Summary
As cells enter mitosis, the genome is restructured to facilitate chromosome segregation, accompanied by dramatic changes in gene expression. However, the mechanisms that underlie mitotic transcriptional regulation are unclear. In contrast to transcribed genes, centromere regions retain transcriptionally active RNA Polymerase II (RNAPII) in mitosis. Here, we demonstrate that chromatin-bound cohesin is necessary to retain elongating RNAPII at centromeres. We find that WAPL-mediated removal of cohesin from chromosome arms during prophase is required for the dissociation of RNAPII and nascent transcripts and failure of this process dramatically alters mitotic gene expression. Removal of cohesin/RNAPII from chromosome arms in prophase is important for accurate chromosome segregation and normal activation of gene expression in G1. We propose that prophase cohesin removal is a key step in reprogramming gene expression as cells transition from G2 through mitosis to G1.
Graphical Abstract

eTOC blurb
During mitosis chromosomes condense and transcription is silenced to facilitate chromosome segregation. However, centromeres are transcribed during mitosis. Perea-Resa find that mitotic cohesin regulation is important for mitotic transcriptional silencing, mitotic centromere transcription, and transcriptional restart in G1. Coupled cohesin/transcription dynamics ensures chromosome segregation and gene expression reprograming across mitosis.
Introduction
During mitosis, interphase chromatin organization is erased as chromosomes are condensed and individualized in preparation for their segregation (Gibcus et al., 2018). As chromosomes condense, the pattern of gene expression is dramatically altered (Liang et al., 2015; Palozola et al., 2017; Prescott and Bender, 1962). Mitotic transcriptional silencing is thought to occur by preventing new initiation through phosphorylation of the general transcription factors, TFIID and TFIIH (Akoulitchev and Reinberg, 1998; Segil et al., 1996), as well as phosphorylation of the C-terminal domain (CTD) of RNA Polymerase II (RNAPII) (Bellier et al., 1997). Additionally, p-TEF-b is activated at mitotic onset to promote the transcriptional run-off of paused RNAPII during prophase (Liang et al., 2015). In contrast to the bulk of the genome, centromeres escape general mitotic transcriptional inhibition and retain elongating RNAPII (Chan et al., 2012). The previously described mechanisms for mitotic transcriptional silencing cannot account for the persistence of active RNAPII at centromeres, suggesting that additional mechanisms regulate the spatial pattern of mitotic transcriptional inhibition.
Cohesin is a major component of interphase and mitotic chromosomes regulating transcription, DNA double-strand break repair, and sister chromatid cohesion (Nasmyth and Haering, 2009). Cohesin is linked to transcription through its interaction with Mediator, which promotes enhancer-promoter interactions (Kagey et al., 2010), the formation of topologically-associated domains (TADs) (Rao et al., 2017), interaction with the super-elongation complex (Izumi et al., 2015), and recruitment of transcriptional activators to transcription factor hotspots (Yan et al., 2013). In interphase cells, cohesin is widely distributed throughout the genome where it regulates both chromosome structure and cohesion between replicated chromatids. During mitotic prophase, the kinases Plk1, Aurora B, and Cdk1 phosphorylate the cohesin-associated proteins Sororin and SA2 to promote cohesin removal from chromosome arms by the cohesin release factor WAPL, in a process termed the ‘prophase pathway’ (Haarhuis et al., 2014). Centromeric cohesin is protected from WAPL-mediated removal by Shugoshin proteins until the metaphase-anaphase transition when cohesin rings are cleaved by the Separase protease (Uhlmann et al., 2000). While it is established that cohesin loops formed in cis regulate chromatin architecture and gene expression during interphase, it is not clear whether the cohesin connecting duplicated sister chromatid also affects gene expression during mitosis.
Here, we use centromere transcription as a model to study the mechanisms of mitotic transcription inhibition. We find that dissociation of RNAPII and cohesin from mitotic chromosomes are temporally and spatially correlated. We show that chromatin-localized cohesin is both necessary and sufficient to retain elongating RNAPII on mitotic chromosomes. Additionally, we demonstrate that static cohesin prevents dissociation of elongating RNPII from both interphase and mitotic chromosomes. Our results indicate that WAPL-mediated removal of cohesin from chromosomes during prophase functions to regulate gene expression in both mitosis and G1 and contributes to accurate chromosome segregation during mitosis.
Results
Cohesin promotes selective retention of RNA Pol II at centromeres in mitosis
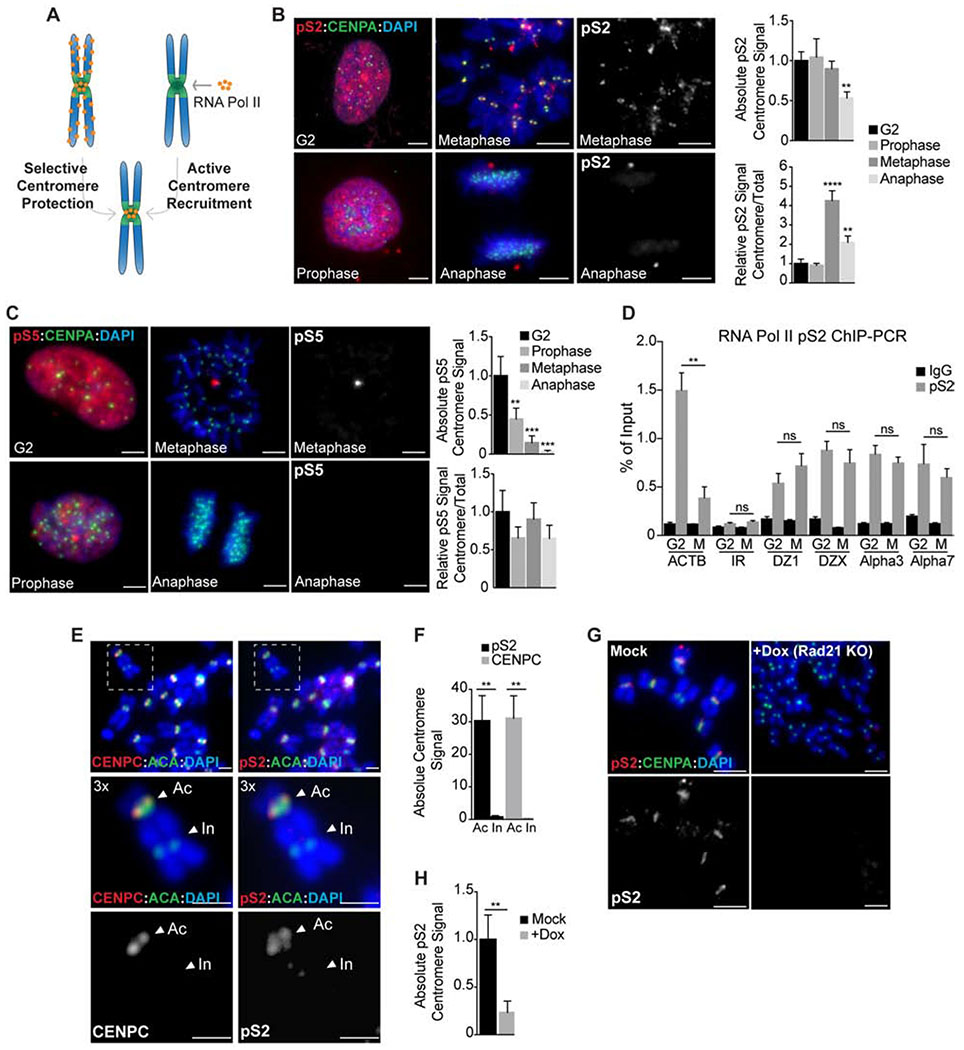
The differential localization of RNAPII during mitosis could be explained by its active recruitment to centromeres or reflect selective protection from a global removal pathway (Fig. 1A). To determine how RNAPII is selectively localized to centromeres during mitosis, we analyzed the distribution of actively-elongating (CTD phospho-Ser 2, pS2) and promoter-paused (CTD phospho-Ser 5, pS5) RNAPII during early stages of mitosis using non-transformed human RPE-1-hTERT cells (Fig. 1B–C). RNAPII pS2 localized to mitotic centromeres from G2 through metaphase, but then became significantly reduced at centromeres in anaphase as sister chromatids separated (Fig. 1B). In contrast, CTD pS5 intensity steadily declined throughout mitosis (Fig. 1C). We observed similar results for RNAPII localization in Xenopus egg extracts (Fig. S1A–C). To understand the differential localization of RNAPII pS2 to mitotic centromeres, we analyzed both the absolute fluorescent intensity at centromeres and its behavior relative to chromosome arms. We found that the absolute levels of RNAPII pS2 at centromeres remained constant from G2 to metaphase (Fig. 1B). Similarly, chromatin immunoprecipitation (ChIP) analysis using RNAPII CTD pS2 antibodies revealed that RNAPII is present at human centromeres at similar levels in G2 and metaphase, but is removed from the β-actin gene during mitosis (Fig. 1D). These results suggest that the relative enrichment of RNAPII at mitotic centromeres occurs through its selective removal from chromosome arms.
Figure 1. Cohesin promotes the selective retention of RNA Pol II at centromeres in mitosis.
A. Either selective chromosome arm removal or active centromere recruitment early in prophase could explain RNA Pol II enrichment at metaphase centromeres. B-C RNAP pS2 and pS5 localization (red) in chromosome spreads from RPE-1 cells cycled from interphase (G2) to anaphase. Anti-CENPA antibodies (green) and DAPI (blue) were used to visualize centromeres and DNA respectively. Scale bars, 10 μm. Right, quantification of absolute and relative centromere signal. **, P < 0.005; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA, Dunnett’s test). D pS2 occupancy at centromeres of chromosomes 1, 3, 7 and X, represented as the % of input recovered following ChIP in G2 or metaphase (M) RPE-1 cells. Occupancy values at the ACTB gene and at a random intergenic region (IR) were included as positive and negative controls, respectively. **, P < 0.005; (ns) no significant differences (t-test). E-F Analysis of CENPC and pS2 localization (red) in metaphase spreads from dicentric MDA-MB 435 cells. ACA (anti-centromere antibodies; green) stains both active (Ac) and inactive (In) centromeres while CENPC and pS2 (red) are only recruited to the active kinetochore region. Scale bars, 5 μm. Quantification of pS2 and CENPC centromere signal is plotted on F. **, P < 0.005 (t-test). G-H Analysis of pS2 localization (red) on metaphase spreads from doxycycline inducible Rad21 KO cells. Anti-CENPA (green) and DAPI (blue) were used to visualize centromeres and DNA, respectively. Scale bars, 10 μm. Quantification of absolute pS2 centromere levels is represented in H. **, P < 0.005 (t-test). In all cases error bars indicate the standard deviation of the mean.
In vertebrate cells, centromeres are specified epigenetically by the presence of histone variants, and are not defined by precise DNA sequences. To determine if the retention of RNAPII on mitotic chromosomes is linked to the active centromere or is a general property of α-satellite DNA, we analyzed RNAPII localization on a dicentric human chromosome. Structurally dicentric chromosomes contain two α-satellite arrays, but form an active centromere on only one array. Consistent with previous results (Chan et al., 2012), RNAPII only localized to the kinetochore-forming array of a dicentric chromosome (Fig. 1E–F). Thus, RNAPII retention is linked to functional centromere activity.
In addition to displaying differences in kinetochore formation, the inactive centromere on dicentric chromosomes was notably separated from its sister partner indicating the absence of cohesion. To evaluate the role of sister chromatid cohesion in retaining RNAPII at centromeres, we depleted the cohesin subunit Rad21 (Fig. S1D–E). Strikingly, Rad21 depletion resulted in a complete loss of elongating RNAPII localization to mitotic chromosomes (Fig. 1G–H) and in reduced levels of centromeric RNAs (Fig. S1F), suggesting that the presence of cohesin helps retain RNAPII at centromeres. Consistent with a close connection between cohesin and RNAPII, we found that the temporal pattern of cohesin removal during prophase closely parallels that of RNAPII from chromatin in both human cells (Fig. S2A) and Xenopus egg extracts (Fig. S2B). We conclude that removal of cohesin and RNAPII from chromosomes during mitosis is temporally correlated and that centromeric cohesin is necessary for RNAPII localization to the mitotic centromere. Thus, cohesin is an excellent candidate factor that could regulate the spatial pattern of RNAPII during mitosis.
