This study reports on the characterization of two ceftazidime-avibactam (CZA)-resistant KPC-producing Klebsiella pneumoniae strains (KP-14159 and KP-8788) sequentially isolated from infections occurred in a patient never treated with CZA. Whole-genome sequencing characterization using a combined short- and long-read sequencing approach showed that both isolates belonged to the same ST258 strain, had altered outer membrane porins (a truncated OmpK35 and an Asp137Thr138 duplication in the L3 loop of OmpK36), and carried novel pKpQIL plasmid derivatives (pIT-14159 and pIT-8788, respectively) harboring two copies of the Tn4401a KPC-3-encoding transposon.
KEYWORDS: CAZ-AVI, KPC, Tn4401, avibactam, carbapenem-resistant Enterobacterales, carbapenemase, double copy, porin alterations
ABSTRACT
This study reports on the characterization of two ceftazidime-avibactam (CZA)-resistant KPC-producing Klebsiella pneumoniae strains (KP-14159 and KP-8788) sequentially isolated from infections occurred in a patient never treated with CZA. Whole-genome sequencing characterization using a combined short- and long-read sequencing approach showed that both isolates belonged to the same ST258 strain, had altered outer membrane porins (a truncated OmpK35 and an Asp137Thr138 duplication in the L3 loop of OmpK36), and carried novel pKpQIL plasmid derivatives (pIT-14159 and pIT-8788, respectively) harboring two copies of the Tn4401a KPC-3-encoding transposon. Plasmid pIT-8788 was a cointegrate of pIT-14159 with a ColE replicon (that was also present in KP-14159) apparently evolved in vivo during infection. pIT-8788 was maintained at a higher copy number than pIT-14159 and, upon transfer to Escherichia coli DH10B, was able to increase the CZA MIC by 32-fold. The present findings provide novel insights about the mechanisms of acquired resistance to CZA, underscoring the role that the evolution of broadly disseminated pKpQIL plasmid derivatives may have in increasing the blaKPC gene copy number and KPC-3 expression in bacterial hosts. Although not self-transferable, similar elements, with multiple copies of Tn4401 and maintained at a high copy number, could mediate transferable CZA resistance upon mobilization.
INTRODUCTION
Carbapenemase-producing Enterobacterales (CPE) have undergone a rapid and global dissemination worldwide and currently represent a major public health problem due to their difficult-to-treat resistance phenotypes (1).
The urgent need for new agents with anti-CPE activity has partially been addressed by the recent introduction of novel β-lactam/β-lactamase inhibitor combinations, among which ceftazidime-avibactam (CZA) was the first to be released for clinical use (2). Avibactam inhibits KPC and OXA-48 carbapenemases, and CZA has represented a major breakthrough for treating infections caused by CPE producing these enzymes (3). However, acquired resistance to CZA has been reported among KPC-producing strains of Klebsiella pneumoniae (KPC-Kp), either following treatment with the drug or even in the absence of previous exposure (4–9). In these cases, resistance was associated with different mechanisms, including missense mutations in the KPC enzyme (e.g., D179Y, L169P, T243M, EL165-166, V240G/A, and H274Y), alterations of the OmpK35 and OmpK36 porins, upregulation of the AcrAB efflux system, and increased expression of KPC- or even SHV-type β-lactamases (10, 11).
We report here on the characterization of two CZA-resistant KPC-Kp strains isolated from a patient in the absence of previous exposure to the drug; these strains carried a nonfunctional OmpK35 and an altered OmpK36 with a Asp-Thr duplication (positions 137 and 138) in the L3 loop and overproduced KPC-3 due an increased blaKPC gene dosage mediated by pKpQIL-like plasmid derivatives harboring a duplicated Tn4401 transposon and maintained at a higher copy number compared to pKpQIL.
RESULTS AND DISCUSSION
Sources of the CZA-resistant KPC-Kp isolates.
