We aimed to assess the rate and predictive factors of bloodstream infection (BSI) due to multidrug-resistant (MDR) Pseudomonas aeruginosa in neutropenic cancer patients. We performed a multicenter, retrospective cohort study including oncohematological neutropenic patients with BSI due to P. aeruginosa conducted across 34 centers in 12 countries from January 2006 to May 2018. A mixed logistic regression model was used to estimate a model to predict the multidrug resistance of the causative pathogens.
KEYWORDS: multidrug resistant, Pseudomonas aeruginosa, bacteremia, bloodstream infection, neutropenia, cancer, risk factors, predictive model
ABSTRACT
We aimed to assess the rate and predictive factors of bloodstream infection (BSI) due to multidrug-resistant (MDR) Pseudomonas aeruginosa in neutropenic cancer patients. We performed a multicenter, retrospective cohort study including oncohematological neutropenic patients with BSI due to P. aeruginosa conducted across 34 centers in 12 countries from January 2006 to May 2018. A mixed logistic regression model was used to estimate a model to predict the multidrug resistance of the causative pathogens. Of a total of 1,217 episodes of BSI due to P. aeruginosa, 309 episodes (25.4%) were caused by MDR strains. The rate of multidrug resistance increased significantly over the study period (P = 0.033). Predictors of MDR P. aeruginosa BSI were prior therapy with piperacillin-tazobactam (odds ratio [OR], 3.48; 95% confidence interval [CI], 2.29 to 5.30), prior antipseudomonal carbapenem use (OR, 2.53; 95% CI, 1.65 to 3.87), fluoroquinolone prophylaxis (OR, 2.99; 95% CI, 1.92 to 4.64), underlying hematological disease (OR, 2.09; 95% CI, 1.26 to 3.44), and the presence of a urinary catheter (OR, 2.54; 95% CI, 1.65 to 3.91), whereas older age (OR, 0.98; 95% CI, 0.97 to 0.99) was found to be protective. Our prediction model achieves good discrimination and calibration, thereby identifying neutropenic patients at higher risk of BSI due to MDR P. aeruginosa. The application of this model using a web-based calculator may be a simple strategy to identify high-risk patients who may benefit from the early administration of broad-spectrum antibiotic coverage against MDR strains according to the local susceptibility patterns, thus avoiding the use of broad-spectrum antibiotics in patients at a low risk of resistance development.
INTRODUCTION
Bloodstream infection (BSI) is an important cause of morbidity and mortality in neutropenic cancer patients. In recent years, an increase in the incidence of BSI caused by Gram-negative bacilli (GNB) has been reported in this population, as has the emergence of antibiotic resistance (1–5).
Pseudomonas aeruginosa has classically been one of the most important causes of severe sepsis and death among cancer patients with neutropenia (6–8). Recent data in patients with hematological malignancies show that BSI carries a poor prognosis and is associated with the highest mortality among different groups of patients with BSIs (9). In part, this may be due to multidrug-resistant (MDR) P. aeruginosa, which has been found at high rates in some series involving patients with hematological malignancies, particularly in Italy (10–15). Importantly, inadequate empirical antibiotic therapy is frequently administered in this scenario, which contributes to poor survival (10–12, 15).
The recent implementation of new treatment modalities, such as highly toxic myelosuppressive therapies, different types of hematopoietic stem cell transplants (HSCT), and the widespread use of other invasive procedures, may have had an impact on the risk of development of antibiotic resistance. Very few studies have examined the risk factors for MDR P. aeruginosa infections in patients with cancer under these new and evolving conditions or in the current era of widespread antimicrobial resistance (16, 17).
Identifying the risk factors for infections due to MDR P. aeruginosa in neutropenic cancer patients could help physicians more rapidly recognize patients at higher risk. Prompt administration of an empirical therapy active against MDR strains in these high-risk patients might benefit their outcomes. In this regard, estimating the probability of antibiotic resistance using a clinical prediction model could be useful for stratifying patients according to their risk. Along this line, Viasus et al. recently reported a score which identified hematological malignancy, nosocomial acquisition, prior treatment with antipseudomonal cephalosporins and quinolones, prior treatment with corticosteroids, and breakthrough BSI during treatment with quinolones and β-lactams other than ertapenem to be independent risk factors for MDR P. aeruginosa BSI in neutropenic patients (18). A limitation of that study was its single-center design, the relatively small number of BSI episodes, and the lack of performance of external validation. Also, the use of a clinical prediction model could help avoid the use of broad-spectrum antibiotics in patients with a low risk of resistance development and, therefore, improve antibiotic stewardship.
