Fosfomycin is gaining renewed interest for treating urinary tract infections. Monitoring fosfomycin resistance is therefore important in order to detect the emergence of novel resistance mechanisms. Here, we used the Rapid Fosfomycin NP test to screen a collection of extended-spectrum-β-lactamase-producing Escherichia coli isolates from Switzerland and found a fosfomycin-resistant isolate in which a novel plasmid-mediated fosfomycin resistance gene, named fosL1, was identified.
KEYWORDS: fosfomycin, plasmid, Escherichia coli, FosL1
ABSTRACT
Fosfomycin is gaining renewed interest for treating urinary tract infections. Monitoring fosfomycin resistance is therefore important in order to detect the emergence of novel resistance mechanisms. Here, we used the Rapid Fosfomycin NP test to screen a collection of extended-spectrum-β-lactamase-producing Escherichia coli isolates from Switzerland and found a fosfomycin-resistant isolate in which a novel plasmid-mediated fosfomycin resistance gene, named fosL1, was identified. The FosL1 protein is a putative glutathione S-transferase enzyme conferring high-level resistance to fosfomycin and sharing between 57% to 63% amino acid identity with other FosA-like family members. Genetic analyses showed that the fosL1 gene was embedded in a mobile insertion cassette and had likely been acquired by transposition through a Tn7-related mechanism. In silico analysis over GenBank databases identified the FosL1-encoding gene in addition to another variant (fosL1 and fosL2, respectively) in two Salmonella enterica isolates from the United States. Our study further highlights the necessity of monitoring fosfomycin resistance in Enterobacteriaceae to identify the emergence of novel mechanisms of resistance.
INTRODUCTION
Fosfomycin is a broad-spectrum phosphonic acid-derived antibiotic that is naturally produced by Streptomyces fradiae (1). Fosfomycin acts by inhibiting the UDP-N-acetylglucosamine-enolpyruvyl transferase (MurA enzyme) that is involved in a preliminary step of peptidoglycan synthesis by covalently bonding to the latter (1). It is increasingly used as a first-line oral agent for uncomplicated urinary tract infections. Fosfomycin has a large spectrum of activity, good diffusion in the urinary tract, and excellent tolerance among patients. It is considered an alternative treatment option to carbapenems against extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Therefore, its use may significantly contribute to prevent the selection of carbapenem-resistant isolates (2).
Fosfomycin resistance may occur by three major ways: (i) punctual mutations in the MurA-encoding gene leading to a decrease of affinity between the enzyme and fosfomycin, (ii) decrease of the fosfomycin uptake caused by mutations in the glpT and uhpT genes encoding a glycerolphosphate permease and a hexose-phosphate system, respectively; and (iii) the production of fosfomycin-modifying enzymes. Those latter enzymes are generally encoded by plasmid-borne genes, and their expression leads to the permanent modification of the fosfomycin molecule by phosphorylation or by addition of a bacillithiol or glutathione group (1).
Glutathione-S-transferases are Mn2+- and K+-dependent enzymes predominantly in the FosA family. The fosA genes are part of a family of nine variants (fosA1 to fosA9) conferring high resistance to fosfomycin once expressed (3–10). The natural reservoir of the fosA genes has been identified, as members of the Enterobacteriaceae family such as Enterobacter spp. (fosA1 and fosA2), Kluyvera spp. (fosA3 and fosA4), and Klebsiella spp. (fosA3 and fosA4) (11, 12). We recently identified a novel fosA variant, namely, fosA8, for which the progenitor is the enterobacterial species Leclercia adecarboxylata (9). The fosA genes are mostly located on conjugative plasmids, in particular, in Escherichia coli, leading to a high level of resistance to fosfomycin. The fosC2 gene, identified as a class 1 integron (13), also encodes a glutathione transferase identified in Enterobacteriaceae. Other acquired glutathione S-transferase genes have been identified in nonenterobacterial species, such as fosD in Staphylococcus rostri, fosG in Achromobacter denitrificans, fosK in Acinetobacter soli, and fosF in Pseudomonas aeruginosa (14, 15) (see Table S1 in the supplemental material).