The two CZA-resistant KPC-Kp investigated here were isolated at the beginning of 2017 in Italy (from a region where KPc-Kp has been endemic since 2010 [12, 13]) from a kidney transplant recipient. In the early posttransplantation course, the patient experienced a urinary tract infection (UTI) caused by a KPC-Kp (isolate KP-14519) which exhibited a pan-drug-resistant phenotype according to the laboratory testing system (Vitek-2 AST-N202 card, interpreted with EUCAST v7.1 breakpoints). The patient was treated with a double-carbapenem (meropenem plus ertapenem) therapy but subsequently developed a breakthrough bloodstream infection (BSI) caused by a KPC-Kp (isolate KP-8788) with the same resistance profile as KP-14519. Despite an increased dosage of the double-carbapenem therapy, bacteremia persisted, and the infection was cleared only after source control (a double nephrectomy) and the addition of tigecycline to the treatment regimen.
Antimicrobial susceptibility of the CZA-resistant KPC-KP isolates.
Antimicrobial susceptibility testing, using reference broth microdilution, confirmed resistance to carbapenems and other β-lactams and revealed resistance also to CZA even though the patient had never been treated with this drug, which had not been available in the hospital formulary at the time of infection. The isolates were also resistant to fluoroquinolones, aminoglycosides, trimethoprim-sulfamethoxazole, and colistin (see Table S1 in the supplemental material).
Characterization of the KP-14519 and KP-8788 isolates.
To investigate the CZA resistance mechanism, both isolates were subjected to whole-genome sequencing (WGS) analysis using a combined short- and long-read sequencing approach. With KP-14519, the hybrid assembly of sequencing data resulted in a draft chromosome plus three circular molecules represented by three plasmids: pKPN-IT-14519, pIT-14519, and ColE-14519. With KP-8788 the hybrid assembly of sequencing data resulted in three circular molecules represented by the chromosome and the two plasmids pKPN-IT-8788 and pIT-8788 (Table 1 , Fig. 1).
TABLE 1.
Features of the K. pneumoniae KP-8788 and KP-14159 plasmidome
| Strain | Plasmid | Size (bp) | Replicon | Resistance genes | Highly related element(s)a (accession no./% query/% identity) |
|---|---|---|---|---|---|
| KP-14159 | pKPN-IT-14159 | 271,781 | IncFIIK7-IncFIBk | aph(3′)-Ia, aadA2, aac(3)-IIa, aac(6′)-Ib3, blaOXA-1, catA1, catB3, dfrA12, dfrA14, qnrB1, sul1 | pKPN-IT-8788 (CP037929/100/99); pKPN-a68 (CP009777/75/100) |
| pIT-14159 | 88,674 | IncFIBk | blaKPC-3, blaKPC-3 | pKpQIL-UK (KY798507/99/100); pIT-FIPP-1 (HG969999/99/100); pKpQIL-234 (KJ146689/99/100) | |
| ColE-14519 | 14,708 | ColE | aac(6′)-Ib | ColE-LS6 (JX442973/100/100) | |
| KP-8788 | pKPN-IT-8788 | 271,781 | IncFIIK7-IncFIBk | aph(3′)-Ia, aadA2, aac(3)-IIa, aac(6′)-Ib3, blaOXA-1, catA1, catB3, dfrA12, dfrA14, qnrB1, sul1 | pKPN-IT-14159 (MN922301/100/99); pKPN-a68 (CP009777/75/100) |
| pIT-8788 | 102,851 | IncFIBk-ColE | blaKPC-3, blaKPC-3, blaTEM-1, aac(6′)-Ib | pKpQIL-UK (KY798507/87/100); pIT-FIPP-1 (HG969999/87/100); pKpQIL-234 (KJ146689/87/100) |
BLAST results indicate the coverage (%) of the query sequence and the corresponding identity (%) at nucleotide level.
FIG 1.
