Antimalarial drugs have long half-lives, so clinical trials to monitor their efficacy require long periods of follow-up to capture drug failure that may become patent only weeks after treatment. Reinfections often occur during follow-up, so robust methods of distinguishing drug failures (recrudescence) from emerging new infections are needed to produce accurate failure rate estimates. Molecular correction aims to achieve this by comparing the genotype of a patient’s pretreatment (initial) blood sample with that of any infection that occurs during follow-up, with matching genotypes indicating drug failure.
KEYWORDS: Bayesian, efficacy, malaria, microsatellite, molecular correction, recrudescence, reinfection, resistance, therapeutic efficacy study
ABSTRACT
Antimalarial drugs have long half-lives, so clinical trials to monitor their efficacy require long periods of follow-up to capture drug failure that may become patent only weeks after treatment. Reinfections often occur during follow-up, so robust methods of distinguishing drug failures (recrudescence) from emerging new infections are needed to produce accurate failure rate estimates. Molecular correction aims to achieve this by comparing the genotype of a patient’s pretreatment (initial) blood sample with that of any infection that occurs during follow-up, with matching genotypes indicating drug failure. We use an in silico approach to show that the widely used match-counting method of molecular correction with microsatellite markers is likely to be highly unreliable and may lead to gross under- or overestimates of the true failure rates, depending on the choice of matching criterion. A Bayesian algorithm for molecular correction was previously developed and utilized for analysis of in vivo efficacy trials. We validated this algorithm using in silico data and showed it had high specificity and generated accurate failure rate estimates. This conclusion was robust for multiple drugs, different levels of drug failure rates, different levels of transmission intensity in the study sites, and microsatellite genetic diversity. The Bayesian algorithm was inherently unable to accurately identify low-density recrudescence that occurred in a small number of patients, but this did not appear to compromise its utility as a highly effective molecular correction method for analyzing microsatellite genotypes. Strong consideration should be given to using Bayesian methodology to obtain accurate failure rate estimates during routine monitoring trials of antimalarial efficacy that use microsatellite markers.
INTRODUCTION
Effective treatment of Plasmodium falciparum malaria infections is essential but is threatened by the spread of drug resistance to front-line antimalarial drugs, including artemisinin-based combination therapies (ACTs). Frequent monitoring of efficacy (1) is therefore necessary to confirm the effectiveness of current drugs and to evaluate alternatives as they become available or necessary. The World Health Organization (WHO) recommends that countries where malaria is endemic routinely retest their currently used antimalarials at least every 2 years using standard patient-based in vivo trials. These are known as therapeutic efficacy studies (TES) (1); they generally have only one “arm” (i.e., the drug being evaluated). This terminology distinguishes TES from regulatory multiarm trials used to evaluate proposed new regimens.
Current first-line antimalarials are ACTs that are composed of an artemisinin derivative and a partner drug; the artemisinin component is short-lived and rapidly clears parasites, while the partner drug has a longer half-life and is responsible for completely, but more slowly, clearing parasites. Ergo, if treatment fails due to resistance of parasites to the partner drug, it may take several weeks for these drug failures to become patent. TES must therefore follow up patients for several weeks posttreatment to ensure failures are detected. The consequence of this requirement for long follow-up periods in TES is that new infections not present at the time of treatment, termed “reinfections” (2), frequently occur during follow-up in moderate- to high-transmission areas. A patient presenting with detectable malaria parasites during follow-up (known as a recurrence or recurrent infection) may have a reinfection, which does not indicate that treatment failed to clear the patient’s initial infection. Thus, recurrent infections trigger molecular testing that leads to their being classified as either (i) P. falciparum clones that infected the patient pretreatment (initial clones) and were subsequently not cleared by treatment (termed recrudescence) or (ii) reinfections that occurred during follow-up (3). In a methodology called either PCR correction or molecular correction, the genotype of the malaria infection at the time of treatment (the initial sample) is compared with the genotype of any recurrence during follow-up. The purpose of this comparison is to distinguish recrudescences from reinfections so that patients with reinfections can be excluded from subsequent analysis, thus producing a “corrected” drug efficacy estimate.
The original WHO and Medicines for Malaria Venture (MMV) consensus methodology was based on the use of the length-polymorphic markers msp-1, msp-2, and glurp (3). An alternative system, microsatellite markers—segments of repeated genetic motifs—has been explored (4–6), a proposed advantage being the lack of immune selection on the ostensibly neutral microsatellite markers (7). In this methodology, researchers genotype microsatellite loci in both initial and recurrent infections and count the matching loci in each patient, i.e., the loci at which at least a single allele is shared between the initial and recurrent infections. They then define a certain number of matches as indicative of recrudescence. In addition to their use in TES, microsatellites have also been commonly used to assess treatment failure in returning travelers in areas of nonendemicity (8–10).
There are two inherent sources of bias in running TES, independent of the genotyping method. (i) Patients who fail to clear their initial infections may have a reinfection that becomes patent before the recrudescent clone reaches a detectable level; ethically, those patients must be treated and so are removed (or “censored”) from the study before the recrudescence can be observed. (ii) Patients who fail to clear their initial infections may have those infections persist at a low level, below the limit of detection of light microscopy (assumed [see below] to be a count of 108 total parasites in the patient), so that parasites are never detected during follow-up; the frequency of this event is influenced by the duration of follow-up in the trial, i.e., the longer the follow-up, the less likely it is to occur.
Classification of recurrences into reinfections and recrudescences also introduces potential bias into estimates of the true failure rate (11–14). Genotyping of blood samples is imperfect and suffers from three key limitations. Firstly, patients are often infected by two or more malaria parasite clones in high-transmission areas, and the lower-density “minority” clones contribute genetic signals that are hard to detect in the amplification process, meaning their low-frequency alleles may fail to be detected in either blood sample (4). Secondly, there are inherent error rates when measuring the genotype of a parasite, for example, through imperfect determination of fragment length or sequencing error. Finally, there is the nontechnical limitation that reinfections can, by chance, share alleles with clones of the initial infection; this is more pervasive in areas of lower genetic diversity (6). These three factors combine to generate several additional factors that need to be considered in the analysis of malaria drug trials. (i) Recrudescent infections can be misclassified as reinfection if alleles of the recrudescent clone were not detected when genotyping the initial infection. (ii) Recrudescent infections can be misclassified as reinfections if a recrudescent clone has a sufficient number of base pair read errors (i.e., at multiple markers) so that it appears to be a reinfection. (iii) A reinfection can be misclassified as recrudescent if it shares (by chance or due to base pair read error) a genetic signal(s) with the clones present at the time of treatment.
Typically, microsatellite data are analyzed by applying a mathematically simple match-counting algorithm that uses an arbitrary threshold for the number of loci that have common alleles between the initial and day-of-failure samples. In these algorithms, if the two samples have matching alleles at, or above, the threshold number of loci, they are classified as recrudescences, and otherwise, as reinfections. Typically, classification of an infection as a recrudescence requires matches at most, if not all, sampled loci (4, 15, 16). This kind of counting algorithm deals with only the unprocessed, raw genetic data and makes no allowance for errors due to the three additional factors described above, resulting in increased risk of misclassification, although more advanced statistical algorithms have been proposed to adjust for these potential biases (6).
A recent publication (17) presented a statistical method based on Bayesian probability to analyze microsatellite data to calculate drug failure rates. The method generates the posterior probability that a recurrent infection is a recrudescence and has subsequently been used to analyze TES data (18–20). The biases listed above mean that a simple method of counting matching microsatellites between samples may never be able to reliably classify a patient as having reinfection or recrudescence. Bayesian analyses constitute a better, more flexible approach capable of dealing with these uncertainties, and the advantages of a Bayesian approach are explained elsewhere (17).
