A clinically relevant risk factor for Clostridioides difficile-associated disease (CDAD) is recent antibiotic treatment. Although broad-spectrum antibiotics have been shown to disrupt the structure of the gut microbiota, some antibiotics appear to increase CDAD risk without being highly active against intestinal anaerobes, suggesting direct nonantimicrobial effects. We examined cell biological effects of antibiotic exposure that may be involved in bacterial pathogenesis using an in vitro germfree human colon epithelial culture model.
KEYWORDS: Clostridioides difficile, enteric organoid/enteroid, germ free, host response, postantibiotic effect, systems biology
ABSTRACT
A clinically relevant risk factor for Clostridioides difficile-associated disease (CDAD) is recent antibiotic treatment. Although broad-spectrum antibiotics have been shown to disrupt the structure of the gut microbiota, some antibiotics appear to increase CDAD risk without being highly active against intestinal anaerobes, suggesting direct nonantimicrobial effects. We examined cell biological effects of antibiotic exposure that may be involved in bacterial pathogenesis using an in vitro germfree human colon epithelial culture model. We found a marked loss of mucosal barrier and immune function with exposure to the CDAD-associated antibiotics clindamycin and ciprofloxacin, distinct from the results of pretreatment with an antibiotic unassociated with CDAD, tigecycline, which did not reduce innate immune or mucosal barrier functions. Importantly, pretreatment with CDAD-associated antibiotics sensitized mucosal barriers to C. difficile toxin activity in primary cell-derived enteroid monolayers. These data implicate commensal-independent gut mucosal barrier changes in the increased risk of CDAD with specific antibiotics and warrant further studies in in vivo systems. We anticipate this work to suggest potential avenues of research for host-directed treatment and preventive therapies for CDAD.
INTRODUCTION
Clostridioides difficile-associated disease (CDAD) is a CDC urgent public health threat (1), with 453,000 incident cases in the United States in 2011 (2). CDAD is estimated to account for more than 44,500 deaths and over $5 billion in related health care costs in the United States each year (3). CDAD treatment failure is increasing due to rising levels of antibiotic-resistant (4) and hypervirulent (5) strains of C. difficile and high rates of persistent and recurrent infections (6). New treatment and prevention strategies are needed. A promising strategy for treating some bacterial infections is host-directed therapy (7). Translation to CDAD therapy requires a better understanding of disease susceptibility.
CDAD pathogenesis requires the outgrowth of the etiologic agent, Clostridioides difficile, in the gastrointestinal tract. While a functional gut microbiome is able to prevent the outgrowth of C. difficile (8), in large part due to bacterially dependent production of secondary bile acids (9), loss of a functional gut microbiome allows for outgrowth of the pathogen. Once sufficient cell density is reached, the bacteria begin secreting toxins (10), particularly TcdB, which is primarily responsible for the disease’s symptoms and pathogenesis (11).
A clinically relevant risk factor for CDAD is recent antibiotic treatment (12). There is substantial evidence supporting a causal link between microbiome disruption by antibiotics and CDAD (reviewed in reference 13); however, the antibiotic effect on the microbiome seems unlikely to be the sole causal mechanism for CDAD. Microbiome signatures of patients with and without CDAD (14) are more pronounced than those arising from antibiotic treatment alone, which produces widely varied changes to microbial community structure (15–17). Notwithstanding the frequency of proteobacteria blooms following antibiotic exposure (18), no shared taxonomic change has been identified in successful fecal microbial transplant donors (19) or recipients (20) with other diseases, and alterations to bacterial load do not correlate with the risk of CDAD (21). Taken together, these data suggest one or more host-dependent contributions to the antibiotic-associated risk of CDAD.
Recent work has shown that host-acting drugs have a significant effect on bacteria (22). The inverse has also been shown; a commonly prescribed antibiotic cocktail alters the mitochondrial function of enterocytes in germfree mice (23), demonstrating the commensal-independent effect of antibacterials on the host gut mucosal barrier in this rodent model. Yet, the effects of CDAD-associated antibiotics on the host—especially the human host—and how these effects might contribute to CDAD are not known. Antibiotics are known to affect human host responses in other settings, such as the well-known alterations in airway inflammation and mucosal barrier behavior induced by macrolides (24). Extensive and lasting effects on gene expression and function in the immune system and multiple organ systems, in association with generation of reactive oxygen species, were observed in mice exposed to ciprofloxacin, one CDAD-associated antibiotic (25). Although previous studies have explored the effects of antibiotics in general on the host in a variety of animal models and tissues—including the gut (23)—isolating the effects of particular antibiotics on host-dependent mechanisms of antibiotic-associated CDAD requires a controlled study assessing these mechanisms for multiple antibiotics with various degrees of CDAD-associated risk in the same experimental context, preferably with models that include elements of the human mucosal barrier response.
RESULTS
CDAD-associated antibiotics induce distinct changes in host gene expression.
Based on reported meta-analyses of community-associated C. difficile infection and antibiotics (26, 27) we selected 3 antibiotics representing the odds ratio spectrum from less than 1 (tigecycline, no risk) to 6 (ciprofloxacin, medium risk) to 20 (clindamycin, highest risk) in order to achieve complete coverage of the CDAD risk landscape. Tigecycline, a derivative of tetracycline that is delivered intravenously, is currently used to treat CDAD (28) and hence is an attractive antibiotic for this study. Although there are currently no epidemiological studies giving an odds ratio to tigecycline, we assigned it a low risk based on its similarity to tetracycline, an antibiotic which showed no increased risk of CDAD in the meta-analyses (27), and its strong safety profile (29) in clinical use, including for CDAD treatment.
We used clindamycin and ciprofloxacin for CDAD-associated antibiotics, as they have the highest risk of CDAD among commonly used antibiotics (26, 27). Although all three are considered broad-spectrum antibiotics, they differ in their mechanisms of action and species specificity. We considered mechanism of action as part of the study design. Tigecycline (low risk) and clindamycin (high risk) share a similar mechanism of action, both targeting bacterial translation machinery. Conversely, ciprofloxacin (intermediate risk) inhibits bacterial DNA replication. Notably, ciprofloxacin has an FDA black-box label arising from adverse human side effects on the nervous and musculoskeletal systems (30), yet it has been used to treat Crohn’s disease, a chronic gastrointestinal disorder, with a reasonable safety profile on the gastrointestinal tract although unclear efficacy (31). The mechanisms for effects of ciprofloxacin on human cells have been implicated in damage to mitochondrial DNA and in alterations in DNA-modifying enzymes, but a clear picture has not emerged (30, 32). Gene expression and phenotypic effects of these antibiotics on human colon mucosal barrier cells will thus help illuminate whether additional off-target effects of these drugs exist and should be further studied.
To test the commensal-independent effects of antibiotics on human tissue, we used a transwell-based in vitro epithelial barrier without bacteria to model a germfree human gut (33, 34). We treated mature mucosal barriers with antibiotics dosed from the basal side, using clinically relevant dose ranges and estimating the maximum concentration of drug in serum (Cmax) in each case from a combination of the FDA inserts (Table 1) and published literature on pharmacokinetics for each drug, which provide Cmax values comparable to those in Table 1 (29, 35–40). The standard dosing for intravenous tigecycline includes a 100-mg loading dose followed by maintenance doses, resulting in Cmax values comparable to those shown in Table 1 (29, 39); however, protocols involving higher daily doses (400 mg every 24 h [q24h]) are also used (41), motivating exploration of high-end dosing ranges.
TABLE 1.