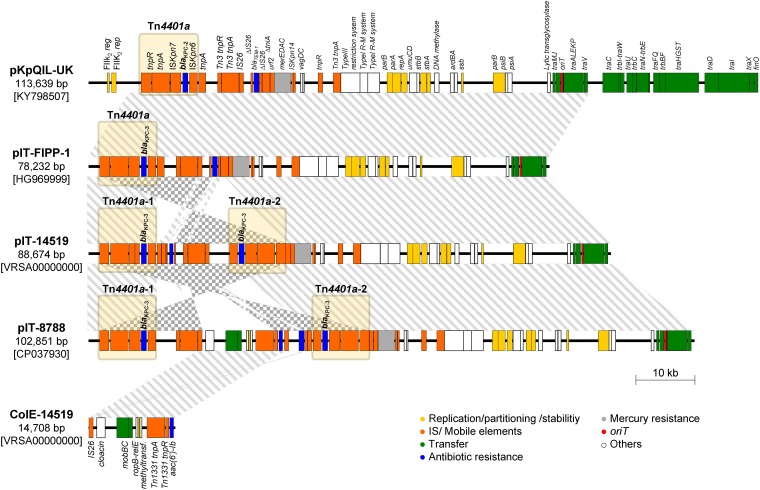
Linear map of pIT-14519 and pIT-8788, two novel KPC-encoding pKpQIL derivative plasmids. A comparison with most similar elements (i.e., pKpQIL-UK and pIT-FIPP-1) is also shown, together with the linear map of ColE-14159 plasmid. The straight lines or the checkered pattern between plasmids indicate a sequence identity ≥99% in the same or in the opposite orientation, respectively. Genes are represented as rectangles with a color-based legend according to their function. Elements belonging to the Tn4401a transposon are grouped with a light yellow rectangle.
In silico analysis of the KP-14519 and KP-8788 genomes confirmed the identification as K. pneumoniae sensu stricto and revealed that both isolates belonged to the clade II lineage of ST258 (14). Alignment of KP-14519 short reads against the chromosome of KP-8788 revealed that the two isolates differed by only a single nucleotide polymorphism (SNP) in the rseB gene, causing a missense mutation (Lys56Glu) in the σE factor regulatory protein.
Investigation of the major porins status showed that (i) OmpK35 was truncated (AA89-STOP) due to a frameshift mutation that was commonly observed among ST258 isolates (15, 16) and (ii) OmpK36 was altered by a two-amino-acid duplication (Asp137Thr138) within the transmembrane β-strand loop 3 (L3), which was previously associated with decreased susceptibility to ertapenem and meropenem (17, 18). Interestingly, this alteration is proximal to the Gly134Asp135 duplication, which was commonly detected among ST258 isolates (15, 16, 19) and shown to result in pore constriction associated with reduced susceptibility to various antimicrobial agents, including CZA (19). Therefore, it is likely that the OmpK36Asp137Thr138 could also result in pore constriction and contribute to decreased CZA susceptibility.
The chromosome of the two isolates also carried (i) a blaSHV-11 gene, typically resident in this K. pneumoniae lineage; (ii) mutations leading to amino acid substitutions in the topoisomerase IV ParC subunit (Ser80Ile) and in the DNA gyrase GyrA subunit (Ser83Ile), known to be associated with fluoroquinolone resistance; and (iii) a nonfunctional mgrB gene due to an original deletion (Δ110/119) similar to a previously described alteration associated with colistin resistance (20).
Plasmidome of KP-14519.
KP-14519 carried three plasmids: pKPN-IT-14519, pIT-14519, and ColE-14519.
Sequence analysis of pKPN-IT-14519 revealed that it was a 271,781-bp IncFIIK7-IncFIBk replicon carrying genes associated with resistance to aminoglycosides [aph(3′)-Ia, aadA2, aac(3)-IIa, aac(6′)-Ib3], β-lactams (blaOXA-1), chloramphenicol (catA1 and catB3), sulfonamides (sul1), trimethoprim (dfrA12 and dfrA14), and fluoroquinolones (qnrB1). This plasmid was similar to other plasmids previously detected in ST258 KPC-Kp strains (Table 1, Fig. 1).