We utilized a computer modelling approach to simulate therapeutic outcome following antimalarial therapy in antimalarial trials. In these simulated data sets, the parasitemia of each patient’s clones is calculated at every time step, and thus, the true status (reinfection or recrudescence) of all recurrent infections is known. Using the simulated data, we then evaluated the ability of the Bayesian algorithm to correctly distinguish reinfections from recrudescences. This allowed us to quantify the accuracy of the method, which has not been possible in vivo; due to imperfect molecular correction techniques, the true failure rate of the population cannot be known. We also compared the performance of the Bayesian algorithm to that of a threshold-based match-counting algorithm and investigated whether the advantages of a Bayesian methodology are realized in the analysis of data from antimalarial TES and whether the approach is truly as robust as postulated in the original paper (17).
This study had three main objectives: first, to evaluate the accuracy of failure rate estimates generated using microsatellite data in conjunction with a match-counting algorithm (as is currently typical); second, to assess the advantages of Bayesian analysis methodology, both in its ability to recover the true failure rate and the diagnostic ability to distinguish recrudescence from reinfections; and third, to check whether the methodologies based on microsatellite loci are robust across drugs with different posttreatment prophylactic profiles (i.e., partner drugs with varying half-lives that determine when reinfections start to occur), across different transmission intensities (which determine rates of reinfection in TES), and in regions with differing levels of genetic diversity at microsatellite loci.
RESULTS
Results were generated for artemether-lumefantrine (AR-LF) and artesunate-mefloquine (AS-MQ) under the assumption of a failing and a nonfailing drug for three scenarios of transmission intensity (methods). Here, we focus on the results for AR-LF, while the results for AS-MQ are fully described in the supplemental material.
Failure rate estimates and comparison to the true failure rate.
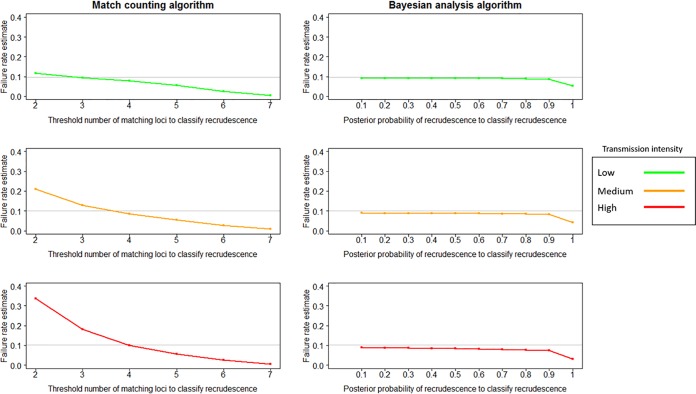
The match-counting algorithm was sensitive to transmission intensity; no threshold value of matching loci at which a recurrence was classified as recrudescent was able to accurately estimate the true failure rate across all transmission scenarios for either failing (Fig. 1) or nonfailing (Fig. 2) AR-LF therapy. Failure rate estimates declined as the threshold increased. Failure rate estimates increased as transmission increased, presumably due to the greater number of reinfections, some of which would be misclassified as recrudescences; this effect was greater at low thresholds, when the probability of such misclassification was greater. A threshold of 4 matching loci produced estimates close to the true failure rate for all nonfailing AR-LF scenarios. For failing AR-LF scenarios, a threshold of 3 matching loci produced the estimate closest to true failure in the low-transmission scenario, and a threshold of 4 matching loci produced the closest estimate in the high-transmission scenario, with the medium-transmission scenario intermediate between the two. However, using a threshold of 3 matching loci in a high-transmission scenario overestimated the failure rate (an estimated failure rate of 0.18 compared to a true failure rate of 0.1). A threshold of 4 matching loci gave an estimate of 0.08 relative to a 0.0997 true failure rate for the failing, medium-transmission scenario and an estimate of 0.077 relative to a true failure rate of 0.0965 for the failing, low-transmission scenario. A threshold of 7 matching loci resulted in extremely large underestimates of failure rates for failing AR-LF therapy: 0.005 relative to a true failure rate of 0.0965 in the low-transmission scenario, 0.008 relative to a true failure rate of 0.0997 in the medium-transmission scenario, and 0.006 relative to a true failure rate of 0.1 in the high-transmission scenario.
FIG 1.
Failure rate estimates obtained using the match-counting algorithm and the Bayesian analysis algorithm for failing AR-LF therapy under low-, medium-, and high-transmission scenarios. The true failure rate is denoted in each plot by a horizontal gray line. For the match-counting algorithm, the threshold for the number of matching loci at which a recurrence was classified as a recrudescence varied between 2 and 7. For the Bayesian analysis, the cutoff for posterior probability at which a recurrence was classified as a recrudescence varied between ≥0.1 and ≥0.9.
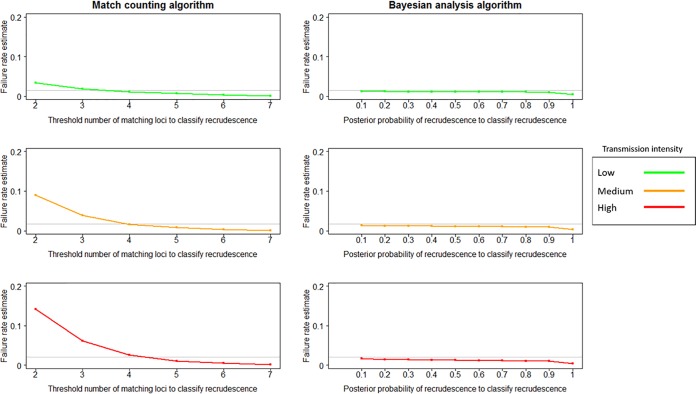
FIG 2.
Failure rate estimates obtained using the match-counting algorithm and the Bayesian analysis algorithm for nonfailing AR-LF therapy under low-, medium-, and high-transmission scenarios. The true failure rate is denoted in each plot by a horizontal gray line. For the match-counting algorithm, the threshold for the number of matches at which a recurrence was classified as a recrudescence varied between 2 and 7. For the Bayesian analysis, the cutoff for posterior probability at which a recurrence was classified as a recrudescence varied between ≥0.1 and ≥0.9.
In contrast to the match-counting method, the Bayesian algorithm recovered the true failure rate at a high degree of accuracy across all transmission settings and for both calibrations of the true drug failure rate (Fig. 1 and 2). Values of the posterior probability of recrudescence, P, were used to distinguish recrudescence from reinfection. Between 0.1 and 0.9 produced good, consistent failure rate estimates with only a slight decline as P increased; using a P value of 1 to classify a recrudescence resulted in a substantial decrease in failure rate estimates. For all nonfailing and failing drug scenarios, treating all infections with P values of ≥0.1 as recrudescences generated a failure rate estimate within 0.01 of the true failure rate.
ROC curves for the Bayesian algorithm.
The general trend was that the area under the receiver operator characteristic (ROC) curve (AUC) decreased as transmission intensity increased (Fig. 3), with values of 0.872 and 0.835 in the failing and nonfailing high-transmission scenarios, respectively—these correspond to a “good” diagnostic test. The AUC was higher for failing AR-LF than nonfailing AR-LF for any given transmission scenario. When the ROC curve was calculated for only high-density recrudescence, the AUC increased to ≥0.968 in all scenarios—an “excellent” diagnostic test.
