Abstract
Pancreatic cancer (PanCa) is a highly lethal disease with a poor 5 year survival rate, less than 7%. It has a dismal prognosis, and more than 50% of cases are detected at an advanced and metastatic stage. Gemcitabine (GEM) is a gold standard chemotherapy used for PanCa treatment. However, GEM-acquired resistance in cancer cells is considered as a major setback for its continued clinical implementation. This phenomenon is evidently linked to de novo lipid synthesis. PanCa cells rely on de novo lipid synthesis, which is a prime event in survival and one of the key drivers for tumorigenesis, cancer progression, and drug resistance. Thus, the depletion of lipogenesis or lipid metabolism can not only improve treatment outcomes but also overcome chemoresistance, which is an unmet clinical need. Toward this effort, our study reports a unique paclitaxel–poly(lactic-co-glycolic acid) (PLGA) nanoparticles (PPNPs) formulation which can target lipid metabolism and improve anticancer efficacy of GEM in PanCa cells. PPNPs inhibit excessive lipid formation and alter membrane stability with compromised membrane integrity, which was confirmed by Fourier transform infrared and zeta potential measurements. The effective interference of PPNPs in lipid metabolic signaling was determined by reduction in the expression of FASN, ACC, lipin, and Cox-2 proteins. This molecular action profoundly enhances efficacy of GEM as evident through enhanced inhibitory effects on the tumorigenic and metastasis assays in PanCa cells. These data clearly suggest that the ablation of lipid metabolism might offer an innovative approach for the improved therapeutic outcome in PanCa patients.
Introduction
Pancreatic cancer (PanCa) remains a second deadliest cancer related disease with an estimated 56,770 new cases and 45,750 deaths in the United States in 2019.1 It is highly lethal cancer with a 5 year mortality rate of 95%. The majority of patients are not eligible for surgery because of the advanced stage of disease or metastasis to other vital organs. At this stage, chemotherapy is highly advised rather radiation or surgery alone. There are few chemotherapy regimens such as Gemzar (gemcitabine), Abraxane (albumin-bound paclitaxel formulation), FOLFOX (combination of folinic acid, fluorouracil, and oxaliplatin), and FOLFIRINOX (combination of fluorouracil, irinotecan, leucovorin, oxaliplatin, and folinic acid)2,3 that have been widely used; however, none of them are efficient to improve overall survival more than 6–12 months. Additionally, cocktail chemotherapy regimen(s) introduce severe systemic toxicities and drug resistance; thus, cancer cells do not respond to these regimens. These events offer recurrence of disease or highly aggressive and metastasis disease which is difficult to tackle. Thus, there is an urgent clinical unmet need to overcome such short fall in effective chemotherapy for PanCa.
The literature strongly shows that glucose and lipid metabolism contribute poor patients’ survival.4−6 The metabolic pathway is one of the distinct features of tumorigenesis and cancer progression.7 During cancer development, the de novo lipid synthesis gets triggered distinct from normal cells, causing an increase in the production of fatty acids.7−9 The synthesis of fatty acids in normal cells is at a lower level.10,11 The increase in demand of energy is supplied by an increase in the enzymes involved in lipogenesis, which further form lipids, glucose, and amino acids, in turn, leading to proliferation and differentiation of tumor cells.12,13 The phospholipids generated after fatty acid synthesis play an integral part in the formation of the cell membrane and some of the them function as signaling molecules in different oncogenic pathways. In fact, some lipids can act as a biomarker for cancer diagnosis as the lipid composition changes from normal cells as compared to cancer cells.7−9,14
In the cytosol, citrate is broken down by adenosine triphosphate (ATP) citrate lyase to form acetyl coenzyme A (acetyl-CoA), which is an important lipid synthesis substrate. Acetyl-CoA carboxylase (ACC) is responsible for conversion of acetyl-CoA to malonyl-CoA. Fatty acid synthase (FASN) further converts malonyl-CoA to palmitic acid and the synthesis of fatty acid proceeds thereon.7−9,14 There have been various findings related to apoptosis and growth arrest of cancer cells, as FASN is inhibited.15,16 Supplementation of a lipid synthesis inhibitor (5-(tetradecyloxy)-2-furoic acid) or ACC/FASN inhibitor (cerulenin and irgasan) can be efficient to reduce the proliferation and increase apoptosis in cancer cells.3,15 In de novo lipid synthesis, sterol regulatory element-binding protein-1 (SREBP-1)14,16 regulates the expression of FASN and ACC, thus facilitating the production of lipids, subsequently endorses proliferation of cancer cells. It has now been increasingly accepted that targeting or modulating lipid metabolism in cancer cells is an emerging therapeutic strategy. To this end, several inhibitors/drugs have been developed and tested in several preclinical and clinical trials (or trials are ongoing). There are number of clinical trials underway to learn the therapeutic benefit with inhibitors blocking lipid metabolism. These include gemcitabine and a combination of disulfiram (NCT02671890), paricalcitol (NCT02030860), and simvastatin (NCT00944463). A recent study reports that an FASN inhibitor, orlistat, with gemcitabine combination not only stimulates cell-cycle arrest and apoptosis through induction of ROS but also promotes gemcitabine uptake and metabolism in PanCa cells.4
Chemotherapy is a standard form of treatment for PanCa. Gemcitabine is the first-line chemotherapy agent which gets converted to disphosphate (dFdCDP) and triphosphate (dFdCTP) intracellularly. Inactivation of ribonucleotide reductase, which is integral for DNA replication and inhibition of DNA by dFdCDP, leads to apoptosis eventually by incorporating itself into DNA.8,9 When the human concentrative nucleoside transporter (hCNT1) expression is at a lower level, there is limited gemcitabine transport in cells. Because gemcitabine is a hydrophilic drug which requires an efficient transport to aid its uptake across the hydrophobic cell membrane.17 Gemcitabine is metabolized by cytidine deaminase which causes the drug to be rapidly cleared, that is, decreased circulation time leading to its reduced therapeutic efficacy.18,19 In order to combat this, elevated doses of gemcitabine have been administered that have caused toxic effects such as nausea and difficulty in breathing. In order to increase its bioavailability, different approaches have been undertaken.19 Additionally, other efflux pumps such as P-glycoprotein (P-gp) or multidrug resistant gene-1/5 (MDR-1 or MRP5) expression can hinder gemcitabine uptake because of elevation of drug-resistant features.18 Treatment efficacy of gemcitabine can be improved with agents that can alter the expression of the transporters18,19 or by increased gemcitabine uptake.20
A clinical trial of Nab-paclitaxel (abraxane, albumin-bound paclitaxel nanoparticle) and gemcitabine proved that the combination was more effective as compared to gemcitabine alone in antitumor activity. Nab-paclitaxel is known to decrease the cytidine deaminase responsible for gemcitabine metabolism and thus improving its half-life within the body.21 Until today, there is no study dealing with lipid metabolism in conjunction with paclitaxel or paclitaxel with gemcitabine to control the PanCa growth. Our laboratory has formulated a unique paclitaxel–poly(lactic-co-glycolic acid) (PLGA) nanoparticles (PPNPs) formulation, containing PLGA, which is an ideal carrier for drug delivery because of its features such as biocompatibility and biodegradation,22 and pluronic F-127 that has the capability to inhibit P-glycoprotein efflux for increasing intracellular drug concentration.23 This study reports that our novel PPNPs nanoformulation effectively altered lipid metabolism in PanCa cells and enhanced therapeutic efficacy of gemcitabine.
