Abstract
This year marks the 40th anniversary of the initial identification of p53 as a transformation-related antigen, which was the result of our effort to identify an antigenically-distinct tumor antigen of a chemically-induced mouse tumor and develop a cancer vaccine. Many researchers at the time viewed this effort as folly. Since then, its characterization has progressed from being an attractive cancer vaccine candidate to recognition as a key player in regulating critical pathways controlling the cell cycle and oncogenesis. Advances in molecular immunology and oncology have enhanced the role of p53 in both fields. It is now apparent that p53 plays a critical role in controlling immune recognition and responses in normal tissues as well as the tumor microenvironment. Together with the advances in clinical implementation of p53-based cancer immunotherapy, they highlight the importance of p53 in many areas of basic and translational cancer research.
For many years the related concepts of tumor-specific antigens (TSAs), cancer vaccines and immunotherapy were considered by many cancer researchers to be the fantasies of a few tumor immunologists. At this moment, when cancer immunotherapy is being applied successfully to treat several types of cancer, it is interesting to look back more than forty years to review how p53, one of the most commonly mutated proteins associated with cancer, was initially identified by tumor immunologists in their quest to identify TSAs and ultimately develop cancer vaccines.
The origins of tumor immunology date back to the early half of the 20th century with the establishment of inbred strains of mice. The demonstrated immunogenicity of experimental-, chemical- and viral-induced tumors of inbred mice, namely sarcomas, led to the concept of TSAs. What was remarkable was that each of the chemically-induced tumors, but not the virally-induced tumors, was immunologically distinct. Immunization of mice with one chemically-induced tumor did not confer immunity to another tumor induced by the same chemical in another mouse of the same inbred mouse strain. Thus arose the idea that chemically-induced tumors of inbred mice expressed individual, distinct tumor-specific transplantation antigens (TSTAs). However, the existence of unique or antigenically distinct TSTAs was not accepted unequivocally until Hellstrom et al. demonstrated using an inbred strain of mice, that following surgical removal of a chemically-induced sarcoma from its host, the tumor could be used to immunize its autologous host and protect it from a subsequent tumor challenge that was otherwise lethal to naive mice of the same strain (1). These results strongly supported the concept of TSTAs and their potential use in developing vaccines for immunotherapy of human cancer. From that point on, it was widely appreciated that further development of cancer immunotherapy required the molecular identification of mouse and human TSTAs.
Tumor immunologists sought an approach to identifying the molecular basis of TSTAs of chemically induced tumors in inbred mice. In the early 1970’s, two pioneers in the field of tumor immunology, Lloyd J. Old at Memorial Sloan Kettering Cancer Center (MSKCC) and Lloyd W. Law at the National Cancer Institute (NCI) independently set out to identify the unique TSTA of a chemically-induced mouse tumor, using two distinct approaches, serology and biochemistry, respectively. An important common factor in their approaches was that both laboratories chose to study the same tumor, the BALB/c Meth A sarcoma. The Meth A sarcoma was originally induced by methycholanthrene (MCA) in a BALB/cSKI mouse, which had been derived from BALB/cJ mice bred at MSKCC, by Old in 1958 and subsequently converted by serial passage in mice into a single cell ascites suspension of tumor cells that could be rapidly and serially transplanted in the peritoneal cavity of mice, similar to transplantable lymphoid tumors (2). Due to its ascites form and ease of transplantation, the Meth A sarcoma became the “work horse” of many studies, albeit with the caveat that the original and low passage specimens of the tumor or normal tissues from the host mouse were available, since cryopreserving tumor cells in liquid nitrogen was introduced years later. Consequently, “genetic antigenic drift” was always a potential problem in identifying any highly restricted antigen expressed by Meth A sarcoma.
The serologically defined Meth A antigen
In the early 1970’s, Lloyd Old established the human tumor serology laboratory at MSKCC with the aim of identifying TSAs of human melanoma using patients’ sera. He also continued his interest in the unique TSTAs of chemically-induced tumors of mice and their potential to reveal the nature of human TSAs. This new research effort followed in the footsteps of earlier collaborations with his colleague, Edward (Ted) Boyce at MSKCC. Together, they had serologically defined numerous cell surface alloantigens of lymphoid and tumor origin expressed on mouse immune and leukemic cells, including the Lyt antigens (now known as CD4 and CD8) using suspensions of these cells in rabbit complement-mediated, antibody-dependent cell cytotoxic (ADCC) assays (2). The underlying and prejudicial concept guiding Old’s new effort was that TSTAs were cell surface antigens and in the case of the chemically-induced tumors, the products of a polymorphic gene family similar in nature to the major histocompatibility complex (MHC) antigens and immunoglobulins rather than the product of random genetic or epigenetic events. If the former was found to be the case, identifying one TSTA could elucidate the nature of other TSTAs and facilitate vaccine development. If the latter scenario was the case, each tumor would have to be analyzed to identify its TSTA, presenting significant logistical difficulties in the approach to cancer vaccine development.
In 1974, Albert DeLeo joined Old’s lab at MSKCC. His background was far removed from tumor immunology. He had received his Ph.D. in biochemistry at Columbia University under David Sprinson and had completed a post-doctoral fellowship at MIT under Boris Magasanik, which in part involved using rabbit antisera to identify the structural gene of glutamine synthetase in K. aerogenes. He also had been an instructor in an elective senior undergraduate course given by Phil Robbins that focused on differences in lactic dehydrogenase isoenzyme expression in transformed and non-transformed cell lines. With this background, he became fascinated with the use of antibodies to identify biochemical differences between normal and cancer cells. This interest led him eventually to Lloyd Old.
Given his background, DeLeo’s research project was to serologically identify murine TSTAs using antibodies present in the antisera of mice hyperimmunized to Meth A and, most importantly, to several new chemically-induced sarcomas of BALB/cJ mice. The first step was to establish a panel of monolayer cell lines derived from 20 MCA-induced sarcomas of BALB/cJ mice (the CMS series) with an emphasis on cryopreserving specimens of the original tumors and their early in vivo passages as well as early in vitro-passaged cell lines.