Antibiotics used in this study
| Antibiotic (dose type) | Concn used (μg/ml) | Odds ratio (95% CI)a | Typical dosageb | Cmax in serum (μg/ml)b | Half-life (h)b |
|---|---|---|---|---|---|
| Tigecycline (standard) | 1 | 0.91 (0.57–1.45) | 100 mg loading, 50 mg q12h, intravenous | 0.87 | 15–44 |
| Tigecycline (high) | 20 | ||||
| Ciprofloxacin (standard) | 5 | 5.65 (4.38–7.28) | 400 mg q12h, oral | 5.4 | 3.5–4 |
| Ciprofloxacin (high) | 200 | ||||
| Clindamycin (standard) | 3 | 20.43 (8.50–49.09) | 600 mg q6h, oral | 10.8 | 2 |
| Clindamycin (high) | 300 |
Basal dosing of colonic epithelial monolayers at low doses near the reported plasma Cmax is expected to replicate some but not all features of colonic epithelial exposure. The lack of significant metabolism in vitro results in exposure to Cmax throughout the experiment, rather than episodically, as in vivo. Furthermore, although the orally dosed antibiotics are absorbed in the intestine, with subsequent strong partitioning into tissue compartments, all 3 antibiotics considered in this study are also known to be partially excreted in feces in humans (35, 36, 42), resulting in apical exposure in vivo. Tigecycline is given intravenously and is partly eliminated via biliary export into the intestine and passage to feces (40). Tigecycline also strongly partitions into tissue compartments (40, 42), with concentrations in the colon tissue exceeding that in plasma (36, 42); whether it exhibits basal-to-apical transport in the colon is not reported in the literature, but the relatively high in vivo basal concentrations suggest that our basal dosing scheme is reasonable for tigecycline. Orally administered ciprofloxacin has variable absorption in the intestine (38) and, with a relatively long half-life, builds up substantial concentrations in blood over several days of treatment. Ciprofloxacin is partly excreted in urine, but also in feces, where its presence may arise in part from systemic circulation via a known basal-to-apical transport route in colon epithelia (35, 38). Thus, for ciprofloxacin, over the course of the 24- to 48-h experiment, we likely exposed the cells in the ciprofloxacin case to both basal and apical antibiotic due to basal-apical transport. Clindamycin is administered orally or systemically and in both cases partitions strongly into tissues; it is also excreted into the intestine via biliary transport (43). Whether it is transported basal-apically in colonic epithelia is unknown, but the basal dosing used here is reasonable given the strong tissue partitioning.
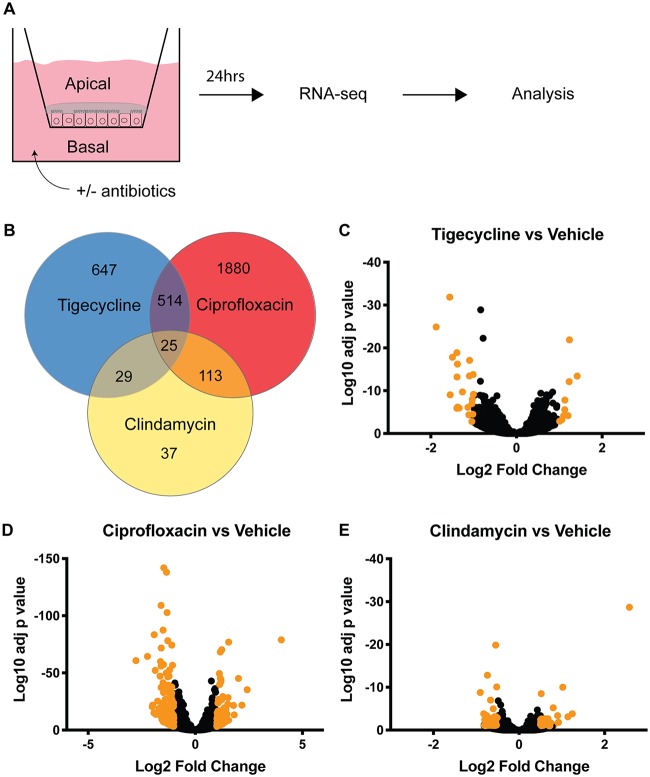
In order to capture extremes of response, Caco2/HT29-MTX monolayers were exposed for 24 h at either standard dose concentrations (equivalent to Cmax) or higher doses, as listed in Table 1, then harvested and lysed. Transcriptome sequencing (RNA-seq) identified gene expression changes under high- and low-exposure conditions, with the largest number of transcriptional changes being driven by ciprofloxacin exposure (Fig. 1B to E). Tigecycline was insoluble in water and was therefore dosed in dimethyl sulfoxide (DMSO). All other antibiotics were dosed in water and compared back to water vehicle. We found DMSO exposure at high concentration had a significant effect on gene expression (Fig. S1), and we therefore performed subsequent analysis using the lower concentrations of both DMSO and tigecycline, listed as “standard” dose type in Table 1.
FIG 1.
Transcriptome sequencing (RNA-seq) identifies differential gene expression changes following 24 h of antibiotic treatment. (A) Schematic of RNA-seq procedure; 6 transwells per condition. (B) Venn diagram and color key of gene expression patterns for high-dose treatment. (C to E) Volcano plots of gene expression changes for high concentrations of indicated antibiotic compared to vehicle. Highlighted points have at least a 2-fold change with an adjusted P value of <0.01.
Unsupervised hierarchical clustering of the 606 genes with statistically significant gene expression changes in at least two exposure groups, using the high concentrations for ciprofloxacin and clindamycin but the low concentration for tigecycline, revealed antibiotic-specific alterations of the gut transcriptome (Fig. S2). This clustering showed various patterns of transcriptional response to exposure. Several transcripts shared similar expression configurations across experimental conditions, some had dose-dependent effects correlating to increasing or decreasing CDAD risk, and still others exhibited more complex behaviors not apparent from the initial clustering. However, the ciprofloxacin-driven expression changes dominated the clustering, highlighting the need for more nuanced computational analysis.
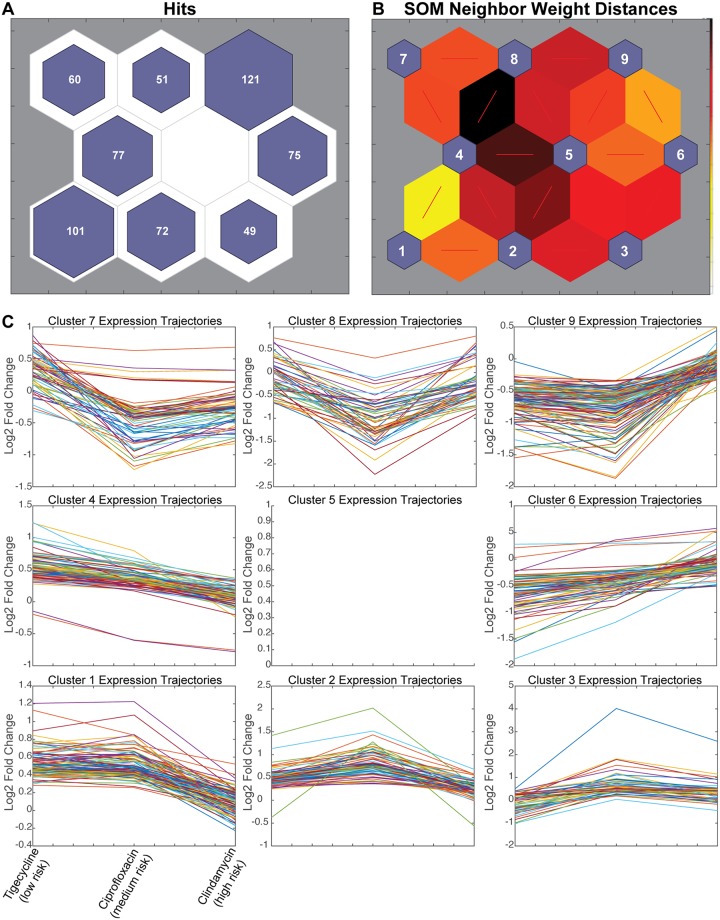
In order to identify genes with transcriptional changes shared between both CDAD-associated antibiotics, we used a self-organizing map (SOM). An SOM is a neural network-based unsupervised clustering technique that groups similar observations together on the SOM neurons. Here, we used the SOM to cluster gene transcript fold changes across antibiotic exposures to identify genes with expression changes associated with CDAD risk. Similarly to other dimensionality-reduction techniques, such as principal-component analysis (PCA), SOMs produce a low-dimensional projection of high-dimensional data that facilitates visualization of patterns. However, unlike PCA or our previous hierarchical clustering, the SOM analysis simultaneously merges two important features, as follows: (i) it incorporates information about the expected number of clusters in the data by defining the number of SOM neurons based on experimental design (number of conditions), and (ii) it allows the data to drive identification of the most informative groups among those clusters (i.e., SOM neurons).