FIG 3.
ROC curves showing the diagnostic ability of the Bayesian analysis method for 3 scenarios of transmission intensity for nonfailing and failing AR-LF therapy. ROC curves and AUC are shown for all recrudescence and for high-density recrudescence. A high-density recrudescence was defined as explained in the text.
Distribution of posterior probability of recrudescence.
Figure 4 shows the distribution of the posterior probabilities of recrudescence for all recurrences, stratified according to the true classification of their recurrence: reinfection, low-density recrudescence, or high-density recrudescence. The distributions were nearly binary in every scenario: nearly all posterior probabilities in the patient population were <0.1 or ≥0.9. Some trends here were intuitive (note that there are different scales on the y axes), i.e., a larger number of reinfections occurred as transmission intensity increased and a larger number of recrudescences occurred in scenarios in which failing drugs were administered. The small number of patients whose infections had estimated probabilities of recrudescence between (but not including) 0.1 and 0.9 was reflected in the minor changes in failure rate estimates as P changed (Fig. 1 and 2).
FIG 4.
Distributions of the posterior probabilities of recrudescence estimated by the Bayesian algorithm for 3 scenarios of transmission intensity for nonfailing and failing AR-LF therapy. A high-density recrudescence was defined as explained in the text.
Most patients whose recurrences had P values of <0.1 were reinfections. Given that ≥0.1 was the choice of P value that produced the most accurate failure rate estimate (Fig. 1 and 2), the cause of the (slight) underestimate of the failure rate was due to the proportion of patients with infections at P values of <0.1 who had, in reality, recrudescent infections. In simulations of failing drugs, at all transmission intensities, ∼5% of recurrent infections that had P values of <0.1 were truly recrudescent infections. For simulations of nonfailing drugs, at all transmission intensities, ∼2.5% of recurrent infections that had P values of <0.1were truly recrudescent infections. Notably most of these were low-density recrudescence; only 0.03% to 0.05% of recurrent infections that had P values of <0.1 were high-density recrudescences in simulations of failing drugs, and 0.02% to 0.06% of recurrent infections that had P values of <0.1 were high-density recrudescences in simulations of nonfailing drugs. There were a small number of recurrent infections with P values of ≥0.1 that were truly reinfections, but in all scenarios, this number was small relative to the number of recurrent infections that had P values of <0.1 and were truly recrudescent.
Consequently, the underestimation of the failure rate that occurs due to truly recrudescent infections having P values of <0.1 was greater than the overestimation due to reinfections having P values of ≥0.1; thus, these reinfections with P values of ≥0.1 were not leading to an overestimation of the failure rate.
Figures 1, 2, and 4 show that overestimation of the failure rate due to misclassification of reinfection as recrudescence did not significantly affect the Bayesian algorithm due to its high specificity; nearly all reinfections have a posterior probability of recrudescence of <0.1. A slight underestimate of the failure rate occurred with all values of P from ≥0.1 to ≥0.9 (inclusive) to classify a recrudescence, due to the algorithm assigning posterior probabilities of <0.1 to a small proportion of infections with low-density recrudescence.
Determinants of posterior probability of recrudescence.
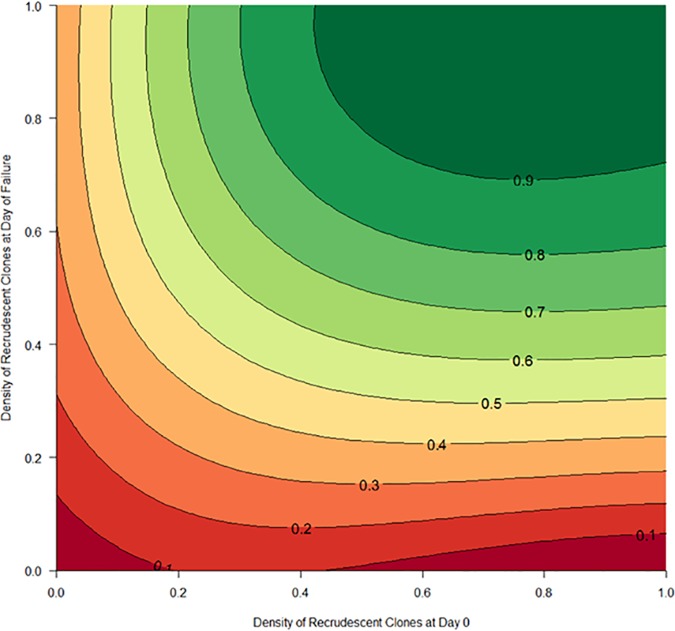
Figure 5 is a contour plot showing the estimated posterior probabilities of recrudescence estimated by the Bayesian algorithm as a function of the densities of the recrudescent clone(s) in the recurrent and initial samples. There was a clear trend for the posterior probability of recrudescence to increase as both densities increased, reinforcing the result illustrated in Fig. 4; the density of recrudescent clones was an important determinant of the posterior probability of recrudescence returned for a given patient. Errors in classification were due almost entirely to the finite sensitivity of genotyping causing some low-density clones to be missed during genotyping.
FIG 5.
Contour plot of the posterior probability of recrudescence estimated by the Bayesian algorithm as a function of the density of recrudescent clones (i.e., the proportion of recrudescent clones in the total recurrent infection biomass) in the initial sample and the recurrent sample. The plot shows the combined data from all 6 scenarios modeled for AR-LF. Each contour line indicates the posterior probability of recrudescence, and the areas between the lines represent the numbers of recurrent infections in the population with those posterior probabilities.
Analysis of artesunate-mefloquine.
We simulated and analyzed AS-MQ in the same manner as for AR-LF. Full results are shown in the supplemental material. The results were very consistent with those for AR-LF. The match-counting algorithm for classifying recurrences as reinfections or recrudescences could not consistently provide accurate failure rate estimates across a variety of scenarios and often resulted in extreme over- or underestimates of the true failure rate, depending on the choice of threshold. The Bayesian analysis method generated failure rate estimates at a high degree of accuracy across all scenarios, although there was an underestimate of 1.6% in the high-transmission failing drug scenario. As with AR-LF, using P values of ≥0.1 to classify an infection as a recrudescence provided the most accurate failure rate estimate for AS-MQ in every scenario.
Very low genetic diversity scenario.
As expected, in the very low genetic diversity scenarios, failure rate estimates increased due to misclassification of reinfection as recrudescence (the first factor identified in the introduction; see the supplemental material). The match-counting algorithm was unable to recover accurate failure rate estimates in any scenario. However, using a high threshold of matching loci to classify a recrudescence (6 or 7) did not lead to overestimates of the failure rate, even in a high-transmission setting, under conditions of very low genetic diversity. Importantly, the Bayesian method recovered accurate failure rate estimates in low genetic diversity scenarios using a P value of ≥0.1 to classify a recrudescence (see the supplemental material).
Patients with subpatent, undetectable parasitemia during follow-up.
We calculated the number of patients who had undetectable, subpatent parasitemia on the final day of follow-up (i.e., a total <108 parasites; either reinfection or recrudescence) and the proportion of these patients who were harboring subpatent recrudescent infections. The results are shown in full in Table S3 in the supplemental material. The results we present above are based on analysis of patients with patent recurrent infections (i.e., those who have detectable parasites during follow-up). In our model, it is possible for a patient to be a true failure (i.e., fail to clear their initial parasite clones) but never have detectable levels of parasites (either recrudescent clones or reinfections) during follow-up (methods). If the number of these patients was large, it would induce bias in our results. However, the proportion of total patients who were true failures but had no recurrent infection was extremely low (between 0 and 0.001 across all scenarios for nonfailing and failing AR-LF therapy), so we can assume the duration of follow-up, at least in our simulations, was sufficiently long to capture nearly all failures and hence to safely draw conclusions about the entire study population.