Experimental Section
Cell Culture
PanCa cell lines (HPAF-II and PANC-1) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in DMEM/F12 or DMEM (HyClone Laboratories, Inc. South Logan, Utah, USA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, GA, USA) and 1% (w/v) penicillin–streptomycin (Gibco, Thermo Fisher Scientific, Grand Island, NY) at 37 °C in a humidified atmosphere containing 5% CO2. Both cell lines were maintained as mycoplasma-free and used within 4–6 months of resuscitation. Cells were trypsinized once they reach 70–80% confluency and seeded as per the required experiments.
Preparation and Characterization of Paclitaxel-Encapsulated PLGA Nanoparticles
Paclitaxel-encapsulated PLGA nanoparticles (PPNPs) were prepared using a nanoprecipitation method as previously described.24 In brief, 200 mg of PVA (Sigma, P8136) was dissolved in 20 mL of Milli-Q to achieve 1% PVA. To this solution, the organic phase containing 90 mg of PLGA (Lactel Polymers, B6010-4) and 10 mg of paclitaxel in 5 mL of acetone was added dropwise and left to stir overnight at 800 rpm. Then, 5 mg of poly(l-lysine) (Sigma, P2636) and 5 mg of the F127 polymer (Sigma, P2443) in 2 mL of Milli-Q were added dropwise to the formulation to achieve a stable coating on nanoparticles. Similarly, PLGA control formulation was generated without paclitaxel in nanoparticle formulation. Particle size and zeta potential measurement were performed based on the principle of dynamic light scattering using a Zetasizer (Nano ZS, Malvern Instruments, Malvern, UK). Measurements were performed in triplicate using 10 μL of the sample and 990 μL of phosphate-buffered saline (PBS). The spherical particle morphology of generated PPNPs was imaged using a JEOL 1210-JEM transmission electron microscope (JEOL Ltd, Tokyo, Japan). In this study, 20 μL of 1 mg/mL PPNP sample solution was placed on a shiny side of 200 mesh formvar-coated copper grid (grid size: 97 μm; Ted Pella Inc, Redding, CA). A 2% w/v of uranyl acetate solution contrast stain solution was used to stain nanoparticles for better visibility. These air-dried nanoparticles on the grid were imaged at 100,000× following our published protocol.25
Rh123 Dye Exclusion Assay
Rh123 dye exclusion assay was performed to test the extent of possibility of the drug to internalize in cells. To examine this phenomenon, 0.5 million PanCa cells were seeded in a 6-well plate and allowed them to attach overnight. Cells were treated with 10 nM PTX or 10 nM PPNPs for 24 h. Then, the cells were washed three times with 1× PBS and incubated with 100 μg of Rh123 dye (1 mg/mL stock solution) for 30 min and imaged under a microscope or Rh123 dye fluorescence in cells was quantified using flow cytometry. For visualization of Rh123 in cells, after incubation with Rh123, cells were given PBS washes and visualized under an AMF4300 EVOS FL Imaging System (Life Technologies, Carlsbad, CA, USA). For semiquantification analysis of Rh123 dye in cells, after incubation with Rh123 dye, cells were trypsinized and centrifuged to remove unbound dye present in culture media and then injected 10,000 cells into a NovoCyte Flow Cytometer (ACEA NovoCyte 1000, ACEA Biosciences, Inc. San Diego, CA, USA). The dye quantification in cell population was measured using an FITC channel (λex: 485 nm and λem: 520 nm).26
Cell Proliferation by xCELLigence
Cell proliferation of PANC-1 and HPAF-II cells was carried out by xCELLigence assay using real-time cell analyzer (RTCA) DP instrument (Roche) to evaluate influence of PPNPs/GEM combination treatment. For this assay, PanCa cell lines (6000 cells/well) were seeded in E plates according to manufacturer’s specifications.27 After 24 h (cells attached to plates), various treatments (10 nM PTX, 10 nM PPNPs, 100 nM GEM, 10 nM PPNPs + 50 nM GEM, and 10 nM PPNPs + 100 nM GEM or controls) were introduced, and the study continued for 65 h. The selection of 10 nM PTX or PPNPs and 100 nM GEM for all in vitro studies was based on our previous study24,28 which suggests that these concentrations are effective and influence on molecular effects in PanCa cells.
Lipid Extraction and FT-IR Spectroscopy
For this study, PanCa cells were replated (0.5 million cells/well) in a 6-well plate in 2 mL of the respective medium. The cells were allowed to attach to plates overnight and treated with 10 nM PTX, 10 nM PPNPs, 100 nM GEM, and 10 nM PPNPs + 100 nM GEM for 24 h. Treatments with PBS and nanoparticles alone (no paclitaxel) were considered as controls. After treatment, cells containing plates were washed three times with 1× PBS, trypsinized, and pelleted down at 2000 rpm using a Sorvall ST 8 Centrifuge (Thermo Fisher Scientific, Suzhou, China). The membrane lipids were extracted from PanCa cell pellets following Bligh and Dyer protocol. To a cell pellet, 250 μL of chloroform was added and vortexed, followed by the addition of 16.8 μL of 6 M HCl and 250 μL of chloroform, again samples were vortexed. About 250 μL of water was added to this solution, vortexed, and centrifugation was carried out for 2000 rpm using an Eppendorf centrifuge 5415C (Brinkmann Instruments, Inc., N. Y., USA) for 5 min. Samples were incubated at 4 °C for 1 h and the lipid portions were collected from the lower phase.29 To examine lipid profile alternation in cancer cells with treatments, we employed Fourier transform infrared (FT-IR) spectroscopy. For this, equal amounts of lipid extracts (based on protein quantification) were deposited on a diamond/ZnSe attenuated total reflection crystal plate and allowed to air-dry. Then, FT-IR spectra were obtained using a PerkinElmer Spectrum 100 FTIR spectrometer (Waltham, MA, USA) in the range from 4000 to 650 cm–1 with a resolution of 2 cm–1. Each spectrum is an average of 32 scans.
Confocal Microscopy
Confocal microscopy was employed in order to test the effect of different treatment groups on the cellular lipid membrane. In this study, PanCa cells were seeded in chamber slides and after 70% confluency, cells were treated with 10 nM PTX, 10 nM PPNPs, 100 nM GEM, and 10 nM PPNPs + 100 nM GEM or control groups for 24 h. After treatment, 0.7 μL of stain was added to each well containing 1 mL of media and incubated for 30 min. Triton X-100 (0.1%) was used to permeabilize cells, followed by washing twice with PBS. Cell Mask deep red plasma membrane stain was used to stain the lipid membranes of cells. Upon DAPI staining, VECTASHIELD Mounting Medium (Vector Labs, Burlingame, CA, USA) with a coverslip was used to mount the slides. The membrane disruption was observed using a laser confocal microscope (Carl Zeiss LSM 710, Oberkochen, Germany).30
Western Blotting
To examine superior inhibitory metabolic effects of PPNPs/GEM combination on cancer cells, we performed immunoblotting. For this study, PANC-1 and HPAF-II were seeded in 100 mm dishes and supplemented treatments: 10 nM PTX, 10 nM PPNPs, 100 nM GEM, and 10 nM PPNPs + 100 nM GEM combination with control and PLGA formulation for 48 h. 2× SDS lysis extraction buffer (Santa Cruz Biotechnology, Santa Cruz, CA) was used to prepare protein lysates. Protein concentration was quantified using Bradford assay. Cell lysate proteins were separated by 10% SDS-PAGE gels, transferred on to a PVDF membrane, followed by blocking with 1% BSA. Then, membranes were probed for protein expression with antibodies: Bax, Bcl-2, FASN, ACC, Cox-2, Lipin, ATP citrate lyase, and β-actin, followed by specific secondary antibodies.31 The bound antibodies were viewed under a UVP Gel Doc system.