Recent studies show that glutathione transferases may be inhibited in vitro by the presence of phosphonoformate (PPF), therefore restoring the susceptibility of the isolate to fosfomycin when supplemented (16). This molecule may be used to differentiate resistance to fosfomycin related to the production of glutathione transferases from those that are not.
We recently performed a screening of fosfomycin resistance among a collection of ESBL-producing E. coli from Switzerland recovered in 2012 (17). Hence, we identified a fosfomycin-resistant isolate harboring a novel fos gene coding for a putative glutathione S-transferase located on a conjugative plasmid.
RESULTS AND DISCUSSION
E. coli isolate R249 belonged to the extraintestinal phylogroup B1 and to a novel sequence type (ST) (adk53, fumC40, gyrB47, icd13, mdh36, purA28, and recA29). It was resistant to broad-spectrum cephalosporins, fluoroquinolones, tetracycline, sulfonamides, kanamycin, tobramycin, chloramphenicol, fosfomycin, and trimethoprim-sulfamethoxazole. It remained susceptible only to cephamycins (cefoxitin), carbapenems, amikacin, colistin, and tigecycline. PCR screening using specific primers followed by sequencing identified the resistance determinants responsible for the overall resistance phenotype, namely, aac(3)-IIa, aac(6′)-Ib-cr, blaCTX-M-15, catB3, sul1, and dfrA17 (Table 1).
TABLE 1.
E. coli strains used in this study and corresponding resistance genotypes
| Strains | MIC (μg/ml) |
Resistance genes identified | |
|---|---|---|---|
| FSFa | FSF+PPF | ||
| R249 | >1,024 | 64 | aac(3)-IIa, aac(6’)-Ib-cr, blaCTX-M-15, catB3, sul1, dfrA17, fosL1 |
| TC-R249b | >1,024 | 32 | fosL1, catB3 |
| TOP10-p249 | >1,024 | 32 | fosL1 |
| J53 | <4 | ND | None |
| TOP10 | <4 | ND | None |
FSF, fosfomycin.
TC, transconjugant.
The MIC of fosfomycin measured by the agar dilution method was 1,024 μg/ml for E. coli R249 and decreased to 64 μg/ml after adding PPF (5 mM) in the supplemented medium. The inhibition of the resistance mechanism by PPF suggested that the fosfomycin resistance pattern was likely related to the production of a glutathione S-transferase, such as those belonging to the FosA family (16). However, PCR amplification of the known fosA and fosC (fosA1-9 and fosC2) remained negative.
E. coli J53 transconjugant (TC-249) was successfully recovered by a mating-out assay, showing high-level resistance to fosfomycin but also to chloramphenicol. PCR assays showed that the corresponding conjugative plasmid coharbored the catB3 gene encoding a chloramphenicol acetyltransferase along with the fosfomycin resistance determinant (Table 1). The conjugation frequency was estimated to be ca. 10−2, therefore quite high but actually corresponding to what was reported for IncX-type plasmids (18). PCR-based replicon typing (PBRT) analysis and Kieser extraction revealed that this plasmid was ca. 40 kb in size and belonged to the IncX1 incompatibility group.