The aim of the present study was to assess the rate and evolution of multidrug resistance among P. aeruginosa isolates causing BSI in neutropenic cancer patients over recent years and to develop a clinical prediction model to estimate the probability of multidrug resistance acquisition in this population. To this end, we used data from a large multicenter, international cohort from 34 centers in 12 countries.
RESULTS
Rate of multidrug resistance.
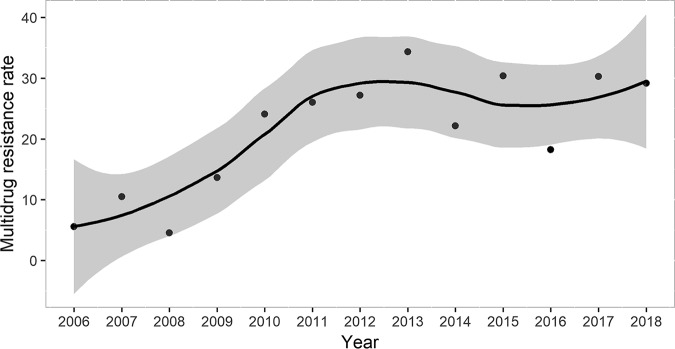
Of a total of 1,217 episodes of BSI due to P. aeruginosa occurring in 1,177 patients, 309 episodes (25.4%) were caused by MDR strains, of which 234 (19.3%) were considered to be extensively drug resistant (XDR). The rate of multidrug resistance by country is detailed in Table 1. It was found to be the highest in Colombia and Argentina, followed by Italy, and it presented the lowest rates in the United Kingdom and Switzerland. Notably, the rate of multidrug resistance among P. aeruginosa isolates increased significantly over the study period (P = 0.033) (Fig. 1). The distribution of the rates of multidrug resistance according to the centers and the number of episodes included is shown in fig. S1 in the supplemental material.
TABLE 1.
Rates of multidrug resistance among Pseudomonas aeruginosa isolates by country
| Country | No. of episodes included | Rate (%) of MDRa P. aeruginosa | 95% confidence interval |
|---|---|---|---|
| Colombia | 19 | 57.89 | 33.50–79.74 |
| Argentina | 47 | 46.81 | 32.11–61.92 |
| Italy | 123 | 40.65 | 31.88–49.87 |
| Chile | 13 | 30.77 | 9.09–61.42 |
| Slovakia | 32 | 25 | 11.46–43.40 |
| Turkey | 114 | 24.56 | 16.98–33.50 |
| Spain | 642 | 23.21 | 19.99–26.67 |
| Brazil | 125 | 19.2 | 12.70–27.20 |
| Lebanon | 22 | 18.18 | 5.18–40.28 |
| Germany | 41 | 12.2 | 4.08–26.20 |
| Switzerland | 28 | 10.71 | 2.26–28.22 |
| United Kingdom | 11 | 9.091 | 0.23–41.27 |
MDR, multidrug resistant.
FIG 1.
Evolution of multidrug resistance rates among Pseudomonas aeruginosa isolates from 2006 to 2018.
Information regarding whether the P. aeruginosa strains were MDR or not was provided for all the isolates. A detailed susceptibility profile was available for 1,156 P. aeruginosa strains. Of them, 18.6% were resistant to cefepime, 21.9% were resistant to ceftazidime, 25.2% were resistant to piperacillin-tazobactam, 23% were resistant to meropenem, 25.4% were resistant to imipenem, 26.7% were resistant to ciprofloxacin, 9.4% were resistant to amikacin, 11.3% were resistant to tobramycin, and 1.2% were resistant to colistin. The activities of fosfomycin, ceftazidime-avibactam, and ceftolozane-tazobactam were tested against 312, 30, and 39 strains, respectively, and the rates of resistance were 10.4%, 0.7%, and 1%, respectively.
Clinical characteristics.