Ectopic cohesin retains active RNA Pol II on metaphase chromosomes
To determine if prophase cohesin removal is required for the eviction of RNAPII from mitotic chromosomes, we generated an Auxin-Inducible Degron (AID) human cell line for the cohesin release factor WAPL (Fig. S2C–D). Addition of auxin (IAA) to WAPL-AID cells resulted in WAPL degradation, cohesin retention, and the persistence of sister chromatid cohesion along the entire length of mitotic chromosomes (Fig. S2D–H). Strikingly, WAPL degradation following auxin addition also resulted in the retention of RNAPII pS2 on the arms of mitotic chromosomes while no effect was detected on pS5 localization (Fig. 2A–C; Fig. S2I,J). Similar results were obtained following WAPL RNAi in human cells (Fig. S2K–M) or WAPL immuno-depletion in Xenopus egg extracts (Fig. S2N–P). In addition, preventing cohesin removal by inhibition of either Aurora B or Plk1 also resulted in retention of elongating RNAPII (pS2) on mitotic chromosome arms in both human cells (Fig. S3A,B) and Xenopus egg extracts (Fig. S3C,D). Retention of RNAPII pS2 in WAPL-depleted cells was dependent on the presence of cohesin as co-depletion of WAPL and Rad21 resulted in a lack of RNAPII pS2 on mitotic chromosomes, similar to the phenotype observed after Rad21 depletion (Fig. S3E,F). Thus, prophase cohesin removal is an important step in the release of elongating RNAPII from mitotic chromosomes.
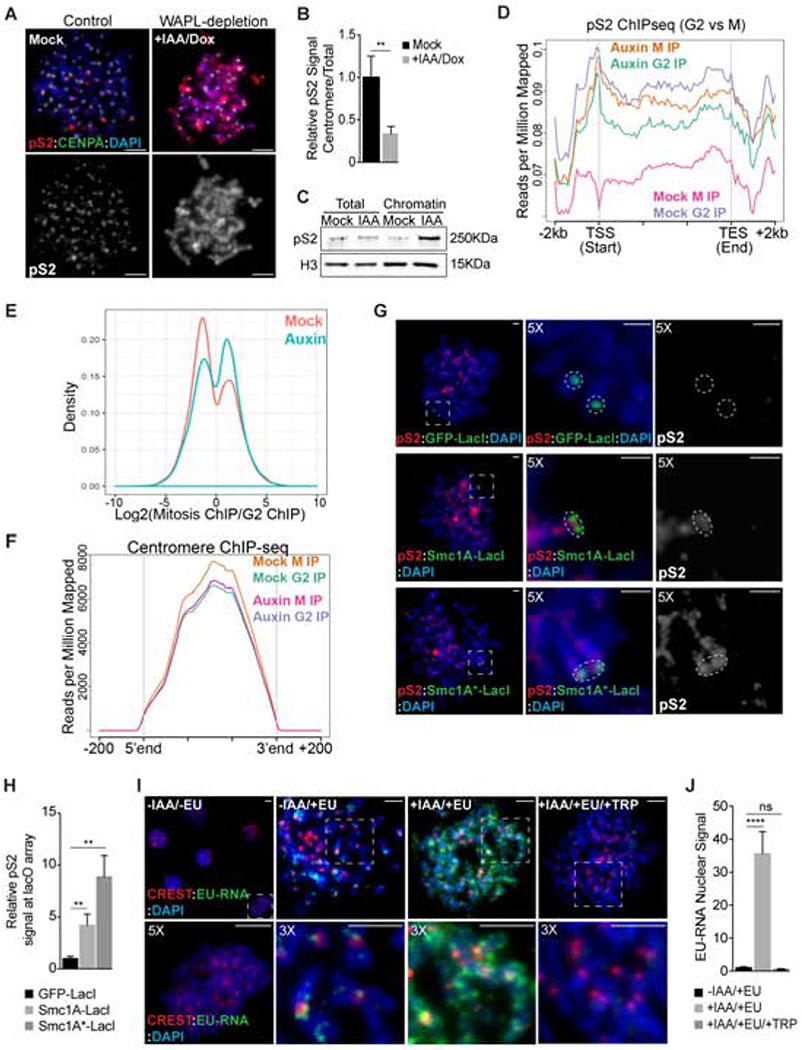
Figure 2. Ectopic cohesin retains RNA Pol II activity on metaphase chromosomes.
A Analysis of pS2 localization (red) in metaphase spreads from WAPL-AID cells. Cells were incubated with DMSO (Mock) or IAA and doxycycline for 48 h to promote WAPL-AID degradation. Anti-CENP-A antibodies (green) and DAPI (blue) were used to visualize centromeres and DNA, respectively. Scale bars, 5 μm. B Quantification of pS2 centromere enrichment in images from A. Data represent relative enrichment as a ratio of centromere to total nuclear signal. Levels were normalized to control cells. C pS2 levels in total or chromatin protein fractions from metaphase WAPL-AID cells measured by Western blotting. Levels of histone H3 provide a loading control. D Metaplot of relative pS2 ChIP-seq coverage across all genes showing RNA-seq expression levels of FPKM > 10. E Histogram of changes in pS2 ChIP-seq occupancy between G2 and mitosis in control (mock) or auxin-treated cells. F Histogram representing pS2 occupancy at centromere gene models between G2 and mitosis in control (mock) or auxin-treated cells. G pS2 localization (red) in metaphase spreads from U2OS cells following the tethering of LacI-tagged proteins to the lacO locus. GFP-LacI, SMC1A(WT)-LacI, or SMC1A*(mt)-LacI fusion proteins were transiently expressed and metaphase chromosome spreads were analyzed 48 h after transfection. Anti-GFP or anti-FLAG antibodies were used to detect the LacI-tagged proteins (green) and DAPI staining to visualize chromosomes (blue). Scale bars, 2 μm. H Quantification of pS2 levels at the lacO array following cell transfection as detailed in G. I-J Analysis of nascent RNA localization on metaphase chromosome spreads from WAPL-AID cells. Cells were incubated in the presence or absence of EU for nascent RNA labeling. EU incorporation was detected by Click-iT chemistry (green). Anti-centromere antibodies (CREST; red) and DAPI staining (blue) were used for centromere and chromosome labelling, respectively. Scale bars, 5 μm. J Quantification of total EU nuclear fluorescence in images from I. Data represent the mean of three independent experiments and values were normalized to control cells (−IAA/+EU). In all cases, error bars indicate the standard deviation of the mean. **, P < 0.005; ****, P < 0.0001; (ns) no significant differences (t-test).
To determine if the cytological loss of RNAPII pS2 from mitotic chromosomes reflects a loss from active genes, we performed genome-wide ChIP-seq with RNAPII pS2 in control and WAPL-depleted cells in G2 and mitosis. Consistent with our immunofluorescence results in control cells, we observed robust RNAPII pS2 ChIP signals at expressed genes in G2 cells that was reduced to background levels during mitosis (Fig. 2D,E, Fig. S3G). In contrast, in WAPL-depleted cells RNAPII pS2 ChIP signal on active genes was high in G2 and remained high in mitosis (Fig. 2D,E, Fig. S3H). Interestingly, RNAPII pS2 ChIP signals at centromere regions were constant between G2 and mitosis and were unaffected by WAPL depletion (Fig. 2F, Fig. S3I,J). Additionally, we detected low levels of RNAPII pS2 on a subset of genes in mitosis that were not bound in G2 (Fig. 2E). Taken together, our data suggest that cohesin removal from chromosome arms during mitotic prophase is a key step in the eviction of elongating RNAPII from chromosomes during mitosis. Thus, cohesin protection at active centromeres explains the selective retention of RNAPII at centromeres during mitosis.
To determine if cohesin is sufficient to promote transcription of an ectopic locus during mitosis, we tethered either wild type Smc1a (a subunit of cohesin) or an ATPase-deficient form of Smc1a that is resistant to WAPL-mediated removal (Elbatsh et al., 2016) to an array of lac operator sequences located at the arm of chromosome 1 in U2OS cells (Janicki et al., 2004). Tethering of Smc1a-LacI led to the recruitment of the cohesin subunit Smc3 (Fig. S4A,B). Treatment with IPTG to disrupt the LacI-LacO interaction retained 50% of cohesin signal, suggesting that the entire cohesin complex is stably recruited to DNA (Fig. S4C,D). Tethering of Smc1-LacI, but not GFP-LacI, resulted in the strong recruitment of elongating RNAPII (pS2) to interphase and mitotic chromosomes (Fig. 2GH; Fig. S4E–F). We also observed a modest recruitment of RNAPII pS5 upon cohesin tethering (Fig. S4G,H). During mitosis, RNAPII recruitment was more efficient when WAPL-resistant cohesin was tethered to chromatin (Fig. 2H). Taken together, our results demonstrate that cohesin removal from euchromatin by the ‘prophase pathway’ helps evict RNAPII from mitotic chromosomes and that retention of cohesin is sufficient to allow RNAPII to persist on chromosomes.
To understand if cohesin retention on mitotic chromosomes results in enzymatically active RNAPII, we next pulse-labeled control and WAPL-AID cells with the RNA label EU during mitosis (Jao and Salic, 2008). Mitotic chromosome spreads from control cells displayed newly-synthesized RNA present primarily at centromeres (Fig. 2I). In contrast, WAPL-AID cells displayed newly-synthesized RNA throughout the chromosomes (Fig. 2I). Importantly, this nascent-RNA signal on mitotic chromosomes in WAPL-AID cells was eliminated by treatment with triptolide, an inhibitor of RNAPII transcription initiation (Titov et al., 2011) (Fig. 2I,J), likely through RNAPII degradation following extended triptolide treatment (Novais-Cruz et al., 2018). Taken together our results demonstrate that removal of cohesin from chromosome arms during prophase is important for removing elongating RNAPII and nascent transcripts and that protection of cohesin at the centromere is necessary for RNAPII localization to the centromere during mitosis.
Cohesin retention alters gene expression reprogramming during the G2-M transition
Failure to remove cohesin from euchromatin in prophase in WAPL-depleted cells results in a broad distribution of active RNAPII on mitotic chromosomes. To understand the impact of mitotic cohesin retention on the transcriptome, we performed RNA sequencing of newly synthesized transcripts (EU labeled) from G2 and mitotically-arrested control and WAPL-depleted cells. Principal Component Analysis (PCA) indicated that experimental replicates clustered and that G2 and mitotic samples were well separated by PC1 in control cells (Fig. 3A). WAPL-depleted cells displayed significantly different gene expression from control cells, consistent with previous work (Haarhuis et al., 2017; Tedeschi et al., 2013). In WAPL-depleted cells, G2 and mitotic cells were not well-separated by PC1, suggesting a similar pattern of gene expression persists between G2 and mitosis. Analysis of the sequence coverage across all genes demonstrated a significant peak at the 5’ end of genes in G2 cells (Fig. 3B, S5A), likely representing paused, promoter-proximal RNAPII. Upon entry into mitosis, this 5’ peak was significantly reduced in both control and WAPL-depleted cells, suggesting that persistent cohesin does not affect the process of RNAPII activation by p-TEF-b to release from its promoter-proximal pause state. Indeed, we found that inhibition of several different steps of transcriptional initiation during mitosis did not affect the distribution of RNAPII on mitotic chromosomes in control cells (Fig. S4I–K). This suggests that persistent cohesin does not promote transcription initiation during mitosis, but retains elongating RNAPII on chromatin.