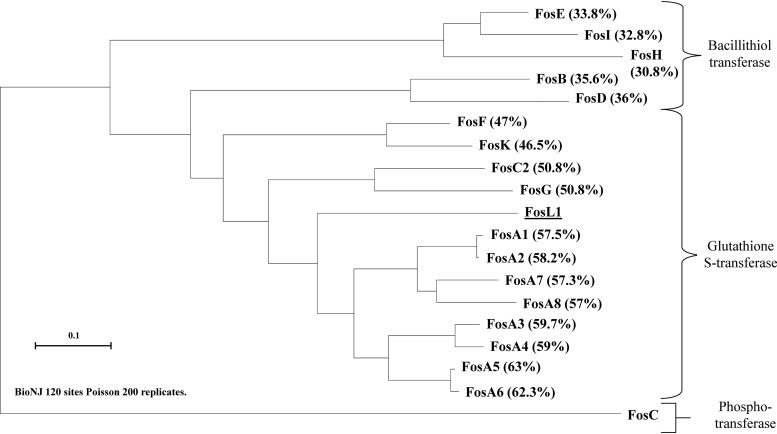
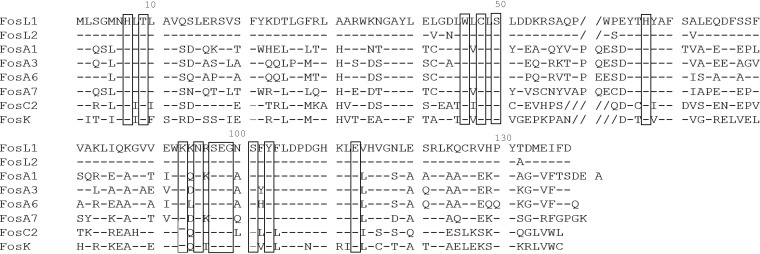
Shotgun cloning using whole-cell DNA of E. coli TC-249 gave recombinant E. coli TOP10 strains resistant to fosfomycin, a resistance also antagonized by the addition of PPF. Restriction profiling using the HindIII enzyme revealed that a ca. 2-kb insert was cloned into the pACYC184 recombinant vector. Sequencing of the DNA insert identified a novel fosfomycin resistance gene, which was named fosL1 (GenBank accession number MN464149) to follow the current nomenclature with the latest fos gene, identified as fosK (15). This gene was 411 bp long coding for a putative protein of 136 amino acids. The FosL1 protein shared 57% and 63% of identity with the FosA1 to FosA8 enzymes and 50% of identity with the FosC2 enzyme (Fig. 1). Alignment of the amino acid sequences of the main Fos proteins showed that FosL1 possessed the conserved amino acid residues involved in the binding of the Mn2+ and K+ cations, a feature shared with other Fos-like glutathione transferases (Fig. 2). To gain insight into the backbone of the fosL1-bearing plasmid, whole-genome sequencing of the E. coli transconjugant DNA but also the E. coli clinical isolate was performed. DNA extraction was performed using a Sigma-Aldrich GenElute bacterial genomic DNA kit. Then, genomic libraries and sequencing were performed using a strategy adapted to the Illumina MiniSeq system as reported previously (9) with 300-bp paired-end reads and a coverage of 50 times. The generated FastQ data were compiled and analyzed using the CLC Genomic Workbench (version 7.5.1; CLC Bio, Aarhus, Denmark), with reads assembled as described previously (9); resulting contigs were uploaded into the Center for Genomic Epidemiology server (http://www.genomicepidemiology.org/). Even though the total assembly process could not close the sequence of the natural plasmid because of repeated sequences, it did confirm that the fosL1 plasmid was indeed the IncX1, with no other plasmid backbone identified from the transconjugant. Corresponding sequences have been deposited under BioProject accession number PRJNA596098, and the whole genome of isolate 249 has been deposited in GenBank under accession number WTUN00000000 (BioSample SAMN13612190).
FIG 1.
Phylogenetic tree obtained for all the identified Fos enzymes, including the bacillithiol and glutathione transferases by distance method using a neighbor-joining algorithm (SeaView version 4 software). Branch lengths are drawn to scale and are proportional to the number of amino acid substitutions with 200 bootstrap replications. The distance along the vertical axis has no significance. Percentages of amino acid identity shared between the FosL1 enzyme and the other Fos enzymes are indicated in brackets.
FIG 2.
Amino acid sequence comparison between FosL1 and different members of glutathione transferases. Boxes bracket amino acids implicated in Mn2+, K+, and fosfomycin bond. Hyphens represent the conserved amino acids with the FosL1 sequence and dashes represent the gaps in the amino acid sequence.