Zeta Potential Measurement of Whole Cells
Zeta potential of cancer cells after treating with 10 nM PTX and PPNPs, 100 nM GEM, and 10 nM PPNPs + 100 nM GEM combination with respect to their control and blank nanoparticle group was measured using a Zetasizer (Nano ZS, Malvern Instruments, Malvern, UK). After 24 h treatment, cells were washed three times with 1× PBS and trypsinized. About 10 μL of the sample was diluted with 990 μL of PBS before measurement. Zeta potential measurements were performed in triplicate.
Statistical Analysis
All the experimental data, statistical analysis, and graphical representations were processed using GraphPad Prism 5 (San Diego, CA) software. The data are shown as an average of representation of triplicates and mean ± sem. Student’s t-test was applied to analyze the difference between groups. *p < 0.05 or #p < 0.05 was considered statistically significant in these tests.
Results
Characterization of PPNPs
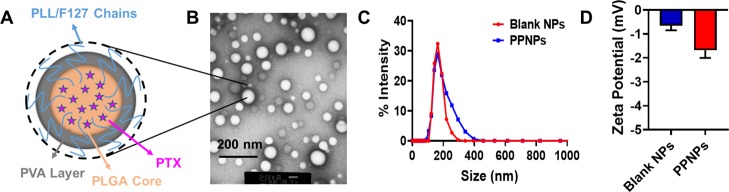
All physicochemical characterization of PPNPs was conducted to confirm the encapsulation of paclitaxel in PLGA NPs and it is consistent with our previous reports.24 In brief, putative structure, transmission electron microscopy image, and dynamic light scattering measurements of PPNPs are presented in Figure 1. The TEM image of PPNPs confirms its uniformity of particle size and its distribution in the dried form. Similarly, the average hydrodynamic particle size of control PLGA NPs and PPNPs was found to be 160 ± 5 and 190 ± 3 nm, respectively. TEM-based particles size appears to be smaller than dynamic light scattering (DLS)-based particle size measurements. This difference between TEM and DLS size variation may be due to the difference in the measurement environment. DLS measurements provide particle size in suspension, which may represent higher because of hydrodynamic volume of polymers and other formulation excipients, while TEM size indicates the particles those were completely in the dried state. Zeta potential was found to be −0.688 and −1.68 mV for these nanoparticles. This confirms that paclitaxel encapsulation did not significantly alter the particle size, distribution, and zeta potential.
Figure 1.
Structure and characterization of paclitaxel-encapsulated poly(lactic-co-glycolic acid) nanoparticles (PPNPs). (A) Schematic structure illustration of PPNPs generated in the solvent evaporation and nanoprecipitation process. (B) Representative transmission electron microscopy image of PPNPs. JEOL 2010-JEM was used to image nanoparticles at a direct magnification of 100,000× with the help of a 2%w/v uranyl acetate EM stain solution. (C,D) Particle size and zeta potential measurements of blank PLGA NPs and PPNPs in a Zetasizer. Data represent the average of triplicate measurements using 10 μL of the sample and 990 μL of PBS.
Chemosensitization Ability of PPNPs
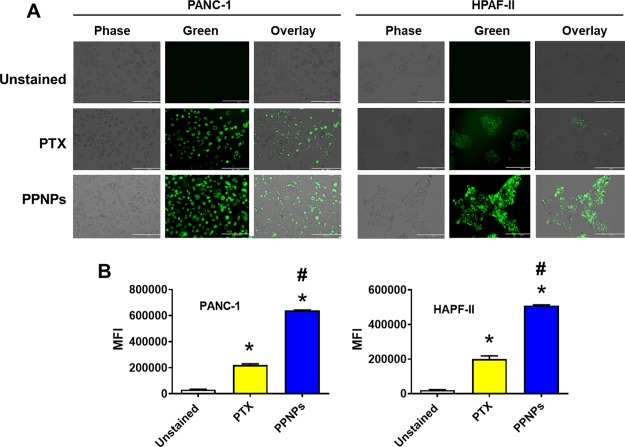
Rh123 dye exclusion assay can be extended to determine the chemosensitization potential of the drug or formulation, which has been previously employed in our laboratory. Rh123 is a red color dye that easily crosses the cell membrane and stains the cytoplasm when cells express low P-gp or multidrug resistance protein.32 On the other hand, there may be less Rh123 stain in cells when cells express more P-gp. This represents that it acts as a probe for a chemosensitizer, that is, more Rh123 access means more drug can be accumulated in cells without pumping out drug through exocytosis or exporters. This phenomenon was evaluated by microscopy and flow cytometry methods.
In the microscopy method, PTX-treated PanCa cells show a minimum uptake of Rh123 because of export of Rh123 due to presence of P-gp/MDR proteins (transporters, responsible for pumping out the drug) on cells (Figure 2A), whereas in PPNP-treated cells, it appears to have significantly more Rh123 dye in cells because of inhibition of P-gp efflux. To further confirm these results, the flow cytometry method was employed. In the flow cytometry method, the mean flow intensity values also suggest an increase in Rh123 stain with PTX and a greater increase with PPNPs (significant) compared to that of control cells and PTX-treated cells (Figure 2B).
Figure 2.
Drug efflux inhibition phenomenon evaluated by Rh123 dye exclusion assay. (A) Cancer cells were treated for 24 h with 10 nM PTX or 10 nM PPNPs and incubated with Rh123 for 30 min before visualizing them under a microscope to obtain representative images. Rh123 showed more accumulation in treatment groups of PPNPs, indicating P-gp and drug export inhibition in this treatment group. (B) Mean fluorescence intensity in an FL1 channel (488 excitation, blue laser, 530 ± 15 nm, FITC/GFP) was measured for treatment groups. PPNPs showed increased fluorescence in both PanCa cells as compared to 10 nM PTX treatment, indicating that PTX in solution gets effluxed because of P-gp. Data are presented as the mean ± SEM (n = 3). *p < 0.05 and #p < 0.05 represent the significant value compared to the control and PTX, respectively.