The architecture of the SOM employed here to map the 606 significant genes is based on increased or decreased gene expression (2 directions) in each of three experimental conditions (i.e., clindamycin, ciprofloxacin, or tigecycline, each compared to its respective control), with an extra neuron for noisy profiles (23 + 1 = 9 neurons). Genes with similar expression patterns cluster in a node, with the number of genes per node indicated (Fig. 2A). Plotting neighbor weight distances allows for the visualization of similarities between nodes (Fig. 2B).
FIG 2.
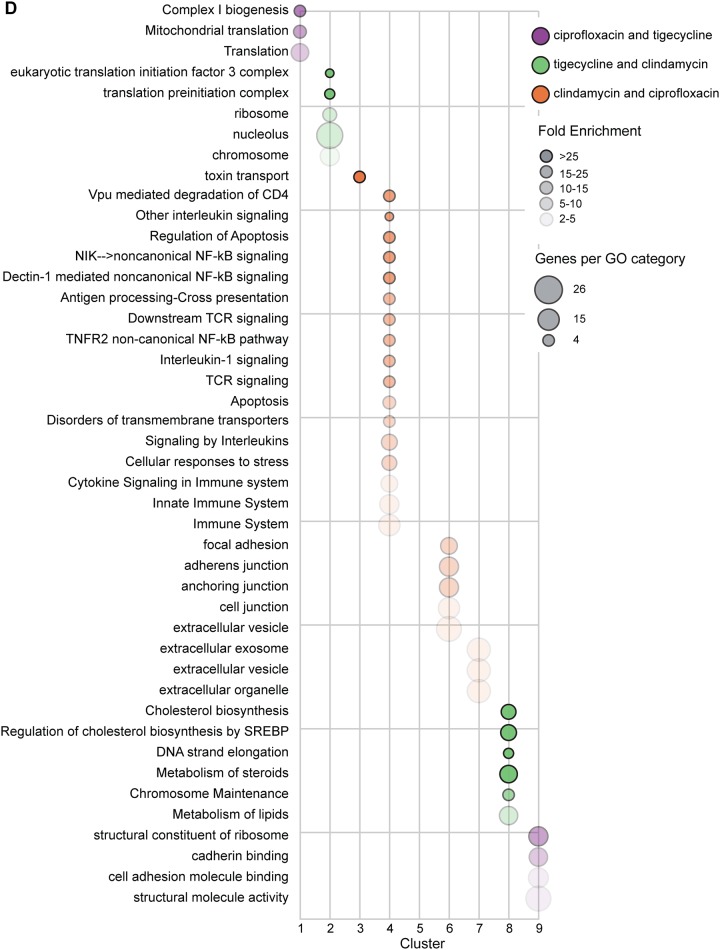
Self-organizing map (SOM) predicts that CDAD-associated antibiotics may alter barrier and immune functions. (A) SOM of 606 genes with statistically significant expression changes by RNA-seq. (B) SOM neighbor weight distances indicate level of similarity between each node pair. (C) Line graphs of all genes in each node, plotted as increasing risk of CDAD on the x axis by standardized fold change on the y axis. (D) Bubble chart showing overrepresented gene ontology (GO) terms for clusters as indicated by the PANTHER overrepresentation test; false-discovery rate (FDR), <0.05.
Each neuron of the SOM captured gene expression responses to antibiotic exposure that grouped according to changing CDAD risk ratios. These patterns could then be investigated by plotting line graphs of the gene fold changes across increasing CDAD risk for each node (Fig. 2C). Two nodes identified gene expression responses that were specifically elevated (node 3) or repressed (node 7) in response to CDAD-associated antibiotic exposure. Another two nodes (4 and 6) captured risk ratio-dependent changes in gene expression responses to CDAD-associated antibiotic exposure, with genes on node 4 being more downregulated and genes on node 6 being more upregulated in antibiotics with higher CDAD risk ratios. Altogether, nodes 3, 4, 6, and 7 capture a set of 261 genes with expression patterns common among ciprofloxacin and clindamycin that indicated a shared pattern of expression unique to the CDAD-associated antibiotics (Fig. 2C), despite different mechanisms of action between ciprofloxacin and clindamycin and similar mechanisms for tigecycline and clindamycin.
In order to identify the biological functions associated with CDAD-associated antibiotic exposure, we performed gene ontology (GO) enrichment analysis (GOEA) of each node (Fig. 2D, Table S1). We would expect nodes that cluster by mechanism of action to be enriched in related GO terms. For instance, the gene expression responses common to tigecycline and clindamycin (nodes 2 and 8) were enriched for the cellular targets of those drugs, translation machinery and chromosome maintenance (Fig. 2D). It is important to note that these targets are considered bacterial cellular components, yet we found they impacted mammalian cells. This finding from the SOM clustering that grouped known target-associated gene expression responses to tigecycline and clindamycin provided an important positive control for interpreting the biological functions associated with the other SOM neurons.
We then analyzed the SOM clusters that captured genes with shared patterns of expression response to CDAD-associated antibiotics (nodes 3, 4, 6, and 7), but distinct from unassociated antibiotics, to generate mechanistic hypotheses of host-dependent mechanisms of CDAD. The GOEA functional annotations of CDAD-associated antibiotic exposure showed an accumulation of cellular toxins in the cell via retrograde secretion (node 3, toxin transport) coupled with a decrease in secretion out of the cell (node 7). We found that as antibiotic-CDAD risk ratios increased, genes associated with immune signaling GO terms were suppressed (node 4), and expression of genes associated with cell-cell and cell-extracellular matrix (ECM) connections was increased (nodes 6 and 7), both in a dose-dependent manner. Node 6 captured genes related to focal adhesion and anchoring junctions, pathways reported to be enhanced in gut wound-healing responses (44), suggesting CDAD-associated antibiotics might result in greater cell stress or death, a hypothesis that can be tested experimentally. The suppression in immune signaling may also alter important barrier functions critical in host-pathogen response (reviewed in references 45), and likewise can be probed in subsequent experiments. Overall, GOEA of these SOMs suggested that exposure to CDAD-associated antibiotics resulted in alterations to transport of extracellular components out of the cell and toxins into the cell and in a reduced immune capacity after only 24 h of exposure.
CDAD-associated antibiotics reduce mucosal barrier and immune functions.
Based on the results of the SOM analysis, we hypothesized that CDAD-associated antibiotic exposure would result in acute and possibly persisting effects of impaired epithelial barrier and impaired innate immune cell function. We tested these SOM predictions experimentally using three complementary levels of in vitro models, as follows: (i) acute effects on epithelial barrier survival and function exposed for 24 h to standard doses of antibiotics listed in Table 1; (ii) chronic (3-day) effect on epithelial exposure to antibiotics and toxin; and (iii) acute effects on innate immune cell function exposed to standard concentrations of the antibiotics listed in Table 1.
