP2, TRIM59, YIPF6 and ITGAV expression were associated with CCA prognosis. In addition to reports that TRIM59 can be knocked out to inhibit the proliferation of CCA [28], other DERNAs were reported for the first time to show differences between tumour and normal tissue and were significantly associated with the prognosis of CCA patients.
Subsequently, we used the iTRAQ quantitative proteomics technique to screen out DEPs between CCA and nontumour tissues. By using the iTRAQ technique, we found 487 DEPs, of which 235 were upregulated and 252 were downregulated. By integrating transcriptome data with proteome data, we found that CASK, ITGAV and APOF were differentially expressed at the mRNA and protein levels. Through survival analysis, these DERNAs can serve as prognostic biomarkers for CCA. Through multiomics joint analysis, it was found that the proteins that were translated from these DERNAs were also differentially expressed between CCA and nontumour tissue. Therefore, these candidate proteins are likely associated with the prognosis of patients and may help us to understand the occurrence and development of CCA from a systematic and comprehensive perspective. Given the above reasons and the results of the literature review, we believe that ITGAV, APOF and CASK may be potential protein prognostic biomarkers and thus conducted further research on them. We used immunohistochemistry to identify the expression profiles of these candidate proteins in CCA tissues. After a preliminary evaluation of the immunohistochemical results by the immunoreactivity score, we found that the differential expression of CASK between CCA and nontumour tissues was significant. We further evaluated CASK expression in 84 CCA patients and explored the association between CASK expression and prognosis in patients with CCA.
Integrin subunit alpha V (ITGAV), a member of the integrin family, has been found to be associated with cell differentiation and metastasis and can serve as a prognostic biomarker for a variety of cancers [29–31]. In our study, we found that ITGAV mRNA expression was significantly upregulated in CCA and that upregulated expression was associated with better prognosis in CCA patients through bioinformatics analysis. Through iTRAQ quantitative proteomics, we found that the expression of ITGAV protein in CCA samples was stronger than that in nontumour samples, which was consistent with the results at the mRNA level. This finding is also consistent with the expression of ITGAV in gastric cancer, but overexpression of ITGAV has been found to be associated with a poor prognosis in gastric cancer patients [29]. Kang et al. reported that lncRNA AY can promote hepatocellular carcinoma metastasis by inducing transcription of ITGAV [32]. ITGAV may promote or inhibit cancer according to the type of tumour [33, 34]. Therefore, further research is needed to clarify its mechanism in the development of carcinoma.
Apolipoprotein F (APOF) is a lipid transfer inhibitor protein that can inhibit the activity of cholesteryl ester transfer protein (CETP) and plays an important role in lipid metabolism [35–37]. Wang et al. found that the expression of APOF at both the mRNA and protein levels in hepatocellular carcinoma was significantly downregulated, and low APOF expression was associated with poor RFS in hepatocellular carcinoma patients [38]. In this study, we found that APOF expression at both the mRNA and protein levels in CCA was significantly downregulated compared with in noncancer tissues. Through survival analysis, we found that the OS of CCA patients with high APOF expression at the mRNA level was significantly lower than that of other CCA patients. The mechanism of APOF in CCA is unclear and may be related to lipid metabolism. Future research is needed to clarify the relationship between APOF in CCA and lipid metabolism, which may provide new ideas for research on CCA.
Peripheral plasma membrane protein CASK (CASK), also known as calcium/calmodulin-dependent serine protein kinase, is a member of the membrane-related guanylate kinase (MAGUK) family, a group of cytoskeletal proteins composed of modular domain arrays [39, 40]. CASK, which has been intensively studied in neurons, is deemed to be an important histone protein in cortical networks and may be involved in the establishment and maintenance of synaptic connections [41–44]. The role of CASK in carcinoma has been reported only in oesophageal cancer, gastric cancer and colorectal cancer [45, 46]. Wang et al. found that CASK expression in oesophageal cancer was upregulated at the mRNA and protein levels [39]. Wei et al. reported that CASK is highly expressed in colorectal cancer and is associated with poor prognosis in colorectal cancer patients [47]. In this study, we first found that CASK was upregulated at the mRNA and protein levels in CCA compared with nontumour tissue by using bioinformatics and proteomics. Through Kaplan–Meier survival analysis, we found that the upregulated CASK expression at both the mRNA and protein levels was significantly related to a better prognosis in CCA patients and that CCA patients with high CASK expression at the protein level had a longer recurrence-free survival rate, which means that low CASK expression is an independent prognostic risk factor in CCA patients. Moreover, univariate and multivariate analyses also showed that low CASK expression was an independent risk factor for recurrence and poor prognosis.