DISCUSSION
Our in silico experiment showed that the Bayesian algorithm generated extremely accurate estimates of the true failure rate across different transmission intensity and drug failure rate scenarios. In contrast, the match-counting algorithm showed high potential for misclassification bias, with no single threshold able to consistently estimate the true failure rate.
Our results highlight the important role that computer modelling approaches can play in evaluating the performance of genotyping-based classification algorithms. This kind of approach is essential for this evaluation because, unlike real field data, we know the true failure rate of drugs in silico and so can readily identify the most accurate and/or robust method of analysis. In contrast, analysis of field data demonstrates that failure rate estimates vary depending on the choice of methodology (e.g., between criteria used to define recurrence as reinfection or recrudescence [11]), but since the real failure rate in a clinical trial is unknown, it is not possible to demonstrate which method is most accurate or robust. A further advantage of simulated data sets is that we can observe the conditions under which a method fails to return a correct classification (Fig. 4 shows an example). We are confident in our conclusions for several reasons.
Our first main conclusion is that, despite its wide use, match counting of microsatellites to distinguish recrudescence from reinfection is not a robust approach because the estimated drug failure rate is highly dependent on the threshold used to define a recrudescence. By definition, the same clone of malaria parasites will have the same genotype in the initial and recurrent samples. However, the observed genotype (described by the microsatellite alleles) may differ due to issues inherent in the genotyping method (failure to detect minority alleles or errors in measuring base pair lengths of alleles)—accounting for this difference is the purpose of including a degree of flexibility in the molecular correction process, i.e., varying thresholds. Use of microsatellites to correct trials in vivo has, up to now, generally relied upon a simplified analysis method, such as the match-counting algorithm described here. Hwang et al. (16) used 8 markers and defined a match at 7 or more loci as a recrudescence. Greenhouse et al. (4) investigated 6 markers and subsequently used 4 to analyze samples, with a match at every locus being required to classify a recurrence as a recrudescence. Mwangi et al. (15) used 5 loci and considered a match at 5 to be a recrudescence, 0 to be a reinfection, and intermediate values to be mixed infections.
The high thresholds generally used to classify a recurrence as a recrudescence (either most or all of the available loci must match to define a recrudescence) likely results in substantial underestimates of failure rates. For the in silico failing AR-LF results presented here, failure rate estimates with a threshold of 2 ranged from 15% in a low-transmission scenario to 50% in a high-transmission scenario relative to the true failure rates of ∼10% (Fig. 1). However, a threshold of 7 provided estimates that ranged between 0.5% and 0.6% relative to the true failure rates of ∼10%. For nonfailing AR-LF (Fig. 2), failure rate estimates with a threshold of 2 ranged from 7% in a low-transmission scenario to 24% in a high-transmission scenario relative to the true failure rates of ∼2%. In other words, the potential bias induced by the choice of a breakpoint for the match-counting algorithm could result in either rejecting an efficacious drug or continuing to use a failing drug, and this is further complicated by the sensitivity of the breakpoint to transmission intensity (Fig. 1 and 2); the same issues are present in using the match-counting algorithm for AS-MQ (see the supplemental material). This is perhaps not surprising. In the context of genetic markers for classification of recurrent infections in TES, microsatellites are very similar to the marker glurp used in the WHO/MMV method, i.e., they are defined only as length polymorphism with no allelic families. This has led some commentators to suggest that glurp is so unreliable that it should simply be omitted from the WHO/MMV method or used only to resolve disparate msp-1 and msp-2 results (11).
The results presented here strongly suggest that stringent thresholds (i.e., requiring all or most loci to have matching alleles) underestimate the failure rate (and overestimate efficacy). With the seven microsatellites used in these simulations, failure rate estimates produced by the match-counting algorithm varied with both the choice of threshold and the transmission intensity, but in all scenarios, a threshold of 5 matching loci underestimated the failure rate; a threshold of either 3 or 4 produced the closest estimate (Fig. 1 and 2). A threshold of 2 would lead to large overestimates of the failure rate. The reason that stringent thresholds underestimated the failure rate is that low-density recrudescence can be overlooked in patients who have a polyclonal initial or recurrent infection. Note that the threshold producing the most accurate estimate increased from 3 to 4 as transmission increased from low to high—this was because in higher-transmission areas there was a greater impact of reinfections incorrectly classified as recrudescence due to sharing alleles by chance. However, this is dependent on the genetic diversity of the markers used.
When a match-counting algorithm for interpreting microsatellite data is used, we strongly suggest that the failure rates obtained with multiple threshold points be reported (for example Plucinski et al. reported failure rate estimates based on thresholds of matching at all loci and matching at all except a single locus [see Table 2 in reference 17]). This reflects the difficulty (in our opinions, the impossibility) of identifying a robust threshold (Fig. 1 and 2) a priori. Additionally, we suggest that stringent thresholds (requiring all or a very high proportion of loci to match) be generally avoided. Inaccuracies of the failure rate estimates using the match-counting algorithm were a concern for failing drugs; a threshold of 4 was a reasonable approach in our nonfailing drug scenarios, as most recurrences were likely to be reinfection and 4 appeared to be a sufficient threshold to prevent overestimation of failure rates due to misclassifying reinfections as recrudescences. Consequently, a feasible approach for using microsatellites in TES would be to use the match-counting algorithm initially, assess the failure rate estimates produced with a range of thresholds, and pass any result that indicates a drug failure rate higher than 5% through a Bayesian algorithm for reanalysis. We note that in our results, the estimates produced by each threshold are sensitive to transmission intensity, but even with a high transmission intensity, a threshold of 4 would not mistakenly indicate that a failing drug was nonfailing (Fig. 1).
Our second main conclusion is that application of the Bayesian algorithm produces relatively accurate and stable estimates of the failure rate in all transmission scenarios for both failing and nonfailing drugs with use of a posterior probability (P) of 0.1. This result is consistent for analysis of AR-LF and AS-MQ and even for AR-LF in a very low genetic diversity setting, where a P value of 0.1 is effective due to the high specificity of the Bayesian algorithm, i.e., misclassification of reinfection as recrudescence is extremely infrequent (Fig. 4; see the supplemental material). However, note that in the very low diversity setting, failure rate estimates increased as transmission intensity increased, and in areas of higher transmission than we simulated here, there may be a risk of accurate classification with this method, though this presupposes that low genetic diversity (characteristic of low-transmission settings) could occur within an area of high transmission.
The type of pharmacokinetic/pharmacodynamic (PK/PD) modelling that we used to generate parasite dynamics posttreatment has been widely validated and used by our group (21, 22), and the approach is being increasingly used by other groups (e.g., reference 23). Results are highly robust for both AR-LF and AS-MQ (i.e., partner drugs with different lengths of posttreatment prophylaxis), different levels of transmission intensity, and different levels of drug failures and return an intuitive result (increase the failure rate) when very low genetic diversity is simulated. We wish to underline the fact that there are a large number of PK/PD calibrations for AR-LF and AS-MQ in the field; we have chosen the parameterizations here (see the supplemental material) based on our previous work and because their role in the current study is solely to generate plausible profiles of parasite dynamics over time (i.e., see Fig. 1 in reference 24) and to obtain genetic data with which we can evaluate different methods of molecular correction. We could describe parasite dynamics using other methods (for example, predetermining a number of clones at a given time and randomly drawing their densities [25]) but chose to use a PK/PD model for increased realism and relative simplicity and to provide the ability for users to calibrate the model to their liking. The crucial part of our methodology is how we calculate detection of microsatellites in blood samples and specifically that it is based on the relative density of alleles in the parasitemia (and thus is dependent on relative clone numbers) and accounts for a sampling limit and inherent errors in reading microsatellite lengths. We are confident that while use of different PK/PD parameters would change a given patient’s parasite dynamic profile, anything but the most novel parameterization would be unlikely to markedly change our results, given that (i) we simulate 10,000 patients and (ii) parameters are varied within the model so that a large range of alternative parameterizations is already at least partly included in our simulations.