Plasmid pIT-14519 was an 88,674-bp IncFIBk replicon highly similar (>99% identity over 99% of the sequence length) to previously characterized pKpQIL-like KPC-encoding plasmids from ST258 KPC-Kp strains, including pIT-FIPP-1 from the Italian ST258 KPC-Kp index strain FIPP-1 (21) (Table 1, Fig. 1). These plasmids are deletion derivatives of the archetypal IncFIIK2 KPC-encoding plasmid pKpQIL (22) lacking parts of the FIIK2 replicon and of the tra locus (Fig. 1). Compared to the above pKpQIL derivatives, however, pIT-14159 was missing blaTEM-1, while it carried an additional copy of Tn4401a (named Tn4401a-2) located within the ΔtniA gene (Fig. 1), which could contribute to increase the blaKPC-3 copy number in the bacterial host. Indeed, evaluation of the blaKPC copy number and of carbapenemase specific activity in a crude cell extract revealed higher values in K. pneumoniae KP-14519 versus those of K. pneumoniae FIPP-1, which correlated with the higher resistance level to MEM, CAZ, and CZA of the former strain (Table 2).
TABLE 2.
Comparison between meropenem, ceftazidime, ceftazidime-avibactam MICs, blaKPC fold changes, and normalized MEM-hydrolyzing activities against various strainsa
| Strain | ST | No. of blaKPC-3 copies/plasmid | blaKPC-carrying plasmid replicon(s) | MIC (μg/ml) |
Mean blaKPC fold change ± SD | Mean normalized MEM sp act ± SDb | Porin status |
Source or reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | CAZ | CZA | OmpK35 | OmpK36 | |||||||
| K. pneumoniae | |||||||||||
| KP-14519 | 258 | 2 | FIB | 1,024 | 4,096 | 32 | 3.47 ± 0.24 | 2.3 ± 0.7 | AA89-STOP | Asp135, Thr136 | This study |
| KP-8788 | 258 | 2 | FIB-ColE | 2,048 | >4,096 | 64 | 6.89 ± 0.26 | 4.4 ± 0.9 | AA89-STOP | Asp135, Thr136 | This study |
| FIPP-1 | 258 | 1 | FIB | 512 | 1,024 | 8 | Refc | Ref | AA122-STOP | Disrupted by IS5-like | 21 |
| E. coli | |||||||||||
| DH10B(pIT-8788) | NAd | 2 | FIB-ColE | 32 | 4,096 | 8 | 6.18 ± 0.16 | 8.1 ± 1.5 | NA | NA | This study |
| DH10B(pIT-FIPP-1) | NA | 1 | FIB | 4 | 128 | 1 | Ref | Ref | NA | NA | 21 |
| DH10B(pIT-01C22) | NA | 1 | FIIK2-FIB | 1 | 128 | 0.5 | –2.00 ± 0.69 | 0.60 ± 0.2 | NA | NA | 21 |
| DH10B | NA | NA | NA | ≤0.125 | 0.25 | 0.25 | NA | NA | NA | NA | Invitrogen |
Comparisons were determined between meropenem (MEM), ceftazidime (CAZ), and ceftazidime-avibactam (CAZ-AVI; 4 μg/ml) MICs, blaKPC fold changes (reported as log2-transformed values), and normalized MEM-hydrolyzing activity against the following strains: (i) K. pneumoniae FIPP-1 harboring plasmid pIT-FIPP-1, highly similar to plasmid pIT-14159 from K. pneumoniae KP-15159 and to p IT-8788 from K. pneumoniae KP-8788, showing a similar porin’s genetic background, and (ii) E. coli DH10B carrying pIT-8788 or comparator pKpQIL-like plasmids (i.e., pIT-FIPP-1 or pIT-001C22).
Evaluation of the MEM-hydrolyzing activity was performed on cell crude extracts. A normalization (ratio) of the enzymatic specific activity was performed against a reference (Ref) strain according to the bacterial species.
Ref, reference. K. pneumoniae FIPP-1 and E. coli DH10B(pIT-FIPP-1) were used as references for the normalization of K. pneumoniae isolates and E. coli DH10B transformants, respectively.