The mechanism by which CASK plays a role in the development of carcinoma is unclear. It has been reported that the interaction and coexpression of CASK and CX43 can affect cell migration [48]. Zhou et al. found that the expression of miR-203 can inhibit the growth and invasion of gastric cancer cells by inhibiting the expression of CASK, which may provide a new idea for explaining how CASK is involved in the development of tumours from the perspective of a competing endogenous RNA (ceRNA) network [46]. Interestingly, when we studied the TCGA database by using the GEPIA web tool, we also found that patients with high CASK expression tended to have poorer OS and RFS than other hepatocellular carcinoma patients. This also indicates that CASK may be located upstream of tumour-related pathways and may modulate the entire network in a complex manner, showing different roles depending on the type of tumour. Therefore, the potential role of CASK in the process of tumourigenesis or tumour development and its mechanism deserve further investigation.
Through bioinformatics prediction, we can more easily find and verify candidate proteins that may be used as prognostic markers of CCA. Moreover, the integrated analysis of multiomics data will help us to explore the mechanism of CCA from a holistic perspective. However, due to a limited funding situation, we were only able to evaluate CASK as a prognostic marker for CCA, and the mechanism of CASK’s role in CCA remains unclear. Further research on this issue will help to elucidate the molecular mechanism of the occurrence or progression of CCA and may provide potential therapeutic targets for the treatment of CCA.
Conclusions
In summary, we used bioinformatics analysis to identify 875 DERNAs and explored their potential functions in relation to CCA. Ten prognostic related DERNAs that have not been previously reported were screened out. By using the iTRAQ technique, we identified 487 DEPs between CCA and nontumour tissues. Multiomics integrated analysis revealed differences at the mRNA and protein levels for CASK, ITGAV and APOF in CCA tissues compared to nontumour tissues. Low expression of CASK at the mRNA level is associated with a poor prognosis in CCA patients. Low CASK expression at the protein level is an independent risk factor for recurrence and poor prognosis in CCA patients. Our study found a reliable method for the screening of biomarkers for CCA and may provide a comprehensive and systematic perspective for the in-depth study of the pathogenesis of CCA.
Supplementary information
Additional file 1. The data of differentially expressed RNAs in GSE32225.
Additional file 2. Volcano plots of the DERNAs.
Additional file 3. Heatmap of the DERNAs.
Acknowledgements
We sincerely thank American Journal Experts (https://www.aje.com/) for its linguistic correcting assistance during the preparation of this manuscript.
Abbreviations
- CCA
Cholangiocarcinoma
- DERNAs
Differentially expressed RNAs
- DEPs
Differentially expressed RNAs
- GEO
Gene Expression Omnibus
- TCGA
The Cancer Genome Atlas
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- iTRAQ
Isobaric tags for relative and absolute quantitation
- CASK
Peripheral plasma membrane protein CASK
- ITGAV
Integrin subunit alpha V
- APOF
Apolipoprotein F
- IHC
Immunohistochemistry
- DAB
Diaminobenzidine
- OS
Overall survival
- RFS
Recurrence free survival
- LC–MS
Liquid chromatography–mass spectrometry
- TMA
Tissue microarray
- SCX
Strong cation exchange
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- GEPIA
Gene Expression Profiling Interactive Analysis
- FC
Fold change
Authors’ contributions
Research design: ZD, LG, WM, YL. Conducted experiments: LG, ZD, JZ. Data collection and analysis: ZD, LG, WF. Writing and editing manuscript: ZD, GS. Revised the manuscript: WM, YL, JY, XL. Funding acquisition: WM, YL. Provided intellectual contribution: XZ, ZP, PY, BB. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation (Grant Number 81872036), and Lanzhou Science and Technology Bureau (Grant Number 2019-4-43), and Science and Technology Project of Chengguan District of Lanzhou City (Grant Number 2019JSCX0092 and 2019RCCX0038), and Lanzhou talent innovation project (Grant Number 2018-RC-13).
Availability of data and materials
The data used to sustain the results of this work are included in the article and its supplementary information files.