We assessed the barrier function of Caco2/HT29-MTX mucosal barriers following 24 h of exposure to standard concentrations of antibiotics (Table 1) dosed in the basal compartment. We found a small but significant increase in cell death in monolayers with ciprofloxacin exposure, which agrees with results of previous work using significantly higher concentrations (46), and the same result with clindamycin, which has not been demonstrated previously (Fig. S3). It is unclear whether this increase is biologically significant, but the upregulation in adhesion-related genes is consistent with a wound-healing response (44). Despite the increase in cell death, none of the antibiotics used affected the physical integrity of the barrier as determined by transepithelial electrical resistance 24 h post exposure (Fig. S4).
To better mimic the 3- to 7-day course of antibiotics in routine in vivo human treatment patterns, we extended the exposure period to a 3-day basal dose, again using the standard concentrations listed in Table 1. We quantified both cell-associated and secreted mucin production, as these strongly influence microbial interactions with the mucosal barrier. Total cell-bound (Fig. 3A) mucin of Caco2/HT29-MTX monolayers was reduced robustly following exposure to both CDAD-associated antibiotics, while mucins in low-risk CDAD exposure groups remained unchanged (Fig. 3A and B). These cell-bound mucins, including MUC1 and MUC4, are critical for protection from intestinal pathogens, as they alert the cell to the presence of invading pathogens through intracellular signal transduction (reviewed in reference 47). Secreted mucins, which provide both a niche for beneficial microbes and a physical obstacle preventing pathogens from accessing the epithelial surface, were reduced with both CDAD-associate antibiotics, although the clindamycin exposure group does not reach statistical significance (Fig. 3B). Together, this reduction in secreted and cell-bound mucins with CDAD-associated antibiotics could provide increased access to the epithelia for C. difficile and its toxins.
FIG 3.
CDAD-associated antibiotics reduce mucosal barrier and immune functions. (A) Cell-bound and (B) secreted mucin quantification. (C) Expression fold changes according to quantitative PCR (qPCR) of muc17 in primary cell-derived 2-dimensional (2D) enteroids relative to vehicle. (D to F) Immune-competent monolayers. (D) tlr4 and (E) il8 gene expression fold change by qPCR with lipopolysaccharide (LPS) stimulation relative to vehicle. (F) Total IL-8 basal secretion with LPS stimulation by enzyme-linked immunosorbent assay (ELISA). Representative data, experiments repeated 2 or 3 times with similar results, n = 3 per experiment. (G) Quantification of phagocytosis and (H) intracellular killing of Escherichia coli by primary macrophages pretreated for 3 days with indicated antibiotics. Primary cell data in open bars, cell line data in filled bars. Representative data, experiments repeated 2 or 3 times with similar results, n = 3 per experiment. Statistical significance was determined by Student’s t test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To assess the effect of our antibiotic panel on primary tissue, we repeated this experiment using primary cell-derived 2-dimensional (2D) enteroids. We assayed for one of the main transmembrane mucin genes in the colon (48), muc17, as it was highly expressed in this donor and was one of the mucin genes that was significantly downregulated in our RNA-seq analysis. We found that expression of muc17 was reduced with ciprofloxacin exposure but not with clindamycin or tigecycline exposure (Fig. 3C); however, we did not assay for other mucin genes in this donor. The inconsistency between cell lines and primary tissue could suggest a reduced role for mucin changes in clindamycin exposure or a donor-specific reduced effect of clindamycin. Other cell-associated mucins (e.g., Muc1 or Muc4) may be contributing to the changes in this primary cell donor shown in Fig. 3A.
To assess the effect of extended, low-dose antibiotic exposure on immune function, we treated an immunocompetent mucosal barrier (Caco2/HT29-MTX monolayers with monocyte-derived dendritic cells added to the basal compartment) for 3 days with each antibiotic, again dosing from the basal side. Interleukin 8 (IL-8) secretion is the primary chemokine implicated in CDAD (5). IL-8 is required for neutrophil recruitment to contain the infection, yet neutrophils are also implicated in progression of disease (49). Thus, a delicate control over dissemination and clearance of neutrophils is likely required for resolution of infection.
We therefore assessed the effect of antibiotics on the ability of immunocompetent mucosal barriers to induce il8 expression and IL-8 secretion following lipopolysaccharide (LPS) stimulation. LPS signals through TLR4 and tlr4 gene expression should increase following its activation, yet tlr4 expression did not increase with LPS stimulation following ciprofloxacin exposure (Fig. 3D). Clindamycin-treated barriers had lower levels of tlr4 relative to those of the vehicle, although this was not significantly lower by Student’s t test than those for tigecycline (Fig. 3D). We found that il8 gene expression (Fig. 3E) is reduced following ciprofloxacin and clindamycin exposure but unchanged with tigecycline in LPS-treated barriers. IL-8 secretion (Fig. 3F) was reduced to a statistically significant extent in all exposure groups. It is likely that the magnitude of change is important in the case of CDAD-associated antibiotics.
To test whether the immune cells are impaired in function, we performed phagocytosis and killing assays using green fluorescent protein-positive (GFP+) Escherichia coli. We found that pretreating macrophages with CDAD-associated antibiotics reduced both the phagocytosis of E. coli (Fig. 3G) and the subsequent killing of phagocytosed E. coli (Fig. 3H). Together, these data support the hypothesis that CDAD-associated antibiotics lead to a loss of immune responsiveness, which one can imagine might contribute to outgrowth of C. difficile.
Antibiotic effects are recapitulated in primary tissue.
Previous work has shown the importance of mucus—primarily made up of mucins—in preventing C. difficile toxins from entering gut cell lines in culture (50). CDAD pathology is driven by the cytotoxic effects of toxins, primarily those of TcdB (5). TcdB enters epithelial cells through receptor-mediated endocytosis (51), although none of the known toxin receptors (NECTIN3, CSPG4, or FZD1/2/7) had significantly altered expression following exposure to any antibiotic we tested. Following acidification of the vacuole, the toxin enters the cytosol, where it glucosylates its GTPase targets, Rho, Rac, and Cdc42. This leads to actin depolymerization, characterized by visible rounding of the cell, and eventually to cell death. The in vitro human mucosal barrier used in this work is affected by TcdB as expected, by cell rounding and areas of cell death, as determined by holes in the monolayer after 48 h of exposure to C. difficile spent medium (Fig. S5), demonstrating the utility of the system for studying TcdB effects.
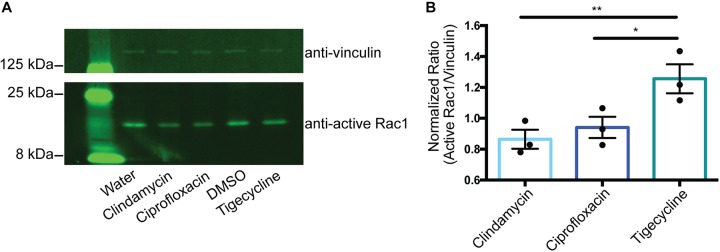
To understand the translation of these altered barrier properties to potential impact in CDAD, we apically exposed mucosal barriers with TcdB (following pretreatment with standard concentrations of antibiotics [Table 1] in the basal compartment) and measured its action by the loss of a cellular target, activated Rac-1, by Western blot analysis. Both CDAD-associated antibiotics had deactivation of Rac1 at 24 h, while tigecycline and controls were still active by quantitative Western blot (Fig. 4A and B), implicating a shared sensitization to C. difficile toxin from CDAD-associated antibiotics with a mechanism independent of commensals.