The main practical drawback of the Bayesian algorithm is the need to run a Bayesian analysis. The methodology is published and available (17), but application requires some experience in programming and Bayesian statistics. The analysis is computationally expensive (see the supplemental material) and may be difficult to run on an average personal computer. However, this should not be allowed to be an impediment, given the importance of accurate malaria drug trials, and one solution to this would be for a central body to offer such analyses as a service or to support application of the algorithm through an Internet-based application.
One problem with the microsatellite-genotyping approach is its inability to detect low-density minority clones (the limit here was set to 25%), a problem common to other markers, such as the WHO markers msp-1, msp-2, and glurp (the peak height cutoff for ignoring a signal as noise is generally between 10% and 20% [11]). The slight underestimates of the failure rate produced by Bayesian analyses occurred primarily because the algorithm is unable to correctly identify all low-density recrudescences (Fig. 4 and 5)—this reflected minority alleles being missed during amplification and sequencing. There is now considerable interest in using deep-sequenced amplicons as markers, because the method allows detection of alleles at very low frequencies (less than 2% of the frequency of the most frequent allele). We are currently investigating these markers, using a strategy analogous to that described above, to investigate their potential role in molecular correction. Even if they prove accurate and robust, it is likely to be several years before they are validated, with a consensus methodology identified and routinely used in trials. Meanwhile, it appears that Bayesian analysis of a suite of microsatellite markers does constitute a robust and accurate method for analysis of malaria drug efficacy trials.
MATERIALS AND METHODS
Study design.
We used existing PK/PD models (21, 22, 26) to simulate parasite intrahost dynamics following treatment in 10,000 patients. We simulated whether original clones were cleared or survived drug treatment. If they survived, we then noted whether the recrudescent clone(s) became patent during follow-up and if/when reinfections occurred and became patent. We allowed clones present at the time of treatment to have different numbers (densities) and assigned microsatellite alleles to each clone in the infection. This allowed us to simulate the genetic information that would occur during routine follow-up in these simulated patients, which reflects the inherent problems in the follow-up and genotyping processes (i.e., inability to detect low-density clones and genotyping errors, as described above).
We ran 12 different scenarios, varying the drug, the failure rate, and the level of transmission intensity. The last factor is quantified as the force of infection (FOI), which is the frequency at which reinfections emerge per person per year. Transmission intensity also affects the initial number of clones in each patient at the time of treatment (commonly known as the multiplicity of infection [MOI] or, equivalently, complexity of infection) and the level of genetic diversity in the population allele structure (see below for details). For each scenario, we used the Bayesian algorithm and the match-counting algorithm to generate estimates of the failure rate. We then compared the estimate of the failure rate with the true failure rate to assess the accuracy of each algorithm. It is important to note that our methodology has two distinct steps. First, the PK/PD model simulates parasite dynamics posttreatment, and we used a series of heuristics to calculate which alleles would be observable at any given time step. This provided data sets that were akin to those obtained in vivo, where each patient’s infection was described by observable alleles in the initial sample and observable alleles of any recurrent sample. Secondly, we applied the match-counting and Bayesian algorithms to these data to obtain failure rate estimates. The simulated data set could then be used to analyze algorithm performance against the true failure rate (known from the simulation).
Computational methodology.
All modelling and subsequent analysis was conducted using the statistical programming language R (version 3.5.1) (27). Figures were produced using base R graphics and the ggplot2 package (28). For hardware details and programming considerations, see the supplemental material.
Trial scenarios.
Twelve TES scenarios were simulated. This article presents results obtained from simulations of AR-LF therapy, with results for AS-MQ presented in the supplemental material. The purpose of simulating two distinct treatments is to reflect the different posttreatment prophylactic durations of the drugs—AS-MQ persists at killing concentrations for longer than AR-LF. We primarily wanted to analyze the use of microsatellite markers for AR-LF treatment for which a Bayesian approach has previously been applied (17), but we also wanted to test if results were consistent for a drug with a longer posttreatment prophylactic period. For each drug, we simulated nonfailing drugs with low failure rates (1 to 2%) and failing drugs with higher failure rates (∼10%). The true failure rate of the drug is determined by the half-maximal inhibitory concentration (IC50) of the drug in the parasite population; the IC50 of each clone is drawn from a distribution of values (see Table S1 in the supplemental material). Note that we arbitrarily changed the mean IC50 values of partner drugs within the model to obtain different levels of treatment failure. We do not imply that the values of IC50 used here are representative of any particular field scenario, but rather, we use them to investigate the accuracy of techniques to analyze clinical trial data in a simulation environment. In our simulations, the true failure rate changes with the MOI (a higher MOI means that there are more clones within a patient that are potentially able to survive treatment, and so, the true failure rate increases), so we altered the mean IC50 between scenarios for failing drugs to keep the true failure rates within a percentage of each other between scenarios. Each drug calibration (i.e., nonfailing AR-LF, failing AR-LF, nonfailing AS-MQ, and failing AS-MQ) were run in low-, medium-, and high-transmission settings. These scenarios incorporated varying distributions of MOI at time of treatment, different frequency distributions of microsatellite alleles (obtained from Angola and based on transmission levels [see the supplemental material, parts 2.3 and 2.4]), and different transmission intensities (quantified by FOI [see the supplemental material, part 2.2]). The calibration of each scenario in terms of MOI, allele frequency, FOI, and mean IC50/true failure rate is presented in the supplemental material. Each scenario simulated 10,000 patients for a 28-day follow-up period for AR-LF and a 42-day follow-up period for AS-MQ.
The data we used for distributions of microsatellite markers came from three sentinel sites in Angola (17, 18), which represent areas with moderate to high transmission and thus relatively high diversity (see the supplemental material, part 2.3). The risk of misclassifying a reinfection as a recrudescence is, intuitively, higher in areas of lower genetic diversity (additional factor iii described in the introduction), so we artificially generated an additional distribution of marker allele frequency with very low genetic diversity by modifying the allele distributions from the low-diversity area (as described in the supplemental material, part 2.4) to investigate the accuracy of failure rate estimates under these conditions.
PK/PD model specifications and output.
We utilized a computer-based mechanistic PK/PD (mPK/PD) model of drug treatment of uncomplicated P. falciparum malaria with either AR-LF or AS-MQ based on previous models (21, 22, 26). The methodology used the drug concentration profile in each patient to calculate the change in parasite counts (parasitemia) of each malaria parasite clone over time following drug treatment; this produced quantitative estimates of parasite dynamics in a patient following treatment (see Fig. S1 in the supplemental material). The drug concentration over time in the patient population for each partner drug is shown in Fig. S2 and S3 in the supplemental material. An alternative approach is to generate parasite dynamics by arbitrarily deciding on a day of recurrence for a patient and then assigning the recurrence as constituting recrudescences and/or reinfections (29). It would then be straightforward to draw the parasite numbers in each clone from a uniform distribution, but a PK/PD model was chosen for the ability to easily test different levels of drug failure, for increased realism, and to allow future users to easily recalibrate the methodology with parameters of their choice. Pharmacokinetic parameters varied between patients, and pharmacodynamic parameters varied between clones when they were drawn from distributions of PK and PD parameters (see Table S1 in the supplemental material for full parameter lists and additional considerations).