Ethics approval and consent to participate
Collection and usage of tissue samples in this work was approved by the Ethics Committee for Human Research, Lanzhou University, and written informed consents were obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanyan Lin and Wenbo Meng contributed equally to this work
Contributor Information
Yanyan Lin, Email: ldyy_linyy@lzu.edu.cn.
Wenbo Meng, Email: mengwb@lzu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-020-01212-z.
References
- 1.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borbath I, Ceratti A, Verslype C, Demols A, Delaunoit T, Deleporte A, et al. Combination of gemcitabine and cetuximab in patients with advanced cholangiocarcinoma: a phase II study of the Belgian Group of Digestive Oncology. J Clin Oncol. 2011;29(4_suppl):245. doi: 10.1200/jco.2011.29.4_suppl.245. [DOI] [PubMed] [Google Scholar]
- 3.Gaudig I, Schwoerer H, Ramadori G. Treatment of cholangiocarcinoma: retrospective analysis of a single-center experience. J Clin Oncol. 2012;30(4_suppl):376. doi: 10.1200/jco.2012.30.4_suppl.376. [DOI] [Google Scholar]
- 4.Suh KS, Roh HR, Koh YT, Lee KU, Park YH, Kim SW. Clinicopathologic features of the intraductal growth type of peripheral cholangiocarcinoma. Hepatology. 2000;31(1):12–17. doi: 10.1002/hep.510310104. [DOI] [PubMed] [Google Scholar]
- 5.Hatzaras I, Choti MA, Schulick RD, Alexandrescu S, Pulitano C, Clary BM, et al. Patterns of recurrence after resection of intrahepatic cholangiocarcinoma: results from a multi-institutional cohort of 449 patients. J Clin Oncol. 2012;30(4_suppl):267. doi: 10.1200/jco.2012.30.4_suppl.267. [DOI] [Google Scholar]
- 6.Park J, Kim M, Kim J, Lee J. Survival time and its prognostic factors in advanced unresected cholangiocarcinoma without chemoradiation. J Clin Oncol. 2009;27(15_suppl):e15681. [Google Scholar]
- 7.Mosnier JF, Kandel C, Cazals-Hatem D, Bou-Hanna C, Gournay J, Jarry A, et al. N-cadherin serves as diagnostic biomarker in intrahepatic and perihilar cholangiocarcinomas. Mod Pathol. 2009;22(2):182–190. doi: 10.1038/modpathol.2008.123. [DOI] [PubMed] [Google Scholar]
- 8.Hu B, Fan H, Lv X, Chen S, Shao Z. Prognostic significance of CXCL5 expression in cancer patients: a meta-analysis. Cancer Cell Int. 2018;18:68. doi: 10.1186/s12935-018-0562-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel AH, Harnois DM, Klee GG, LaRusso NF, Gores GJ. The utility of CA 19-9 in the diagnoses of cholangiocarcinoma in patients without primary sclerosing cholangitis. Am J Gastroenterol. 2000;95(1):204–207. doi: 10.1111/j.1572-0241.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Li J, Xia Y, Gong R, Wang K, Yan Z, et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol. 2013;31(9):1188–1195. doi: 10.1200/JCO.2012.41.5984. [DOI] [PubMed] [Google Scholar]
- 11.Hatzaras I, Schmidt C, Muscarella P, Melvin WS, Ellison EC, Bloomston M. Elevated CA 19-9 portends poor prognosis in patients undergoing resection of biliary malignancies. HPB (Oxford). 2010;12(2):134–138. doi: 10.1111/j.1477-2574.2009.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardini A, Corti B, Fiorentino M, Altimari A, Ercolani G, Grazi GL, et al. Expression of connective tissue growth factor is a prognostic marker for patients with intrahepatic cholangiocarcinoma. Dig Liver Dis. 2005;37(4):269–274. doi: 10.1016/j.dld.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Bonney GK, Craven RA, Prasad R, Melcher AF, Selby PJ, Banks RE. Circulating markers of biliary malignancy: opportunities in proteomics? Lancet Oncol. 2008;9(2):149–158. doi: 10.1016/S1470-2045(08)70027-5. [DOI] [PubMed] [Google Scholar]
- 14.Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57(7):1501–1508. doi: 10.1093/jxb/erj168. [DOI] [PubMed] [Google Scholar]