FIG 4.
CDAD-associated antibiotics reduce mucosal barrier function in primary tissue. (A) Immunoblots against active Rac1 following 24 h of apical TcdB exposure in primary cell-derived monolayers pretreated with standard concentrations of antibiotics (Table 1) for 48 h. (B) Quantification of blots, with Rac1:vinculin normalized to vehicle control. n = 3, paired t test; *, P < 0.05; **, P < 0.01.
DISCUSSION
Here, we demonstrate the convergent changes to in vitro germfree human gut models from two separate CDAD-associated antibiotics in the absence of commensal bacteria. CDAD has been suggested to be a multiphase system, in which germination, outgrowth, and toxin production each have distinct signals upon which they activate (52). Substantial work has shown that antibiotics contribute to CDAD by changing commensal structure and by removing inhibition on both germination and outgrowth (53). Yet, the specific structural definition of a CDAD-inhibitory gut microbiome remains elusive.
Our data suggest a potential mechanism by which an already outgrown but microbiome-controlled population of C. difficile might be able to take hold and produce toxins following the host changes of CDAD-associated antibiotics, namely loss of mucin barrier, increased sensitivity to toxin, and reduced innate immune response. We found that both CDAD-associated antibiotics led to increased toxin transport (Fig. 2D) and concomitant sensitivity to toxin B (Fig. 4).
Increased relative abundance of proteobacteria is associated with CDAD and has been proposed to be a risk factor (54). Proteobacterial bloom following antibiotic treatment might be accounted for by the loss of E. coli and LPS responsiveness we observed. This in vitro study is limited, however, by the relatively short time frame for investigation of effects. Additionally, the simplicity of the model, which lacks microbes and peripheral organs, does not capture the complex reciprocal interplay between the shifting microbial populations and the mucosal barrier. Specifically, the presence of microbes contributes to mucus production in mouse models (55). New microfluidic microphysiological systems are emerging that enable coculture of anaerobic microbial populations with colonic mucosal barriers, and when these become validated and accessible, the combined effects on host and microbe populations can be assessed more fully.
Our work suggests that further study of how CDAD-associated antibiotics might influence CDAD pathology through commensal-independent effects on human tissue may yield insights important for clinical practice.
Antibiotics with high risk for CDAD may prime the host to be less prepared for combating C. difficile infection and pathogenesis by reducing barrier function and immune cell capability and increasing toxin sensitivity, suggesting possible implications for host-directed prophylactic or CDAD treatment therapies. Encouragingly, host-directed antioxidant treatment concomitant with ciprofloxacin therapy showed mitigation of effects on host tissues in a mouse model (25).
There are several limitations to this pilot study. This work was done in a germfree, in vitro setting. As such, its results may not be generalizable to in vivo settings. In addition to the simplicity of the cell populations used, we added antibiotics only to the basal culture medium, which may not fully recapitulate the combined apical and basal exposure known to occur in vivo, as all 3 of the antibiotics in the study are partially excreted in feces. While ciprofloxacin is known to be basal-apically transported in colon epithelia, thus likely mitigating the lack of deliberate apical exposure in vitro, whether such transport occurs for tigecycline and clindamycin is unknown. This model also lacks reciprocal effects of microbe-host interactions that occur in vivo. The impact of antibiotics on the gut bacteria—and their impact on the host—cannot be extrapolated from this work and will need to be studied in the future.
An additional limitation to this study is that we focused on the effects of antibiotics with the highest and lowest risks, and therefore did not test effects of commonly used classes, such as beta-lactams. Further work is needed to understand the commensal-independent, host-dependent effects of this and other classes of antibiotics on gut barrier function.
MATERIALS AND METHODS
Tissue culture: cell lines.
Caco2 (clone C2BBe1, passage 48 to 58; ATCC, Manassas, VA) and HT29-MTX (passage 20 to 30; Sigma-Aldrich, St. Louis, MO) were maintained in Dulbecco’s modified Eagle medium (DMEM) (Gibco, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA), 1% GlutaMax (Gibco), 1% nonessential amino acids (NEAA, Gibco), and 1% penicillin-streptomycin (Pen-Strep). Both cell lines were passaged twice postthawing before their use for transwell seeding. Briefly, the apical side of a transwell membrane was coated with 50 mg/ml collagen type I (Corning Inc., Corning, NY) overnight at 4°C. Caco2 at 80 to 90% confluence and HT29-MTX at 90 to 95% confluence were harvested using 0.25% trypsin-EDTA and mechanically broken up into single cells. A 9:1 ratio of C2BBe1 to HT29-MTX was seeded onto 12-well 0.4-mm-pore polyester transwell inserts (Corning, Tewksbury, MA) at a density of 105 cells/cm2. Seeding medium contained 10% heat-inactivated FBS, 1× GlutaMax, and 1% P-S in advanced DMEM (Gibco). Seven days postseeding, the medium was switched to a serum-free gut medium by replacing FBS with insulin-transferrin-sodium selenite (ITS; Roche, Indianapolis, IN), and the epithelial cultures were matured for another 2 weeks. P-S was left out of the medium during experimental procedures.
Tissue culture: primary cells.
Colon organoids (enteroids) used in this study were established and maintained as previously described (56, 57). Endoscopic tissue biopsy specimens were collected from the ascending colon of deidentified individuals at either Boston Children’s Hospital or Massachusetts General Hospital with the donors’ informed consent. Methods were carried out in accordance to the standards of the Institutional Review Board of Boston Children’s Hospital (protocol number IRB-P00000529) and the Koch Institute Institutional Review Board Committee, as well as those of the Massachusetts Institute of Technology Committee on the Use of Humans as Experimental Subjects. Tissue was digested in 2 mg · ml−1 collagenase I (catalog [cat.] no. 07416; Stemcell Technologies, Vancouver, BC, Canada) for 40 min at 37°C, followed by mechanical dissociation, and isolated crypts were resuspended in growth factor-reduced Matrigel (cat. no. 356237; Becton, Dickinson) and polymerized at 37°C. Organoids were grown in expansion medium (EM) consisting of advanced DMEM/F12 supplemented with L-WRN conditioned medium (65% vol/vol, cat. no. CRL-3276; ATCC,), 2 mM GlutaMax (cat. no. 35050-061; Thermo Fisher), 10 mM HEPES (cat. no. 15630-080; Thermo Fisher), penicillin-streptomycin (Pen-Strep) (cat. no. 15070063; Thermo Fisher), 50 ng · ml−1 murine epidermal growth factor (EGF) (cat. no. PMG8041; Thermo Fisher), N2 supplement (cat. no. 17502-048; Thermo Fisher), B-27 supplement (cat. no. 17502-044; Thermo Fisher), 10 nM human [Leu15]-gastrin I (cat. no. G9145; Sigma), 500 μM N-acetyl cysteine (cat. no. A9165-5G; Sigma), 10 mM nicotinamide (cat. no. N0636; Sigma), 10 μM Y27632 (cat. no. 1293823; Peprotech), 500 nM A83-01 (cat. no. 2939; Tocris), 10 μM SB202190 (cat. no. 1523072; Peprotech), and 5 nM prostaglandin E2 (cat. no. 72192; Stemcell Technologies) at 37°C and 5% CO2. Organoids were passaged every 7 days by incubating in Cell Recovery Solution (cat. no. 354253; Corning) for 40 min at 4°C, followed by mechanical dissociation and reconstitution in fresh Matrigel at a 1:4 ratio.