NA, not applicable.
Plasmid ColE-14519 was a 14,708-bp replicon of the ColE family, carrying an aac(6′)-Ib aminoglycoside resistance determinant (Fig. 1) and was identical to plasmids commonly detected among ST258 isolates (15, 16) (Table 1).
Plasmidome of KP-8788.
KP-8788 carried two plasmids: pKPN-IT-8788 and pIT-8788. The former was virtually identical to pKPN-IT-14519, differing only by a synonymous substitution in the Tn3 transposase gene (Table 1). The latter plasmid was a 102,851-bp IncFIBk-ColE multireplicon apparently resulting from the cointegration of pIT-14519 and ColE-14519 derived from an IS26-mediated homologous recombination event. The other major differences between pIT-14519 and pIT-8788 consisted of the inversion of the backbone segment between the proximal IRs of Tn4401a-1 and Tn4401a-2 and the presence of a blaTEM-1 β-lactamase gene in pIT-8788 (Fig. 1).
Interestingly, evaluation of the blaKPC copy number and of carbapenemase specific activity in a crude cell extract revealed higher values for K. pneumoniae KP-8788 versus those of K. pneumoniae KP-14519 and K. pneumoniae FIPP-1, which correlated with the higher resistance level to MEM, CAZ, and CZA exhibited by the former strain (Table 2). Altogether, these results suggested that cointegration of the ColE-plasmid with the pKpQIL derivative resulted in a higher plasmid copy number compared to pIT-14519.
To further evaluate the potential role of different pKpQIL derivatives in affecting CZA susceptibility, an Escherichia coli DH10B transformant carrying pIT-8788 was constructed and compared to previously obtained E. coli DH10B strains carrying plasmids pIT-FIPP-1 (a pKpQIL derivative with partial deletion of FIIK2 replicon and of the tra locus and carrying a single copy of Tn4401) and pIT-01C22 (an original pKpQIL plasmid carrying a single copy of Tn4401) (21). Evaluation of the blaKPC copy number, carbapenemase expression, and antibiotic resistance in these strains revealed that pIT-8788 was associated with higher blaKPC copy numbers, carbapenemase specific activities, and resistance levels to MEM, CAZ, and CZA in comparison to strains carrying the other plasmids (Table 2). Although DH10B(pIT-8788) was still susceptible to CZA, the MIC (8 μg/ml) was close to the resistance breakpoint and 8- to 16-fold higher than those of the DH10B strains carrying the other plasmids. Altogether, these findings supported a significant role of pIT-8788 in conferring CZA resistance.
Conjugal transfer experiments from either K. pneumoniae KP-8788 or E. coli DH10B(pIT-8788) to an E. coli J53 recipient were unsuccessful, consistent with the partial deletion of the tra locus in pIT-8788. However, the presence of a complete transfer origin (oriT) within the IncFIIK plasmid backbone and of mob genes provided by the ColE element potentially make pIT-8788 mobilizable by helper plasmids. Further experiments are needed to further evaluate the transferability potential of pIT-8788.
Concluding remarks.
Characterization of KP-14519 and KP-8788, two CZA-resistant KPC-Kp belonging to the same ST258 clade II clonal lineage isolated from infections occurred in a kidney transplant recipient, provided some new insights into the mechanisms of acquired CZA resistance in KPC-Kp.