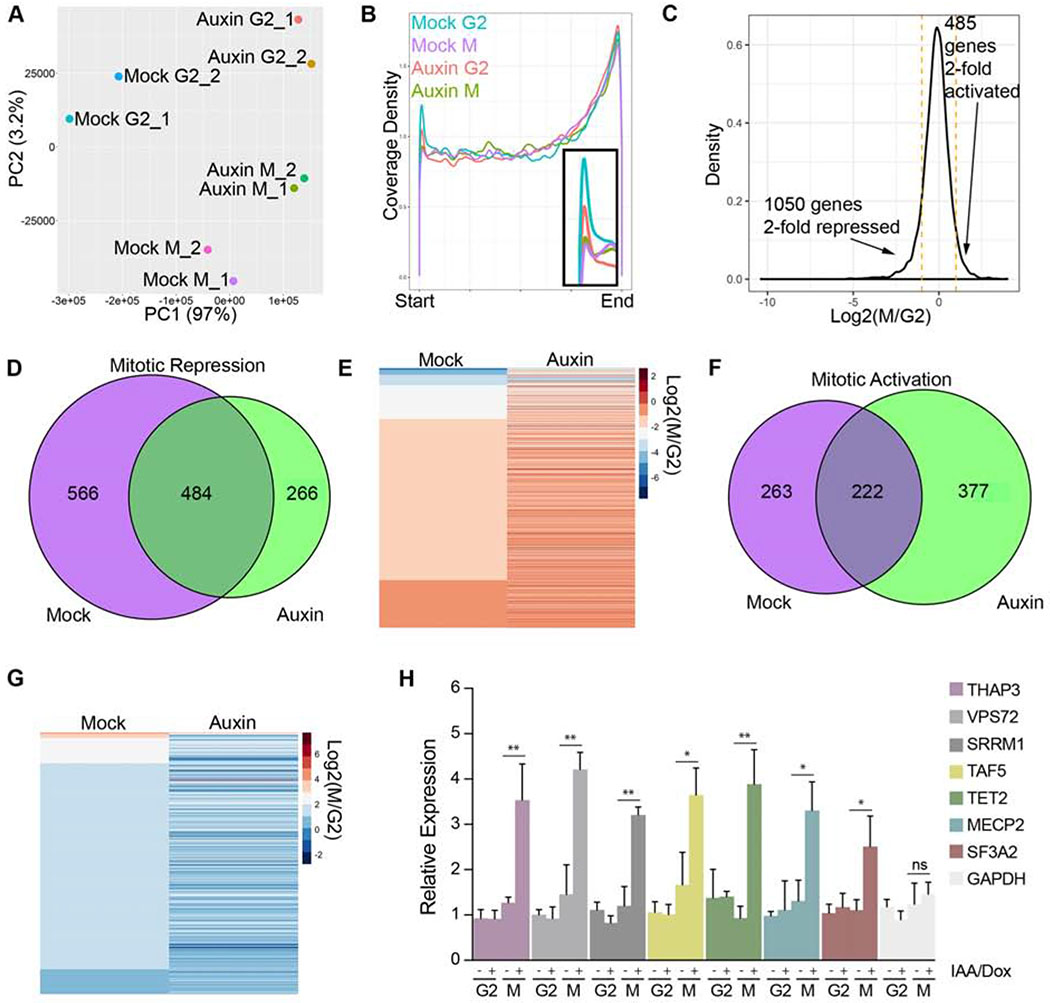
Figure 3. Cohesin retention alters gene expression reprogramming in the G2-M transition.
A Principal Component Analysis (PCA) of nascent gene expression in G2 and mitosis from replicate cultures of mock or auxin-treated WAPL-AID cells. B Metaplot of relative sequencing coverage across all genes expressed at FPKM > 10. C Histogram of changes in gene expression between G2 and mitosis in control cells. D Venn diagram of overlap of mitotically-repressed genes in control and auxin-treated WAPL-AID cells. E Heatmap of mitotically repressed genes in control and auxin-treated WAPL-AID cells. F Venn diagram of mitotically activated genes in control and WAPL-AID cells. G Heatmap of mitotically activated genes in control and auxin-treated WAPL-AID cells. H qRT-PCR validation of EU-labeled transcription in control and WAPL-depleted cells in G2 and mitosis. Error bars represent the SD of the mean. *, P < 0.05; **, P < 0.005 (t-test). (ns) no significant differences (t-test).
In control cells, we found that hundreds of genes were transcriptionally activated or repressed during mitosis (Fig. 3C), consistent with previous work (Liang et al., 2015). However, of the 1050 genes that were > 2-fold repressed in control cells, less than half of these genes were repressed in WAPL-depleted cells (Fig. 3C–E, S5B–F). Thus, persistent cohesin results in decreased mitotic transcriptional repression. Reciprocally, of the 485 genes that were > 2-fold transcriptionally activated in control cells, less than half of these genes were activated in WAPL-depleted cells (Fig. 3C,F–G, S5D). Consistent with sequencing data, qRT-PCR results showed increased transcript levels of several mitotically-repressed genes in WAPL-depleted cells (Fig. 3H).
Elongating RNAPII localizes to centromere regions in G2 and M and this localization is not affected by WAPL-depletion. To monitor expression of centromere sequences, we mapped sequencing reads to recently developed sequence models for human centromeres (Miga et al., 2014) and used the mapped reads to create transcript models for centromere RNAs. We found that RNAs mapping to centromere transcript models showed little change between G2 and mitosis in control cells and were unaffected by WAPL depletion (Fig. S5G–I). However, we found that depletion of Rad21 resulted in a significant reduction in a-satellite transcripts by Q-RT-PCR (Fig. S1F). Taken together, these results suggest that the ability to remove cohesin from chromosomes is important for the cell cycle dynamics of RNAPII and that persistent cohesin results in dampened transcriptional changes between G2 and mitosis.
Cohesin retention impairs the release of elongating RNA Pol II from chromosomes
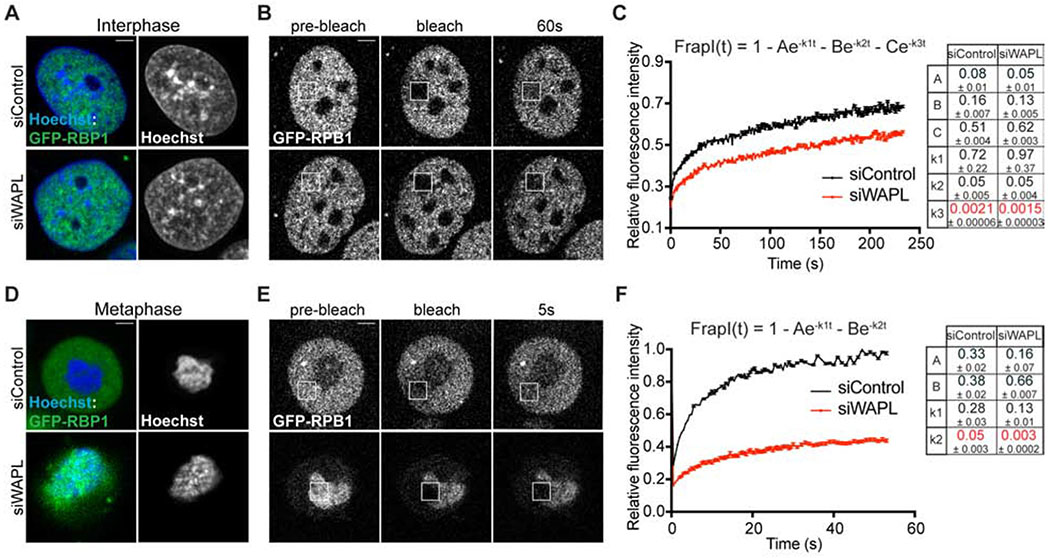
Our genetic and transcriptional inhibition experiments show that static cohesin specifically retains elongating RNAPII on mitotic chromosomes without affecting transcription initiation. RNA-seq shows that WAPL-depleted cells lose transcriptional dynamics between G2 and M. To determine how persistent cohesin affects the dynamics of RNAPII association with chromosomes, we performed fluorescent recovery after photobleaching (FRAP) analysis in GFP-RPB1 knock-in cells (Steurer et al., 2018). In control cells during interphase, the fluorescent recovery of GFP-RPB1 fit to a three-component model representing the different chromatin-bound RNAPII populations during transcription: initiation (A, k1), promoter-paused (B, k2), and elongating states (C, k3) (Fig. 4A–C and Fig. S6A,C), consistent with prior work (Darzacq et al., 2007; Steurer et al., 2018). Diffusion did not contribute to RNAPII FRAP recovery, suggesting that most RNAPII is chromatin-bound (Fig. S6E–H)(Darzacq et al., 2007; Steurer et al., 2018). In contrast to controls, WAPL-depleted interphase cells showed significantly slower fluorescent recovery of GFP-RPB1 (Fig. 4A–C). Modeling of this curve behavior indicated that this difference was primarily due to a higher fraction of elongating RNAPII, 44% vs 51%, and a ~ 35 % lower dissociation rate of elongating RNAPII (k3) (Fig. 4A–C, Fig. S6C). As a consequence, the pool of RNAPII available for promoter binding is reduced as suggested by a lower pseudo-on rate (A) (Fig. 4C, Fig. S6C). During mitosis, control cells displayed a rapid fluorescent recovery of GFP-RPB1. Analysis of the turnover dynamics indicated that a two-component model could explain the fast fluorescence turnover primarily based on the presence of freely diffusing (non-chromatin bound) RNAPII (Fig. 4D–F, Fig. S6B,I). In contrast, WAPL-depleted cells displayed a much slower, diffusion-independent recovery with a behavior resembling that of interphase, elongating RNAPII dynamics (Fig. 4D–F, Fig. S6J). In summary, analysis of GFP-RBP1 fluorescent recovery demonstrates that cohesin removal promotes the release of elongating RNAPII from chromatin in interphase and mitotic cells.
Figure 4. Cohesin retention impairs elongating RNA Pol II release from chromosomes.
GFP-RPB1 knock-in fibroblasts were used for the analysis of RNA Pol II dynamics by FRAP. A, D Co-staining of GFP-RPB1 (green) and DNA (Hoechst, blue) in live interphase (A) or mitotic (D) cells 48 h after transfection with either siControl or siWAPL siRNAs. B, E Cells were bleached 48 h after transfection with either siControl or siWAPL siRNAs and imaged for fluorescence recovery. Images of the indicated times are shown for representative cells in interphase (B) and mitosis (E). Scale bars, 3 μm. C, F Relative fluorescence intensity between bleached (squares) and non-bleached reference areas are plotted in time. Measurements from interphase and mitotic cells were fitted to three- or two-component models, respectively. The equation and a table containing the values estimated for each parameter are shown to the right. n ≥ 14 cells were used for each condition from three independent experiments. In all cases, error bars indicate the standard error of the mean (sem).
Ectopic mitotic transcription results in Aurora-B and Shugoshin1 mislocalization and anaphase chromosome segregation defects.