For 2D enteroid studies, at day 7 post passaging, colon organoids were collected, Matrigel was dissolved with Cell Recovery Solution for 40 min at 4°C, followed by incubation of Matrigel-free organoids in trypsin (cat. no. T4549; Sigma) at 37°C for 5 min. Organoids were mechanically dissociated into single cells, resuspended in EM without nicotinamide and with 2.5 μM thiazovivin (cat. no. 3845; Tocris) in the place of Y27632, and seeded onto 24-well 0.4-μm-pore polyester transwell inserts (cat. no. 3493; Corning) coated with a mixture of 200 μg/ml type 1 collagen and 1% Matrigel at a density of 1 × 105 cells/transwell. After 3 to 4 days of incubation, monolayers were confluent and differentiation was initiated. For differentiation, apical medium was replaced with advanced DMEM/F12 plus HEPES, GlutaMax, and Pen-Strep, and basal medium was replaced with differentiation medium (DM), which is EM without L-WRN conditioned medium, nicotinamide, prostaglandin E2, and Y27632 but supplemented with 100 ng ml−1 human recombinant noggin (cat. no. 120-10C; Peprotech) and 20% R-spondin-conditioned medium (cat. no. SCC111; Sigma). Transepithelial electrical resistance (TEER) measurements were performed using the EndOhm-12 chamber with an EVOM2 meter (World Precision Instruments). At day 8 post seeding, the 2D enteroids were washed to remove Pen-Strep and used for further experimentation.
Monocyte-derived dendritic cells were used as the immune component of the gut when indicated. Briefly, peripheral blood mononuclear cells (PBMCs) were processed from Leukopak (Stemcell Technologies). Monocytes were isolated from PBMCs using the EasySep human monocyte enrichment kit (cat. no. 19058; Stemcell Technologies) and were differentiated in RPMI medium (Gibco) supplemented with 10% heat-inactivated FBS (Gibco), 50 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; BioLegend, San Diego, CA), 35 ng/ml IL-4 (BioLegend), and 10 nM retinoic acid (Sigma). After 7 days of differentiation (at day 19 or 20 post epithelial cell seeding), immature dendritic cells were harvested using phosphate-buffered saline (PBS; Gibco) and seeded onto the basal side of the gut transwells in the absence of Pen-Strep 1 day prior to the start of experiment. Macrophages were derived similarly, but with macrophage colony-stimulating factor (M-CSF; BioLegend) at 500 ng/ml.
RNA preparation and qPCR.
RNA was prepared using a PureLink RNA minikit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. DNA removal was done on a column with PureLink DNase (Invitrogen). cDNA synthesis using a High-Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA) according to the product insert. Quantitative PCR (qPCR) was completed using TaqMan assays with Fast Advanced master mix (Applied Biosystems) per the manufacturer’s guidelines.
3′-DGE library preparation.
RNA samples were quantified and quality assessed using a Fragment Analyzer (Advanced Analytical). Total RNA (20 ng) was used for library preparation with ERCC spike-in control mix A (10−6 final dilution; Ambion). All steps were performed on a Tecan EVO 150 instrument. 3′-digital gene expression (DGE) custom primers 3V6NEXT-bmc#1 to 3V6NEXT-bmc#24 were added to a final concentration of 1.2 μM (5′-/5Biosg/ACACTCTTTCCCTACACGACGCTCTTCCGATCT[BC6]N10T30VN-3′, where 5Biosg = 5′ biotin, [BC6] = 6-bp barcode specific to each sample/well, and N10 = unique molecular identifiers; Integrated DNA Technologies). After addition of the oligonucleotides, samples were denatured at 72°C for 2 min, followed by addition of SmartScribe reverse transcriptase (RT) per the manufacturer’s recommendations along with template-switching oligo5V6NEXT (12 μM, 5′-iCiGiCACACTCTTTCCCTACACGACGCrGrGrG-3′, where iC = iso-dC, iG = iso-dG, and rG = RNA G) and incubation at 42°C for 90 min, followed by inactivation at 72°C for 10 min. Following the template-switching reaction, cDNA from 24 wells containing unique well identifiers was pooled and cleaned using RNA AMPure beads at 1.0×. cDNA was eluted with 90 μl of water, followed by digestion with exonuclease I at 37°C for 45 min and inactivation at 80°C for 20 min. Single-stranded cDNA was then cleaned using RNA AMPure beads at 1.0× and eluted in 50 μl of water. Second-strand synthesis and PCR amplification were done using the Advantage 2 polymerase mix (Clontech) and the SINGV6 primer (10 pmol, 5′-/5Biosg/ACACTCTTTCCCTACACGACGC-3′; Integrated DNA Technologies). PCR was performed for 12 cycles, followed by cleanup using regular solid-phase reversible immobilization (SPRI) beads at 1.0×, and cDNA was eluted with 20 μl of elution buffer. Successful amplification of cDNA was confirmed using the Fragment Analyzer. Illumina libraries were then produced using standard Nextera tagmentation, substituting the P5NEXTPT5-bmc primer (25 μM, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCG*A*T*C*T*-3′, where * indicates phosphorothioate bonds; Integrated DNA Technologies) in place of the normal N500 primer.
Final libraries were cleaned using SPRI beads at 1× and quantified using the Fragment Analyzer and qPCR before being loaded for paired-end sequencing using the Illumina NextSeq 500 platform.
Sequencing data analysis.
Postsequencing quality control on each of the libraries was performed to assess coverage depth, enrichment for mRNA (exon/intron and exon/intergenic density ratios), fraction of rRNA reads, and the number of detected genes using bespoke scripts. The sequencing reads were mapped to the hg38 reference genome using star/2.5.3a. Gene expression counts were further estimated using ESAT version 1 (58).
Phagocytosis and intracellular killing assays.
Escherichia coli GFP (ATCC 25922GFP) was grown to the early log phase in LB medium plus ampicillin, then washed and resuspended in RPMI without antibiotic at the appropriate density. PBMC-derived macrophages were treated with low doses (Table 1) of indicated antibiotics for 3 days in RPMI with heat-inactivated FBS (Atlanta Biologicals). Antibiotic was removed, and antibiotic-free RPMI with GFP+ E. coli was added at a multiplicity of infection (MOI) of 10:1. Extracellular E. coli was washed off after 24 h. For phagocytosis, at 30 min postinfection, cells were fixed and permeabilized, stained with DAPI (4′,6-diamidino-2-phenylindole, Thermo Fisher) and ActinRed 555 ReadyProbes (Molecular Probes, Life Technologies, Carlsbad, CA) stained, and imaged. The percentage of macrophages with at least one GFP+ bacterium was calculated from fluorescence microscopy images. For the intracellular survival assay, macrophages were lysed in water, and supernatants were plated for CFU.
Self-organizing map.
Gene expression fold changes between the controls and the antibiotic-treated cultures were considered a function of CDAD risk and were normalized to be between 0 and 1 across the 3 antibiotics used. Genes without expression fold changes across all 3 conditions were omitted. The map was initialized with a 2-dimensional 3 × 3 square grid and implemented using the MATLAB (MathWorks, Natick, MA) R2017b Neural Network Toolbox.
Gene ontology enrichment analysis.
Gene ontology enrichment on all GO terms was performed using the free online PANTHER overrepresentation test (59–61). The false-discovery rate (FDR) was set to <0.05.
Data representation and statistical analysis.
Prism 8 software (GraphPad Software, La Jolla, CA) was used to graph all data except that for SOMs. Statistical tests of measurements were used from the Prism suite as noted in the figure legends. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Viability/cytotoxicity analysis.
Viability of monolayers post-antibiotic exposure was assessed using the Viability/Cytotoxicity Assay for Animal Live & Dead Cells kit (Biotium, Fremont, CA) according to the package insert. The ratio of red to green cells was measured using ImageJ.
Secreted and cell-bound mucin quantification by Alcian blue colorimetric assay.