The number of initial clones in each patient was drawn from a distribution of MOI that depended on local transmission intensity (MOI ranges between 1 and 5 [see Fig. S4 in the supplemental material]); the starting parasitemia of each clone was drawn from a log-uniform distribution between 1010 and 1011. We describe parasitemia in terms of parasite counts rather than parasite densities. We do this because the models track changes in parasite counts over time, and we do not parameterize patients in a way that would allow us to easily convert counts to parasite densities (i.e., patients do not have parameters for blood volume, white blood cell [WBC] count or red blood cell count, etc.), nor would including these parameters aid the mechanistic simulation of the model or improve the accuracy of the results. For reference, assuming a patient with 4.5 liters of blood and a WBC count of 8,000/μl of blood, parasitemias of 1010 and 1011 would correspond to densities of 2,222 parasites/μl of blood and 22,222 parasites/μl of blood, respectively, according to the WHO counting procedure (30). Previous modelling approaches used 1012 parasites as the upper limit of parasitemia; this level of parasitemia is likely to be lethal or at least exceed the maximum parasite density exclusion criterion in a clinical trial (typically 100,000 parasites/μl); hence, we used 1011 as the upper limit for any single clone at the time of treatment. The number of reinfections that occurred during follow-up was determined by the parameter FOI, which we used as our measure of transmission intensity (see the supplemental material, part 2.2). The days on which reinfections occurred were drawn from a Poisson distribution whose mean was the FOI. Reinfections were assumed to emerge from the liver at a count of 105 parasites (31, 32) (see the supplemental material). PK parameters were varied between patients, and PD parameters were varied between malaria parasite clones, so that each patient and clone responded differently to treatment (see Table S1 in the supplemental material for a list of PK/PD parameter means and associated coefficients of variation [CV]). The growth rates of all clones were assumed to be identical and were set to 1.15/day as in previous modelling work (26, 33); this is equivalent to a parasite multiplications rate of 10 per 48-h cycle. The simulation assumed that if the total parasitemia (i.e., the sum of the parasitemias of all clones) in a patient at any time reached 1012, then density-dependent effects, such as fever, acted to control and stabilize the parasitemia, effectively setting the growth rate of every clone in that patient to 0. Aside from this density-dependent effect, we did not attempt to model patient-acquired immunity, as accurately modelling such acquisition is notoriously difficult. It is likely to affect recrudescent and reinfecting clones equally, so we would not expect it to alter how recurrent infections are classified. We did not model parasite sequestration (see reference 34 for justification). The output of this PK/PD model was, for each patient, the exact number of parasites of each malaria parasite clone (whether that clone was an initial infection or a reinfection) at each time-step of the model (in days) (see Fig. S1 for an example). A patient in a real TES would be removed from the trial and retreated when a recurrence occurred, but no such ethical imperative exists in silico, so we tracked the patients the full length of follow-up, with the advantage that we could determine if any initial clones were still present on the final day of follow-up, even though in vivo that patient might have been removed from the trial (right censored) earlier due to a recurrence caused by reinfection.
Modelling microsatellite genotyping and detectability of alleles.
Genotypes were assigned to every clone (both initial infections and reinfections) at seven microsatellite markers: 313, 383, TA1, polya, PfPK2, 2490, and TA109. Alleles at each marker were defined by their lengths (in base pairs) (see details in the supplemental material, part 2.3).
The genotype of the initial malaria infection of each patient was taken on the day of treatment. This genotype signal was a composite of all the clones present in the initial infection and was determined by the technical accuracy and sensitivity of genotyping (see the introduction and below). Each patient was then checked for recurrent parasites during follow-up in a typical clinical trial schedule, i.e., at days 3, 7, 14, 21, and 28 for AR-LF and additionally at days 35 and 42 for AS-MQ.
On all the days of follow-up except day 3, a recurrence was identified if the sum of the parasitemias of all the clones in a patient exceeded 108, which we assumed was the minimum parasitemia at which detection by light microscopy was possible (35). This corresponds to a parasite density of roughly 22 parasites/μl of blood, assuming a patient with 4.5 liters of blood and 8,000 WBC/μl. If total parasitemia was less than 108, then recurrent parasites would not be observed by microscopy (and thus, the patient would not be genotyped on that day). On day 3, if total parasitemia exceeded 108 but was <25% of the total parasitemia of the initial sample, the patient continued in the trial; if parasites were present at >25% of initial parasitemia, the patient was classed as an early treatment failure, according to WHO procedure (1). Genotyping of initial and recurrent samples was then simulated using the following 3-stage protocol.
First, we included a sampling limit. A finite volume of blood was available for genotyping. A parasite clone would not be detected if its density was so low that no parasites were included in the blood sample analyzed. Thus, the density and volume of the processed blood sample defined the limit of detection. We assumed this limit to be 108 (i.e., no clone present in fewer than 108 parasites would be detected [see the supplemental material, part 3, for calculation and justification]).
Secondly, the majority allele for each microsatellite was the allele with the highest parasitemia (if multiple clones shared alleles at a marker, the allelic signal for that marker was the sum of the parasitemias of the clones). We assumed that for an allele to be detected, the parasitemia of that allele must be ≥25% of the parasitemia of the majority allele; this reflects the sensitivity of microsatellite genotyping to infer low-frequency alleles.
Finally, we included the chance that the length of each microsatellite might be misread due to genotyping errors such as stutter bands (7). The chance of an error of plus or minus length x was assumed to be described by the following geometric distribution (described in reference 17): 0.8 × (0.2)x.
The output of these simulations was, for each patient, the microsatellite alleles (quantified by their lengths in base pairs for each locus) at each of the seven loci observed in the initial sample and at any recurrent infection in that patient. A small example (100 patients) is shown in the supplemental material. These are exactly the data recorded in standard TES (and are used as input for the Bayesian algorithm in vivo [17, 18]), so they formed the basis for our PCR correction and failure rate estimates.
Terminology of results.
Our terms “recurrent infection/recurrence,” “recrudescence,” and “reinfection” are consistent with the WHO terminology (2).
We frequently use the additional term “true failure” to describe the failure rate that we know occurred during our simulations (and that is unknown in a real TES). We determined whether each patient was a true failure based on parasitemia. A patient was a true failure if on the final day of follow-up (day 28 for AR-LF and day 42 for AS-MQ) he/she still harbored any parasites from any initial clone. The true failure rate is the frequency of these patients across the entire population. Our model tracked patients over the full length of follow-up, and thus, our “true failure” classification captured patients who would, in a real trial, have been removed earlier in the trial with a recurrent infection classified as a reinfection (and whose recrudescent clones would then not be observed).
A key advantage of our in silico approach is its ability to interrogate the Bayesian algorithm, i.e., to investigate diagnostic ability and determine under which circumstances it would misclassify recurrences.