- 15.Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144(4):829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh S, Tang S-J, Sreenarasimhaiah J, Lara LF, Siddiqui A. The clinical utility and limitations of serum carbohydrate antigen (CA19-9) as a diagnostic tool for pancreatic cancer and cholangiocarcinoma. Dig Dis Sci. 2011;56(8):2491–2496. doi: 10.1007/s10620-011-1709-8. [DOI] [PubMed] [Google Scholar]
- 21.Yan XQ, Zhang W, Zhang BX, Liang HF, Zhang WG, Chen XP. Inactivation of Smad4 is a prognostic factor in intrahepatic cholangiocarcinoma. Chin Med J (Engl). 2013;126(16):3039–3043. [PubMed] [Google Scholar]
- 22.Tang Z, Yang Y, Zhang J, Fu W, Lin Y, Su G, et al. Quantitative Proteomic Analysis and Evaluation of the Potential Prognostic Biomarkers in Cholangiocarcinoma. J Cancer. 2019;10(17):3985–3999. doi: 10.7150/jca.29354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer. 2016;16(8):525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun YV, Hu Y-J. Integrative analysis of multi-omics data for discovery and functional studies of complex human diseases. Adv Genet. 2016;93:147–190. doi: 10.1016/bs.adgen.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vantaku V, Dong J, Ambati CR, Perera D, Donepudi SR, Amara CS, et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin Cancer Res. 2019;25(12):3689–3701. doi: 10.1158/1078-0432.CCR-18-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu D-Y, Qu R-X, Lu T-R, Wu H-Y. Cancer bioinformatics for updating anticancer drug developments and personalized therapeutics. Rev Recent Clin Trials. 2017;12(2):101–110. doi: 10.2174/1574887112666170209161444. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, Zhang B, Meng D, Ge C. Comprehensive analysis of DNA methylation and gene expression profiles in cholangiocarcinoma. Cancer Cell Int. 2019;19(1):352. doi: 10.1186/s12935-019-1080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen H, Zhang J, Zhang Y, Feng Q, Wang H, Li G, et al. Knockdown of tripartite motif 59 (TRIM59) inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR signalling pathway. Gene. 2019;698:50–60. doi: 10.1016/j.gene.2019.02.044. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Chen H, Jiang Z, Lin Y, Wang X, Xiang J, et al. Integrin subunit alpha V promotes growth, migration, and invasion of gastric cancer cells. Pathol Res Pract. 2019;215(9):152531. doi: 10.1016/j.prp.2019.152531. [DOI] [PubMed] [Google Scholar]
- 30.Logan NJ, Camman M, Williams G, Higgins CA. Demethylation of ITGAV accelerates osteogenic differentiation in a blast-induced heterotopic ossification in vitro cell culture model. Bone. 2018;117:149–160. doi: 10.1016/j.bone.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waisberg J, De Souza Viana L, Affonso Junior RJ, Silva SRM, Denadai MVA, Margeotto FB, et al. Overexpression of the ITGAV gene is associated with progression and spread of colorectal cancer. Anticancer Res. 2014;34(10):5599–5607. [PubMed] [Google Scholar]
- 32.Kang CL, Qi B, Cai QQ, Fu LS, Yang Y, Tang C, et al. LncRNA AY promotes hepatocellular carcinoma metastasis by stimulating ITGAV transcription. Theranostics. 2019;9(15):4421–4436. doi: 10.7150/thno.32854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viana LS, Affonso RJ, Silva SR, Denadai MV, Matos D, Souza C, et al. Relationship between the expression of the extracellular matrix genes SPARC, SPP1, FN1, ITGA5 and ITGAV and clinicopathological parameters of tumor progression and colorectal cancer dissemination. Oncology. 2013;84(2):81–91. doi: 10.1159/000343436. [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Lee CH, Bae JT, Nam KT, Moon DB, Hwang OK, et al. Inhibition of skin carcinogenesis by suppression of NF-κB dependent ITGAV and TIMP-1 expression in IL-32γ overexpressed condition. J Exp Clin Cancer Res. 2018;37(1):293. doi: 10.1186/s13046-018-0943-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Driscoll DM, Morton RE. Molecular cloning and expression of lipid transfer inhibitor protein reveals its identity with apolipoprotein F. J Biol Chem. 1999;274(3):1814–1820. doi: 10.1074/jbc.274.3.1814. [DOI] [PubMed] [Google Scholar]