Apical medium (secreted mucin) or supernatant from the lysed cell pellet (cell-bound mucin) was stained for 2 h at room temperature with 1% Alcian blue (Electron Microscopy Sciences) at a ratio of 1:4 with the sample. Dye-treated mucin was sedimented by a 30-min centrifugation, followed by two wash steps in wash buffer (290 ml 70% ethanol, 210 ml 0.1 M acetic acid, and 1.2 g MgCl2). Dye-treated mucin was resuspended in 10% SDS, and absorbance at 620 nm was read on a plate reader. Calculations were made based on a known standard prepared in parallel.
Western blotting.
Western blotting was performed under reducing conditions using iBlot 2 dry blotting system (Invitrogen) standard procedures. Primary antibodies were incubated at 4°C overnight and diluted as follows in Odyssey blocking buffer: rabbit monoclonal anti-Vinculin antibody (EPR8185; abcam, Cambridge, MA) at 1:3,000, mouse monoclonal anti-Rac1 antibody (23A8; abcam) at 1:750, and purified mouse Anti-Rac1 antibody (clone 102/Rac1; BD Transduction Laboratories, San Jose, CA) at 1:750. For detection, goat anti-rabbit or anti-mouse IR800-conjugated secondaries at 1:8,000 (LI-COR Lincoln, NE) were incubated for 30 min at room temperature in Odyssey blocking buffer (Tris-buffered saline [BS]) with 0.1% Tween 20. Imaging of the membrane used an LI-COR Odyssey imager with settings as follows: 24-μm resolution and a high-quality laser intensity of 2.0 on the 800 channel.
Chemokine quantification.
Secreted IL-8 was measured by Quantikine enzyme-linked immunosorbent assay (ELISA) human IL-8/CXCL8 immunoassay (R&D Systems, Minneapolis, MN) per the manufacturer’s guidelines.
Data availability.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (62) and are accessible under GEO series accession number GSE135383.
Supplementary Material
ACKNOWLEDGMENTS
J.C.K. was supported by an administrative supplement to grant NIH R01-EB021908. D.K.B. was supported by the Research Beyond Borders Program of Boehringer Ingelheim Pharmaceuticals. This work was supported in part by Koch Institute Support (Core) Grant P30-CA14501 from the National Cancer Institute and by NIEHS grant P30-ES002109. We thank the members of the L.G.G. and D.A.L. labs for their scientific input.
J.C.K. and L.G.G. conceived the project. J.C.K. designed the experiments. J.C.K. acquired and analyzed the data, with technical support from J.V. and C.W. J.C.K. and D.K.B. performed self-organizing map analysis, with critical validation from D.A.L. J.C.K. drafted the original manuscript. All authors contributed to manuscript revisions. L.G.G. and D.A.L. provided oversight and leadership for the project. L.G.G. provided grant support for all study materials and reagents.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.CDC. 2015. 2015 annual report for the Emerging Infections Program for Clostridium difficile Infection. CDC, Atlanta, GA. [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai K, Gupta SB, Dubberke ER, Prabhu VS, Browne C, Mast TC. 2016. Epidemiological and economic burden of Clostridium difficile in the United States: estimates from a modeling approach. BMC Infect Dis 16:303. doi: 10.1186/s12879-016-1610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MTG, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 5.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePestel DD, Aronoff DM. 2013. Epidemiology of Clostridium difficile infection. J Pharm Pract 26:464–475. doi: 10.1177/0897190013499521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R. 2018. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov 17:35–56. doi: 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theriot CM, Bowman AA, Young VB. 2015. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:825–816. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. 2015. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. mBio 6:S88–10. doi: 10.1128/mBio.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eze P, Balsells E, Kyaw MH, Nair H. 2017. Risk factors for Clostridium difficile infections—an overview of the evidence base and challenges in data synthesis. J Global Health 7:010417. doi: 10.7189/jogh.07.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theriot CM, Young VB. 2015. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu Rev Microbiol 69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Lugli GA, Mancabelli L, Turroni F, Duranti S, Mangifesta M, Viappiani A, Ferrario C, Maggio M, Lauretani F, de Vos W, van Sinderen D, Meschi T, Ventura M. 2016. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep 6:25945–25912. doi: 10.1038/srep25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhalodi AA, van Engelen TSR, Virk HS, Wiersinga WJ. 2019. Impact of antimicrobial therapy on the gut microbiome. J Antimicrob Chemother 74:i6–i15. doi: 10.1093/jac/dky530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langdon A, Crook N, Dantas G. 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panda S, Khader El I, Casellas F, López Vivancos J, García Cors M, Santiago A, Cuenca S, Guarner F, Manichanh C. 2014. Short-term effect of antibiotics on human gut microbiota. PLoS One 9:e95476-7. doi: 10.1371/journal.pone.0095476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin N-R, Whon TW, Bae J-W. 2015. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Vermeire S, Joossens M, Verbeke K, Wang J, Machiels K, Sabino J, Ferrante M, Van Assche G, Rutgeerts P, Raes J. 2016. Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. J Crohns Colitis 10:387–394. doi: 10.1093/ecco-jcc/jjv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song Y, Garg S, Girotra M, Maddox C, Rosenvinge von EC, Dutta A, Dutta S, Fricke WF. 2013. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One 8:e81330-11. doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Contijoch EJ, Britton GJ, Yang C, Mogno I, Li Z, Ng R, Llewellyn SR, Hira S, Johnson C, Rabinowitz KM, Barkan R, Dotan I, Hirten RP, Fu S-C, Luo Y, Yang N, Luong T, Labrias PR, Lira S, Peter I, Grinspan A, Clemente JC, Kosoy R, Kim-Schulze S, Qin X, Castillo A, Hurley A, Atreja A, Rogers J, Fasihuddin F, Saliaj M, Nolan A, Reyes-Mercedes P, Rodriguez C, Aly S, Santa-Cruz K, Peters L, Suárez-Fariñas M, Huang R, Hao K, Zhu J, Zhang B, Losic B, Irizar H, Song W-M, Di Narzo A, Wang W, Cohen BL, DiMaio C, Greenwald D, Itzkowitz S, Lucas A, Marion J, Maser E, Ungaro R, Naymagon S, Novak J, Shah B, Ullman T, Rubin P, George J, Legnani P, Telesco SE, Friedman JR, Brodmerkel C, Plevy S, Cho JH, Colombel J-F, Schadt EE, Argmann C, Dubinsky M, Kasarskis A, Sands B, Faith JJ. 2019. Gut microbiota density influences host physiology and is shaped by host and microbial factors. Elife 8:337–326. doi: 10.7554/eLife.40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 55:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgun A, Dzutsev A, Dong X, Greer RL, Sexton DJ, Ravel J, Schuster M, Hsiao W, Matzinger P, Shulzhenko N. 2015. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut 64:1732–1743. doi: 10.1136/gutjnl-2014-308820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamaoki J, Kadota J, Takizawa H. 2004. Clinical implications of the immunomodulatory effects of macrolides. The American J Medicine Supplements 117:5–11. doi: 10.1016/j.amjmed.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 25.Kalghatgi S, Spina CS, Costello JC, Liesa M, Morones-Ramirez JR, Slomovic S, Molina A, Shirihai OS, Collins JJ. 2013. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in mammalian cells. Sci Transl Med 5:192ra85. doi: 10.1126/scitranslmed.3006055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown KA, Khanafer N, Daneman N, Fisman DN. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 57:2326–2332. doi: 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshpande A, Pasupuleti V, Thota P, Pant C, Rolston DDK, Sferra TJ, Hernandez AV, Donskey CJ. 2013. Community-associated Clostridium difficile infection and antibiotics: a meta-analysis. J Antimicrob Chemother 68:1951–1961. doi: 10.1093/jac/dkt129. [DOI] [PubMed] [Google Scholar]