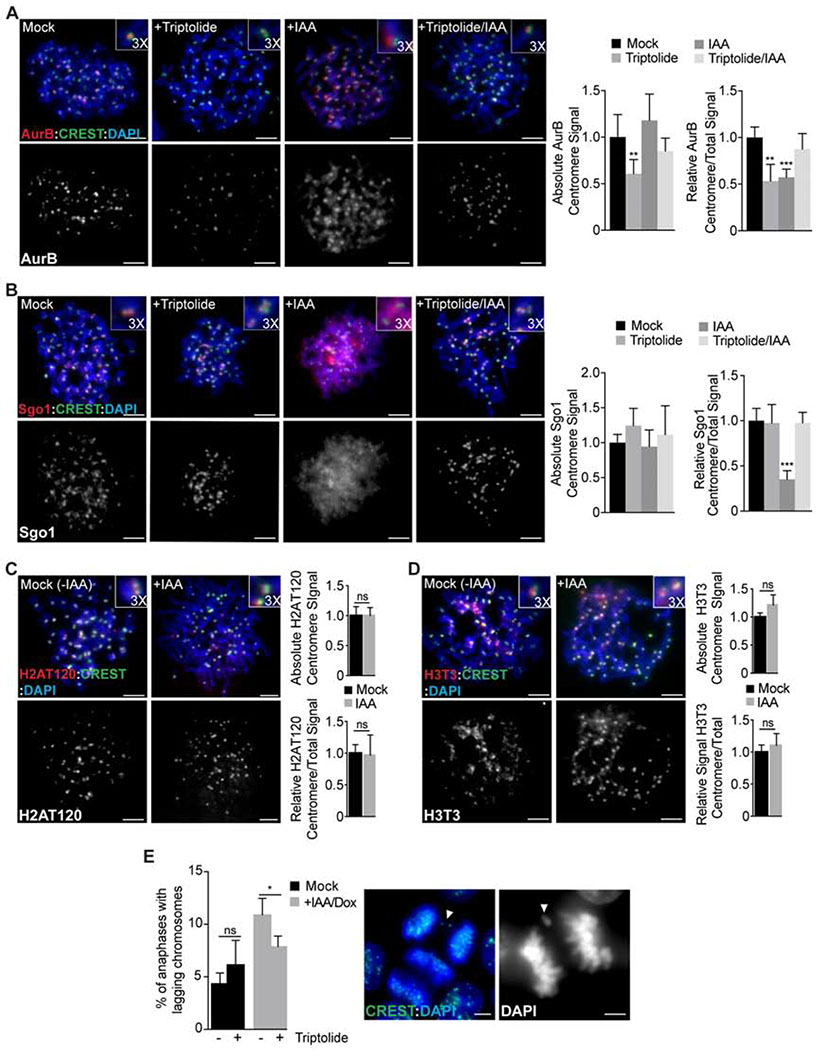
Failure to remove cohesin in WAPL-depleted cells results in retention of elongating RNAPII and nascent transcripts on mitotic chromosomes (Fig. 2). However, it is not clear if retention of RNAPII on mitotic chromosomes affects mitotic progression. In mitosis, RNAPII and chromosomal RNAs are typically restricted to centromeres (Chan et al., 2012; Johnson et al., 2017). Interestingly, both the CPC and Sgo1 interact with centromeric RNA (Perea-Resa and Blower, 2018). The CPC is normally localized to the inner centromere through an interaction of Survivin with Histone H3 phosphorylated at T3 (H3pT3) by Haspin kinase and an interaction with Sgo1, which is targeted to centromeres by H2A phosphorylated at T120 (H2ApT120) by Bub1 kinase (Tsukahara et al., 2010; Wang et al., 2010; Yamagishi et al., 2010). WAPL-depletion causes retention of cohesin on mitotic chromosome arms, loss of Chromosome Passenger Complex (CPC) and Shugoshin 1 (Sgo1) enrichment at the inner centromere, and an increase in anaphase chromosome segregation defects (Gandhi et al., 2006; Haarhuis et al., 2013; Kueng et al., 2006; Shintomi and Hirano, 2009; Tedeschi et al., 2013). To determine how ectopic mitotic transcription influences mitosis, we evaluated CPC/Sgo1 localization and chromosome segregation in WAPL-depleted cells. Consistent with previous results, Aurora-B and Sgo1 were less enriched at the inner centromere region in WAPL knockout cells (Fig. 5A,B). However, WAPL-depletion did not alter the levels of H2ApT120 or H3pT3 at the inner centromere (Fig. 5C,D). To determine if transcription contributes to CPC and Sgo1 localization, we inhibited transcription in late G2 cells using triptolide, which resulted in decreased levels and enrichment of Aurora B at the centromere, but had no effect on Sgo1 localization in control cells (Fig. 5A,B). Surprisingly, transcriptional inhibition in WAPL-depleted cells restored inner centromere enrichment of both Aurora-B and Sgo1 (Fig. 5A,B). We conclude that mislocalization of Aurora-B and Sgo1 in WAPL-depleted cells is caused by ectopic mitotic transcription and that mitotic transcriptional silencing on chromosome arms is important for concentrating Sgo1 and the CPC at the inner centromere at metaphase.
Figure 5. Ectopic RNA Pol II activity on metaphase chromosomes impairs Aurora B and Shugoshin 1 localization causing chromosome segregation defects.
A-B Analysis of Aurora B (AurB, A) and Shugoshin 1 (Sgo1, B) localization (red) in metaphase chromosome spreads from control (mock) WAPL-AID cells or following treatment with Triptolide, IAA/Dox or IAA/Dox + Triptolide. Quantification of absolute and relative centromere signal is showed on the right. Error bars indicate the standard deviation of the mean. **, P < 0.005; ***, P < 0.001 (one-way ANOVA, Dunnett’s test). C-D Analysis of H2AT120 (C) and H3T3 (D) localization (red) in metaphase chromosome spreads from control (mock) or auxin-treated WAPL-AID cells. Quantification of absolute and relative centromere signal is showed on the right. Error bars represent the standard deviation of the mean. (ns) no significant differences (t-test). E % of anaphases showing lagging chromosomes measured in control (mock) WAPL-AID cells or after treatment with Triptolide, IAA/Dox or IAA/Dox + Triptolide. Error bars indicate the standard deviation of the mean. *, P < 0.05; (ns) no significant differences (t- test). Example of anaphases showing lagging chromosomes are showed on the right. In all cases, anti-centromere antibodies (CREST; green) and DAPI staining (blue) were used for centromere and chromosome labelling, respectively. Scale bars, 5 μm.
To determine if ectopic mitotic transcription contributes to the chromosome segregation defects observed in WAPL-depleted cells, we analyzed anaphase chromosome segregation. Consistent with previous work, we found that WAPL depletion results in a significant increase in lagging chromosomes during anaphase (Fig. 5E)(Haarhuis et al., 2013; Tedeschi et al., 2013). Transcriptional inhibition using Triptolide treatment from late G2 to mitosis did not increase anaphase segregation defects in control cells (Fig. 5E). Interestingly, transcriptional inhibition in WAPL-depleted cells resulted in a statistically significant decrease in anaphase segregation defects. Collectively, our results suggest that ectopic mitotic transcription contributes to the anaphase segregation defects in WAPL-depleted cells through the altered localization of mitotic factors including Aurora-B and Sgo1.
Cohesin removal though the prophase pathway is required for G1 gene expression and cell cycle progression
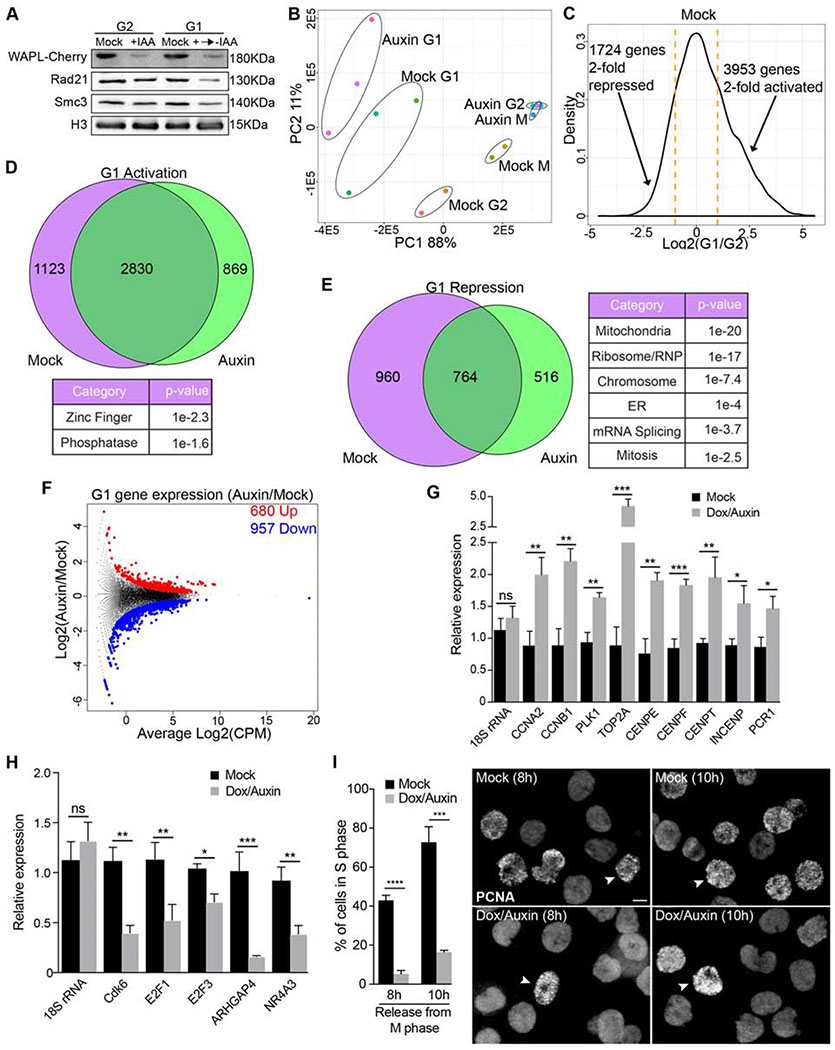
Failure to remove cohesin from prophase chromosomes in WAPL-depleted cells results in dramatically altered gene expression in mitosis (Fig. 3). In addition, failure to remove chromosome arm cohesin results in increased Separase-mediated cleavage of chromatin-bound cohesin at the metaphase-anaphase transition (Tedeschi et al., 2013), resulting in less total cohesin in G1. To understand how failure of the prophase pathway affects gene expression in G1, we synchronized control and WAPL-depleted cells in mitosis and released them into G1 in the absence of auxin and in the presence of EU to label newly synthesized transcripts. WAPL protein levels did not recover to control levels in the 2 hours after release into mitosis (Fig. 6A). Consistent with previous results, we found that WAPL-depletion resulted in reduction of cohesin subunits Rad21 and Smc3 specifically in G1 cells (Fig. 6A). PCA analysis of gene expression showed that WAPL-depleted G1 cells exhibited significantly different gene expression than control G1 cells (Fig. 6B). In control cells, we observed major changes in gene expression between G2 and G1 cells (Fig. 6C). Of the 3953 genes 2-fold activated in G1 control cells, 1123 were not activated in WAPL-depleted cells (Fig. 6C,D). Of the 1724 genes that were 2-fold repressed in control cells, 960 were not repressed in WAPL-depleted cells (Fig. 6C,E). Additionally, a direct comparison of G1 gene expression in control and WAPL-depleted cells revealed hundreds of genes either up- or downregulated in WAPL-depleted cells (Fig. 6F, Table S1). Interestingly, G1 genes upregulated in WAPL-depleted cells included many core mitotic regulators (e.g. CCNA2, CCNB1) (Fig. 6F,G). Additionally, G1 genes downregulated in WAPL-depleted cells included many proteins involved in DNA replication and cell cycle progression (e.g. E2F1, E2F3, Cdk6) (Fig. 6F,H). Collectively, these results demonstrate that dynamic cohesin is important for repressing G2-expressed genes and activating G1-expressed genes.
Figure 6. Cohesin dynamics is required for G2/G1 gene expression reprogramming.
A Analysis of protein levels by WB in control (mock) and auxin-treated WAPL-AID cells arrested in G2 or released into G1. B Principal Component Analysis (PCA) of nascent gene expression in G2, mitosis (M) and G1 from replicate cultures of mock or auxin-treated WAPL-AID cells. C Histogram of changes in gene expression between G2 and G1 in control cells. D Venn diagram of overlap of G1-activated genes in control and auxin-treated WAPL-AID cells. Results from GO analysis are displayed below. E Venn diagram of overlap of G1-repressed genes in control and auxin-treated WAPL-AID cells. Results from GO analysis are displayed on the right. F Volcano plot representing upregulated and downregulated newly-synthesized transcripts in WAPL-depleted cells in G1. G-H Validation of RNAseq data by qRT-PCR using EU-RNA from G1 control (mock) or auxin-treated WAPL-AID cells. Relative expression is plotted for upregulated genes (G) or downregulated genes (H) in auxin-treated WAPL-AID cells using 18S rRNA levels as a reference. In all cases, error bars indicate the standard deviation of the mean. *, P < 0.05; **, P < 0.005; ***, P < 0.001; (ns) no significant differences (t-test). I Analysis of G1 progression by PCNA staining. The % of cells in S phase (left) was measured in control and WAPL-depleted cells released from metaphase arrest for the indicated times. Error bars indicate the standard deviation of the mean. ***, P < 0.001; ****, P < 0.0001 (t-test). Representative images with arrow heads highlighting a cell in S phase (right). Scale bar, 10 μm.
To determine if G1 gene expression changes in WAPL-depleted cells affected cell cycle progression, we monitored entry into S-phase by staining for proliferating cell nuclear antigen (PCNA). WAPL-depleted cells show a significant delay in S-phase entry compared to control cells (Fig. 6I). Thus, cohesin dynamics are a key component of the molecular mechanism that reprograms gene expression through mitosis to ensure proper cell progression.