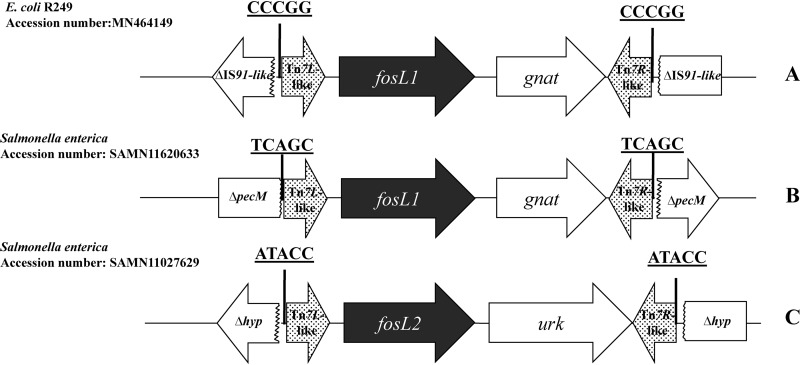
Analysis of the sequences surrounding the fosL1 gene showed that it was part of a 1,597-bp-long cassette along with a putative N-acetyl-transferase (Fig. 3A). Noteworthy, 5-bp direct repeat sequences (CCCGG) were identified at each extremity of this cassette, suggesting that the acquisition of that latter had occurred through a transposition mechanism, although no transposase gene was identified in this cassette. In silico analysis using the BLAST alignment tool showed that the sequences directly adjacent to the direct repeats and therefore corresponding to the cassette boundaries were highly similar to those found at the extremities of Tn7-like transposons, namely, the Tn7L/R sequences. The latter sequences have been shown to play a major role in the mobilization of Tn7-like elements (19, 20). Further analysis of adjacent sequences flanking this fosL1-containing cassette identified sequences of an IS91-like element on both extremities. In fact, it may be speculated that this IS91-like element had been truncated by the fosL1 cassette, probably by a Tn7-like-mediated transposition mechanism with an undefined transposase activity likely acting in trans.
FIG 3.
Schematic structures of the different mobile insertion cassettes harboring the fosL gene. (A) Genetic structure identified in R249 (this study). (B and C) Genetic structures identified in silico in Salmonella species isolates. Tn7L/R, sequences showing high nucleotide identity with the Tn7 extremities recognized by the transposases of the transposon Tn7; gna, gene encoding a putative acetyltransferase; IS91-like, insertion sequence truncated by the insertion of the fosL1 cassette between nucleotides 150 and 151; pecM, gene encoding a histone acetyltransferase HPA2 and related acetyltransferases (protein ID BBB37423.1); urk, gene encoding a putative uridine kinase; hyp, hypothetical gene.
Altogether, these data showed that the fosL1 gene is located in a mobile insertion cassette (mic). Those genetic structures were first described in a Bacillus cereus isolate (21). They are small genetic elements containing one or several open reading frames that present imperfect inverted repeats (IRs) in their extremities. They are not capable of mobility by themselves, since they lack a transposase gene but they can be effectively mobilized in trans by a transposase that is able to recognize its extremities. Those mics were previously described containing resistance genes such as the plasmid-mediated quinolone resistance qnrS2 or more recently the colistin resistance gene mcr-5 (22–24).
In silico analysis using the NCBI database identified the exact same fosL1 gene in a Salmonella enterica isolate recovered from the United States (accession number EAW2818841.1) (Fig. 3B). This isolate was sequenced by the National Subtyping Network for Foodborne Disease Surveillance established by the CDC (25). In that case, the fosL1 gene was also part of a mic-type structure sharing 99% nucleotide identity, differing by only three nucleotide substitutions in the Tn7L-like sequence. That element was also bracketed by 5-bp-long direct repeats (TCAGC) (Fig. 3B). Furthermore, another gene encoding a FosL1-like enzyme was also identified in silico in the genome of another S. enterica from the United States (GenBank accession number SAMN11027629). This enzyme that we named FosL2 shares 96% amino acid identity (only five amino acid substitutions, namely, L41V, D43N, P61S, A70V, and T130A) with FosL1. The fosL2 gene was also located in a mic element; however, it was different from the one bearing the fosL1 gene, being 1,491 bp long and bracketed by 5-bp-long direct repeats (Fig. 3C). Using the PlasmidFinder tool (26), the fosL1 and fosL2 genes identified in the Salmonella sp. isolates were found to be located on IncQ1 and IncP-like plasmids, respectively.