- 28.Herpers BL, Vlaminckx B, Burkhardt O, Blom H, Biemond Moeniralam HS, Hornef M, Welte T, Kuijper EJ. 2009. Intravenous tigecycline as adjunctive or alternative therapy for severe refractory Clostridium difficile infection. Clin Infect Dis 48:1732–1735. doi: 10.1086/599224. [DOI] [PubMed] [Google Scholar]
- 29.Rello J. 2005. Pharmacokinetics, pharmacodynamics, safety and tolerability of tigecycline. J Chemother 17(Suppl 1):12–22. doi: 10.1179/joc.2005.17.Supplement-1.12. [DOI] [PubMed] [Google Scholar]
- 30.Marchant J. 2018. When antibiotics turn toxic. Nature 555:431–433. doi: 10.1038/d41586-018-03267-5. [DOI] [PubMed] [Google Scholar]
- 31.Townsend CM, Parker CE, MacDonald JK, Nguyen TM, Jairath V, Feagan BG, Khanna R. 2019. Antibiotics for induction and maintenance of remission in Crohn’s disease. Cochrane Database Syst Rev 2:CD012730. doi: 10.1002/14651858.CD012730.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badal S, Her YF, Maher LJ. III, 2015. Nonantibiotic effects of fluoroquinolones in mammalian cells. J Biol Chem 290:22287–22297. doi: 10.1074/jbc.M115.671222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsamandouras N, Chen WLK, Edington CD, Stokes CL, Griffith LG, Cirit M. 2017. Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. AAPS J 19:1499–1512. doi: 10.1208/s12248-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen WLK, Edington C, Suter E, Yu J, Velazquez JJ, Velazquez JG, Shockley M, Large EM, Venkataramanan R, Hughes DJ, Stokes CL, Trumper DL, Carrier RL, Cirit M, Griffith LG, Lauffenburger DA. 2017. Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol Bioeng 7:383–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowes S, Simmons NL. 2002. Multiple pathways for fluoroquinolone secretion by human intestinal epithelial (Caco-2) cells. Br J Pharmacol 135:1263–1275. doi: 10.1038/sj.bjp.0704560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubino CM, Ma L, Bhavnani SM, Korth-Bradley J, Speth J, Ellis-Grosse E, Rodvold KR, Ambrose PG, Drusano GL. 2007. Evaluation of tigecycline penetration into colon wall tissue and epithelial lining fluid using a population pharmacokinetic model and Monte Carlo simulation. Antimicrob Agents Chemother 51:4085–4089. doi: 10.1128/AAC.00065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flaherty JF, Rodondi LC, Guglielmo BJ, Fleishaker JC, Townsend RJ, Gambertoglio JG. 1988. Comparative pharmacokinetics and serum inhibitory activity of clindamycin in different dosing regimens. Antimicrob Agents Chemother 32:1825–1829. doi: 10.1128/aac.32.12.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brumfitt W, Franklin I, Grady D, Hamilton-Miller JM, Iliffe A. 1984. Changes in the pharmacokinetics of ciprofloxacin and fecal flora during administration of a 7-day course to human volunteers. Antimicrob Agents Chemother 26:757–761. doi: 10.1128/aac.26.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbour A, Schmidt S, Ma B, Schiefelbein L, Rand KH, Burkhardt O, Derendorf H. 2009. Clinical pharmacokinetics and pharmacodynamics of tigecycline. Clin Pharmacokinet 48:575–584. doi: 10.2165/11317100-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann M, DeMaio W, Jordan RA, Talaat R, Harper D, Speth J, Scatina J. 2007. Metabolism, excretion, and pharmacokinetics of [14C]tigecycline, a first-in-class glycylcycline antibiotic, after intravenous infusion to healthy male subjects. Drug Metab Dispos 35:1543–1553. doi: 10.1124/dmd.107.015735. [DOI] [PubMed] [Google Scholar]
- 41.Baron J, Cai S, Klein N, Cunha B. 2018. Once daily high dose tigecycline is optimal: tigecycline PK/PD parameters predict clinical effectiveness. J Clin Med 7:49–47. doi: 10.3390/jcm7030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodvold KA, Gotfried MH, Cwik M, Korth-Bradley JM, Dukart G, Ellis-Grosse EJ. 2006. Serum, tissue and body fluid concentrations of tigecycline after a single 100 mg dose. J Antimicrob Chemother 58:1221–1229. doi: 10.1093/jac/dkl403. [DOI] [PubMed] [Google Scholar]
- 43.Dhawan VK, Thadepalli H. 1982. Clindamycin: a review of fifteen years of experience. Rev Infect Dis 4:1133–1153. doi: 10.1093/clinids/4.6.1133. [DOI] [PubMed] [Google Scholar]
- 44.Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, Johansen JV, Li Y, Madsen CD, Nakamura T, Watanabe M, Nielsen OH, Schweiger PJ, Piccolo S, Jensen KB. 2018. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22:35–49.e7. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkes J, Viswanathan VK, Savkovic SD, Hecht G. 2003. Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut 52:439–451. doi: 10.1136/gut.52.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herold C, Ocker M, Ganslmayer M, Gerauer H, Hahn EG, Schuppan D. 2002. Ciprofloxacin induces apoptosis and inhibits proliferation of human colorectal carcinoma cells. Br J Cancer 86:443–448. doi: 10.1038/sj.bjc.6600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. 2008. Mucins in the mucosal barrier to infection. Mucosal Immunol 1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Putten JPM, Strijbis K. 2017. Transmembrane mucins: signaling receptors at the intersection of inflammation and cancer. J Innate Immun 9:281–299. doi: 10.1159/000453594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jose S, Madan R. 2016. Neutrophil-mediated inflammation in the pathogenesis of Clostridium difficile infections. Anaerobe 41:85–90. doi: 10.1016/j.anaerobe.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olson A, Diebel LN, Liberati DM. 2013. Effect of host defenses on Clostridium difficile toxin–induced intestinal barrier injury. J Trauma Acute Care Surg 74:983–990. doi: 10.1097/TA.0b013e3182858477. [DOI] [PubMed] [Google Scholar]
- 51.Papatheodorou P, Zamboglou C, Genisyuerek S, Guttenberg G, Aktories K. 2010. Clostridial glucosylating toxins enter cells via clathrin-mediated endocytosis. PLoS One 5:e10673. doi: 10.1371/journal.pone.0010673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hryckowian AJ, Pruss KM, Sonnenburg JL. 2017. The emerging metabolic view of Clostridium difficile pathogenesis. Curr Opin Microbiol 35:42–47. doi: 10.1016/j.mib.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abt MC, Pamer EG. 2014. Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol 29:16–22. doi: 10.1016/j.coi.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seekatz AM, Young VB. 2014. Clostridium difficile and the microbiota. J Clin Invest 124:4182–4189. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson M. 2015. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, Ingber DE. 2018. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep 8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, Almeqdadi M, Wu K, Oberli MA, Sánchez-Rivera FJ, Park YK, Liang X, Eng G, Taylor MS, Azimi R, Kedrin D, Neupane R, Beyaz S, Sicinska ET, Suarez Y, Yoo J, Chen L, Zukerberg L, Katajisto P, Deshpande V, Bass AJ, Tsichlis PN, Lees J, Langer R, Hynes RO, Chen J, Bhutkar A, Jacks T, Yilmaz ÖH. 2017. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol 35:569–576. doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A, Mikkelsen TS. 2014. Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv 10.1101/003236. [DOI]
- 59.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The Gene Ontology Consortium. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, The Gene Ontology Consortium . 2000. Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (62) and are accessible under GEO series accession number GSE135383.