Discussion
Role of cohesin dynamics on gene expression reprogramming
During mitotic entry, interphase chromatin structure is erased and gene expression is dramatically altered (Gibcus et al., 2018; Liang et al., 2015; Palozola et al., 2017). However, the molecular mechanisms controlling this process are not clear. Our results demonstrate that the prophase pathway of cohesin removal is a key factor in rewiring transcription upon mitotic entry and mitotic exit. Removal of cohesin from chromosome arms facilitates the release of elongating RNAPII, while cohesin retention at centromeres is required for persistence of elongating RNAPII at centromeres of metaphase chromosomes. As cells exit mitosis, transcription is rapidly reactivated (Palozola et al., 2017; Teves et al., 2018) in a manner that is temporally correlated with cohesin loading. However, it was unclear if cohesin is required for transcription regulation following mitosis. WAPL-depletion results in a dramatic failure to properly repress G2-expressed genes important for mitosis and activate G1-expressed genes important for DNA replication and repair. Mis-regulation of G1 gene expression could result from several possible causes. First, during prophase WAPL-mediated release of cohesin from chromosome arms safeguards cohesin from Separase decay (Tedeschi et al., 2013), which may facilitate the rapid deposition of cohesin on chromosomes in telophase and early G1 (Watrin et al., 2006). Cohesin cleavage in WAPL-depleted cells could result in a lack of cohesin available in G1 cells, leading to a failure to activate or repress transcription. Second, failure of the prophase pathway could lead to a failure to erase the G2 gene expression program during mitosis and result in inappropriate expression of mitotic genes in G1. Interestingly, cohesin remains bound to binding sites of transcription factor clusters in mitosis while transcription factors are evicted, suggesting a role for cohesin in cell memory (Yan et al., 2013). Prophase cohesin removal could be important for resetting expression of a subset of genes during mitosis. Finally, although we removed auxin from WAPL-depleted cells upon release into G1, WAPL protein levels are not restored in G1 WAPL-depleted cells. A lack of dynamic cohesin in G1 could result in changes in gene expression independently of events that occurred during mitosis. In addition to mitosis, gene expression shows an extensive reprogramming following cell differentiation during development. Mutations of genes encoding the cohesin subunits SMC1A, SMC3, and RAD21, or their regulators NIPBL and HDAC8, have been identified in the Cornelia de Lange syndrome (CdLS), a cohesinopathy causing severe developmental disorders (Banerji et al., 2017). Importantly, cells from CdLS patients do not exhibit defects in mitotic sister chromatid cohesion, but do show significantly altered transcriptional profiles (Mannini et al., 2015), supporting a central role for cohesin and its regulation rewiring gene expression during development.
Role of cohesin in centromere transcription
Transcription has been observed at endogenous, neo-, and artificial centromeres in many different organisms (reviewed in: (Perea-Resa and Blower, 2018)). Centromere transcription promotes changes to centromeric chromatin structure that are important for deposition of new CENP-A during interphase (Bobkov et al., 2018; Chen et al., 2015; McNulty et al., 2017; Molina et al., 2016; Nakano et al., 2008; Swartz et al., 2019). Strikingly, retention of elongating RNAPII at centromeres has been observed in metaphase chromosomes of human, fly and frog cells (Blower, 2016; Chan et al., 2012; Molina et al., 2016; Rosic et al., 2014), and several roles for mitotic centromere transcription have been proposed (Blower, 2016; Chan et al., 2012; Liu et al., 2015). However, a recent study where transcription was inhibited specifically in mitotically arrested cells has questioned the role of mitosis-specific transcription for accurate chromosome segregation (Novais-Cruz et al., 2018). Consistent with this study, we find that inhibition of transcription in very late G2 and mitosis does not result in anaphase chromosome segregation defects (Fig. 5E). WAPL-depletion results in an increase in anaphase chromosome segregation defects and a failure to concentrate Aurora B and Sgo1 at metaphase centromeres (Haarhuis et al., 2013; Kueng et al., 2006; Tedeschi et al., 2013). Interestingly, both Aurora B and Sgo1 proteins have promiscuous RNA-binding activity (Jambhekar et al., 2014; Liu et al., 2015). We show that failure to concentrate Sgo1 and Aurora B at the inner centromere in WAPL-depleted cells is a result of ectopic transcription, possibly mediated by an interaction of Sgo1 and the CPC with nascent transcripts. WAPL knockout cells exhibit chromosome segregation defects (Haarhuis et al., 2013; Tedeschi et al., 2013) that are partially reversed by transcription inhibition in late G2 and mitosis. We speculate that restricting mitotic transcription to the centromere promotes accurate chromosome segregation by reinforcing the localization of the CPC and Sgo1 to the inner centromere region. In early mitosis, the CPC contributes to cohesin removal from chromosome arms, which likely removes a CPC recruitment signal from euchromatin and contributes to CPC/Sgo1 concentration at the inner centromere.
Regulation of transcription by cohesin in mitosis
Cohesin is a major constituent of genome architecture and regulates many aspects of gene transcription during interphase. Global analyses of cohesin distribution by ChIP-seq have demonstrated a correlation between the cohesin complex and active transcription (Busslinger et al., 2017; Schaaf et al., 2013). However, during early mitosis most cohesin and cohesin-mediated TADs are removed and it was not clear if chromatin-bound cohesin also regulated transcription at this cell stage. Our results reveal cohesin as one key factor determining the distribution of elongating RNA Pol II (pS2) on mitotic chromosomes. Consistent with previous studies (Chan et al., 2012), we find specific retention of actively elongating RNAPII pS2 while promoter paused RNA RNAPII pS5 is largely absent from metaphase chromosomes. Additionally, mitotic inhibition of transcription initiation does not affect the localization of RNAPII pS2 to centromeres and only elongating RNAPII is retained on mitotic chromosomes in WAPL-depleted cells. Collectively, these data support the absence of transcription initiation during mitosis limiting the role of cohesin to latter steps of transcription regulation. Interestingly, several studies suggest cohesin facilitates the transition of RNA Pol II from promoter-paused to the actively elongating state (Izumi et al., 2015; Schaaf et al., 2013). In addition, cohesin promotes transcription termination in yeast (Gullerova and Proudfoot, 2008). However, our RNAPII pS2 ChIP-seq and EU-seq results do not reveal extension of transcription beyond transcription termination sites (Fig. 2D and not shown), suggesting that static cohesin does not influence transcription termination in human cells. Our FRAP results reveal increased levels of chromatin-bound elongating RNAPII when cohesin is retained on mitotic chromosomes. We also observe a similar effect in interphase cells, but the effect is more subtle because transcription initiation is active during this cell stage. Thus, our work indicates that cohesin removal promotes release of elongating RNAPII from mitotic chromatin. Static cohesin could influence elongating RNAPII by changing the transcription elongation rate or by directly tethering elongating RNAPII to chromatin. A slower release of elongating RNAPII from chromatin is likely to alter aspects of the transcription cycle by limiting the availability of free RNAPII to initiate transcription.
Here, we demonstrated that the removal of cohesin from chromosomes during prophase facilitates mitotic RNAPII dynamics and contributes to transcription inhibition during mitosis and further reactivation in G1. Our results demonstrate that cohesin removal is coupled to transcriptional remodeling as a component of the process that restructures the genome in preparation for mitotic chromosome segregation. Our work suggests that one function of the ‘prophase pathway’ of cohesin removal is to reprogram gene expression during mitosis.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michael Blower (blower@molbio.mgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
hTERT RPE-1 (retina epithelium eye) cells were cultured in DMEM:F12 medium supplemented with 10% fetal bovine serum (FBS, Sigma), sodium bicarbonate (0.25%), and 1% penicillin/streptomycin (Corning). HeLa S3 cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. AID-WAPL cell line generation is described below. HeLa-Cas9 cells expressing Rad21 guide RNAs were cultured in DMEM supplemented with tetracycline-free FBS and doxycycline (Sigma D3447, DMSO) added as previously reported to induce Cas9 expression (McKinley and Cheeseman, 2017). MRC-5 fibroblast cells stably expressing GFP-RPB1 were kindly provided by A.B. Houtsmuller (Steurer et al., 2018) and cultured in DMEM:F12 with 10% FBS and 1% penicillin/streptomycin. lacO-array-containing U2OS cells is a kind gift from D. Spector and were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. MDA-MB 435 cells were grown in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. In all cases cells were incubated at 37° in the presence of 5% CO2.
Xenopus egg extracts
Wild-type female Xenopus laevis frogs were obtained from Nasco (www.enasco.com). Mature eggs were obtained by injecting frogs with hCG as described below.
METHOD DETAILS
Cell culture
For G2 arrest RO-3306 (Sigma SML0569, DMSO) was added to a 10μm final concentration for 16h. Cells were washed with 1xPBS and fresh medium added to release cells into mitosis for the indicated times. Metaphase arrest was induced adding nocodazole (Sigma M1404, DMSO) to 0.1μM for 16h. In EU-labelling experiments cells were first synchronized at G1/S using 2mM thymidine (Sigma T1895, water) for 16h, released into fresh medium for 3h and then arrested at G2 or metaphase using RO-3306 or nocodazole. In both cases 5-EU substrate (Abcam ab146642, water) was added to a 0.25mM final concentration for 12h. For G1 samples, cells arrested in nocodazole were washed and released into fresh medium for 2h in the presence of 5-EU (0.25mM). Aurora-B and Plk1 inhibition was performed adding barasertib or BI-2536 (Selleckchem, DMSO) to a final concentration of 1 and 0.1μm respectively. Inhibition of Cdk7 and Cdk9 kinase activities was induced adding THXZ1 or LDC000067 at 10μm (ApexBio, DMSO) respectively to metaphase arrested cells for 1h. 1μm Triptolide (Sigma T3652, DMSO) was used to inhibit RNAPII transcription initiation in metaphase arrested cells for 12h (Fig. 2I; Fig. 5). RNAPII degradation is likely induced as previously reported (Wang et al., 2011). In lacO competition assays, IPTG (Sigma I6758, water) was added to a 1mM final concentration for 1h. AID-WAPL proteolytic decay was induced incubating HeLa S3 cells in medium supplemented with 1 μg/ml doxycycline (Sigma D3447, DMSO) and 0.5 mM IAA (Indole-3 Acetic Acid sodium salt, Sigma I5148) for 48h.
For G1/S progression analysis, control and auxin-treated AID-WAPL cells arrested in nocodazole were release in fresh medium −/+ Doxycycline/IAA for the indicated times. IF using anti-PCNA antibodies was performed as previously described (Xiao et al., 2007) and the % of cells in S phase (equally distributed foci) evaluated in each time point (n ≥ 200).
Generation of WAPL-AID cells
To create WAPL-AID cells we first created a repair template that contains a central cassette of mAID-mCherry-P2A-Neomycin (pMB1152). This construct was created using InPhusion cloning from (pMK294, Addgene #72832)(Natsume et al., 2016). We then synthesized 750bp homology regions flanking the WAPL stop codon and fused these to the AID selection cassette using InPhusion cloning. The complete repair template was created by using the InPhusion reaction as a PCR template. PCR product was purified and cotransfected into HeLa S3 cells with a plasmid expressing a WAPL sgRNA and Cas9 (cloned into pX330, Addgene# 42230) (pMB1155). Cells were allowed to grow for 3 days then were split into 500ug/ml Neomycin for ~14 days. Single cell colonies were isolated using cloning rings. Single colonies were initially screened for nuclear mCherry fluorescence, then by Western blot with antibodies against Xenopus WAPL that cross react with human WAPL. Potential homozygous knock-in cell lines were then screened by PCR with primers that recognize both the endogenous stop codon or primers that recognize the knock-in junction.
siRNA and DNA transfection
ON-TARGETplus Human WAPL siRNA-SMARTpool (Dharmacon, 23063) were transfected into RPE-1 or HeLa cells using the RNAiMAX transfection reagent (Thermofisher 13778030). 20μM siRNAs aliquots were incubated with transfection agent in Optimem medium at room temperature for 20 minutes and added to cells grown in medium without antibiotics. siGENOME non-targeting siRNA (Dharmacon, D-001210) was transfected in parallel as a negative control. The effects of siRNAs were evaluated 48 hours after transfection.