PPNPs Treatment Efficiently Enhances GEM Treatment Efficacy in PanCa Cells
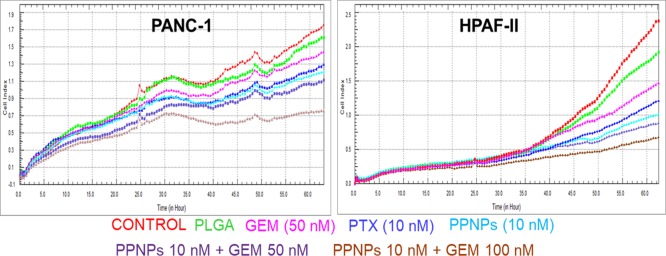
To examine if lipid metabolism inhibition with PPNP treatment can be translated into therapeutic benefit, functional assays such as cytotoxicity were performed in GEM combination using HPAF-II and PANC-1 cells (Figure 3). Proliferation is an inherent property of cancer cells. To investigate the effect of PPNPs/GEM combination treatment on PanCa cells, a real-time xCELLigence assay was conducted (Figure 3). This assay provides the cell index which is basically the electrical impedance due to the presence of cells that indicate the growth of cells. The cell index was continuously hiked in the control, PLGA NPs, and GEM-treated groups, while PPNPs and PPNPs/GEM combination treatment group effectively reduced the cell index growth. The overall cell index growth was greatly reduced by PPNPs/GEM combination treatment. However, the effect of PPNPs (10 nM)/GEM (100 nM) combination is more pronounced than PPNPs (10 nM)/GEM (50 nM). Therefore, this data confirm that PPNPs (10 nM)/GEM (100 nM) exhibited a significant growth control over all other treatments.
Figure 3.
PPNPs/GEM combination inhibits proliferation of PanCa cells. Effect of PPNPs/GEM on proliferation of PanCa cells was determined using xCELLigence. 10 nM PTX, 100 nM GEM, 10 nM PPNPs, 10 nM PPNPs/50 nM GEM, and 10 nM PPNPs/100 nM GEM were used for treatment with respect to the control and blank PLGA formulation.
Spectral Analysis Confirms Reduction in Lipids with PPNP Treatment
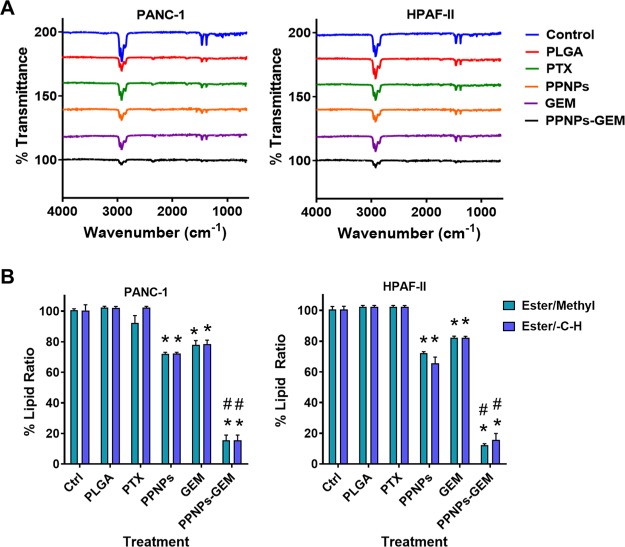
FT-IR spectra of the extracted lipids from PanCa cell lines treated with various groups are presented in Figure 4. The control cells exhibited two strong transmittance peaks in the region 3000–2800 and 1740 cm–1 which are characteristic of the lipid acyl chain (v-CH2 or v-CH3) on unsaturated carbon and ester carbonyl (vibrational stretching), respectively. Such a functional distinct characteristic peak was reported to lipids in the literature.29 With treatments (PTX and GEM), a slight decrease in lipid transmittance peaks was observed. However, the PPNPs and combination group of PPNPs (10 nM) and GEM (100 nM) showed greater impact on the lipid structures as evident from the intensity of the spectra in these peak regions. This result is indicating a significant role of PPNPs in inhibition of lipid production in pancreatic cells to enhance GEM efficacy.
Figure 4.
PPNPs and PPNPs/GEM combination lower cellular lipid contents in PanCa cells. (A) FT-IR spectra of cellular lipids of PanCa cells that were obtained after treatment with various groups. The lipid contents were extracted using Bligh and Dyer protocol and lipids. The spectral data represent an average of 32 scans. (B) Graphical representation of the methyl (ester moiety) and −CH group (ester moiety) of cellular lipids upon combination treatment. Data are indicated as the mean ± SEM (n = 6). *p < 0.05 and #p < 0.05 represent the significant value of treatment groups compared to the control and the PPNPs/GEM group compared to other treatment groups, respectively.
Confocal Microscopy
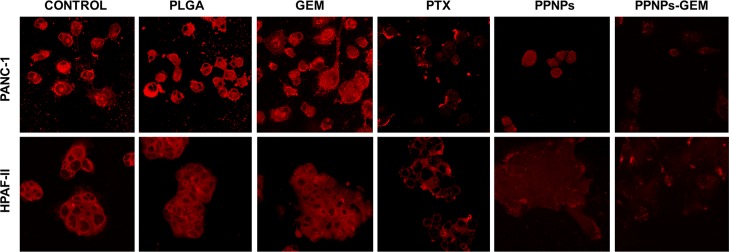
FT-IR spectral analysis is an indication of superior effects of PPNPs/GEM combination on the lipid structure in cancer cells. However, the abovementioned study only refers to extracted cell membrane lipids but do not represent intact cell membranes of cancer cells. Therefore, a confocal microscopy study was performed to delineate this aspect in whole cells. Cell Mask deep red plasma membrane stain is a dye that specifically illuminates the plasma membrane upon incubation. This dye was used to observe visual changes in the membrane with various treatments (Figure 5). It was observed that with PTX and GEM drug treatments, there was a decrease in lipid production (less lipid stain) on cell membranes compared to the control groups (PBS or PLGA). PPNPs treatment inhibited almost all lipid production on the cell membrane, but the cell morphology is round, whereas in the case of PPNPs/GEM combination treatment, there was no lipid staining which indicates a complete disruption of lipid membranes and/or inhibition of lipid production.
Figure 5.
PPNPs and PPNPs/GEM combination treatment disrupt plasma lipid membrane integrity in PanCa cells. Representative confocal microscopy images of PanCa cells after treatment with PBS/no treatment, blank PLGA NPs, 10 nM PTX, 100 nM GEM, 10 nM PPNPs, and 10 nM PPNPs/100 nM GEM. Membrane integrity was shown by the Deep Red plasma membrane known to stain the plasma membrane. Decrease in the intensity of the staining indicating disruption of the membrane due to treatment.
PPNPs Alter Zeta Potential (Charge) on PanCa Cell Membranes
To evaluate the combination treatment enhancement effects, we focused on studying whole cell zeta potential after treatments. Zeta potential is defined as the electrostatic potential near the surface of a particle/material which is corrected by charges of the opposite sign in the environment. Chemotherapeutic agents and cell membrane signals often may disturb the cellular membrane and their integrity that can alter the surface zeta potential of the cancer cells.33−36 Thus, in this study, the surface charge of the lipid membrane that exists on HPAF-II and PANC-1’s cellular membranes was determined after treatment with various groups and compared with the control (Figure 6). The cancer cells themselves present an initial zeta potential of −7.65 ± 0.4 (PANC-1) and −6.79 ± 0.3 (HPAF-II). There was no significant change noticed in the zeta potentials upon treatment with PTX and GEM. However, a noticeable change in zeta potential was observed with PPNPs and PPNPs + GEM treatments in HPAF-II and PANC-1 PanCa cells. Phosphatidylserine is a phospholipid which is present on the inner leaflet of the lipid membrane. Before undergoing apoptosis, this phospholipid is seen on the extracellular leaflet, giving the signal that it is ready to undergo cell death.37 Our treatment of PPNPs and PPNPs/GEM allows for flipping of phosphatidylserine from the inner to outer leaflet.