In the years preceding the development of hybridomas and monoclonal antibodies, defining the antigenic specificity of conventionally prepared mouse antisera was the realm of serology and its use of absorption techniques to essentially convert a polyvalent serum into one that was monovalent against a defined target. Using a method initially developed by Eda Bloom in Old’s laboratory (3), a microADCC assay was modified using in vitro-cultured sarcoma cells as targets and fetal rabbit serum as a complement to identify cell surface antigens expressed by the sarcoma cell lines and control, low passage normal lung fibroblasts. Surprisingly, nearly all the antisera, including those defining lymphoid alloantigens, such as the Lyt antigens and several leukemia-associated antigens showed cross-reactivity in the microADCC assay against the sarcomas and late-passage mouse lung fibroblasts, which are spontaneously transformed after several in vitro passages, but are not transformed during early passages. As a result, determining the alloantigen expression on the sarcomas and normal fibroblasts needed to be done in the classical manner by absorption analysis using the antisera and target cells that defined the allogenetic specificities of the antisera. This analysis led to the observation that 1) regardless of the immunogen used, essentially all conventionally prepared mouse antisera contained complement-dependent cytotoxic antibodies recognizing the murine leukemia virus (MuLV) gp70 envelope protein and 2) the BALB/c sarcomas, with the significant exceptions of Meth A and two newly induced sarcomas, CMS4 and CMS5, expressed MuLV and gp70. Interestingly, normal fibroblasts of C57BL/6J but not BALB/cJ origin expressed gp70 in the absence of virion expression. Consequently, cell lines derived from Meth A and two newly induced sarcomas, CMS4 and CMS5, which remained MuLVneg for more than ten in vitro passages, were used for further study.
After demonstrating that Meth A, CMS4 and CMS5 expressed individual, non-cross-reacting distinct TSTAs in in vivo tumor rejection assays, hyperimmune antitumor sera in BALB/cJ mice were prepared. To generate these antisera, mice were immunized subcutaneously a minimum of three times with irradiated in vivo-grown Meth A ascites cells or fragments of CMS4 and CMS5 tumor prior to lethal challenges with non-irradiated, in vivo-grown tumor fragments or cells in order to avoid the use of culture media and trypsin and potential cross reactivity with “tissue culture” antigens. Tumor challenges were repeated with increasing tumor dosages over the course of many months. Throughout this process, sera were obtained from the mice and tested for specificity against the in vitro-grown tumor cell line established from the immunizing tumor in microADCC assays. In this manner, restricted antibody reactivities against Meth A, CMS4 or CMS5 were detected in these antisera, with Meth A antisera having the highest titer, followed by CMS4 and with CMS5 antisera being relatively weak in comparison. The Meth A and CMS4 antisera were extensively analyzed by absorption techniques using panels of cell lines established from other tumors of the CMS series, as well as many established and newly isolated tumors that had been induced with chemical carcinogens, radiation or viruses in BALB/c and other inbred mouse strains and a panel of normal tissues of BALB/c mice. The exciting conclusion of these results, constituting a major breakthrough in the field, was that the Meth A and CMS4 antisera contained two non-cross-reacting, serologically defined unique TSAs that descriptively corresponded to the individually distinct TSTAs of these two chemically-induced sarcomas (4).
The serologically defined Meth A antigen and the identification of p53
The obvious next step was the molecular identification of the serologically defined Meth A and CMS4 TSAs using the hyperimmune antisera. To this end, a collaboration was initiated with Lloyd Law and Ettore Appella at the National Cancer Institute. Lloyd Law was a legend in the field of tumor immunology; Appella was a well-established protein chemist and biochemist. In their approach to identifying the Meth A TSTA, they had developed a method to purify cell-free extracts of tumors and tested purified Meth A fractions for their ability to induce immunity to Meth A in in vivo tumor rejection assays. Unfortunately, absorption of the Meth A antisera with their biochemically purified Meth A fractions did not block the activity of the Meth A antisera in the micro-ADCC assay. The next step, performed by Garrett Dubois, a postdoctoral fellow in Appella’s lab, was immunoprecipitation of radiolabeled Meth A cell-free extracts with the Meth A antisera followed by SDS-PAGE analysis. These assays showed that the antisera recognized a predominant 50 kDa protein present in Meth A as well as in other tumor cell extracts, but not detected in normal fibroblast extracts. To our great disappointment, absorption of the antisera with Meth A sarcoma cells did not block the immunoprecipitating antibodies. We now recognized that we were most likely dealing with two antibody specificities and possibly two distinct molecular entities. At this point, Gilbert Jay, a molecular virologist at NCI, joined us as we expanded the SDS-PAGE analysis, which led to confirmation that the band at ~ 50 kDa in the gels was not an IgG-related artifact, and that antibodies in the Meth A/CMS4 antisera were detecting a 53 kDa protein. We probed a wide range of murine tumors and leukemias of chemical and viral origin as well as spontaneous tumors and normal tissues for the presence of the p53 kDa species. Both antisera yielded similar results, but we exclusively used the Meth A antisera because it was easier to prepare and had a higher titer. Diverting from his past practice of naming serologically defined, but molecularly undefined tumor antigens with the combination of “G” to honor Ludwig Gross, the virologist who identified MuLV, and/or the tumor target used to define the antigen (such as the GIX, GERLD and Meth A antigens), Old suggested that a more molecular description akin to that used with MuLV-related antigens (i.e. gp70, p15, p30) be used for the new antigen. In this manner, the first description of the 53kDa antigen, now designated p53, was reported in 1979 (5). Our main conclusions were that p53 was of cellular origin and its expression was transformation related, regardless of the mechanism of cellular transformation. All the chemical, viral or spontaneously induced transformed mouse cells we tested expressed p53.