These data highlight that FosL-like resistance determinants are already disseminated among enterobacterial isolates in different parts of the world, and their presence on various plasmid supports may be the consequence of a functional mobility of the mic-fosL elements.
Interestingly, a previous study described a mobile insertion cassette containing a fosD-like gene coding for bacillithiol transferase acquired in the chromosome of a Bacillus cereus isolate. The extremities of that mic structure showed significant homology with the inverted repeats of insertion sequence IS231 (IS4 family) (27).
Very recently, we identified another novel fosA variant, namely, fosA8 (9), identified from an E. coli isolate. The progenitor of the fosA8 gene was identified, being the enterobacterial species Leclercia adecarboxylata. In the present study, we did not find any putative progenitor species for fosL when searching the GenBank databases. Our finding constitutes further evidence that a variety of Fos-like determinants are currently circulating in Enterobacteriaceae, and particularly, in E. coli, being transferable sources of acquired resistance among clinical isolates. Their identification may not simply rely on PCR experiments using primers designed from the previously described fosA-fosC genes, since sequences of those two recently identified genes are too divergent compared to the former. Since fosfomycin is currently considered a critical antimicrobial molecule (used as first-line therapy in many situations but also as a last-resort option when dealing with infections caused by some carbapenem-resistant isolates), strategies should be implemented to identify those resistance determinants and limit their spread when it is still possible.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Isolate R249 was recovered in 2012 from a patient’s urine sample in Switzerland (17). The azide-resistant E. coli J53 strain was used as recipient for the conjugation experiments. The plasmid-free E. coli TOP10 strain (Thermo Fisher, Zug, Switzerland) was used as recipient of the genomic bank generated by the shotgun cloning experiment.
Antimicrobial susceptibility testing and determination of fosfomycin resistance.
Antimicrobial susceptibility testing was performed according to the standard disc diffusion methods using Muller-Hinton (MH) agar plates (Bio-Rad, Cressier, Switzerland) according to the CLSI recommendations (28). Fosfomycin resistance determination was performed using the Rapid Fosfomycin NP test (29) followed by MIC determination by agar dilution using MH agar plates supplemented with glucose-6-phosphate (G6P) at 25 μg/ml and with concentrations of fosfomycin (Sigma-Aldrich, Switzerland) ranging from 4 to 2,048 μg/ml.
Molecular analyses.
Screening of resistance determinants was performed by PCR using specific primers. Multilocus sequence type was obtained using the Center for Genomic Epidemiology server (MLST 1.8) (30). Phylogroup determination was performed using the PCR-based Clermont method (31).
Plasmid analysis.
Transferability of the fosfomycin resistance determinant was assessed by a mating-out assay. Briefly, isolate R249 and E. coli strain J53 were separately inoculated overnight in LB broth. The samples were then mixed at a ratio of 10:1 (recipient/donor) for 5 h and plated on LB agar plates supplemented with azide (100 μg/ml), G6P (25 μg/ml), and fosfomycin (50 μg/ml). Antimicrobial susceptibility testing was performed on the E. coli transconjugant in order to identify putative coresistance associated. Kieser extraction (32) followed by gel electrophoresis analysis were performed for the resulting E. coli transconjugant strain (TC-249) in order to estimate the size of the transferred plasmid. E. coli strain 50192 carrying four plasmids with known sizes (7 kb, 48 kb, 66 kb, and 154 kb) was used as molecular marker.
PCR-based replicon typing (PBRT) was performed to identify specific replicase genes eventually differentiating plasmid incompatibility groups (33). Replicon typing of in silico-identified plasmid was performed using the web-based tool PlasmidFinder version 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) (26).
Shotgun cloning.