Figure 6.
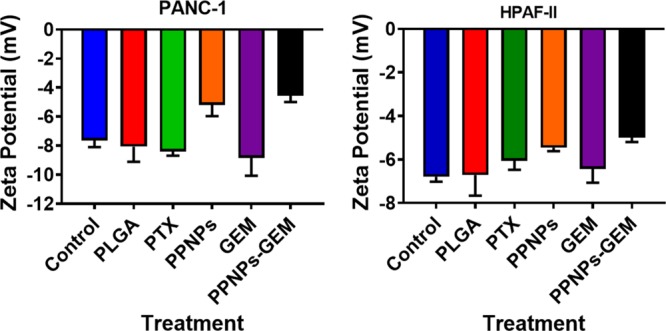
PPNPs/GEM treatment alters surface charge on PanCa cells. Zeta potential measurements were performed on whole cell suspension in PBS after 24 h treatment of PanCa cells with 10 nM PTX, 100 nM GEM, 10 nM PPNPs, and 10 nM PPNPs/100 nM GEM. No treatment and blank PLGA NPs served as the respective control groups. PPNPs/GEM exhibits neutralization of the negatively charged functional membrane. Data are represented as mean ± SEM (n = 3).
PPNPs Treatments Deregulate FASN-Mediated Lipid Metabolism
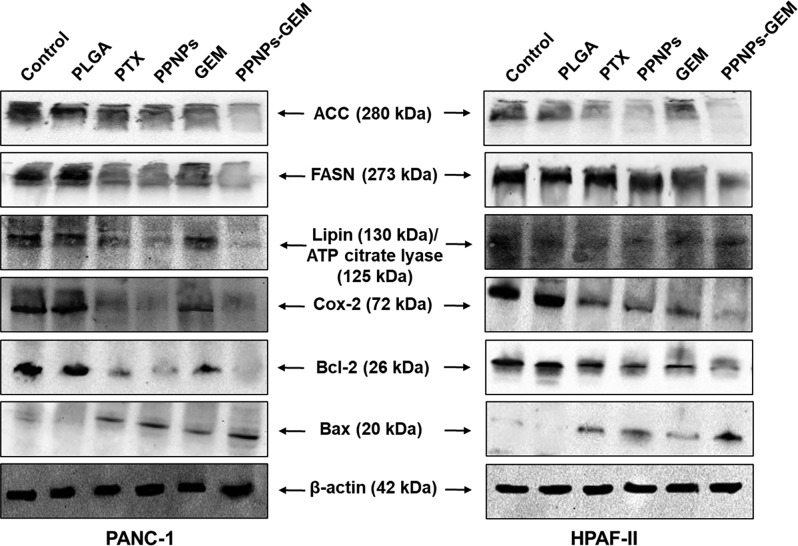
PPNPs/GEM combination treatment leads to alteration in lipid structures, which were determined by FT-IR and confocal microscopy. To further delineate molecular effects of the combination therapy on lipid metabolism, the immunoblotting method was followed. Bax (a pro-apoptotic protein) protein expression was found to be upregulated more effectively with combination treatment as compared to the control and alone drugs. Bcl-2 (an anti-apoptotic protein) protein expression was decreased in the groups treated with drugs alone, while combination treatment resulted in a significant decrease (Figure 7). Protein expression of the lipid metabolism pathway such as ACC, FASN, and Cox-2 was confirmed to be downregulated with PPNPs alone and PPNPs/GEM combination.38
Figure 7.
Molecular effect of PPNPs and PPNPs/GEM on proteins associated with lipid membrane metabolism in PanCa cells. Cell lysates were collected after 48 h treatment with 10 nM PTX, 100 nM GEM, 10 nM PPNPs, and 10 nM PPNPs/100 nM GEM. PBS and blank PLGA NPs served as the respective control groups. Panel showing decrease in ACC, FASN, Lipin, ATP citrate lyase, Cox-2, and Bcl-2 expression and upregulation of Bax (apoptosis regulator) with PPNPs and PPNPs and GEM combination.
Discussion
Gemcitabine is an FDA approved first-line chemotherapeutic agent for PanCa; however, its poor uptake and acquired resistance show only marginal benefits in patients’ survival.18,19 Gemcitabine requires safe entry into the cell and becomes the biologically active triphosphate form by phosphorylation. Such a passive entry and accumulation of gemcitabine always depend on mutational deficiency of nucleoside transporters. To achieve therapeutic concentration at tumors, a huge dose of gemcitabine (∼1000 mg/m2) needs to be administered because of its poor half-life that leads to remarkable systemic toxicities (kidney and liver dysfunctions). In spite, tumor uptake of the drug is suboptimal due to high desmoplasia in pancreatic tumors that elevate the issues related to drug resistance. To increase the uptake and therapeutic ability, there is a need for a novel strategy in order to deliver the drug toward tumor site. Paclitaxel or its FDA-approved nanoformulation (Abraxane) induces apoptosis in cancer cells at low nanomolar concentrations. Abraxane considered as the first line of therapy in advanced breast, lung, and pancreatic cancers along with gemcitabine or other chemotherapy agents. Several therapeutic combinations are under clinical evaluation. Paclitaxel treatment primarily stabilizes tubulin polymerization, thus reducing tumor growth. Paclitaxel nanoparticles can also facilitate gemcitabine internalization into tumors by softening tumor tissue.39
The literature supports that strategic inhibition of lipid metabolism or FASN in cancer cells promotes synergy with chemotherapy.40 There is an evidence through the Cancer Genome Atlas that among many pathways’ metabolic pathways (lipid metabolism) is strongly linked to poor gemcitabine response in PanCa patients.4,5 This eventually lowers gemcitabine uptake, thus offering resistance and survival nature in cancer cells. A recent study advices that strategic inhibition of FASN with orlistat evidently decreases gemcitabine metabolic activity and enhances the uptake by cancer cells.4 Similarly, various lipid metabolism pathway inhibitors promote apoptosis in cancer cells and a lower dose of gemcitabine is required for effective therapy.
The current standard of therapy for PanCa is Nab-paclitaxel combination with gemcitabine. This combination offers mean survival time 13.3 months.41 There are a number of reasons reported for such improved therapeutic benefits, including enhanced level of gemcitabine uptake in tumor, decrease of desmoplasia by nab-paclitaxel in tumors, and by activating tumor angiogenesis that enhanced targetability of GEM at tumor site. However, there is no evidence that paclitaxel plus gemcitabine combination may regulate lipid metabolism. In this attempt, we used our own paclitaxel nanoparticle formulation (Figure 1). This is because our group has constructed a unique nanoparticle formulation, that is, PPNPs which can be further functionalized for targeted delivery to pancreatic tumors. A recent report from our group demonstrated that PPNPs can efficiently reduce tumor growth in orthotopic tumor xenograft mouse models.24 Thus, in this study, we delineated its effect on the lipid metabolism pathway in PanCa cells (Figures 4–7).