Plasmid DNA transfection was performed in all cases using the Lipofectamine 3000 reagent (Thermofisher, L3000001). DNA (2.5-5 μg) and transfection reagent were incubated in Optimem medium at room temperature for 20 minutes and then added to the cell medium. The expression of transfected DNA was analyzed 48-72 hours after transfection.
Egg extracts from Xenopus laevis
CSF-arrested Xenopus egg extracts were prepared and utilized for immunofluorescence as described previously (Hannak and Heald, 2006). Sperm nuclei were cycled through interphase by the addition of 0.6 mM CaCl2 for 90 minutes, then driven back into mitosis by adding an equal volume of CSF-arrested extract for the indicated times. AuroraB and Plk1 inhibition was performed adding barasertib or BI-2536 to a final concentration of 10 and 1μm respectively.
WAPL inmunodepletion was performed using a customized in-house antibody (see below) coupled to protein A dynabeads (Life technologies) and added at 0.125μm/μl egg extract. IgG-coupled beads were used at same concentration in parallel as control. Reactions were incubated at 4° for 1h before being used for immunofluorescence or western blot.
Recombinant protein and Antibody generation
A N-terminal fragment corresponding to 1-466 amino acids (~ 46 KDa) of Xenopus WAPL protein was cloned to express a GFP-WAPL-6xHis fusion protein. For CENPC expression a fragment containing 381-712 amino acids (~ 32 KDa) of Xenopus CENPC protein was used. In both cases, expression was performed in BL21 Rosetta cells as previously described (Jambhekar et al., 2014) and purified using NiNTA and Superdex S200. Peak fractions from S200 were incubated with Precision Protease (GE Healthcare, 27084301) overnight at 4° to excise GFP. Cleaved WAPL/CENPC was bound to HiTrap Heparin and eluted with a linear gradient of NaCl (from 150 mM to 1 M). Fractions containing WAPL/CENPC were loaded into acrylamide gel and a 46/33 KDa band excised and used for antibody generation in rabbit or guinea pig respectively (Cocalico Biologicals). Antibodies were purified from serum by affinity purification using the same recombinant proteins employed for animal injection and the concentration estimated measuring absorbance at 280nm. Specificity was tested by WB using total protein samples from Xenopus egg extracts revealing bands corresponding to the full length of WAPL or CENPC proteins.
Immunofluorescence
A list of antibodies used in this study is displayed in Supplemental Table 1. For chromosome spreads 500μl of cells (2x105cells/ml) were centrifuged in a cytospin (1800rpm, 10min). Slides were fixed in 1xPBS containing 4% PFA for 10min followed by permeabilization in 1xPBS with 0.5% Triton X-100 for 20min and washed twice with 1xPBS. Slides were blocked in 1xPBS supplemented with 10% ultrapure BSA (Invitrogen, AM2616) for 10min before primary and secondary antibody incubations at 37°. For pS2 and pS5 immunostaining an unfixed KCM protocol was used instead (Chan et al., 2012). Cells (2x105cells/ml) were swelled in KCl (65mM) at 37° for 10min before cytospin. Slides were incubated in KCM buffer supplemented with 10% ultrapure BSA for 10min prior to primary and secondary antibody incubations performed at 4° for 1h. Slides were finally washed with KCM, fixed in 4% PFA KCM and mounted with DAPI containing solution (Sigma, DUO82040). For EU-labelled RNA visualization the Click-iT RNA Imaging Kit was used following the manual specifications (Invitrogen, C10329). Alexa Fluor 488 azide was employed in the Click-iT reaction performed with fixed cells previously incubated with or without 0.25mM EU.
Xenopus egg extracts were fixed in 1xPBS with 4% PFA for 10min prior to centrifugation (10200rpm, 20min) through a cushion buffer containing 1xBrB, 30% glycerol and 0.1% Triton-X-100. Coverslips were washed once in 1xPBS prior to blocking in 1xPBS containing 0.2% Tween and 10% BSA. Incubation of primary antibodies were performed at 4° overnight and secondary at room temperature during 1h. DAPI containing medium was used for mounting.
In all cases samples were storage at 4° under dark conditions until image acquisition.
Western Blotting
Total protein from human cells was extracted incubating cells in lysis buffer (150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA, 5 mM MgCl2, 1% NP 40, 0.5 mM DTT, 1 mM PMSF) supplemented with protease inhibitors (Roche). Protein concentrations were calculated by Bradford and 10 mg were typically loaded for PAGE-SDS. Proteins were transferred to nitrocellulose membranes (Amersham Protran 0.45 μm NC) using Tris/Glycine buffer with 20% methanol for 1h at room temperature or overnight at 4°. Membranes were then blocked in 1x TBST supplemented with 5% skim milk powder (Millipore). Antibodies were diluted in TBST 5% milk and incubated at 4° overnight (primary) or 1 hour (secondary). Membranes were finally developed using Western Lighting Plus ECL (Perkin Elmer) and scanned in a Chemidoc MP imager (BioRad). Total protein from Xenopus extracts were obtained following standard protocols and employed in western blot as described above.
Chromatin-associated protein fractions from human cells or Xenopus egg extracts were purified following previously described protocols (MacCallum et al., 2002; Mendez and Stillman, 2000).
RNA purification
Total RNA from human cells (~ 106 cells) was extracted using Trizol (Invitrogen, 15596026). Cells were collected and washed in 1xPBS prior to resuspension in 1ml Trizol. 250μl of chloroform was then added for protein extraction following RNA precipitation with isopropanol. RNA pellet was finally washed in 70% ethanol and resuspended in nuclease-free water (Ambion, AM9932). RNA levels were measured using a Biodrop and storage at −20°.
EU-labelled RNA was purified from total RNA extracted from G2, metaphase (M) or G1 arrested cells incubated with 0.25mM EU for 12 hours (G2 and M) or 2h (G1). For EU-RNAseq we previously removed the rRNA from 5μg total RNA using Ribo-Zero rRNA Removal Kit (Illumina) following the manual specifications. EU-labelled RNA was then recovered from depleted RNA (~ 500 ng) using the Click-iT RNA Capture Kit (Invitrogen, C10365). Renilla and Luciferase custom biotinylated probes were used as spike in controls in the EU-RNAseq experiment. Templates DNA were generated by PCR and in-vitro transcribed using the Hiscribe T7 kit (NEB E2030) including biotinylated UTP. Probes were purified by LiCl precipitation and added after the Click-iT reaction at 0.25ng/μl and 0.025 ng/μl final concentration respectively. Purified EU-RNA was used for either cDNA synthesis or library preparation (see below).
Library preparation
ChIPseq from G2 and M cells was performed using DNA recovered from pS2 IP and ~ 10 ng of sonicated INPUTs as starting material (see below for ChIP protocol details). Libraries were prepared using the NEBNext ChIP-Seq kit (NEB6240S) in combination with the NEBNext Multiplex Oligo Set (NEB7335L) following the manual specifications. Amplified libraries were submitted for QC and sequencing to Genewiz.
EU-RNA from G2, metaphase and G1 cells treated or not with Auxin and doxycycline were analyzed in duplicated (Auxin vs Mock). For library construct we used EU-labelled RNA purified from ribo-depleted samples as described above and the NEBNext Ultra Directional RNA Library Prep Kit (NEB) following the manual specifications. Amplified libraries sizes were evaluated by loading into an 8% native PAGE and gel bands excised and purified overnight in a NH4Ac/SDS (0.45 M/0.045%) solution. Libraries were finally precipitated with isopropanol and Glycogen (Thermofisher, 10814010), dissolved in nuclease-free water and submitted to the MGH-sequencing facility for QC and Illumina sequencing.
RT-qPCR
Total or EU-labelled RNA (~ 500 ng) was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad, 1708890) following the protocol specifications. cDNA was then diluted and used as template in 15 μl final volume RT reactions containing IQ-Sybr Green reagent (Bio-Rad, 1708880). Water and -RT samples were included as negative controls. A list of primers used in these experiments is displayed in supplemental table 2.
ChIP
Chromatin immunoprecipitation assay was performed essentially following the protocol reported by Myers (v011014) (21). Briefly, G2 and metaphase arrested RPE-1 cells were collected and fixed in 1xPBS containing 1% formaldehyde for 10 minutes. Glycine was added to a final concentration of 0.125M to stop cross-linking and cells washed in cold 1xPBS. Intact nuclei from ~ 5x106 cells were extracted with 1ml of Farnham lysis buffer and finally resuspended into 300μl of RIPA buffer. Nuclei were fragmented using a Qsonica sonicator for 5 minutes (30 seconds on/off, 40%). The efficiency of fragmentation was measured as the enrichment of ~ 300 bp fragments, tested on an agarose gel, and the DNA quantified in a Biodrop. To perform immunoprecipitations Dynabeads M-280 Sheep Anti-Rabbit (Invitrogen 11203D) were coupled to 5 μg of pS2 or IgG rabbit antibodies and incubated with 25 μg of fragmented chromatin samples overnight at 4°. Beads were then washed with LiCl wash buffer and eluted in 200 μl of IP elution buffer. Cross-link was reversed by incubating beads at 65° for 1 hour, supernatant (DNA) recovered after centrifugation and incubated at 65° for 14h. In parallel, input samples (25 μg of fragmented chromatin) were incubated at 65° for 14h and then purified using QIAquick PCR Purification Kit (QIAGEN). Serial dilutions of Input and recovered DNA samples were used as template in 15 μl RT reactions containing iQ-Sybr Green reagent (Bio-Rad). In all the assays water was included as negative control and data represented as the percentage of input recovered in each condition. The analysis of an intergenic region (IR) was included as a negative control. The primers employed in this assay are listed in supplemental table 2.
For ChIP-seq, DNA recovered from pS2 IP and ~ 10 ng of sonicated INPUTs were used for library preparation (see above).
Plasmids and constructs
For expression of SMC1A-LacI fusion proteins we used the pGK215 vector (Addgene 45110) (22) where GFP encoding DNA was replaced by the full-length cDNA encoding the SMC1A protein (pMB1208). A point mutation was then introduced to express a modified SMC1A version (SMC1A*) containing an amino acid substitution (L1128V)(pMB1209) that generates a WAPL-resistant cohesin ring (23). In both cases a C-terminal FLAG was added for further detection. The SV40 promoter was finally cloned to drive the expression of GFP-, SMC1A- or SMC1A*-LacI fusion proteins in mammalian cells. All DNA amplifications were performed using the Q5 High Fidelity polymerase (NEB). For constructs related to the AID-WAPL cell line see above.
For protein expression in bacteria DNA fragments encoding WAPL (1-466aa) and CENPC (381-712aa) from Xenopus laevis were cloned into the pET30a-GFP plasmid to express GFP/6xHis-tagged fusion proteins.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image acquisition, analysis and plotting
All images were acquired using the microscope set-up described previously (Jambhekar et al., 2014). In all cases images were captured from at least three independent experiments considering 10-20 cells per replicate. This numbers were significantly increased in the experiments with the double RAD21/WAPL mutant cells. For quantitative analysis all images were captured using identical conditions and are displayed following identical settings. Prior to the analysis all images were background-subtracted using Image J software. To delimit inner centromere CENPA, CENPC and ACA antibodies were employed to stain kinetochores and a 2.5μm diameter circle including both sister kinetochores was drawn. Total chromatin signal was measured using DAPI staining as reference and relative centromere measurement were calculated normalizing average centromere to average total nuclear signal. In lacO-tethering experiments an equal size circle was drawn to delimit the lacO area detected by tethering GFP-, SMC1A- or SMC1A*-LacI fusion proteins. DAPI signal was used as reference to measure total chromatin levels and to normalize and calculate average relative signal values at lacO.
Statistical analyses and plotting of image measurements were performed using Prism 7 software. Differences between two groups of data were analyzed by a single-sample t-test. For multiple group analyses one-way ANOVA was performed and Dunnett’s comparisons test considered.