The baseline and clinical characteristics of all 1,217 P. aeruginosa BSI episodes are reported in Table 2. The great majority of episodes occurred in patients with hematological malignancies (75.3%), with acute leukemia (44.7%) being the most frequent underlying disease. Lung cancer (29.6%) was the most common malignancy among patients with solid tumors. Profound neutropenia (<0.1 × 109/liter) was present in 61.5% of the cases, and 23.8% of the cases were in HSCT recipients. An endogenous source (37.4%) and pneumonia (25.6%) were the most frequent sources of BSI. More than one-third of the patients (33.9%) presented with septic shock. More than 50% of the patients had received antibiotics in the previous month.
TABLE 2.
Baseline and clinical characteristics of neutropenic cancer patients with Pseudomonas aeruginosa bloodstream infectiona
| Characteristic | Value for patients with: |
P value | ||
|---|---|---|---|---|
| Non-MDR P. aeruginosa infection (n = 908) | MDR P. aeruginosa (n = 309) | Study population (n = 1,217) | ||
| Mean (SD) age (yr) | 58.9 (16.2) | 54.4 (15.5) | 57.8 (16.2) | <0.001 |
| No. (%) of male patients | 577 (63.5) | 174 (56.3) | 751 (61.7) | 0.028 |
| No. (%) of patients with: | ||||
| Hematological disease | 641 (70.6) | 276 (89.3) | 917 (75.3) | <0.001b |
| Acute leukemia/myelodysplastic syndrome | 287 (31.6) | 164 (53) | 451 (37) | 0.001 |
| Lymphoma | 235 (25.8) | 71 (22.9) | 306 (25.1) | 0.336 |
| Multiple myeloma/Waldenström disease | 59 (6.4) | 15 (4.8) | 74 (6) | |
| Other | 60 (6.6) | 26 (8.4) | 46 (3.7) | |
| HSCT | 182 (26.6) | 108 (35.0) | 290 (23.8) | |
| Allogeneic HSCT | 97 (10.6) | 80 (25.8) | 177 (14.5) | |
| Autologous HSCT | 85 (9.3) | 28 (9) | 113 (9.2) | |
| GVHD | 49 (5.3) | 29 (9.3) | 78 (6.4) | |
| Solid tumor | 267 (29.4) | 33 (10.6) | 300 (24.6) | <0.001b |
| Lung cancer | 79 (8.7) | 10 (3.2) | 89 (7.3) | |
| Lower gastrointestinal tract tumor | 28 (3) | 2 (0.6) | 30 (2.4) | |
| Urinary tract cancer | 24 (2.6) | 5 (15.1) | 29 (2.3) | |
| Breast cancer | 28 (3) | 0 | 28 (2.3) | |
| Head and neck tumor | 22 (2.4) | 4 (0.3) | 26 (2.1) | |
| Other | 86 (9.4) | 12 (3.8) | 98 (8.05) | |
| Comorbidities | 453 (52.1) | 133 (45.7) | 586 (50.5) | 0.067 |
| Diabetes mellitus | 75 (8.2) | 11 (3.5) | 86 (7) | 0.009 |
| Chronic heart disease | 106 (11.6) | 44 (14.2) | 150 (12.3) | 0.236 |
| Chronic obstructive pulmonary disease | 79 (8.7) | 21 (6.7) | 100 (8.2) | 0.387 |
| Chronic liver disease | 25 (2.7) | 11 (3.5) | 36 (2.9) | 0.566 |
| Chronic renal disease | 26 (2.8) | 6 (1.9) | 32 (2.6) | 0.528 |
| Profound neutropenia (<0.1 × 109/liter) | 526 (59.7) | 202 (66.9) | 728 (61.5) | 0.032 |
| High-risk MASCC index score (<21 points) | 551 (67.2) | 213 (74.7) | 764 (69.1) | <0.001 |
| Grade III-IV mucositis | 111 (12.4) | 58 (19.1) | 169 (14.1) | 0.005 |
| Previous corticosteroid therapy (within 1 mo) | 456 (51.3) | 176 (58.1) | 632 (53) | 0.048 |
| Prior fluoroquinolone prophylaxis (within 1 mo) | 98 (10.9) | 97 (31.7) | 195 (16.2) | <0.001 |
| Prior antibiotic therapy (within 1 mo) | 414 (46.5) | 251 (81.8) | 665 (55.6) | <0.001 |