The cellular membranes are primary impediment and the first line of guard for the cell from their environments to survive. This barrier limits the exchange of charged molecules or ions across the cellular membrane and creates the cell membrane potential. Such a cell membrane potential can act as a distinguishable characteristic for cancerous cells compared to normal cells. The literature indicates that cancerous cells exhibit a higher degree of membrane negativity (a lower cellular membrane potential) over normal cells because of their rapid membrane changes, transformation, and fluidity.36 Another study demonstrated that a greater negative membrane potential was observed for metastases (−35 mV) and primary lesions (−25 mV) while quasi neutral for noncancer cells (0 mV).42 This advice change in the cellular membrane potential can possibly determine its fate. Higher negative membrane potential ultimately determines cancer cell’s zeta potential. Chemotherapies can regulate such a phenomenon and alter the membrane potential. This is evident from our zeta potential values of cell membranes treated with PPNPs and PPNPs/GEM combination treatment (Figure 6). All these events demonstrate that PPNPs act as a anticancer drug that offers apoptosis via modulation lipid metabolic pathways.
Drug uptake can be estimated through Rh123 dye exclusion assay. Rh123 is a probe used for P-gp studies32 and here it mimicked the role of gemcitabine. The PPNP treatment group showed increased intensity with regard to uptake of Rh123, confirming that our nanoformulation has the ability to inhibit P-gp efflux of drugs (Figure 2) (gemcitabine) and, in turn, increase its intracellular concentration, thus improving its therapeutic efficacy (Figure 3). Our results suggest some lipid modifications as the spectral intensity decreases for characteristic lipid functional groups (Figure 4). Membrane dysregulation was monitored by measuring the membrane charge and integrity of the membrane with regard to the combination therapy. The cell membrane is made up of different phospholipids which are responsible for the charge on the surface. Phosphatidylserine is one of the major phospholipids present in the membrane component, and it undergoes flipping, further leading to apoptosis.37 The combination treatment quickens this flipping process, in turn, signaling the apoptotic molecules. Alteration in the membrane was also confirmed by confocal microscopy with the help of a dye that stains the plasma membrane. Gemcitabine in combination with our formulation showed decreased intensity of the membrane stain. These results give us evidence that our combination of PPNPs have the capability to alter the lipid membrane to improve therapeutic efficacy of GEM (Figure 5).
In many cancers, dysregulation of lipid metabolism is often linked to poor prognosis. Previous studies have shown that inhibition of FASN has led to apoptosis in prostate,43 breast,44 and pancreatic4 cancers. Sun et al.,45 demonstrated that SREBP-1, a master regulator of lipid metabolism (through FASN and ACC), can ablate the tumorigenesis and progression of PanCa. In this investigation, we performed immunoblotting analysis to show the molecular effects of PPNPs and PPNPs/GEM combination on certain key regulators which are known to be upregulated in PanCa.4 These include FASN, ACC, Lipin, ATP citrate lyase, and Cox-2. A significant downregulation of these proteins was noticed with PPNPs and combination treatment compared with controls (Figure 7). SREBP-1 regulates the activity of FASN and ACC. The rate-limiting step in synthesis of fatty acids is mediated by ACC. Our data showed a decrease in expression of FASN, a major enzyme responsible for elongation of fatty acids and, in turn, causes progression of lipogenesis (Figure 7).
PLGA-based drug formulations are widely evaluated in a number of clinical applications. This includes delivery of a number of therapeutic molecules including paclitaxel. To the best of our knowledge, the PPNPs delivery with gemcitabine combination treatment would be the first report being used to examine the lipid metabolism mechanism in PanCa. We also noticed that SREBP-1-associated lipogenesis inhibition was achieved with PPNPs and PPNPs/GEM combination to control the cell growth and induction of apoptosis.
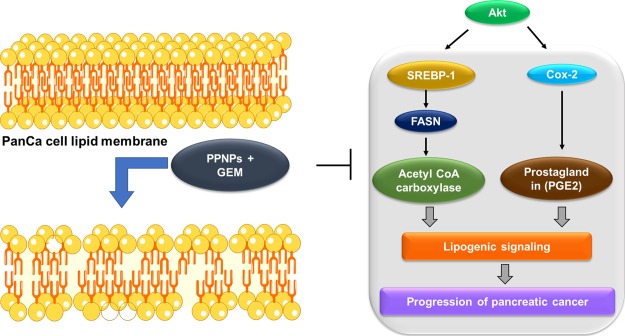
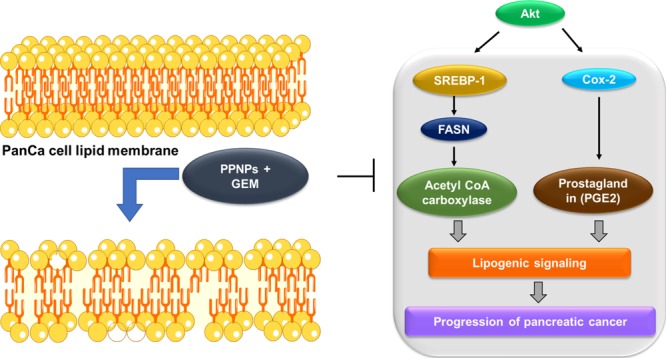
Altogether, our study suggests that PPNPs and PPNPs/GEM combination can effectively inhibit the progression of cancer cells via ablation of the de novo lipid synthesis and induce apoptosis in cancer cells (Scheme 1).
Scheme 1. Schematic Representation of Paclitaxel Nanoformulation and Gemcitabine Combination Impairs de novo Lipid Synthesis in PanCa.
Conclusions
This study reports a new concept that the PPNP platform offers improved therapeutic benefit with gemcitabine in PanCa. The data demonstrate that PPNPs and PPNPs/GEM combination efficiently reduced proliferation and lipid membrane structures of cancer cells. This improved response was confirmed by inhibitory potential of lipid metabolism and enhanced pro-apoptotic activity. The outcomes of this study provide a novel molecular mechanism for enhanced chemotherapeutic efficacy of paclitaxel–gemcitabine combination in PanCa. However, it remains a puzzle how to deliver both drugs (paclitaxel and gemcitabine) in one nanoplatform to further boost their therapeutic efficacy for PanCa treatment.
Acknowledgments
The authors wish to acknowledge the support by the National Institutes of Health (R01 CA210192, R01 CA206069, R01 CA204552) and partial support from the Herb Kosten Foundation, and Faculty Start-up fund from UTRGV to M.M.Y., M.J., and S.C.C.
Author Contributions
A.S. and P.K.B.N. contributed equally to this work. A.S., P.K.B.N., and S.S. performed experiments. A.S., P.K.B.N., S.S., B.B.H., M.J., S.C.C., and M.M.Y. curated study and participated in discussion and writing the manuscript. M.M.Y and S.C.C. conceived the idea and were crucially involved throughout the study. B.B.H., M.J., M.M.Y, and S.C.C. evaluated data, participated in the discussion of results, and edited and reviewed the manuscript.
The authors declare no competing financial interest.