As expected, given the lack of previous exposure to the drug, no mutations known to be associated with CZA resistance were detected in the KPC enzyme expressed by these isolates. Resistance to CZA was apparently related with alterations of the major porins OmpK35 and OmpK36, in combination with overproduction of the KPC-3 enzyme due to increased blaKPC gene dosage. Although similar mechanisms have already been reported to be associated with reduced susceptibility to CZA (11, 23), some findings with the studied isolates are original. In particular, the increased blaKPC gene dosage in this case was related to the presence of a duplicated Tn4401a carried on single pKpQIL-like plasmids (pIT14519 and pIT-8788), while previous studies reported the presence of the gene located on two different plasmids or the presence of multiple copies of blaKPC without a complete characterization of the genetic supports (11, 23, 24). A similar phenomenon, which to our best knowledge was previously reported in mutants selected in vitro with meropenem-vaborbactam (25), is a matter of concern since these plasmids could represent a mechanism of transferable CZA resistance. In fact, although the plasmids were not self-transferable due to a partial deletion of the tra locus, they could be mobilized in the presence of helper plasmids, such as other IncF-type plasmids (i.e., carrying a highly similar transfer locus) which are widely detected among successful clones of K. pneumoniae (e.g., ST512) (15, 16). This is particularly worrisome in an epidemiological context where K. pneumoniae clones carrying alterations of the OmpK35 and/or OmpK36 porins (such as those of clonal group 258, ST11, ST15, and ST395) are frequent (15, 16). In fact, these clones may represent a genetic background potentially conducive to the emergence of clinical resistance to CZA upon transfer of similar plasmids.
Finally, our findings also revealed a further mechanism to increase the resistance level mediated by KPC-encoding plasmids, consisting in the cointegration of a pKpQIL-like derivative carrying the duplicated Tn4401a with a ColE replicon which resulted in a mosaic plasmid maintained at an even higher copy number. Since this plasmid cointegrate was found in an isolate obtained following failure of the double-carbapenem treatment, it could be speculated that it might have been selected in vivo following the exposure to carbapenems, to which the patient was subjected.
MATERIALS AND METHODS
Bacterial isolates and in vitro susceptibility testing.
K. pneumoniae KP-14519 and KP-8788 were isolated from a urine culture and a blood culture, respectively, of a patient who had received a kidney transplant. Bacterial identification was carried out using matrix-assisted laser desorption ionization–time of flight mass spectrometry (bioMérieux, Marcy l’Etoile, France) and confirmed by WGS data. Confirmatory antimicrobial susceptibility testing was carried out with broth microdilution using lyophilized custom plates (Micronaut-S MHK; Merlin Diagnostika GmbH, Berlin, Germany) and interpreted according to the EUCAST clinical breakpoints (EUCAST breakpoint tables version 10.0, 2020 [www.eucast.org/clinical_breakpoints/]). Broad-range concentrations of MEM (0.125 to 4096 μg/ml), CAZ (0.125 to 4,096 μg/ml), and CZA (CAZ, 0.0625 to 128 μg/ml, with avibactam at the fixed concentration of 4 μg/ml) were tested using reference broth microdilution according to CLSI (M100) guidelines (26) and antibiotic powders obtained from Sigma-Aldrich (St. Louis, MO).
Whole-genome sequencing and bioinformatics.
Total bacterial DNA was isolated from KP-8788 and KP-14519 using the phenol-chloroform method (27) and subjected to WGS on an Illumina MiSeq platform (Illumina, San Diego, CA), as previously described (28). WGS was also performed by the Oxford Nanopore MinION system (Oxford Nanopore Technologies, Oxford, UK), and de novo hybrid assemblies were generated using Unicycler v0.4.6 (29). Genome annotation and comparisons, as well as sequences physical maps, were performed as previously described (21, 28). Annotation of IS elements was performed using the ISFinder database (http://www-is.biotoul.fr/). In silico identification of antimicrobial resistance genes, plasmid replicons, and bacterial and plasmid sequence types (ST) were carried out using dedicated tools available at the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). SNPs on core genome were evaluated using snippy (https://github.com/tseemann/snippy), as previously described (30).
Gene transfer experiments.
Electrotransformation experiments were carried out using competent E. coli DH10B (Invitrogen; Waltham, MA) and K. pneumoniae KP-8788 total DNA. Transformants were selected on Mueller-Hinton agar (MHA) supplemented with 8 μg/ml ceftazidime. Transfer of blaKPC gene was always confirmed by real-time PCR (31). Conjugal transfer experiments were carried out as previously described, using K. pneumoniae KP-8788 as donor and both E. coli MKD-135 (rifampin resistant) and E. coli J53 (sodium azide resistant) as recipients (32). Transconjugants were selected on MHA supplemented with 8 μg/ml ceftazidime plus 100 μg/ml rifampin (E. coli MKD-135) or 150 μg/ml sodium azide (E. coli J53).