The follow-up studies of p53 by our group yielded additional, very important insights into the nature of p53. First, trypsin digestion of p53 from chemically-induced tumors, but not virally-induced tumors showed distinct peptides, implying that genetic alterations in p53, namely mutations, occurred during chemical carcinogenesis, but not viral oncogenesis. Second, the elevated expression of p53 in normal tissues or cells with high proliferative indices, namely testis cells and mitogen-activated lymphocytes, indicated a relationship between p53 expression and proliferation (6). The role of p53 in proliferation was confirmed later in collaboration with Ed Mercer and Renato Baserga at Temple University (Philadephia, PA), when they demonstrated that microinjection of a p53 monoclonal antibody that had been generated at MSKCC by Wolfgang Dippold (7) into non-transformed BALB/c 3T3 cells blocked serum-stimulated cell growth and proliferation (8).
Regarding the serologically defined Meth A antigen, we had to conclude that the IgM defining the tumor-specific Meth A antigen in the micro-ADCC assay had a specificity distinct from that of the immunoprecipitating IgG antibodies recognizing p53 in Meth A and other tumors. We still considered the possibility that p53 might be a polymorphic gene product and that the serologically defined Meth A and CMS4 TSAs were unique determinants expressed on p53 molecules in these tumors. This was at a time, however, when the nature of the T cell receptor and the mechanisms of antigen processing and presentation in the context of class I MHC molecules were relatively unknown. Consequently, our subsequent research efforts were not focused on relating p53 to the elusive nature of highly restricted TSTAs of chemically induced tumors. p53 was later identified as an SV40 large T-antigen binding protein (9-11), a tumor suppressor gene product (12) and a frequently mutated gene in cancer (13). In the ensuing 40 years, each year the complexity and importance of the roles of p53 in oncogenesis and literally all aspects of cell biology intensified (14).
Early efforts to develop p53 peptide-based vaccines
In the 1990’s, the antigenic character of p53 was revisited as the field of T-cell immunology was able to demonstrate how antigenic determinants derived from p53, by then shown to be localized in the nucleus, came to be presented on the cell surface in the context of class I MHC molecules for T cell recognition and potentially function in tumor rejection. Figure 1A shows a timeline for p53-based vaccines, beginning with the identification of the serologically defined Meth A antigen and then forward to the clinical introduction of p53-based immunotherapy. Given the advances in T-cell immunology, Old’s group showed that the Meth A p53 mutation at codon 234 (M to I) could be incorporated as a 9-mer mutant p53232-240 peptide that was able to bind to H-2Kd molecules. A vaccine consisting of the incomplete Freund’s adjuvant and the Meth A mutant p53232-240 peptide was then used to induce H-2Kd-restricted, Meth A mutant p53232-240 peptide-specific cytotoxic T-cell lymphocyte (CTL) cells in mice with subsequent rejection of Meth A sarcoma (15).
Figure 1. Spectrum of p53-based immunotherapy of cancer.
A) Timeline for application of p53 Meth A antigen for cancer immunotherapy. There was a significant delay between the initial discovery in 1979 and its application beginning in 1993. B) A wide spectrum of vaccines has been introduced for clinical use that involve dendritic cells, viruses or chemical adjuvants as vehicles for p53-based immunizations that are aimed at inducing anti-p53 immune responses in cancer patients.
In 1986, DeLeo relocated to the University of Pittsburgh Cancer Institute (UPCI), newly established under the direction of Ronald Herberman, a well-established tumor immunologist who had first characterized NK cells. In the 1990’s, UPCI became a center for the use of dendritic cells (DCs) as antigen-presenting cells for inducing anti-tumor immune responses in murine systems and potential clinical applications as a vehicle for cancer vaccines. In collaboration with Appella, the Meth A p53 mutation at codon 234 was targeted using a DC-based Meth A peptide vaccine. We showed that the vaccine was able to induce in vitro and in vivo Meth A p53232-240 mutant peptide-specific CTL and, furthermore, produce effective immunity against the Meth A sarcoma in BALB/c mice. More interestingly, the DC-based wild-type sequence p53232-240 peptide vaccine also was shown to be able to induce in vitro and in vivo a wild type sequence p53232-240 peptide-specific CTL and immunity to CMS4 sarcoma, which did not express the p53 234 mutation (16). Additional studies focused on various vehicles to deliver the Meth A mutant p53232-240 epitope. These included using a Meth A epitope cDNA plasmid to either coat gold nanoparticles for “gene gun” immunizations or to transfect DCs to produce a cellular-based Meth A cDNA vaccine. In sum, the results of these studies demonstrated the potential translational use of DC-based p53 vaccines to target p53 mutations or non-mutated, wild-type sequence or “self” p53 epitopes for cancer therapy and, perhaps, prevention.
Further studies in our laboratories of p53-based immunotherapy
By the mid-1990’s, several laboratories had identified CTL-defined wild type sequence human p53 peptide epitopes, which made targeting p53 attractive for developing potentially broadly applicable cancer vaccines (Figure 1B). Transferring the murine DC/p53 peptide vaccine findings to a human tumor system, namely, squamous cell carcinoma of the head and neck (SCCHN), we demonstrated the in vitro efficacy of DC pulsed with the HLA-A2-restricted wild type sequence p53264-272 peptide to induce peptide-specific CD8+ T cells from lymphocytes obtained from peripheral blood of some, but not all, HLA-A2+ normal donors and SCCHN patients tested (17). These CTLs recognized HLA-A2+ tumors expressing p53 molecules with mutations outside of the p53264-272 epitope. In vitro, using the autologous SCCHN patient/tumor PCI-13 system available at UPCI, DCs pulsed with the wild type sequence p53264-272 peptide induced anti-PCI-13 CTL from the patient’s lymphocytes as well. However, there was difficulty in in vitro re-stimulation of p53 264-272-specific-CD8+ CTL from lymphocytes of most normal donors and SCCHN patients. To this end, Doug Loftus in Appella’s laboratory designed a panel of variant peptides of the p53264-272 LLGRNSFEV peptide with substitutions of amino acids at various residues. The variant peptides LLGRNTFEV and LLGRNSWEV were determined to be immunogenic using lymphocytes from donors non-responsive to the parental peptide and they induced anti-p53 peptide CTL recognizing HLA-A2+ tumor cells presenting the wild type sequence LLGRNSFEV p53264-272 epitope and making it an ideal candidate for clinical vaccine use. Subsequently, the modified p53264-272 peptide together with the HLA-A2-restricted149-157 peptide and the Th-helper-defined HLA-DR p53 peptide we identified (18) were used in a Phase I clinical trial of a DC-based multiepitope DC/p53 vaccine for treating SCCHN (19). The immunization protocol was determined to be safe and associated with a promising clinical outcome, decreased Treg levels, and modest vaccine-specific immunity.