References
- Siegel R. L.; Miller K. D.; Jemal A. Cancer statistics, 2019. Ca-Cancer J. Clin. 2019, 69, 7–34. 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Passadouro M.; Faneca H. Managing Pancreatic Adenocarcinoma: A Special Focus in MicroRNA Gene Therapy. Int. J. Mol. Sci. 2016, 17, 718. 10.3390/ijms17050718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassouni B.; Li Petri G.; Liu D. S. K.; Cascioferro S.; Parrino B.; Hassan W.; Diana P.; Ali A.; Frampton A. E.; Giovannetti E. Pharmacogenetics of treatments for pancreatic cancer. Expert Opin. Drug Metab. Toxicol. 2019, 15, 437–447. 10.1080/17425255.2019.1620731. [DOI] [PubMed] [Google Scholar]
- Tadros S.; Shukla S. K.; King R. J.; Gunda V.; Vernucci E.; Abrego J.; Chaika N. V.; Yu F.; Lazenby A. J.; Berim L.; Grem J.; Sasson A. R.; Singh P. K. De Novo Lipid Synthesis Facilitates Gemcitabine Resistance through Endoplasmic Reticulum Stress in Pancreatic Cancer. Cancer Res. 2017, 77, 5503–5517. 10.1158/0008-5472.can-16-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiss K.; Parhofer K. G.; Heinemann V.; Haas M.; Laubender R. P.; Holdenrieder S.; Schulz C.; Boeck S. Glucose and lipid metabolism in patients with advanced pancreatic cancer receiving palliative chemotherapy. Anticancer Res. 2013, 33, 287–292. [PubMed] [Google Scholar]
- Zhou C.; Qian W.; Li J.; Ma J.; Chen X.; Jiang Z.; Cheng L.; Duan W.; Wang Z.; Wu Z.; Ma Q.; Li X. High glucose microenvironment accelerates tumor growth via SREBP1-autophagy axis in pancreatic cancer. J. Exp. Clin. Cancer Res. 2019, 38, 302. 10.1186/s13046-019-1288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami Y.; Rebelo A.; Kleeff J. Lipid Metabolism and Lipid Droplets in Pancreatic Cancer and Stellate Cells. Cancers 2017, 10, 3. 10.3390/cancers10010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczynski J.; Hebanowska A.; Sledzinski T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World J. Gastroenterol. 2014, 20, 2279–2303. 10.3748/wjg.v20.i9.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.; Geng F.; Cheng X.; Guo D. Lipid Metabolism Reprogramming and its Potential targets in cancer. Cancer Commun. 2018, 38, 27. 10.1186/s40880-018-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E.; Schulze A.; Zechner R.; Walther T. C.; Farese R. V. Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Huang J. The expanded role of fatty acid metabolism in cancer: new aspects and targets. Precis. Clin. Med. 2019, 2, 183–191. 10.1093/pcmedi/pbz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz V.; Fajas L. Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene 2010, 29, 4369–4377. 10.1038/onc.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray U.; Roy S. S. Aberrant lipid metabolism in cancer cells - the role of oncolipid-activated signaling. FEBS J. 2018, 285, 432–443. 10.1111/febs.14281. [DOI] [PubMed] [Google Scholar]
- Mounier C.; Bouraoui L.; Rassart E. Lipogenesis in cancer progression (review). Int. J. Oncol. 2014, 45, 485–492. 10.3892/ijo.2014.2441. [DOI] [PubMed] [Google Scholar]
- Nishi K.; Suzuki K.; Sawamoto J.; Tokizawa Y.; Iwase Y.; Yumita N.; Ikeda T. Inhibition of Fatty Acid Synthesis Induces Apoptosis of Human Pancreatic Cancer Cells. Anticancer Res. 2016, 36, 4655–4660. 10.21873/anticanres.11016. [DOI] [PubMed] [Google Scholar]
- Sokolowska E.; Presler M.; Goyke E.; Milczarek R.; Swierczynski J.; Sledzinski T. Orlistat Reduces Proliferation and Enhances Apoptosis in Human Pancreatic Cancer Cells (PANC-1). Anticancer Res. 2017, 37, 6321–6327. 10.21873/anticanres.12083. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Wang X.; Sun W.; Cheng K.; Qin H.; Han X.; Lin Y.; Wang Y.; Lang J.; Zhao R.; Zheng X.; Zhao Y.; Shi J.; Hao J.; Miao Q. R.; Nie G.; Ren H. Precision design of nanomedicines to restore gemcitabine chemosensitivity for personalized pancreatic ductal adenocarcinoma treatment. Biomaterials 2018, 158, 44–55. 10.1016/j.biomaterials.2017.12.015. [DOI] [PubMed] [Google Scholar]
- Rajabpour A.; Rajaei F.; Teimoori-Toolabi L. Molecular alterations contributing to pancreatic cancer chemoresistance. Pancreatology 2017, 17, 310–320. 10.1016/j.pan.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Amrutkar M.; Gladhaug I. P. Pancreatic Cancer Chemoresistance to Gemcitabine. Cancers 2017, 9, 157. 10.3390/cancers9110157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilzad-Kohan H.; Sani S.; Boroujerdi M. Calcitriol Reverses Induced Expression of Efflux Proteins and Potentiates Cytotoxic Activity of Gemcitabine in Capan-2 Pancreatic Cancer Cells. J. Pharm. Pharmaceut. Sci. 2017, 20, 295–304. 10.18433/j37w7r. [DOI] [PubMed] [Google Scholar]
- Frese K. K.; Neesse A.; Cook N.; Bapiro T. E.; Lolkema M. P.; Jodrell D. I.; Tuveson D. A. nab-Paclitaxel potentiates gemcitabine activity by reducing cytidine deaminase levels in a mouse model of pancreatic cancer. Canc. Discov. 2012, 2, 260–269. 10.1158/2159-8290.cd-11-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvantalab S.; Drude N. I.; Moraveji M. K.; Guvener N.; Koons E. K.; Shi Y.; Lammers T.; Kiessling F. PLGA-Based Nanoparticles in Cancer Treatment. Front Pharmacol. 2018, 9, 1260. 10.3389/fphar.2018.01260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello J. C. d.; Moraes V. W.; Watashi C. M.; da Silva D. C.; Cavalcanti L. P.; Franco M. K.; Yokaichiya F.; de Araujo D. R.; Rodrigues T. Enhancement of chlorpromazine antitumor activity by Pluronics F127/L81 nanostructured system against human multidrug resistant leukemia. Pharmacol. Res. 2016, 111, 102–112. 10.1016/j.phrs.2016.05.032. [DOI] [PubMed] [Google Scholar]