Evaluation of carbapenemase activity.
Meropenem-hydrolyzing specific activity (used as proxy for KPC expression) was determined by a spectrophotometric assay on cell crude extracts as previously described (33).
Evaluation of blaKPC copy number.
The blaKPC copy number was evaluated by a relative qPCR quantification, using the ΔΔCT method (34). For each isolate, the CT value of blaKPC was subtracted from the CT value of rpsL (ΔCT), that was used as the internal control (20). The fold change (2–ΔΔCT) was calculated using K. pneumoniae FIPP-1 and E. coli DH10B(pIT-FIPP-1) as reference strains. Results were reported as log2-transformed values of the fold change.
Data availability.
All genome sequences related to this study have been registered under the BioProject number PRJNA526630. The complete genome sequences of K. pneumoniae KP-8788 and plasmids pKPN-IT-8788 and pIT-8788 were deposited under GenBank accession numbers CP037928, CP037929, and CP037930, respectively. The genome sequences of K. pneumoniae KP-14519, including the complete circular plasmids pKP-IT-14159 and ColE-14159, were deposited under WGS accession number VRSA00000000. The complete sequence of pKPN-IT-14159 was deposited under GenBank accession number MN922301.
Supplementary Material
ACKNOWLEDGMENTS
The results of this work were partially presented at the 46th National Congress of the Italian Society of Microbiology, September 2018, in Palermo, Italy (poster P031).
This study was partially supported by a research grant (PRIN) funded by the Italian Ministry of Education, University, and Research (20177J5Y3P_004).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 21:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shirley M. 2018. Ceftazidime-avibactam: a review in the treatment of serious Gram-negative bacterial infections. Drugs 78:675–692. doi: 10.1007/s40265-018-0902-x. [DOI] [PubMed] [Google Scholar]
- 3.Papp-Wallace KM. 2019. The latest advances in β-lactam/β-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert Opin Pharmacother 20:2169–2184. doi: 10.1080/14656566.2019.1660772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. 2018. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother 62:e02497-17. doi: 10.1128/AAC.02497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann AC. 2017. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. 2017. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother 61:e02534-16. doi: 10.1128/AAC.02534-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. doi: 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galani I, Antoniadou A, Karaiskos I, Kontopoulou K, Giamarellou H, Souli M. 2019. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin Microbiol Infect 25:763.e5–763.e8. doi: 10.1016/j.cmi.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Wang J, Wang R, Cai Y. 2019. Resistance to ceftazidime-avibactam and underlying mechanisms. J Glob Antimicrob Resist doi: 10.1016/j.jgar.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Nelson K, Hemarajata P, Sun D, Rubio-Aparicio D, Tsivkovski R, Yang S, Sebra R, Kasarskis A, Nguyen H, Hanson BM, Leopold S, Weinstock G, Lomovskaya O, Humphries RM. 2017. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother 61:e00989-17. doi: 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, AMCLI-CRE Survey Participants, Pantosti A, Pagani L, Luzzaro F, Rossolini GM. 2013. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill 18:20489 https://www.eurosurveillance.org/content/10.2807/ese.18.22.20489-en. [PubMed] [Google Scholar]
- 13.Rossolini GM. 2015. Extensively drug-resistant carbapenemase-producing Enterobacteriaceae: an emerging challenge for clinicians and healthcare systems. J Intern Med 277:528–531. doi: 10.1111/joim.12350. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, Limbago BM. 2015. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, EuSCAPE Working Group, ESGEM Study Group, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Fernández A, Miriagou V, Papagiannitsis CC, Giordano A, Venditti M, Mancini C, Carattoli A. 2010. An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother 54:4178–4184. doi: 10.1128/AAC.01301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wise MG, Horvath E, Young K, Sahm DF, Kazmierczak KM. 2018. Global survey of Klebsiella pneumoniae major porins from ertapenem non-susceptible isolates lacking carbapenemases. J Med Microbiol 67:289–295. doi: 10.1099/jmm.0.000691. [DOI] [PubMed] [Google Scholar]