Fluorescence recovery after photobleaching (FRAP)
All images were acquired using a Ti-2 Eclipse microscope (Nikon) and a previously described GFP-RPB1 knock-in cell line (2) 48h after transfection with siControl or siWapl siRNAs as detailed above. All images were acquired at 37° using a temperature-controlled Tokai-Hit culture dish system. Incubation with Hoechst 33342 (Molecular Probes, 1:5000) for 2h was used for life cell DNA staining. A square of 3x3 microns was imaged during 2 s at 1 s/frame before stimulation for photobleaching using a 488-nm laser at 40% power during 2 s. Time-lapse imaging was performed in two phases: a fast acquisition at 500 ms/frame during 30 s following by a 1 s/frame phase for 3 min and 30 s. For fast recovery analysis images were taken at 300 ms/frame during 30 s followed by a 1 s/frame phase of 30 s. Images were background subtracted and fluorescence intensity evaluated at bleached areas using Fiji software. Measurements were normalized to values obtained before bleaching (t = 0) and related to an unbleached area used as a reference. All recovery graphs show relative fluorescence intensity (RFI) in time (s). In diffusion-dependence tests half nuclei or squares of 3x3 microns were stimulated and imaged as described above. Images were background subtracted and fluorescence intensity evaluated at the indicated time frames in the bleached/unbleached transition zone by line scanning as described previously (Mueller et al., 2008). RFI values were plotted versus the distance (microns) to an arbitrary point set at the edge of the bleached area.
Normalized FRAP data were fit to two or three component, reaction-dominant models for recovery (Sprague et al., 2004) using the non-linear least-squares method in R. We determined the appropriate number of components for each model by examining the distribution of residuals for each fit and by calculating the Akaike Information Criteria (AIC) and Bayes-Schwarz Information Criteria (BIC) for each model as described (Darzacq et al., 2007). AIC and BIC tests were performed using R.
ChIP-seq data analysis
ChIP-seq reads were aligned to the hg38 genome, hg38 centromere models, or hg19 genome using Bowtie2 (Langmead and Salzberg, 2012). Metagene plots were produced using ngs.plot (Shen et al., 2014). To produce metagene plots of expressed genes we analyzed RNAPII pS2 ChIP-seq coverage on all genes expressed at a FPKM >10 in G2 EU-seq experiment. Comparable results were observed when analyzing ChIP-seq coverage on all genes or genes expressed at a FPKM>1. ChIP-seq peaks were called in human centromere models using MACS2 with default settings (Zhang et al., 2008). To produce metagene plots for human centromere ‘genes’ we used bedtools (Quinlan and Hall, 2010) to intersect centromere transcript models created using Cufflinks with centromere ChIP-seq peaks called using MACS2. We created metagene plots for all transcripts that intersected with a ChIP-seq peak.
EU-RNAseq analysis
Fastq files from the Illumina HiSeq were collapsed to unique reads using a custom Perl script. Reads were aligned to the human genome (hg38) or human centromere sequences (Miga et al., 2014) (released with hg38) using TopHat2. Reads mapping the Gencode gene models were counted using the cuffdiff function in Cufflinks (Trapnell et al., 2012). For analysis of centromere transcripts, we used all reads (from 8 libraries) - mapped to the centromere models as input for Cufflinks to build transcripts. We then counted mapping reads against the Cufflinks gene models. All calculations were performed on FPKM normalized values. For analysis of genes we analyzed all genes with a FPKM value > 10 in all G2 samples and > 0 in all mitosis samples. For analysis of enhancers we analyzed all enhancers with a FPKM value > 1 in all G2 samples and > 0 in all mitosis samples. For analysis of centromere transcripts, we did not impose a FPKM cutoff prior to analysis. Genome browser images were produced in R using the Gviz package. All other plots and statistical tests were prepared in R. Direct comparison of G1 gene expression differences was performed using edgeR (Robinson et al., 2010) and read counts generated using htseq-count (Anders et al., 2015).
We attempted to use Renilla and Firefly spike-in RNAs to normalize our EU-RNA-Seq data. However, in one sample the Firefly and Renilla spike-in sequences diverged dramatically and were not usable, suggesting a gene specific effect as observed for spike-in sequences in previous studies (Consortium, 2014; Risso et al., 2014). Additionally, we performed Q-RT-PCR validation experiments of additional EU-labeled biological replicates. In all cases validation analysis corresponded to expression values/ratios obtained using bulk FPKM normalization and not to normalization obtained by spike-in normalization. As a result all our library comparisons were performed with FPKM normalized samples.
To produce metaplots of sequencing coverage across genes we adapted the metaPlotR software (Olarerin-George and Jaffrey, 2017). We created gene models for each human gene expressed at FPKM > 10, including introns, using R. We then converted our mapped reads into bedgraph files using BEDTools. These files were used as input for metaPlotR software. Metaplots were produced using ggplot in R.
DATA AND CODE AVAILABILITY
All sequencing data associated with this study has been deposited in GEO under the Superseries accession: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139856
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| pS2 (Rabbit) | Abcam | Ab5095 |
| pS5 (Rabbit) | Abcam | Ab5131 |
| CENPA (Mouse) | Abcam | Ab13939 |
| CENPC (Rabbit) | Gascoigne et al., 2011 | NA |
| CENPC (Guinea pig) | In house, see methods | NA |
| Anti-Centromere antibody ACA (Human) | Antibodies Inc. | 15-234-0001 |
| H3 (Rabbit) | Abcam | Ab1791 |
| GFP (Mouse) | Abcam | Ab1218 |
| FLAG (Mouse) | Sigma | F3165 |
| CREST (Human) | Inmunovision | HCT-100 |
| RAD21 (Rabbit) | Abcam | Ab922 |
| SMC3 (Rabbit) | Abcam | Ab9263 |
| Cherry (Rabbit) | Abcam | Ab167453 |
| WAPL (Rabbit) | In house, see methods | NA |
| WAPL (Rabbit) | Abcam | Ab70741 |
| AuroraB (Rabbit) | Abcam | Ab2254 |
| Sgo1 (Mouse) | Abcam | Ab58023 |
| H2A pT120 (Rabbit) | Active Motif | 39391 |
| H3 pT3 (Rabbit) | Millipore Sigma | 07-424 |
| IgG (Rabbit) | Jackson ImmunoResearch | 011-000-003 |
| PCNA (Human) | Xiao et al., 2007 | NA |
| Bacterial and Virus Strains | ||
| Bacteria strain: BL21 Rosetta cells | Promega | Cat#L1195 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant protein: GFP-WAPL (1-466)-6xHis | This paper | NA |
| Recombinant protein: GFP-CENPC (381-712)-6xHis | This paper | NA |
| Precision Protease | GE Healthcare | Cat#27084301 |
| RO-3306 | Sigma | Cat#SML0569 |
| Nocodazole | Sigma | Cat#M1404 |
| Thymidine | Sigma | Cat#T1895 |
| 5-EU | Abcam | Cat#Ab146642 |
| Barasertib | Selleckchem | Cat#S1147 |
| BI-2536 | Selleckchem | Cat#S1109 |
| THXZ1 | ApexBio | Cat#B4736 |
| LDC000067 | ApexBio | Cat#B4754 |
| Triptolide | Sigma | Cat#T3652 |
| IPTG | Sigma | Cat#I6758 |
| Doxycycline | Sigma | Cat#D3447 |
| IAA (Indole-3 Acetic Acid) | Sigma | Cat#I5148 |
| RNAiMAX | Thermofisher | Cat#13778030 |
| Lipofectamine 3000 | Thermofisher | Cat#L3000001 |
| IQ-Sybr Green reagent | Bio-Rad | Cat#1708880 |
| Glycogen | Thermofisher | Cat#10814010 |
| Critical Commercial Assays | ||
| Click-iT RNA Imaging Kit | Invitrogen | Cat#C10329 |
| Click-iT RNA Capture Kit | Invitrogen | Cat#C10365 |
| NEBNext ChIP-Seq kit | NEB | Cat#NEB6240S |
| NEBNext Ultra Directional RNA Library Prep Kit | NEB | Cat#E7420S |
| Ribo-Zero rRNA Removal Kit | Illumina | Cat#MRZH116 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#1708890 |
| QIAquick PCR Purification Kit | QUIAGEN | Cat#28106 |
| Deposited Data | ||
| Raw sequencing data (ChIP-seq; nascent RNA-seq) | This paper | GEO: GSE139856 |
| Experimental Models: Cell Lines | ||
| GFP-RPB1 | Laboratory of A.B. Houtsmuller | NA |
| hTERT RPE-1 | Laboratory of B. Chadwick | CVCL_4388 |
| HeLa S3 | Laboratory of P. Kaufman | CVCL_0058 |
| HeLa-Cas9 | Laboratory of I. Cheeseman | NA |
| U2OS (lacO-array) | Laboratory of D. Spector | NA |
| MDA-MB 435 | Laboratory of R.A. Weinberg | CVCL_0417 |
| WAPL-AID (HeLa S3) | This paper | NA |
| Experimental Models: Organisms/Strains | ||
| Xenopus laevis | Nasco | LM00531 |
| Oligonucleotides | ||
| List of oligonucleotides used in this study provided in Table S2 | ||
| Recombinant DNA | ||
| Plasmid: mAID-mCherry-P2A-Neomycin (pMB1152) | This paper | NA |
| Plasmid: WAPL sgRNA in pX330 (pMB1155) | This paper | NA |
| Plasmid: pGK215 | Addgene | Cat#45110 |
| Plasmid: pGK215 (SMC1A; pMB1208) | This paper | NA |
| Plasmid: pGK215 (SMC1A*; pMB1209) | This paper | NA |
| Software and Algorithms | ||
| Statistical analysis and plotting: Prism 7 | Graphpad | NA |
| FRAP data analysis: Fiji | ||
| R | www.r-project.org | NA |
| Bowtie2 | Langmead and Salzberg, 2012 | NA |
| Ngs.plot | Shen et al., 2012 | NA |
| Bedtools | Quinlan and Hall, 2010 | NA |
| Tophat/Cufflinks | Trapnell et al. 2012 | NA |
| Htseq-count | Anders et al., 2015 | NA |
| metaPlotR | Olarerin-George and Jaffrey, 2017 | NA |
| EdgeR | Robinson et al., 2010 | NA |
| MACS | Zhang et al. 2008 | NA |
| Other | ||
| siRNA: WAPL siRNA-SMARTpool | Dharmacon | Cat#23063 |
| siRNA: siGENOME non-targeting siRNA | Dharmacon | Cat#D-001210 |
| DNA staining: Hoechst 33342 | Molecular Probes | Cat#H3570 |
TABLE WITH EXAMPLES FOR AUTHOR REFERENCE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-Snail | Cell Signaling Technology | Cat#3879S; RRID: AB_2255011 |
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat#T9026; RRID: AB_477593 |
| Rabbit polyclonal anti-BMAL1 | This paper | N/A |
| Bacterial and Virus Strains | ||
| pAAV-hSyn-DIO-hM3D(Gq)-mCherry | Krashes et al., 2011 | Addgene AAV5; 44361-AAV5 |
| AAV5-EF1a-DIO-hChR2(H134R)-EYFP | Hope Center Viral Vectors Core | N/A |
| Cowpox virus Brighton Red | BEI Resources | NR-88 |
| Zika-SMGC-1, GENBANK: KX266255 | Isolated from patient (Wang et al., 2016) | N/A |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Streptococcus pyogenes: M1 serotype strain: strain SF370; M1 GAS | ATCC | ATCC 700294 |
| Biological Samples | ||
| Healthy adult BA9 brain tissue | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat#UMB1455 |
| Human hippocampal brain blocks | New York Brain Bank | http://nybb.hs.columbia.edu/ |
| Patient-derived xenografts (PDX) | Children’s Oncology Group Cell Culture and Xenograft Repository | http://cogcell.org/ |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MK-2206 AKT inhibitor | Selleck Chemicals | S1078; CAS: 1032350-13-2 |
| SB-505124 | Sigma-Aldrich | S4696; CAS: 694433-59-5 (free base) |
| Picrotoxin | Sigma-Aldrich | P1675; CAS: 124-87-8 |
| Human TGF-β | R&D | 240-B; GenPept: P01137 |
| Activated S6K1 | Millipore | Cat#14-486 |
| GST-BMAL1 | Novus | Cat#H00000406-P01 |
| Critical Commercial Assays | ||
| EasyTag EXPRESS 35S Protein Labeling Kit | Perkin-Elmer | NEG772014MC |
| CaspaseGlo 3/7 | Promega | G8090 |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE63473 |
| B-RAF RBD (apo) structure | This paper | PDB: 5J17 |
| Human reference genome NCBI build 37, GRCh37 | Genome Reference Consortium | http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/ |
| Nanog STILT inference | This paper; Mendeley Data | http://dx.doi.org/10.17632/wx6s4mj7s8.2 |
| Affinity-based mass spectrometry performed with 57 genes | This paper; and Mendeley Data | Table S8; http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Experimental Models: Cell Lines | ||
| Hamster: CHO cells | ATCC | CRL-11268 |
| D. melanogaster: Cell line S2: S2-DRSC | Laboratory of Norbert Perrimon | FlyBase: FBtc0000181 |
| Human: Passage 40 H9 ES cells | MSKCC stem cell core facility | N/A |
| Human: HUES 8 hESC line (NIH approval number NIHhESC-09-0021) | HSCI iPS Core | hES Cell Line: HUES-8 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain BC4011: srl-1(s2500) II; dpy-18(e364) III; unc-46(e177)rol-3(s1040) V. | Caenorhabditis Genetics Center | WB Strain: BC4011; WormBase: WBVar00241916 |
| D. melanogaster: RNAi of Sxl: y[1] sc[*] v[1]; P{TRiP.HMS00609}attP2 | Bloomington Drosophila Stock Center | BDSC:34393; FlyBase: FBtp0064874 |
| S. cerevisiae: Strain background: W303 | ATCC | ATTC: 208353 |
| Mouse: R6/2: B6CBA-Tg(HDexon1)62Gpb/3J | The Jackson Laboratory | JAX: 006494 |
| Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J | The Jackson Laboratory | RRID: IMSR_JAX:008471 |
| Zebrafish: Tg(Shha:GFP)t10: t10Tg | Neumann and Nuesslein-Volhard, 2000 | ZFIN: ZDB-GENO-060207-1 |
| Arabidopsis: 35S::PIF4-YFP, BZR1-CFP | Wang et al., 2012 | N/A |
| Arabidopsis: JYB1021.2: pS24(AT5G58010)::cS24:GFP(-G):NOS #1 | NASC | NASC ID: N70450 |
| Oligonucleotides | ||
| siRNA targeting sequence: PIP5K I alpha #1: ACACAGUACUCAGUUGAUA | This paper | N/A |