Immunogenicity and Immunoediting of HLA-A201+-restricted p53 epitopes in human cancer
Several of our studies demonstrated that the HLA-A2 p53264-272 epitope was the target of immunoediting in SCCHN. Specifically, the HLA-A2+ SCCHN patients who had tetramer+ p53264-272 CD8+ T cells or in vitro-inducible p53264-272 CD8+ T lymphocytes in their peripheral blood also had tumors that either expressed mutations within this epitope or at codon 273, thereby evading their T lymphocyte activity (20). In other studies of anti-p53264-272 CTL in SCCHN patients, we determined that patients with a high frequency of p53 tetramer+ CD8+ T cells in their peripheral circulation had a naive phenotype, while those with a lower frequency of p53 tetramer+ CD8+ T cells had mature and terminally differentiated phenotypes, the effector phenotypes. In addition, the frequency of p53 tetramer+ CD8+ T cells negatively correlated with p53 expression and tumor stage. This suggested that the CTLs in responsive patients could have eliminated tumor cells capable of processing and presenting the targeted epitope, resulting in the immunoselection and outgrowth of “epitope-loss” tumors (21).
We also studied the immunogenicity of HLA-2-restricted, CTL-defined p53 epitopes that were identified in tumors of HLA-A2+ SCCHN patients that expressed mutations in non-anchor codon missense mutations. Of the six p53 mutant peptide epitopes studied, the p53217-225 peptide bearing the codon mutation Y220C, that had been further modified by Appella to eliminate disulfide linkages, was found to be immunogenic in vitro and the induced p53 Y220C peptide-specific CTL recognized an HLA-A2+ SCCHN tumor cell line expressing the p53 Y220C mutation (22). Our analysis of p53 mutations detected the p53 Y220C in 6/50 (12%) of HLA‐A2+ SCCHN tumors. This unexpectedly high frequency of the p53 Y220C mutation in HLA‐A2+ SCCHN suggested at the time that a p53 mutation-specific vaccine could be expected to induce anti‐tumor immune responses in these HLA‐A2+ patients and be more widely applicable than envisioned at that time for any given p53 missense mutation.
Concordant with our p53 CTL-based studies and the increasing interest in many laboratories in development and clinical introduction of p53-based vaccines, DeLeo revisited the Meth A sarcoma model. This time the question was whether administering the DC/wild type sequence p53232-240 peptide-based vaccine in the preventive and/or therapy setting could prevent chemical carcinogenesis in mice. The results were very discouraging. They indicated that the DC/p53 peptide vaccine induced anti-p53 CTLs in the mice soon after MCA exposure, but the response deteriorated as tumor growth progressed. This finding is consistent with the demonstrated immunosuppressive effects of MCA as well as progressive tumor growth, and with what we now recognize as the tumor immunosuppressive/pro-inflammatory activities of loss of function p53 as well. Tumor induction was more rapid and their growth rate was higher in the immunized mice than in the control mice. The analysis of the tumors of immunized, MCA-treated mice showed classic evidence of immunoediting. To avoid CTL recognition, the outgrowing tumors either lost the expression of H-2Kd or expressed mutations within the p53232-240 epitope (23).
Past and current approaches to the development of p53-based immunotherapy
A. Peptide-epitope p53 vaccines
The effort to identify the repertoire of p53 peptide epitopes available for vaccine use was pioneered by Melief and co-workers starting from their initial identification in the 1990’s of the human p53274-272 as an HLA-A2-restricted wild type sequence p53 peptide capable of being identified by CD8+ T cells (24). Their efforts have progressed to constructing vaccines using various combinations of overlapping p53 peptides that represented most of the p53 molecules encoded by exons 5–8. Their preclinical and clinical studies have included the more recent clinical use of the p53-SLP vaccine. The clinical-grade peptides (nine peptides 20–30 amino acids in length with an overlap of 5–14 amino acids) were admixed with the adjuvant and the vaccine was administered to metastatic colorectal cancer patients in combination with Pegintron (IFN-α) and chemotherapy (25). They found the combination treatment safe and efficient, as significantly more p53-specific T cells were produced compared to treatment with p53-SLP alone. These results underscore the therapeutic potential of p53 vaccines in combination with immune stimuli. Concordant with this approach was the personalized immunization protocol initiated by Carbone et al. in which vaccines consisting of a patient’s peripheral blood lymphocytes pulsed with 17-mer peptides incorporating the p53 mutations identified in that patient’s tumor were administered and immune responses studied (26).