| Prior piperacillin-tazobactam therapy (within 1 mo) | 98 (10.8) | 101 (32.7) | 199 (16.4) | <0.001 |
| Prior antipseudomonal carbapenem therapy (within 1 mo) | 98 (10.8) | 103 (33.3) | 201 (16.5) | <0.001 |
| Prior antipseudomonal cephalosporin therapy (within 1 mo) | 72 (7.9) | 26 (8.4) | 98 (8.1) | 0.80 |
| Prior/current ICU admission | 78 (8.6) | 49 (15.9) | 127 (10.5) | 0.001 |
| Previous hospitalization (within 3 mo) | 553 (61.5) | 191 (62.6) | 744 (61.8) | 0.782 |
| Nosocomial acquisition | 177 (19.5) | 40 (12.9) | 694 (57.0%) | <0.001 |
| Urinary catheter | 122 (13.8) | 84 (28.1) | 206 (17.4) | <0.001 |
| Intravascular catheter | 626 (68.9) | 282 (91.6) | 908 (74.7) | <0.001 |
| Central venous catheter | 452 (49.7) | 164 (53) | 692 (56.8) | |
| Axillary temp of ≥38°C | 797 (88.6) | 285 (92.5) | 1,082 (88.9) | 0.062 |
| Septic shock at presentation | 271 (29.9) | 140 (45.5) | 411 (33.9) | <0.001 |
| Ecthyma gangrenosum | 33 (3.7) | 18 (5.9) | 51 (4.2) | 0.135 |
| Polymicrobial bloodstream infection | 177 (19.5) | 40 (12.9) | 217 (17.8) | 0.012 |
| High-risk bloodstream infection | 420 (52.2) | 141 (48.5) | 561 (51.2) | 0.308 |
| Source of bloodstream infection | ||||
| Endogenous source | 351 (38.7) | 104 (33.7) | 455 (37.4) | 0.022 |
| Pneumonia | 226 (24.9) | 85 (27.5) | 311 (25.6) | |
| Intravascular catheter infection | 74 (8.2) | 38 (12.3) | 112 (9.2) | |
| Neutropenic enterocolitis | 60 (6.6) | 11 (3.5) | 71 (5.8) | |
| Skin and soft tissue infection | 46 (5.1) | 24 (7.7) | 70 (5.7) | |
| Other abdominal | 50 (5.5) | 8 (2.5) | 58 (4.7) | |
| Urinary tract infection | 37 (4.1) | 14 (4.5) | 51 (4.1) | |
| Perianal abscess | 26 (2.8) | 8 (2.5) | 34 (2.8) | |
| Unknown | 11 (1.2) | 5 (1.6) | 16 (1.3) | |
| Otherc | 27 (3.0) | 12 (3.9) | 39 (3.2) | |
MDR, multidrug resistant; HSCT, hematopoietic stem cell transplant; GVHD, graft-versus-host disease; MASCC, Multinational Association for Supportive Care in Cancer; ICU, intensive care unit.
Comparison of solid tumor versus hematological disease.
Other consists of mucositis, n = 24; odontogenic, n = 9; sinusitis, n = 4; otitis, n = 2.
Antibiotic treatment and outcomes.
The early and overall case fatality rates for the entire cohort were 27.8% and 40.1%, respectively, and they were particularly high in patients with high-risk BSI (33.9% and 48.7%, respectively). To assess the impact of antimicrobial resistance on the patients’ outcomes, we analyzed the rates of adequateness of empirical antibiotic therapy only in the 1,000 monomicrobial episodes. In this cohort, early and overall case fatality rates were 28.0% and 40.4%, respectively. Overall, 187 patients (18.7%) received inadequate initial empirical antibiotic therapy, of which 131 (70.1%) had an infection due to an MDR strain (P < 0.001). Also, the rates of persistent BSI (19.2% versus 7.4%, P < 0.001), early case fatality rates (38.6% versus % 22.8%, P < 0.001), and overall case fatality rates (56.2% versus 32.6%, P < 0.001) were significantly higher in patients infected with MDR strains than in those infected with susceptible strains.
Clinical prediction tool for multidrug resistance.