Total DNA of E. coli TC-249 was extracted using the GenElute bacterial genomic DNA kit (Sigma-Aldrich, Switzerland). Then, DNA from both isolate R249 and the recombinant plasmid pACYC184 were digested by the restriction enzyme HindIII. Generated fragments were ligated using a T4 DNA ligase (Thermo Fischer, Switzerland) and transferred into the E. coli TOP10 strain by heat shock. Putative transformants containing the fosfomycin resistance determinant were selected on LB agar supplemented with G6P (25 μg/ml) and fosfomycin (50 μg/ml). Recombinant clones were analyzed by PCR followed by Sanger sequencing using specific primers at each extremity of the cloning site of plasmid pACYC184.
Data availability.
DNA sequencing has been deposited in GenBank under accession number MN464149. Corresponding sequences have been deposited as BioProject accession PRJNA596098; the whole genome of isolate 249 has been deposited in GenBank under accession number WTUN00000000 and BioSample SAMN13612190.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the University of Fribourg, the Swiss National Science Foundation (project FNS-310030_188801), and the Laboratoire Européen Associé INSERM Emerging Antibiotic Resistance in Gram-Negative Bacteria.
L.P. and P.N. designed the study. M.-C.D. provided the material. N.K. and L.P. performed the experiments. N.K., L.P., and P.N. wrote the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Silver LL. 2017. Fosfomycin: mechanism and resistance. Cold Spring Harb Perspect Med 7:a025262. doi: 10.1101/cshperspect.a025262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiker A, Clarke L, Doi Y, Shields RK. 2019. Fosfomycin for treatment of multidrug-resistant pathogens causing urinary tract infection: a real-world perspective and review of the literature. Diagn Microbiol Infect Dis 95:114856. doi: 10.1016/j.diagmicrobio.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Partridge SR, Hall RM. 2005. Gene cassettes potentially encoding fosfomycin resistance determinants. Antimicrob Agents Chemother 49:860–861. doi: 10.1128/AAC.49.2.860-861.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Miao V, Kwong W, Xia R, Davies J. 2011. Identification of a novel fosfomycin resistance gene (fosA2) in Enterobacter cloacae from the Salmon River, Canada. Lett Appl Microbiol 52:427–429. doi: 10.1111/j.1472-765X.2011.03016.x. [DOI] [PubMed] [Google Scholar]
- 5.Tseng SP, Wang SF, Kuo CY, Huang JW, Hung WC, Ke GM, Lu PL. 2015. Characterization of fosfomycin resistant extended-spectrum β-lactamase-producing Escherichia coli isolates from human and pig in Taiwan. PLoS One 10:e0135864. doi: 10.1371/journal.pone.0135864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura G, Wachino J, Sato N, Kimura K, Yamada K, Jin W, Shibayama K, Yagi T, Kawamura K, Arakawa Y. 2014. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione-S-transferases. J Clin Microbiol 52:3175–3179. doi: 10.1128/JCM.01094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Xu X, Guo Q, Wang P, Wang W, Wang M. 2015. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett Appl Microbiol 60:259–264. doi: 10.1111/lam.12366. [DOI] [PubMed] [Google Scholar]
- 8.Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. 2017. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother 61:e00410-17. doi: 10.1128/AAC.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Vuillemin X, Kieffer N, Mueller L, Descombes M-C, Nordmann P. 2019. Identification of FosA8, a plasmid-encoded fosfomycin resistance determinant from Escherichia coli and its origin in Leclercia adecarboxylata. Antimicrob Agents Chemother 63:e01403-19. doi: 10.1128/AAC.01403-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ten Doesschate T, Abbott IJ, Willems RJL, Top J, Rogers MRC, Bonten MM, Paganelli FL. 2019. In vivo acquisition of fosfomycin resistance in Escherichia coli by fosA transmission from commensal flora. J Antimicrob Chemother 74:3630–3632. doi: 10.1093/jac/dkz380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito R, Mustapha MM, Tomich AD, Callaghan JD, McElheny CL, Mettus RT, Shanks RMQ, Sluis-Cremer N, Doi Y. 2017. Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. mBio 8:e00749-17. doi: 10.1128/mBio.00749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito R, Pacey MP, Mettus RT, Sluis-Cremer N, Doi Y. 2018. Origin of the plasmid-mediated fosfomycin resistance gene fosA3. J Antimicrob Chemother 73:373–376. doi: 10.1093/jac/dkx389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother 54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yatsuyanagi J, Saito S, Harata S, Suzuki N, Ito Y, Amano K, Enomoto K. 2004. Class 1 integron containing metallo-β-lactamase gene blaVIM-2 in Pseudomonas aeruginosa clinical strains isolated in Japan. Antimicrob Agents Chemother 48:626–628. doi: 10.1128/aac.48.2.626-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitanaka H, Wachino J, Jin W, Yokoyama S, Sasano MA, Hori M, Yamada K, Kimura K, Arakawa Y. 2014. Novel integron-mediated fosfomycin resistance gene fosK. Antimicrob Agents Chemother 58:4978–4979. doi: 10.1128/AAC.03131-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito R, Tomich AD, McElheny CL, Mettus RT, Sluis-Cremer N, Doi Y. 2017. Inhibition of fosfomycin resistance protein FosA by phosphonoformate (Foscarnet) in multidrug-resistant Gram-negative pathogens. Antimicrob Agents Chemother 61:e01424-17. doi: 10.1128/AAC.01424-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller L, Cimen C, Poirel L, Descombes MC, Nordmann P. 2019. Prevalence of fosfomycin resistance among ESBL-producing Escherichia coli isolates in the community, Switzerland. Eur J Clin Microbiol Infect Dis 38:945–949. doi: 10.1007/s10096-019-03531-0. [DOI] [PubMed] [Google Scholar]
- 18.Dénervaud Tendon V, Poirel L, Nordmann P. 2017. Transferability of the mcr-1 colistin resistance gene. Microb Drug Resist 23:813–814. doi: 10.1089/mdr.2016.0191. [DOI] [PubMed] [Google Scholar]
- 19.Hauer B, Shapiro JA. 1984. Control of Tn7 transposition. Mol Gen Genet 194:149–158. doi: 10.1007/bf00383510. [DOI] [PubMed] [Google Scholar]
- 20.Peters JE, Craig NL. 2001. Tn7: smarter than we thought. Nat Rev Mol Cell Biol 2:806–814. doi: 10.1038/35099006. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Braathen P, Léonard C, Mahillon J. 1999. MIC231, a naturally occurring mobile insertion cassette from Bacillus cereus. Mol Microbiol 32:657–668. doi: 10.1046/j.1365-2958.1999.01388.x. [DOI] [PubMed] [Google Scholar]
- 22.Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis 14:231–237. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snesrud E, Maybank R, Kwak YI, Jones AR, Hinkle MK, McGann P. 2018. Chromosomally encoded mcr-5 in colistin-nonsusceptible Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e00679-18. doi: 10.1128/AAC.00679-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieffer N, Nordmann P, Millemann Y, Poirel L. 2019. Functional characterization of a miniature inverted transposable element at the origin of mcr-5 gene acquisition in Escherichia coli. Antimicrob Agents Chemother 63:e00559-19. doi: 10.1128/AAC.00559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force . 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7:382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Palmenaer D, Vermeiren C, Mahillon J. 2004. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol Microbiol 53:457–467. doi: 10.1111/j.1365-2958.2004.04146.x. [DOI] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing, 27th ed M100-S27 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 29.Nordmann P, Poirel L, Mueller L. 2019. Rapid detection of fosfomycin resistance in Escherichia coli. J Clin Microbiol 57:e01531-18. doi: 10.1128/JCM.01531-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz Pontén T, Ussery DW, Aarestrup FM, Lund OJ. 2012. Multilocus sequence typing of total genome sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 32.Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 33.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequencing has been deposited in GenBank under accession number MN464149. Corresponding sequences have been deposited as BioProject accession PRJNA596098; the whole genome of isolate 249 has been deposited in GenBank under accession number WTUN00000000 and BioSample SAMN13612190.