B. Hot spot p53 mutation-based immunotherapy
Recent preclinical findings at National Cancer Institute in the Rosenberg lab have certainly refocused attention on targeting p53 for cellular-based cancer immunotherapy. From extensive molecular analysis of the T-cell receptor repertoire in tumor infiltrating lymphocytes, a panel of class I and class II MHC-restricted peptide epitopes, defined as “neoantigens”, were identified, which included frequently occurring p53 “hot spot” mutations, such as p53 R175H and p53 Y220C mutations (27). Most importantly, this approach demonstrated that peptides containing these p53 “hot spot” mutations could be processed and presented as MHC complexes by a variety of types of human tumor cells. Their methodology and subsequent findings overcome many of the previous limitations concerning p53 immunotherapy development that were restricted to certain HLA class I specificities and make p53 a much more attractive target than previously envisioned for use in widely applicable cancer vaccines. This latter scenario is certainly exemplified by the recent results by Malekzadeh et al. in developing the potential of adoptive cell therapy that targets p53 hot spot mutations (28). Relative to this point are factors, namely stability, that result from mutations at various sites in p53 that apparently can influence their immune recognition (29, 30).
C. Small molecular weight compounds (SMWCs) to reverse p53 loss of function
In the 1990’s, approaches to neutralizing the loss-of-function of mutated p53 molecules developed using small molecular weight compounds (SMWCs) were first reported. Starting with identification of CP-31398 and followed by PRIMA-1, an extensive panel of SMWCs that directly or indirectly alter mutant p53 molecules and induce tumor cell death have been identified (31). These agents represent an ideal pharmacological approach to a widely applicable and cost-effective p53-based cancer therapy. While obviously circumventing the numerous subtleties confronting p53-based immunotherapy, an underlying caveat of the p53 SMWC therapy is that to date, none of the more extensively studied compounds can absolutely distinguish wild type p53 from mutant p53 molecules. The problem is quite evident from their demonstrated toxicity to activated normal T cells, which express elevated levels of wild type p53, as we observed decades ago. Our own experience in this regard was in evaluating the ability of CP-31398 and PRIMA-1 to enhance p53-based immunotherapy of MCA-treated mice in a prevention and/or therapy setting. The results clearly showed that SMWCs interfered with both the activation of T cells and the induction of anti-tumor immunity (32). Nutlin and its related compounds represent another SMWC class that disrupt the p53–MDM2 interaction, leading to activation of the p53 pathway in normal and tumor cells. As shown in preclinical murine models, nutlin can both block tumor growth and also promote effective anti-tumor immune responses. However, in a dose-dependent manner, it can also be toxic to normal immune cells (33).
D. p53 gene replacement leads to p53-based immunotherapy.
An early focus in the development of p53-based cancer therapy was on gene replacement. Various viral or plasmid vectors, such as adenovirus and oncolytic viruses were modified to incorporate the wild type p53 coding gene to redirect the tumor cell toward senescence and cell death (34-36). The adenoviral/p53 constructs were also incorporated into vaccines, such as DC-based immunization, pioneered by Carbone and colleagues, which were aimed at gene replacement as well as inducing anti-tumor immunity (37). Either administered “naked” or in complex with adjuvants, numerous adenoviral and oncolytic viral constructs have been shown to be clinically safe and are clinically used in China with reasonable success against a wide range of cancers and currently are undergoing extensive advanced trials here in the USA (35,38,39).
A more recent related gene replacement approach involves scL-53, a nanocomplex made of a cationic liposome coated with an anti-transferrin receptor single-chain antibody fragment (scL) that selectively delivers a wild-type human p53 cDNA into tumor but not normal cells via the elevated levels of transferrin receptor on tumors. The scL-53 nanocomplex has been shown to have significant anti-tumor activity in a number of pre-clinical models (40). Of further interest, it also induces anti-tumor immunity. Administration of scL-53 induced a T-cell-mediated immune response but also induced expression of tumor cell PD-L1, which could be neutralized by anti-PD-L1 antibody. The results of recent clinical trials indicated anti-tumor activity in a subset of patients when administered alone or in combination with docetaxel, making scL-53 an exciting new approach to p53-based therapy that incorporates gene replacement and immunotherapy.
New horizons for p53-based cancer immunotherapy
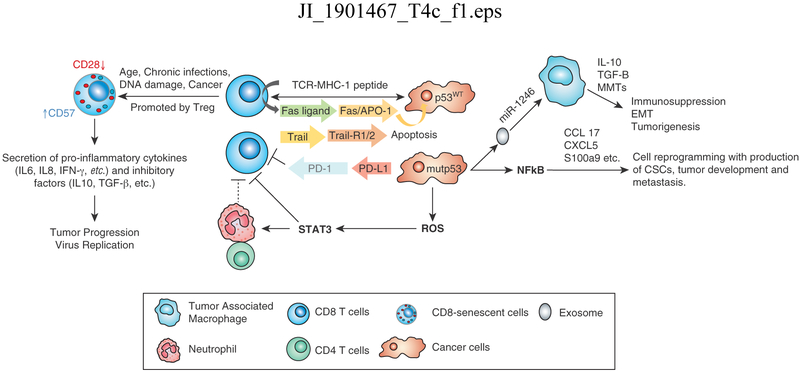
Since its identification as a tumor antigen, immunological-based p53 research has been focused on developing vaccines for immunotherapy. The investigation of the role of p53 in controlling the cell cycle led to the demonstration of its involvement in controlling numerous cellular signaling pathways, including NF-kB, that play key roles in regulating innate and acquired immune responses and controlling inflammation, tolerance and immune suppression (Munoz-Fontana 2016) (41). TLR genes expressed on lymphocytes, dendritic cells and macrophages are p53 targets and changes in their regulation are associated with a range of autoimmune diseases, while p53 also regulates NK cell ligands, influencing their efficacy in anti-tumor response. It is now apparent that wt p53 upregulates the MHC I antigen presentation pathway and DD1α while downregulating PD-1 and the apoptosis receptors Fas/APO-1 and TRAILR1/2 (42-45). In tumor cells lacking p53 or expressing mutp53, MHC I expression is low and PD-L1 is high, resulting in inhibition of the CTL upon binding to the PD-1 receptor. In tumor cells harboring p53mut, upregulation of just the NF-kB signaling pathway, one of many that are influenced by p53, results in increased cytokine production and activation of immune cells. In addition, its activation of ROS induces the Jak-STAT signaling pathway, resulting in reduction of CD8+ T cells plus an increase in immune cell frequencies (46). Furthermore, mutp53 cells shed miR-1246-enriched exosomes with a concomitant uptake by macrophages, triggering an anti-inflammatory immunosuppression state (47). A variety of intracellular and extracellular signals induce senescent immune cells. CD8+CD28− T cells have been identified in multiple tumors and contribute to tumor immune suppression and resistance to immunotherapy through several cytokines and changes in Δ133p53 and p53β isoform expression (48, 49) (Figure 2). Therefore, loss of p53 function not only promotes cellular transformation by altering control of the cell cycle but also facilitates tumor growth by enhancing pro-inflammatory conditions and suppressing components of the innate and adaptive immune systems in the tumor microenvironment. The latter clearly suggests that functional p53 is involved in controlling immune responses; p53-based cancer therapies must maintain a delicate balance in order to promote tumor suppression while concurrently avoiding enhanced autoimmune activities in the tumor microenvironment.