For these analyses, we separated true failures into high- and low-density recrudescence. The performance of PCR correction is likely to depend on its ability to detect genetic signals from low-density clones. The detection limit for low-level genetic signals in our simulation was 25% (to reflect current genotyping sensitivities, as described above), so it is useful to compare the methodologies when patients have high-density recrudescence (recrudescing clones are present at >25% in both initial and recurrent samples) and low-density clones. Technically, a high-density recrudescence was defined as occurring when three conditions were met: (i) if there was a mixed infection of new and recrudescent clones on the day of recurrence, recrudescent clones must have been >25% of the total infection (more specifically, the sum of the parasitemias of all recrudescent clones on the day of recurrence must have been >25% of the sum of the parasitemias of all the clones on the day of recurrence); (ii) clones that recrudesced must constitute at least 25% of the initial infection (more specifically, the sum of the parasitemias of all recrudescent clones on the day of recurrence must have been >25% of the total parasitemias of all the clones in the initial sample); and (iii) the total number of parasites in recrudescing clones on the day of recurrence must have been ≥108 (to be consistent with the sampling limit defined above). If any one of these conditions was not met, then the failure was defined as low density. In this manner, we determined the true classification of each recurrence as a reinfection, high-density recrudescence, or low-density recrudescence.
Match-counting algorithm.
A match-counting algorithm compared the numbers of microsatellite loci that had at least a single allele shared between the initial and recurrent samples (termed a matching locus). Typically, the use of microsatellite markers in vivo requires a high number of matching loci to classify an infection as recrudescent (either all loci or with a single locus permitted not to match [4, 15, 16]). Here, with the 7 loci modeled, we varied the threshold number of matching loci required to classify a recrudescence to determine the impact of this choice of threshold on failure rate estimates. This is a counting algorithm, where a recurrent infection is defined as a recrudescence when the number of matching loci is greater than or equal to a specified threshold. Six threshold values were analyzed for the method: 2, 3, 4, 5, 6, and 7 matching loci (e.g., if a recurrent infection had 3 matching loci with the initial infection, the recurrence was classified as a recrudescence with a threshold of 2 or 3 loci but as a reinfection with the other thresholds).
Bayesian analysis method.
We used the Bayesian analysis method described in reference 17 to interpret our simulated results and to obtain posterior probabilities of recrudescence for each patient. In brief, the Bayesian algorithm uses a Markov chain Monte Carlo approach to sample from the posterior probability of recrudescence for each sample, with the ratio of likelihoods of a reinfection versus a recrudescence derived from the frequencies of the observed alleles. The algorithm jointly estimates several key parameters, such as the genotyping error rate, and accounts for missing data by sampling hidden alleles. The data input into the Bayesian algorithm in our simulations were the same as those in analysis of in vivo trial data, i.e., the microsatellite profiles of initial and recurrent infections in each patient, as shown in the supplemental material.
The Bayesian analysis was then used to define a recurrence as being a recrudescence when the posterior probability of recrudescence in a patient exceeded a value P, where P lies between 0 and 1.
Note that the Bayesian algorithm was applied to our simulated data sets in the same way it is applied to in vivo data (described in reference 17). Crucially, this means that the priors for all parameters are uninformative—we were not calculating any given parameter in the mPK/PD framework and then using that parameter as a prior for the Bayesian algorithm (which would clearly invalidate the results). Note, though, that posterior estimates from reference 17 were used to inform the chance of allele length being misread in the mPK/PD model, as described above.
Assessment of algorithm accuracy.
Both the match-counting algorithm and Bayesian analysis classified a recurrent infection as either reinfection or recrudescence depending on the choice of threshold (for the match-counting algorithm) or posterior probability, P (for the Bayesian analysis). These classifications were then used to generate failure rate estimates for the simulated TES using survival analysis (the WHO-recommended method [1]) with the R packages survival (36) and survminer (37). The failure estimates for both methods were then compared with the true failure rate to assess their accuracy.
The distribution of the posterior probability of recrudescence calculated using the Bayesian algorithm was plotted for each scenario, with recurrences stratified into their true status: low-density recrudescence, high-density recrudescence, or reinfection. ROC curves were constructed using the posterior probability at which an infection would be classified as a recrudescence (from 0 to 1). The AUC was used to quantify the diagnostic ability of the method (38), with an AUC of >0.8 considered to be a good test and an AUC of >0.9 considered to be an excellent test.
We evaluated the ability of the Bayesian algorithm to detect low-density recrudescence by calculating the posterior probability of recrudescence estimated by the Bayesian algorithm for each recurrent infection and categorizing each infection as reinfection, low-density recrudescence, or high-density recrudescence, as described above.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by The Medical Research Council (grants G1100522 and MR/L022508/1), the Bill and Melinda Gates Foundation (grant 1032350), and the Malaria Modeling Consortium (grant UWSC9757). M.P. was supported by the U.S. President’s Malaria Initiative. The funding sources had no role in study design, collection, analysis, or interpretation of data, the writing of the report, or the decision to submit the paper for publication.
We thank Simon Wagstaff and Andrew Bennett of the scientific computing department at the Liverpool School of Tropical Medicine for providing access to the high-performance computing facilities used to generate the results described here. We also thank five staff members from the Centers for Disease Control and Prevention for their thoughtful commentary on the manuscript.
The findings and conclusions in this report are ours and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
S.J. wrote and conducted the simulations, analyzed the results, and wrote the first draft of the manuscript. M.P. wrote the Bayesian algorithm, analyzed the results, and edited the manuscript. E.M.H. wrote the simulations, and edited the manuscript. K.K. wrote the simulations and edited the manuscript. I.M.H. conceived the project, analyzed the results, and edited the manuscript.
We declare no conflict of interest exists.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2009. Methods for surveillance of antimalarial drug efficacy. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.World Health Organization. 2016. Global technical strategy for malaria 2016–2030. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.World Health Organization. 2008. Methods and techniques for clinical trials on antimalarial drug efficacy: Genotyping to identify parasite populations. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 4.Greenhouse B, Myrick A, Dokomajilar C, Woo JM, Carlson EJ, Rosenthal PJ, Dorsey G. 2006. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am J Trop Med Hyg 75:836–842. doi: 10.4269/ajtmh.2006.75.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyachieo A, Van Overmeir C, Laurent T, Dujardin J-C, D’Alessandro U. 2005. Plasmodium falciparum genotyping by microsatellites as a method to distinguish between recrudescent and new infections. Am J Trop Med Hyg 73:210–213. doi: 10.4269/ajtmh.2005.73.210. [DOI] [PubMed] [Google Scholar]
- 6.Greenhouse B, Dokomajilar C, Hubbard A, Rosenthal PJ, Dorsey G. 2007. Impact of transmission intensity on the accuracy of genotyping to distinguish recrudescence from new infection in antimalarial clinical trials. Antimicrob Agents Chemother 51:3096–3103. doi: 10.1128/AAC.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. 1999. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 119:113–125. doi: 10.1017/S0031182099004552. [DOI] [PubMed] [Google Scholar]