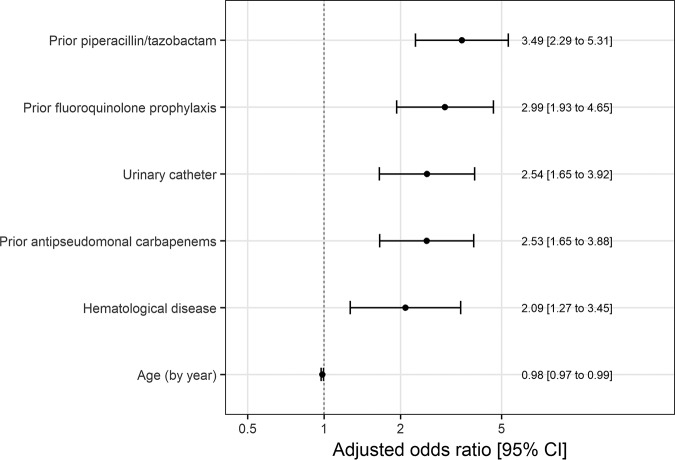
The variables included in the final model were age (continuous variable), underlying disease (hematological malignancy versus solid tumor), fluoroquinolone prophylaxis, prior therapy with piperacillin-tazobactam, prior therapy with antipseudomonal carbapenems, the presence of a urinary catheter, and center (Fig. 2). The percentage of the time that each factor appeared in all the estimated models is shown in Table S1. All the variables included in the model were found to be associated with multidrug resistance, except for older age, which was found to protect against multidrug resistance development.
FIG 2.
Odds ratio and 95% confidence intervals for multidrug resistance predictors included in the final model.
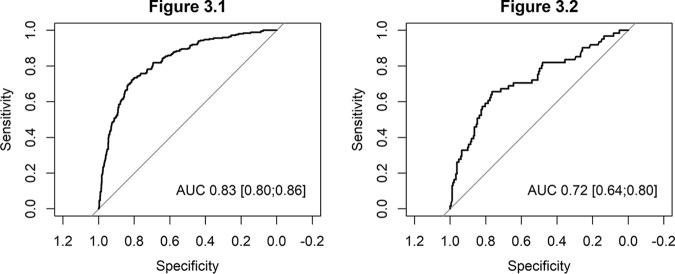
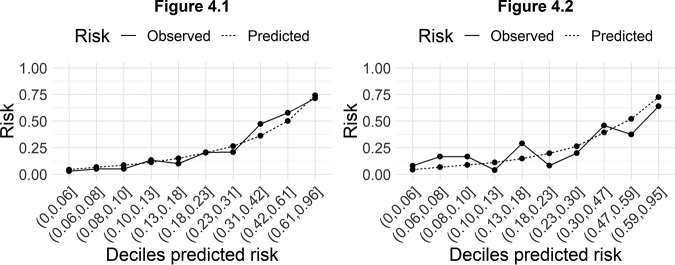
The predictive model obtained in the derivation cohort had excellent discrimination, with an area under the receiver operating characteristic (ROC) curve (AUC) of 0.82 (95% confidence interval [CI], 0.79 to 0.85) (Fig. 3, left). The observed probability corresponded well to the predicted probability, both on average and over the whole range of predictions. A linear regression model had an intercept at 0 and a slope of 1 for the relation between observed and predicted multidrug resistance (Fig. 4, left).
FIG 3.
(Left) Area under the curve of the predictive model of multidrug resistance in patients with Pseudomonas aeruginosa bloodstream infection in the derivation cohort. (Right) Area under the curve of the predictive model of multidrug resistance in patients with Pseudomonas aeruginosa bloodstream infection in the validation cohort.
FIG 4.
(Left) Observed versus predicted risk of multidrug-resistant Pseudomonas aeruginosa bloodstream infection, stratified by deciles of predicted risk, in the derivation cohort. (Right) Observed versus predicted multidrug-resistant Pseudomonas aeruginosa bloodstream infection, stratified by deciles of predicted risk, in the validation cohort.
Internal validation also showed a fair discrimination, with an AUC of 0.72 (95% CI, 0.63 to 0.80) (Fig. 3, right) and good agreement between prediction and observation (Fig. 4, right).
We developed an intuitive online tool to calculate the risk of multidrug resistance using the clinical prediction model that we estimated (http://ubidi.shinyapps.io/ironic). Whether the tool is suitable for use as an intervention to support treatment decisions should be evaluated externally and locally (19). The explanation of how to use the tool is provided in the supplemental material.