Figure 2. Effects of wild type (wt) p53 and mutant (mut) p53 expressed in tumor cells on anti-tumor immune responses and inflammation.
Wt p53 upregulates MHC class I antigen presentation pathway and apoptosis receptors Fas/APO-1 and TRAILR1/2. In tumor cells lacking p53 or expressing p53mut, expression of MHC class I is low and PD-L1 high resulting in inhibition of the CTL upon binding to PD-1 receptor, whereas p53mut promotes upregulation of the NF-kB signaling, increased cytokine production and immune cell activation. In addition, activation of ROS induces the Jak-STAT signaling pathway, resulting in reduction of CD8+ T cells and increased immune cell frequencies. Furthermore, mut p53 cells shed miR-1246-enriched exosomes with a concomitant uptake by macrophages triggering immunosuppression. A variety of intracellular and extracellular signals induce senescent immune cells. CD8+CD28− T cells contribute to immune suppression and resistance to immunotherapy through several cytokines and changes in Δ133p53 and p53β expression.
Interestingly, is also apparent that the other two members of the p53 gene family, p73 and p63, also are active in tumor suppression. The fact that they interact with p53 and their roles in oncogenesis are not dependent on mutagenesis makes them also potentially attractive targets for cancer interventions, including immunotherapy (50, 51). Given the nearly revolutionary impact that immune checkpoint regulation, which is controlled by p53 is having on the clinical implementation of cancer immunotherapy, p53-based immunotherapy is poised for a dramatic breakthrough in the quest for successful cancer treatments (52).
CONCLUSIONS
This year marks the 40th anniversary of our initial identification of p53 as a transformation-related antigen. Our effort to identify an antigenically distinct tumor antigen from a chemically-induced mouse tumor and develop a cancer vaccine, which by today’s level of technology might seem to have been a somewhat naïve approach, has yielded a wealth of knowledge in many areas of cancer research and cell biology that is still a work in progress. Its characterization has evolved from that of a potential and attractive cancer vaccine candidate to a key gene product in regulating critical pathways controlling cellular proliferation and growth and ultimately oncogenesis. Studies in molecular immunology and oncology over the past 40 years have elucidated the roles of p53 in tumor suppression and regulation of the immune response to oncogenic growth. It is apparent that p53 is centrally important in controlling immune recognition and immune responses in normal tissue as well as the tumor microenviroment. With each passing year it becomes more apparent that development of p53-based therapies is very important in controlling this disease (53, 54). These findings together with advances in the development and clinical implementation of p53-based immunotherapy for cancer have justified the efforts viewed by many in cancer research 40 years ago as a fantasy. The results are beyond any immunologist’s wildest dreams.
Acknowledgements
Many investigators have contributed to various aspects of p53 immunotherapy research and we apologize that some could not be cited or discussed in further detail due to space constraints. We thank George Leiman in the Laboratory of Cell Biology for editorial assistance.
This work was supported, in part, by the Intramural Research Program of the National Institutes of Health, the National Cancer Institute.