- Massey A. E.; Sikander M.; Chauhan N.; Kumari S.; Setua S.; Shetty A. B.; Mandil H.; Kashyap V. K.; Khan S.; Jaggi M.; Yallapu M. M.; Hafeez B. B.; Chauhan S. C. Next-generation paclitaxel-nanoparticle formulation for pancreatic cancer treatment. Nanomedicine 2019, 20, 102027. 10.1016/j.nano.2019.102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yallapu M. M.; Gupta B. K.; Jaggi M.; Chauhan S. C. Fabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cells. J. Colloid Interface Sci. 2010, 351, 19–29. 10.1016/j.jcis.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Chowdhury P.; Nagesh P. K. B.; Hatami E.; Wagh S.; Dan N.; Tripathi M. K.; Khan S.; Hafeez B. B.; Meibohm B.; Chauhan S. C.; Jaggi M.; Yallapu M. M. Tannic acid-inspired paclitaxel nanoparticles for enhanced anticancer effects in breast cancer cells. J. Colloid Interface Sci. 2019, 535, 133–148. 10.1016/j.jcis.2018.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setua S.; Khan S.; Yallapu M. M.; Behrman S. W.; Sikander M.; Khan S. S.; Jaggi M.; Chauhan S. C. Restitution of Tumor Suppressor MicroRNA-145 Using Magnetic Nanoformulation for Pancreatic Cancer Therapy. J. Gastrointest. Surg. 2017, 21, 94–105. 10.1007/s11605-016-3222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.; Setua S.; Kumari S.; Dan N.; Massey A.; Hafeez B. B.; Yallapu M. M.; Stiles Z. E.; Alabkaa A.; Yue J.; Ganju A.; Behrman S.; Jaggi M.; Chauhan S. C. Superparamagnetic iron oxide nanoparticles of curcumin enhance gemcitabine therapeutic response in pancreatic cancer. Biomaterials 2019, 208, 83–97. 10.1016/j.biomaterials.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Gasper R.; Vandenbussche G.; Goormaghtigh E. Ouabain-induced modifications of prostate cancer cell lipidome investigated with mass spectrometry and FTIR spectroscopy. Biochim. Biophys. Acta 2011, 1808, 597–605. 10.1016/j.bbamem.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Kashyap V. K.; Wang Q.; Setua S.; Nagesh P. K. B.; Chauhan N.; Kumari S.; Chowdhury P.; Miller D. D.; Yallapu M. M.; Li W.; Jaggi M.; Hafeez B. B.; Chauhan S. C. Therapeutic efficacy of a novel betaIII/betaIV-tubulin inhibitor (VERU-111) in pancreatic cancer. J. Exp. Clin. Canc. Res. 2019, 38, 29. 10.1186/s13046-018-1009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury P.; Nagesh P. K. B.; Khan S.; Hafeez B. B.; Chauhan S. C.; Jaggi M.; Yallapu M. M. Development of polyvinylpyrrolidone/paclitaxel self-assemblies for breast cancer. Acta Pharm. Sin. B 2018, 8, 602–614. 10.1016/j.apsb.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twentyman P. R.; Rhodes T.; Rayner S. A comparison of rhodamine 123 accumulation and efflux in cells with P-glycoprotein-mediated and MRP-associated multidrug resistance phenotypes. Eur. J. Canc. 1994, 30, 1360–1369. 10.1016/0959-8049(94)90187-2. [DOI] [PubMed] [Google Scholar]
- Wonderlin W. F.; Woodfork K. A.; Strobl J. S. Changes in membrane potential during the progression of MCF-7 human mammary tumor cells through the cell cycle. J. Cell. Physiol. 1995, 165, 177–185. 10.1002/jcp.1041650121. [DOI] [PubMed] [Google Scholar]
- Dobrzyńska I.; Szachowicz-Petelska B.; Sulkowski S.; Figaszewski Z. Changes in electric charge and phospholipids composition in human colorectal cancer cells. Mol. Cell. Biochem. 2005, 276, 113–119. 10.1007/s11010-005-3557-3. [DOI] [PubMed] [Google Scholar]
- Zhu X.; Hondroulis E.; Liu W.; Li C.-z. Biosensing approaches for rapid genotoxicity and cytotoxicity assays upon nanomaterial exposure. Small 2013, 9, 1821–1830. 10.1002/smll.201201593. [DOI] [PubMed] [Google Scholar]
- Hondroulis E.; Zhang R.; Zhang C.; Chen C.; Ino K.; Matsue T.; Li C.-Z. Immuno nanoparticles integrated electrical control of targeted cancer cell development using whole cell bioelectronic device. Theranostics 2014, 4, 919–930. 10.7150/thno.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai T. J.; Toombs J. E.; Minna J. D.; Brekken R. A.; Udugamasooriya D. G. Identification of lipid-phosphatidylserine (PS) as the target of unbiasedly selected cancer specific peptide-peptoid hybrid PPS1. Oncotarget 2016, 7, 30678–30690. 10.18632/oncotarget.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Ji F.; Di W.; Chen H.; Wan Y. Activation of acetyl-coenzyme A carboxylase is involved in Taxol-induced ovarian cancer cell death. Oncol. Lett. 2011, 2, 543–547. 10.3892/ol.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.; Brachmann C.; Liu X.; Pierce D. W.; Dey J.; Kerwin W. S.; Li Y.; Zhou S.; Hou S.; Carleton M.; Klinghoffer R. A.; Palmisano M.; Chopra R. Albumin-bound nanoparticle (nab) paclitaxel exhibits enhanced paclitaxel tissue distribution and tumor penetration. Canc. Chemother. Pharmacol. 2015, 76, 699–712. 10.1007/s00280-015-2833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.; Zhang C. J.; Zhu N.; Du K.; Yin Y. F.; Tan X.; Liao D. F.; Qin L. Lipid metabolism and carcinogenesis, cancer development. Am. J. Cancer. Res. 2018, 8, 778–791. [PMC free article] [PubMed] [Google Scholar]
- Saito T.; Ishido K.; Kudo D.; Kimura N.; Wakiya T.; Nakayama Y.; Hakamada K. Combination therapy with gemcitabine and nab-paclitaxel for locally advanced unresectable pancreatic cancer. Mol. Clin. Oncol. 2017, 6, 963–967. 10.3892/mco.2017.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl S.; Leber R.; Rinner B.; Schaider H.; Lohner K.; Zweytick D. Human lactoferricin derived di-peptides deploying loop structures induce apoptosis specifically in cancer cells through targeting membranous phosphatidylserine. Biochim. Biophys. Acta 2015, 1848, 2918–2931. 10.1016/j.bbamem.2015.07.018. [DOI] [PubMed] [Google Scholar]
- Zadra G.; Ribeiro C. F.; Chetta P.; Ho Y.; Cacciatore S.; Gao X.; Syamala S.; Bango C.; Photopoulos C.; Huang Y.; Tyekucheva S.; Bastos D. C.; Tchaicha J.; Lawney B.; Uo T.; D’Anello L.; Csibi A.; Kalekar R.; Larimer B.; Ellis L.; Butler L. M.; Morrissey C.; McGovern K.; Palombella V. J.; Kutok J. L.; Mahmood U.; Bosari S.; Adams J.; Peluso S.; Dehm S. M.; Plymate S. R.; Loda M. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 631–640. 10.1073/pnas.1808834116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu T. H.; Bard J.-M.; Carbonnelle D.; Chaillou C.; Huvelin J.-M.; Bobin-Dubigeon C.; Nazih H. Lithocholic bile acid inhibits lipogenesis and induces apoptosis in breast cancer cells. Cell. Oncol. 2018, 41, 13–24. 10.1007/s13402-017-0353-5. [DOI] [PubMed] [Google Scholar]
- Sun Y.; He W.; Luo M.; Zhou Y.; Chang G.; Ren W.; Wu K.; Li X.; Shen J.; Zhao X.; Hu Y. SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumor Biol. 2015, 36, 4133–4141. 10.1007/s13277-015-3047-5. [DOI] [PubMed] [Google Scholar]