DISCUSSION
Using data from a large international cohort, we have developed a clinical predictive model that allows us to accurately identify neutropenic cancer patients at high risk of BSI due to MDR P. aeruginosa. This clinical tool may benefit these patients by improving the administration of adequate empirical antibiotic treatment, and it may also help optimize the efficacy of antibiotic stewardship programs.
Of particular concern, we found an overall high rate of multidrug resistance among P. aeruginosa isolates, and importantly, a significant increase was observed over time. These findings are in line with other reports that focused on patients with hematological malignancies (11–13, 15, 17), although most of those studies were conducted in the same geographical area. The emergence of resistance among P. aeruginosa isolates causing infection in neutropenic patients is worrisome, since the administration of inadequate empirical antibiotic therapy severely impairs patient outcomes (11, 12, 15). Indeed, we found significantly higher early and overall mortality rates in patients with MDR P. aeruginosa BSI. In addition, in a recent study focused on patients with acute leukemia and BSI, inadequate empirical antibiotic therapy was the only modifiable risk factor independently associated with mortality in patients with MDR P. aeruginosa BSI (20). Therefore, identifying patients at risk of infection due to resistant strains is imperative in order to administer broad-spectrum empirical antibiotics based on local susceptibility patterns and improve patient outcomes. The development of a predictive model could be helpful in assessing and stratifying this risk, and the use of a straightforward web-based calculator would facilitate the prompt application of the predictive model in an easy way at the bedside.
The most important factors associated with the development of antibiotic resistance in our predictive model were exposure to β-lactam antibiotics, such as piperacillin-tazobactam and antipseudomonal carbapenems, and, more importantly, the use of fluoroquinolone prophylaxis. The use of broad-spectrum antipseudomonal β-lactams is frequent in cancer patients, who may present repeated chemotherapy-induced episodes of febrile neutropenia. Nevertheless, these antibiotics and, in particular, carbapenems should be used reasonably, and the duration of empirical antibiotic treatments can be safely shortened, particularly in asymptomatic patients, regardless of their neutrophil count, as we recently demonstrated in a randomized clinical trial (21). Other researchers have also suggested that exposure to fluoroquinolones is a risk factor for infection due to MDR Gram-negative bacilli in cancer patients (16, 17, 22, 23). Hakki et al. recently reported the association between fluoroquinolone prophylaxis and breakthrough BSI with P. aeruginosa strains that are not susceptible to meropenem, probably due to mutations increasing efflux pump activity (16). In addition, fluoroquinolone exposure has been associated with an increased risk of Clostridioides difficile and methicillin-resistant Staphylococcus aureus infection (24, 25). This is of special concern, since the use of universal prophylaxis with fluoroquinolones in neutropenic patients is still routine practice in some institutions. In the absence of current evidence of its impact on mortality, this practice should be seriously reconsidered (26).
The presence of a urinary catheter has previously been reported to be an independent risk factor for MDR GNB BSI in cancer patients (27). This finding could be hypothetically explained by the association between the use of urinary catheters and the increased risk of urinary tract infections. Even though the rate of BSI that originated in the urinary tract in our study was found to be low, its diagnosis could have been limited in our patients, whose inflammatory response and symptoms would have been decreased due to their neutropenia, therefore leading to a low number of urine cultures being performed.
The main strength of the present study is the large number of participating centers from 12 countries around the world. This confers a clear advantage related to a larger sample size and more generalizable results. Moreover, to estimate the clinical prediction model, we used a robust methodology, including multiple imputations to account for missing data, bootstrapping to minimize overfitting, and a validation process. Also, the center effect was addressed by including this variable in the model. However, there are some limitations that should be acknowledged. This was a retrospective study, so the main limitation of the data is related to the potential effects of unmeasured variables and residual confounding. Also, different antimicrobial susceptibility testing methods and different interpretive criteria were used among the different centers, and breakpoints changed during the study period. In addition, the model was validated with data that, while not used to estimate the model, were derived from the same sample, so real external validation is required and is anticipated in the near future. Finally, since this model is specific for MDR P. aeruginosa, its clinical utility will be limited to patients who are found to have a BSI due to P. aeruginosa for which the susceptibility testing results are pending.