Abbreviations used in this article:
- TSA
tumor specific antigen
- TSTA
tumor specific transplantation antigen
- MCA
methycholanthrene
- ADCC
antibody-dependent cytotoxic assay
- MuLV
murine leukemia virus
- SMWC
small molecular weight compound
- scL
single chain antibody fragment
References
- 1.Hellström KE, Hellström I, and Brown JP. 1978. Unique and common tumor-specific transplantation antigens of chemically induced mouse sarcomas. Int. J. Cancer 21: 317–322. [DOI] [PubMed] [Google Scholar]
- 2.Old LJ, Boyse EA, Clarke DA and Carswell E. 1962. Antigenic properties of chemically induced tumors. Ann. N. Y. Acad. Sci 101: 80–106. [Google Scholar]
- 3.Bloom ET 1970. Quantitative detection of cytotoxic antibodies against tumor-specific antigens of murine sarcomas induced by 3-methylcholanthrene. J. Natl. Cancer Inst 45: 443–53. [PubMed] [Google Scholar]
- 4.DeLeo AB, Shiku H, Takahashi T, John M and Old LJ. 1977. Cell surface antigens of chemically induced sarcomas of the mouse. I. Murine leukemia virus-related antigens and alloantigens on cultured fibroblasts and sarcoma cells: description of a unique antigen on BALB/c Meth A sarcoma. J. Exp. Med 146: 720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLeo AB, Jay G, Appella E, Dubois GC, Law LW and Old LJ. 1979. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc. Natl. Acad. Sci. U. S. A 76: 2420–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jay G, DeLeo AB, Appella E, DuBois GC, Law LW, Khoury G and Old LJ. 1980. A common transformation-related protein in murine sarcomas and leukemias. Cold Spring Harbor Sym. Quant. Biol 44: 659–664. [DOI] [PubMed] [Google Scholar]
- 7.Dippold WG, Jay G. DeLeo AB, Khoury G and Old LJ. 1981. p53 transformation-related protein: detection by monoclonal antibody in mouse and human cells. Proc. Natl. Acad. Sci. U. S. A 78: 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer WE, Nelson D, DeLeo AB, Old LJ and Baserga R. 1982. Microinjection of monoclonal antibody to protein p53 inhibits serum-induced DNA synthesis in 3T3 cells. Proc. Natl. Acad. Sci. U. S. A 79: 6309–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linzer DI and Levine AJ. 1979. Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17: 43–52. [DOI] [PubMed] [Google Scholar]
- 10.Lane DP, and Crawford LV. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278: 261–263. [DOI] [PubMed] [Google Scholar]
- 11.Kress M, May E, Cassingena R and May P. 1979. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol 31: 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay CA, Hinds PW and Levine AJ. 1989. The p53 proto-oncogene can act as a suppressor of transformation. Cell 57: 1083–1093. [DOI] [PubMed] [Google Scholar]
- 13.Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hosteller R, Cleary K, Signer SH, Davidson N, Baylin S, Devilee P, et al. 1989. Mutations in the p53 gene occur in diverse human tumour types. Nature 342: 705–708. [DOI] [PubMed] [Google Scholar]
- 14.Levine AJ 2019The many faces of p53: something for everyone. J. Mol. Cell. Biol 11: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi Y, Chen YT and Old LJ. 1994. A mouse mutant p53 product recognized by CD4+ and CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A 91:3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayordomo JI, Loftus DJ, Sakamoto H, De Cesare CM, Appasamy PM, Lotze MT, Storkus WJ, Appella E and DeLeo AB. 1996. Therapy of murine tumors with p53 wild-type and mutant sequence peptide-based vaccines J. Exp. Med 183: 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chikamatsu K, Nakano K, Storkus WJ, Appella E, Lotze MT, Whiteside TL and DeLeo AB. 1999. Generation of anti-p53 cytotoxic T lymphocytes from human peripheral blood using autologous dendritic cells. Clin. Cancer Res 6: 1281–1288. [PubMed] [Google Scholar]
- 18.Ito D, Visus C, Hoffmann TK, Balz V, Bier H, Appella E, Whiteside TL, Ferris RL and DeLeo AB. 2006. The wild-type sequence (wt) p53(25–35) peptide induces HLA-DR7 and HLA-DR11-restricted CD4+ Th cells capable of enhancing the ex vivo expansion and function of anti-wt p53(264–272) peptide CD8+ T cells. J. Immunol 177: 6795–6803. [DOI] [PubMed] [Google Scholar]
- 19.Schuler PJ, Harasymczuk M, Visus C, Deleo A, Trivedi S, Lei Y, Argiris A, Gooding W, Butterfield LH, Whiteside TL, and Ferris RL. 2014. Phase I dendritic cell p53 peptide vaccine for head and neck cancer. Clin. Cancer Res 20: 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann TK, Nakano K, Elder EM, Dworacki G, Finkelstein SD, Appella E, Whiteside TL and DeLeo AB. 2000. Generation of T cells specific for the wild-type sequence p53(264–272) peptide in cancer patients: implications for immunoselection of epitope loss variants. J. Immunol 165: 5938–5944. [DOI] [PubMed] [Google Scholar]
- 21.Albers AE, Qian X, Kaufmann AM, Mytilineos D, Ferris RL, Hoffmann TK and DeLeo AB. 2018. Phenotype of p53 wild-type epitope-specific T cells in the circulation of patients with head and neck cancer. Sci. Rep 8: 10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito D, Visus C, Hoffmann TK, Balz V, Bier H. Appella E, Whiteside TL, Ferris RL and DeLeo AB. 2007. Immunological characterization of missense mutations occurring within cytotoxic T cell-defined p53 epitopes in HLA-A*0201+ squamous cell carcinomas of the head and neck. Int. J. Cancer 120: 2618–2624. [DOI] [PubMed] [Google Scholar]
- 23.Cicinnati VR, Dworacki G, Albers A, Beckebaum S, Tüting T, Kaczmarek E and DeLeo AB. 2005. Impact of p53-based vaccines on primary chemically induced tumors. Int. J.Cancer 113: 961–970. [DOI] [PubMed] [Google Scholar]
- 24.Houbiers JG, Nijman HW, van der Burg SH, Drijfhout JW, Kenemans P, van de Velde CJ, Brand A, Momburg F, Kast WM and Melief CJ. 1993. In vitro induction of human cytotoxic T lymphocyte responses against peptides of mutant and wild type p53. Eur. J. Immunol 23: 2072–2077. [DOI] [PubMed] [Google Scholar]
- 25.Dijkgraaf EM, Santegoets SJ, Reyners AK, Goedemans R, Nijman HW, van Poelgeest MI, van Erkel AR, Smit VT, Daemen TA, van der Hoeven JJ, et al. 2015. A phase 1/2 study combining gemcitabine, Pegintron and p53 SLP vaccine in patients with platinum-resistant ovarian cancer. Oncotarget 6: 32228–32243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carbone DP, Ciernik IF, Kelley MJ, Smith MC, Nadaf S, Kavanaugh D, Maher VE, Stipanov M, Contois D, Johnson BE, et al. 2005. Immunization with mutant p53- and K-ras-derived peptides in cancer patients: immune response and clinical outcome. J. Clin. Oncol 23: 5099–5107. [DOI] [PubMed] [Google Scholar]