- 19.Wong JLC, Romano M, Kerry LE, Kwong HS, Low WW, Brett SJ, Clements A, Beis K, Frankel G. 2019. OmpK36-mediated carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat Commun 10:3957. doi: 10.1038/s41467-019-11756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group . 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papagiannitsis CC, Di Pilato V, Giani T, Giakkoupi P, Riccobono E, Landini G, Miriagou V, Vatopoulos AC, Rossolini GM. 2016. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J Antimicrob Chemother 71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 22.Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2010. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaibani P, Re MC, Campoli C, Viale PL, Ambretti S. 2019. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: epidemiology and genomic characterization. Clin Microbiol Infect doi: 10.1016/j.cmi.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Zhang P, Shi Q, Hu H, Hong B, Wu X, Du X, Akova M, Yu Y. 2020. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect 26 doi: 10.1016/j.cmi.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Sun D, Rubio-Aparicio D, Nelson K, Dudley MN, Lomovskaya O. 2017. Meropenem-vaborbactam resistance selection, resistance prevention, and molecular mechanisms in mutants of KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 61:e01694-17. doi: 10.1128/AAC.01694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. CLSI supplement M100S Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 27.Sambrook J, MacCallum P, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 28.Giani T, Arena F, Pollini S, Di Pilato V, D’Andrea MM, Henrici De Angelis L, Bassetti M, Rossolini GM, Pseudomonas aeruginosa Working Group . 2018. Italian nationwide survey on Pseudomonas aeruginosa from invasive infections: activity of ceftolozane/tazobactam and comparators, and molecular epidemiology of carbapenemase producers. J Antimicrob Chemother 73:664–671. doi: 10.1093/jac/dkx453. [DOI] [PubMed] [Google Scholar]
- 29.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arena F, Di Pilato V, Vannetti F, Fabbri L, Antonelli A, Coppi M, Pupillo R, Macchi C, Rossolini GM. 2020. Population structure of KPC carbapenemase-producing Klebsiella pneumoniae in a long-term acute-care rehabilitation facility: identification of a new lineage of clonal group 101, associated with local hyperendemicity. Microb Genom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coppi M, Antonelli A, Giani T, Spanu T, Liotti FM, Fontana C, Mirandola W, Gargiulo R, Barozzi A, Mauri C, Principe L, Rossolini GM. 2017. Multicenter evaluation of the RAPIDEC CARBA NP test for rapid screening of carbapenemase-producing Enterobacteriaceae and Gram-negative nonfermenters from clinical specimens. Diagn Microbiol Infect Dis 88:207–213. doi: 10.1016/j.diagmicrobio.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Riccobono E, Di Pilato V, Di Maggio T, Revollo C, Bartoloni A, Pallecchi L, Rossolini GM. 2015. Characterization of IncI1 sequence type 71 epidemic plasmid lineage responsible for the recent dissemination of CTX-M-65 extended-spectrum β-lactamase in the Bolivian Chaco region. Antimicrob Agents Chemother 59:5340–5347. doi: 10.1128/AAC.00589-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. 2013. FIM-1, a new acquired metallo-β-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob Agents Chemother 57:410–416. doi: 10.1128/AAC.01953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome sequences related to this study have been registered under the BioProject number PRJNA526630. The complete genome sequences of K. pneumoniae KP-8788 and plasmids pKPN-IT-8788 and pIT-8788 were deposited under GenBank accession numbers CP037928, CP037929, and CP037930, respectively. The genome sequences of K. pneumoniae KP-14519, including the complete circular plasmids pKP-IT-14159 and ColE-14159, were deposited under WGS accession number VRSA00000000. The complete sequence of pKPN-IT-14159 was deposited under GenBank accession number MN922301.