- 36.Lagor WR, Brown RJ, Toh S-A, Millar JS, Fuki IV, de la Llera-Moya M, et al. Overexpression of apolipoprotein F reduces HDL cholesterol levels in vivo. Arterioscler Thromb Vasc Biol. 2009;29(1):40–46. doi: 10.1161/ATVBAHA.108.177105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morton RE, Liu Y, Izem L. ApoF knockdown increases cholesteryl ester transfer to LDL and impairs cholesterol clearance in fat-fed hamsters. J Lipid Res. 2019;60(11):1868–1879. doi: 10.1194/jlr.RA119000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang YB, Zhou BX, Ling YB, Xiong ZY, Li RX, Zhong YS, et al. Decreased expression of ApoF associates with poor prognosis in human hepatocellular carcinoma. Gastroenterol Rep (Oxf). 2019;7(5):354–360. doi: 10.1093/gastro/goz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Lu J, Yang C, Wang X, Cheng L, Hu G, et al. CASK and its target gene Reelin were co-upregulated in human esophageal carcinoma. Cancer Lett. 2002;179(1):71–77. doi: 10.1016/S0304-3835(01)00846-1. [DOI] [PubMed] [Google Scholar]
- 40.Dimitratos SD, Woods DF, Bryant PJ. Camguk, Lin-2, and CASK: novel membrane-associated guanylate kinase homologs that also contain CaM kinase domains. Mech Dev. 1997;63(1):127–130. doi: 10.1016/S0925-4773(97)00668-0. [DOI] [PubMed] [Google Scholar]
- 41.Hsueh YP, Yang FC, Kharazia V, Naisbitt S, Cohen AR, Weinberg RJ, et al. Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J Cell Biol. 1998;142(1):139–151. doi: 10.1083/jcb.142.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaConte LEW, Chavan V, Liang C, Willis J, Schönhense E-M, Schoch S, et al. CASK stabilizes neurexin and links it to liprin-α in a neuronal activity-dependent manner. Cell Mol Life Sci. 2016;73(18):3599–3621. doi: 10.1007/s00018-016-2183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardner KL, Sanford JL, Mays TA, Rafael-Fortney JA. CASK localizes to nuclei in developing skeletal muscle and motor neuron culture models and is agrin-independent. J Cell Physiol. 2006;206(1):196–202. doi: 10.1002/jcp.20449. [DOI] [PubMed] [Google Scholar]
- 44.Gao R, Piguel NH, Melendez-Zaidi AE, Martin-de-Saavedra MD, Yoon S, Forrest MP, et al. CNTNAP2 stabilizes interneuron dendritic arbors through CASK. Mol Psychiatry. 2018;23(9):1832–1850. doi: 10.1038/s41380-018-0027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akrami H, Moradi B, Borzabadi Farahani D, Mehdizadeh K. Ibuprofen reduces cell proliferation through inhibiting Wnt/β catenin signaling pathway in gastric cancer stem cells. Cell Biol Int. 2018;42(8):949–958. doi: 10.1002/cbin.10959. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, Xu G, Yin C, Jin W, Zhang G. Down-regulation of miR-203 induced by Helicobacter pylori infection promotes the proliferation and invasion of gastric cancer by targeting CASK. Oncotarget. 2014;5(22):11631–11640. doi: 10.18632/oncotarget.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei JL, Fu ZX, Fang M, Zhou QY, Zhao QN, Guo JB, et al. High expression of CASK correlates with progression and poor prognosis of colorectal cancer. Tumour Biol. 2014;35(9):9185–9194. doi: 10.1007/s13277-014-2179-3. [DOI] [PubMed] [Google Scholar]
- 48.Márquez-Rosado L, Singh D, Rincón-Arano H, Solan JL, Lampe PD. CASK (LIN2) interacts with Cx43 in wounded skin and their coexpression affects cell migration. J Cell Sci. 2012;125(Pt 3):695–702. doi: 10.1242/jcs.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The data of differentially expressed RNAs in GSE32225.
Additional file 2. Volcano plots of the DERNAs.
Additional file 3. Heatmap of the DERNAs.
Data Availability Statement
The data used to sustain the results of this work are included in the article and its supplementary information files.