- 27.Deniger DC, Pasetto A, Robbins PF, Gartner JJ, Prickett TD, Paria BC, Malekzadeh P, Jia L, Yossef R, M Langhan M, et al. 2018. T-cell responses to TP53 “hotspot” mutations and unique neoantigens expressed by human ovarian cancers. Clin. Cancer Res 24: 5562–5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo NP, et al. 2019. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Invest 129: 1109–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theobald M, Ruppert T, Kuckelkorn U, Hernandez J, Häussler A, Ferreira EA, Liewer U, Biggs J, Levine AJ, Huber C, et al. 1998. The sequence alteration associated with a mutational hotspot in p53 protects cells from lysis by cytotoxic T lymphocytes specific for a flanking peptide epitope. J. Exp. Med 188: 1017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shamalov K, Levy SN, Horovitz-Fried M and Cohen CJ. 2017. The mutational status of p53 can influence its recognition by human T-cells. Oncoimmunology 6: e1285990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanz G, Singh M, Peuget S and Selivanova G. 2019. Inhibition of p53 inhibitors: progress, challenges and perspectives. J. Mol. Cell. Biol 11: 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Liu L, Gomez-Casal R, Wang X, Hayashi R, Appella E, Kopelovich L and DeLeo AB. 2016. Targeting cancer stem cells with p53 modulators. Oncotarget 7: 45079–45093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo G, Miao M, Xiao W, Celis E, and Cui Y. 2017. Local Activation of p53 in the Tumor Microenvironment Overcomes Immune Suppression and Enhances Antitumor Immunity. Cancer Res. 77: 2292–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth J, Dittmer D, Rea D, Tartaglia J, Paoletti E and Levine AJ. 1996. p53 as a target for cancer vaccines: recombinant canarypox virus vectors expressing p53 protect mice against lethal tumor cell challenge. Proc. Natl. Acad. Sci. U. S. A 93: 4781–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang WW, Fang X, Mazur W, French BA, Georges RN, Roth JA. 1994. High-efficiency gene transfer and high-level expression of wild-type p53 in human lung cancer cells mediated by recombinant adenovirus. Cancer Gene Ther. 1: 5–13 [PubMed] [Google Scholar]
- 36.Bressy C, Hastie E and Grdzelishvili VZ. 2017. Combining Oncolytic Virotherapy with p53 Tumor Suppressor Gene Therapy. Mol. Ther. Oncolytics 5: 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keedy V, Wang W, Schiller J, Chada S, Slovis B, Coffee K, Worrell J, Thet LA, Johnson DH and Carbone DP. 2008. Phase I study of adenovirus p53 administered by bronchoalveolar lavage in patients with bronchioloalveolar cell lung carcinoma: ECOG 6597. J. Clin. Oncol 26: 4166–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiappori AA, Williams CC, Gray JE, Tanvetyanon T, Haura EB, Creelan BC, Thapa R, Chen DT, Simon GR, Bepler G, et al. 2019. Randomized-controlled phase II trial of salvage chemotherapy after immunization with a TP53-transfected dendritic cell-based vaccine (Ad. p53-DC) in patients with recurrent small cell lung cancer. Cancer Immunol. Immunother 68: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang WW, Li L, Li D, Liu J, Li X, Li W, Xu X, Zhang MJ, Chandler LA, Lin H, et al. 2018. The First Approved Gene Therapy Product for Cancer Ad-p53 (Gendicine): 12 Years in the Clinic. Hum Gene Ther. 29: 160–179. [DOI] [PubMed] [Google Scholar]
- 40.Moore EC, Sun L, Clavijo PE, Friedman J, Harford JB, Saleh AD, Van Waes C, Chang EH and Allen CT. 2018. Nanocomplex-based TP53 gene therapy promotes anti-tumor immunity through TP53- and STING-dependent mechanisms. Oncoimmunology 7: e1404216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muñoz-Fontela C, Mandinova A, Aaronson SA and Lee SW. 2016. Emerging roles of p53 and other tumour-suppressor genes in immune regulation. Nat. Rev. Immunol 16: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braun MW and Iwakuma T. 2016. Regulation of cytotoxic T-cell responses by p53 in cancer. 2016. Transl. Cancer Res 5: 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu RC, Hwu P, and Radvanyi LG. 2012. New insights on the role of CD8(+) CD57(+) T-cells in cancer. Oncoimmunology 1: 954–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon KW, Byun S, Kwon E, Hwang SY, Chu K, Hiraki M, Jo SH, Weins A, Hakroush S, Cebulla A, et al. 2015. Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science 349:1261669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikuno Uehara I and Tanaka N. 2018. Role of p53 in the Regulation of the Inflammatory Tumor Microenvironment and Tumor Suppression. Cancers 10: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellenstein MD, and de Visser KE. 2018. Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity 48:399–416. [DOI] [PubMed] [Google Scholar]
- 47.Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG and Harris CC. 2018. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun 9: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huff WX, Kwon JH, Henriquez M, Fetcko K and Dey M. 2019. The Evolving Role of CD8+CD28− Immunosenescent T Cells in Cancer Immunology. Int. J. Mol. Sci 20 pii: E2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mondal AM, Horikawa I, Pine SR, Fujita K, Morgan KM, Vera E, Mazur SJ, Appella E, Vojtesek B, Blasco MA, et al. 2013. p53 isoforms regulate aging- and tumor-associated replicative senescence in T lymphocytes. J. Clin. Invest 123: 5247–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Busuttil V, Droin N, McCormick L, Bernassola F, Candi E, Melino G and Green DR. 2010. NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc. Natl. Acad. Sci. U. S. A 107:18061–18066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkatanarayan A, Raulji P, Norton W and Flores ER. 2016. Novel therapeutic interventions for p53-altered tumors through manipulation of its family members, p63 and p73. Cell Cycle 15: 164–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, et al. 2015. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst 108: djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kastenhuber ER and Lowe SW. 2017. Putting p53 in Context. Cell 170: 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeng H, Terabe M and Berzofsky JA. 2018. Cancer vaccines: translation from mice to human clinical trials. Curr. Opin. Immunol 51: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]