- 8.Malvy D, Torrentino-Madamet M, L'Ollivier C, Receveur M-C, Jeddi F, Delhaes L, Piarroux R, Millet P, Pradines B. 2017. Plasmodium falciparum recrudescence two years after treatment of an uncomplicated infection without return to an area where malaria is endemic. Antimicrob Agents Chemother 62:e01892-17. doi: 10.1128/AAC.01892-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo G, L'Episcopia M, Menegon M, Souza SS, Dongho BGD, Vullo V, Lucchi NW, Severini C. 2018. Dihydroartemisinin-piperaquine treatment failure in uncomplicated Plasmodium falciparum malaria case imported from Ethiopia. Infection 46:867–870. doi: 10.1007/s15010-018-1174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plucinski MM, Huber CS, Akinyi S, Dalton W, Eschete M, Grady K, Silva-Flannery L, Mathison BA, Udhayakumar V, Arguin PM, Barnwell JW. 2014. Novel mutation in cytochrome B of Plasmodium falciparum in one of two atovaquone-proguanil treatment failures in travelers returning from same site in Nigeria. Open Forum Infect Dis 1:ofu059. doi: 10.1093/ofid/ofu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messerli C, Hofmann NE, Beck HP, Felger I. 2017. Critical evaluation of molecular monitoring in malaria drug efficacy trials and pitfalls of length-polymorphic markers. Antimicrob Agents Chemother 61:e01500-16. doi: 10.1128/AAC.01500-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh PS, Erlich HA, Higuchi R. 1992. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl 1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- 13.Farnert A, Arez AP, Babiker HA, Beck HP, Benito A, Bjorkman A, Bruce MC, Conway DJ, Day KP, Henning L, Mercereau-Puijalon O, Ranford-Cartwright LC, Rubio JM, Snounou G, Walliker D, Zwetyenga J, do Rosario VE. 2001. Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Trans R Soc Trop Med Hyg 95:225–232. doi: 10.1016/s0035-9203(01)90175-0. [DOI] [PubMed] [Google Scholar]
- 14.Juliano JJ, Gadalla N, Sutherland CJ, Meshnick SR. 2010. The perils of PCR: can we accurately ‘correct’ antimalarial trials? Trends Parasitol 26:119–124. doi: 10.1016/j.pt.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mwangi JM, Omar SA, Ranford-Cartwright LC. 2006. Comparison of microsatellite and antigen-coding loci for differentiating recrudescing Plasmodium falciparum infections from reinfections in Kenya. Int J Parasitol 36:329–336. doi: 10.1016/j.ijpara.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Hwang J, Alemayehu BH, Reithinger R, Tekleyohannes SG, Takele T, Birhanu SG, Demeke L, Hoos D, Melaku Z, Kassa M, Jima D, Malone JL, Nettey H, Green M, Poe A, Akinyi S, Udhayakumar V, Kachur SP, Filler S. 2013. In vivo efficacy of artemether-lumefantrine and chloroquine against Plasmodium vivax: a randomized open label trial in central Ethiopia. PLoS One 8:e63433. doi: 10.1371/journal.pone.0063433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plucinski MM, Morton L, Bushman M, Dimbu PR, Udhayakumar V. 2015. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite genotyping. Antimicrob Agents Chemother 59:6096–6100. doi: 10.1128/AAC.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davlantes E, Dimbu PR, Ferreira CM, Florinda Joao M, Pode D, Felix J, Sanhangala E, Andrade BN, Dos Santos Souza S, Talundzic E, Udhayakumar V, Owens C, Mbounga E, Wiesner L, Halsey ES, Martins JF, Fortes F, Plucinski MM. 2018. Efficacy and safety of artemether-lumefantrine, artesunate-amodiaquine, and dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria in three provinces in Angola, 2017. Malar J 17:144. doi: 10.1186/s12936-018-2290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plucinski MM, Dimbu PR, Macaia AP, Ferreira CM, Samutondo C, Quivinja J, Afonso M, Kiniffo R, Mbounga E, Kelley JS, Patel DS, He Y, Talundzic E, Garrett DO, Halsey ES, Udhayakumar V, Ringwald P, Fortes F. 2017. Efficacy of artemether-lumefantrine, artesunate-amodiaquine, and dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum malaria in Angola, 2015. Malar J 16:62. doi: 10.1186/s12936-017-1712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plucinski MM, Talundzic E, Morton L, Dimbu PR, Macaia AP, Fortes F, Goldman I, Lucchi N, Stennies G, MacArthur JR, Udhayakumar V. 2015. Efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for treatment of uncomplicated malaria in children in Zaire and Uige Provinces, Angola. Antimicrob Agents Chemother 59:437–443. doi: 10.1128/AAC.04181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodel EM, Kay K, Hayes DJ, Terlouw DJ, Hastings IM. 2014. Optimizing the programmatic deployment of the anti-malarials artemether-lumefantrine and dihydroartemisinin-piperaquine using pharmacological modelling. Malar J 13:138–118. doi: 10.1186/1475-2875-13-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kay K, Hastings IM. 2013. Improving pharmacokinetic-pharmacodynamic modeling to investigate anti-infective chemotherapy with application to the current generation of antimalarial drugs. PLoS Comput Biol 9:e1003151. doi: 10.1371/journal.pcbi.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dini S, Zaloumis S, Cao P, Price RN, Fowkes FJI, van der Pluijm RW, McCaw JM, Simpson JA. 2018. Investigating the efficacy of triple artemisinin-based combination therapies for treating Plasmodium falciparum malaria patients using mathematical modeling. Antimicrob Agents Chemother 62:e01068-18. doi: 10.1128/AAC.01068-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaki T, Parry A, Winter K, Hastings I. 2013. Analysing malaria drug trials on a per-individual or per-clone basis: a comparison of methods. Stat Med 32:3020–3038. doi: 10.1002/sim.5706. [DOI] [PubMed] [Google Scholar]
- 25.Ken-Dror G, Hastings IM. 2016. Markov chain Monte Carlo and expectation maximization approaches for estimation of haplotype frequencies for multiply infected human blood samples. Malar J 15:430. doi: 10.1186/s12936-016-1473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter K, Hastings IM. 2011. Development, evaluation, and application of an in silico model for antimalarial drug treatment and failure. Antimicrob Agents Chemother 55:3380–3392. doi: 10.1128/AAC.01712-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 28.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 29.Dahal P, Guerin PJ, Price RN, Simpson JA, Stepniewska K. 2019. Evaluating antimalarial efficacy in single-armed and comparative drug trials using competing risk survival analysis: a simulation study. BMC Med Res Methodol 19:107. doi: 10.1186/s12874-019-0748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization. 2016. Malaria parasite counting, malaria microscopy standard operating procedure—MM-SOP-09. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 31.Stepniewska K, White NJ. 2008. Pharmacokinetic determinants of the window of selection for antimalarial drug resistance. Antimicrob Agents Chemother 52:1589–1596. doi: 10.1128/AAC.00903-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnham P. 1966. Malaria parasites and other Haemosporidia. Blackwell Scientific Publications Ltd., Oxford, United Kingdom. [Google Scholar]
- 33.Simpson JA, Watkins ER, Price RN, Aarons L, Kyle DE, White NJ. 2000. Mefloquine pharmacokinetic-pharmacodynamic models: implications for dosing and resistance. Antimicrob Agents Chemother 44:3414–3424. doi: 10.1128/aac.44.12.3414-3424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S, Kay K, Hodel EM, Chy S, Mbituyumuremyi A, Uwimana A, Menard D, Felger I, Hastings I. 2019. Improving methods for analyzing antimalarial drug efficacy trials: molecular correction based on length-polymorphic markers msp-1, msp-2, and glurp. Antimicrob Agents Chemother 63:e00590-19. doi: 10.1128/AAC.00590-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siahaan L. 2018. Laboratory diagnostics of malaria. IOP Conf Ser: Earth Environ Sci 125:012090. doi: 10.1088/1755-1315/125/1/012090. [DOI] [Google Scholar]
- 36.Therneau T. 2015. A package for survival analysis in S, version 2.38. https://CRAN.R-project.org/package=survival.
- 37.Alboukadel Kassambara MK. 2018. survminer: drawing survival curves using ‘ggplot2’. R package version 0.4.3. https://CRAN.R-project.org/package=survminer.
- 38.Zweig MH, Campbell G. 1993. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39:561–577. doi: 10.1093/clinchem/39.4.561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.