| Primers for XX, see Table SX | This paper | N/A |
| Primer: GFP/YFP/CFP Forward: GCACGACTTCTTCAAGTCCGCCATGCC | This paper | N/A |
| Morpholino: MO-pax2a GGTCTGCTTTGCAGTGAATATCCAT | Gene Tools | ZFIN: ZDB-MRPHLNO-061106-5 |
| ACTB (hs01060665_g1) | Life Technologies | Cat#4331182 |
| RNA sequence: hnRNPA1_ligand: UAGGGACUUAGGGUUCUCUCUAGGGACUUAGGGUUCUCUCUAGGGA | This paper | N/A |
| Recombinant DNA | ||
| pLVX-Tight-Puro (TetOn) | Clonetech | Cat#632162 |
| Plasmid: GFP-Nito | This paper | N/A |
| cDNA GH111110 | Drosophila Genomics Resource Center | DGRC:5666; FlyBase:FBcl0130415 |
| AAV2/1-hsyn-GCaMP6- WPRE | Chen et al., 2013 | N/A |
| Mouse raptor: pLKO mouse shRNA 1 raptor | Thoreen et al., 2009 | Addgene Plasmid #21339 |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Weighted Maximal Information Component Analysis v0.9 | Rau et al., 2013 | https://github.com/ChristophRau/wMICA |
| ICS algorithm | This paper; Mendeley Data | http://dx.doi.org/10.17632/5hvpvspw82.1 |
| Other | ||
| Sequence data, analyses, and resources related to the ultra-deep sequencing of the AML31 tumor, relapse, and matched normal. | This paper | http://aml31.genome.wustl.edu |
| Resource website for the AML31 publication | This paper | https://github.com/chrisamiller/aml31SuppSite |
Highlights.
Mitotic centromere transcription requires cohesin
Prophase cohesin removal releases elongating RNA Pol II and nascent RNA from chromatin
Chromatin release of RNA Pol II/nascent RNA facilitates accurate chromosome segregation
The prophase pathway ensures G2/G1 gene expression reprograming across mitosis
Acknowledgements
We thank Jurgen Marteijn for kindly providing the GFP-RPB1 cell line and Paul Kaufman for providing the HeLa S3 Tir1 cell line. We thank Judith Sharp and Radhika Subramanian for helpful discussions. This work was supported by a grant from the NIH/NIGMS to M.D.B. (GM122893) and support to I.M.C. from the Harold G. & Leila Y. Mathers Charitable Foundation and the NIH/National Institute of General Medical Sciences (R35GM126930), and an American Cancer Society post-doctoral fellowship to L.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests.
The authors declare that they have no competing interests.
References
- Akoulitchev S, and Reinberg D (1998). The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes & development 12, 3541–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji R, Skibbens RV, and Iovine MK (2017). How many roads lead to cohesinopathies? Dev Dyn 246, 881–888. [DOI] [PubMed] [Google Scholar]
- Bellier S, Dubois MF, Nishida E, Almouzni G, and Bensaude O (1997). Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Molecular and cellular biology 17, 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD (2016). Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkov GOM, Gilbert N, and Heun P (2018). Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. The Journal of cell biology 217, 1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, and Peters JM (2017). Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, and Wong LH (2012). Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proceedings of the National Academy of Sciences of the United States of America 109, 1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O’Neill RJ, et al. (2015). Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription. Developmental cell 34, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium S.M.-I. (2014). A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol 32, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, and Singer RH (2007). In vivo dynamics of RNA polymerase II transcription. Nature structural & molecular biology 14, 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbatsh AMO, Haarhuis JHI, Petela N, Chapard C, Fish A, Celie PH, Stadnik M, Ristic D, Wyman C, Medema RH, et al. (2016). Cohesin Releases DNA through Asymmetric ATPase-Driven Ring Opening. Molecular cell 61, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, and Hirano T (2006). Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Current biology : CB 16, 2406–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. (2018). A pathway for mitotic chromosome formation. Science 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullerova M, and Proudfoot NJ (2008). Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132, 983–995. [DOI] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, and Rowland BD (2014). Cohesin and its regulation: on the logic of X-shaped chromosomes. Developmental cell 31, 7–18. [DOI] [PubMed] [Google Scholar]
- Haarhuis JH, Elbatsh AM, van den Broek B, Camps D, Erkan H, Jalink K, Medema RH, and Rowland BD (2013). WAPL-mediated removal of cohesin protects against segregation errors and aneuploidy. Current biology : CB 23, 2071–2077. [DOI] [PubMed] [Google Scholar]
- Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, Krijger PHL, Teunissen H, Medema RH, van Steensel B, et al. (2017). The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell 169, 693–707 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, and Heald R (2006). Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nature protocols 1, 2305–2314. [DOI] [PubMed] [Google Scholar]
- Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, Rajagopalan R, Venditti CP, Gripp K, Samanich J, et al. (2015). Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nature genetics 47, 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, Emerman AB, Schweidenback CT, and Blower MD (2014). RNA stimulates Aurora B kinase activity during mitosis. PloS one 9, e100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. (2004). From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, and Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proceedings of the National Academy of Sciences of the United States of America 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WL, Yewdell WT, Bell JC, McNulty SM, Duda Z, O’Neill RJ, Sullivan BA, and Straight AF (2017). RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, and Peters JM (2006). Wapl controls the dynamic association of cohesin with chromatin. Cell 127, 955–967. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Woodfin AR, Slaughter BD, Unruh JR, Box AC, Rickels RA, Gao X, Haug JS, Jaspersen SL, and Shilatifard A (2015). Mitotic Transcriptional Activation: Clearance of Actively Engaged Pol II via Transcriptional Elongation Control in Mitosis. Molecular cell 60, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Qu Q, Warrington R, Rice A, Cheng N, and Yu H (2015). Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Molecular cell 59, 426–436. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, and Hirano T (2002). ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Molecular biology of the cell 13, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, F CL, Cucco F, Amato C, Quarantotti V, Rizzo IM, Krantz ID, Bilodeau S, and Musio A (2015). Mutant cohesin affects RNA polymerase II regulation in Cornelia de Lange syndrome. Sci Rep 5, 16803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, and Cheeseman IM (2017). Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of p53-Dependent Responses to Cell-Cycle Defects. Developmental cell 40, 405–420 e402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SM, Sullivan LL, and Sullivan BA (2017). Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Developmental cell 42, 226–240 e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J, and Stillman B (2000). Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Molecular and cellular biology 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, and Kent WJ (2014). Centromere reference models for human chromosomes X and Y satellite arrays. Genome research 24, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, and Earnshaw WC (2016). Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nature communications 7, 13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F, Wach P, and McNally JG (2008). Evidence for a common mode of transcription factor interaction with chromatin as revealed by improved quantitative fluorescence recovery after photobleaching. Biophys J 94, 3323–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, and Masumoto H (2008). Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Developmental cell 14, 507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, and Haering CH (2009). Cohesin: its roles and mechanisms. Annual review of genetics 43, 525–558. [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, and Kanemaki MT (2016). Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell reports 15, 210–218. [DOI] [PubMed] [Google Scholar]
- Novais-Cruz M, Alba Abad M, van IWF, Galjart N, Jeyaprakash AA, Maiato H, and Ferras C (2018). Mitotic progression, arrest, exit or death relies on centromere structural integrity, rather than de novo transcription. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarerin-George AO, and Jaffrey SR (2017). MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33, 1563–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palozola KC, Donahue G, Liu H, Grant GR, Becker JS, Cote A, Yu H, Raj A, and Zaret KS (2017). Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358, 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Resa C, and Blower MD (2018). Centromere Biology: transcription goes on stage. Molecular and cellular biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, and Bender MA (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Experimental cell research 26, 260–268. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171, 305–320 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, and Dudoit S (2014). Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic S, Kohler F, and Erhardt S (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. The Journal of cell biology 207, 335–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, and Dorsett D (2013). Genome-wide control of RNA polymerase II activity by cohesin. PLoS genetics 9, e1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, and Heintz N (1996). Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes & development 10, 2389–2400. [DOI] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, and Nestler E (2014). ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, and Hirano T (2009). Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes & development 23, 2224–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, and McNally JG (2004). Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J 86, 3473–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, Chang J, Dealy S, Pothof J, van Cappellen WA, et al. (2018). Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proceedings of the National Academy of Sciences of the United States of America 115, E4368–E4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz SZ, McKay LS, Su KC, Bury L, Padeganeh A, Maddox PS, Knouse KA, and Cheeseman IM (2019). Quiescent Cells Actively Replenish CENP-A Nucleosomes to Maintain Centromere Identity and Proliferative Potential. Developmental cell 51, 35–48 e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. (2013). Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, An L, Bhargava-Shah A, Xie L, Darzacq X, and Tjian R (2018). A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. (2011). XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 7, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T, Tanno Y, and Watanabe Y (2010). Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature 467, 719–723. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, and Nasmyth K (2000). Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375–386. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, and Higgins JM (2010). Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lu JJ, He L, and Yu Q (2011). Triptolide (TPL) inhibits global transcription by inducing proteasome-dependent degradation of RNA polymerase II (Pol II). PloS one 6, e23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, and Peters JM (2006). Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Current biology : CB 16, 863–874. [DOI] [PubMed] [Google Scholar]
- Xiao C, Sharp JA, Kawahara M, Davalos AR, Difilippantonio MJ, Hu Y, Li W, Cao L, Buetow K, Ried T, et al. (2007). The XIST noncoding RNA functions independently of BRCA1 in X inactivation. Cell 128, 977–989. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, and Watanabe Y (2010). Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243. [DOI] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. (2013). Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis ofChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data associated with this study has been deposited in GEO under the Superseries accession: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE139856