In conclusion, the prevalence of multidrug resistance among P. aeruginosa isolates causing BSI in neutropenic cancer patients is an alarming emerging problem. The reasonable use of broad-spectrum β-lactams and, in particular, carbapenems is strongly recommended in order to limit the development of resistance. In addition, the use of universal fluoroquinolone prophylaxis in neutropenic patients should be reconsidered in the current era of increasing antimicrobial resistance. Even though it needs external validation, the proposed prediction model achieves good discrimination and calibration, allowing the risk of BSI due to MDR P. aeruginosa to be estimated in this high-risk population. The application of a predictive model using a web-based calculator would be a simple strategy to identify those patients at the highest risk of infection due to MDR strains who may benefit from broad-spectrum antibiotic coverage, according to the local susceptibility patterns, and it could also help avoid the use of broad-spectrum antibiotics in patients with a low risk of resistance development.
MATERIALS AND METHODS
Study design, patients, and setting.
This study is part of the IRONIC project, a multicenter, international, retrospective cohort study of adult (age, ≥18 years) neutropenic oncohematological patients, including hematopoietic stem cell transplant (HSCT) recipients, diagnosed with at least one episode of P. aeruginosa BSI from 1 January 2006 to 31 May 2018. Subsequent episodes caused by P. aeruginosa occurring in the same patient were included in the study if the interval between them was >1 month.
For this study, all episodes of P. aeruginosa BSI included in the IRONIC database were included. Patients were recruited retrospectively from 34 centers in 12 countries: Spain (n = 14 centers), Turkey (n = 4), Brazil (n = 3), Italy (n = 3), Argentina (n = 2), Germany (n = 2), Chile (n = 1), Colombia (n = 1), Lebanon (n = 1), Slovakia (n = 1), Switzerland (n = 1), and the United Kingdom (n = 1). The number of patients recruited at each center is provided in the supplemental material. The study was conducted in accordance with the STROBE recommendations, and the protocol has been published elsewhere (28).
The protocol of the study was approved by all the appropriate regulatory agencies and local research ethics committees. The need for informed consent and information sheets was waived by the ethics committees because of the retrospective nature of the study.
Definitions.
Neutropenia was defined as an absolute neutrophil count of <0.5 × 109/liter. The Multinational Association for Supportive Care in Cancer (MASCC) score was calculated as described elsewhere (29). A BSI was considered low risk when the infection originated in the urinary tract or was secondary to a vascular catheter infection or to gut translocation (endogenous source). Episodes of BSI originating from other sources were considered high-risk BSI (30). Antimicrobial therapy administered before susceptibility results were available was considered empirical therapy. Empirical antibiotic therapy was considered adequate when it included at least one antibiotic active in vitro against the P. aeruginosa strain causing the infection. A BSI was considered persistent if the blood cultures were positive after the first 48 h of adequate antibiotic therapy. An early case fatality was defined as death from any cause within 7 days of BSI onset. Overall 30-day case fatality was defined as death from any cause within 30 days of BSI onset.
Microbiological studies.
Clinical samples were processed at the microbiology laboratory of each participating center in accordance with standard operating procedures. P. aeruginosa was identified using standard microbiological techniques at each center. In vitro susceptibility was determined according to the EUCAST recommendations in the great majority of centers (31). In the Lebanese center and in one center from Argentina, the CLSI cutoffs were used, and in the center from the United Kingdom, BSAC recommendations were used before 2016 (32). P. aeruginosa isolate phenotypes were stratified in accordance with recent standard definitions (33). We determined an isolate to be an MDR P. aeruginosa isolate when it was not susceptible to at least one agent in three or more of the following antimicrobial categories: aminoglycosides, antipseudomonal carbapenems, antipseudomonal fluoroquinolones, antipseudomonal cephalosporins, antipseudomonal penicillins plus β-lactamase inhibitors, monobactams, fosfomycin, and polymyxins. Moreover, we determined an isolate to be an extensively drug resistant (XDR) P. aeruginosa isolate when it was not susceptible to at least one agent in all but two or fewer of these antimicrobial categories.
Statistical analysis.
The original cohort was randomly split into a derivation cohort that included 80% of the patients and a validation cohort that consisted of the rest of the patients.
The set of candidate risk factors to be included in the model was extracted from the IRONIC case report form, and it mainly included sociodemographic variables, underlying conditions (hematological malignancy versus solid tumor and comorbidities), administration of immunosuppressants and antibiotics within the last 30 days, indwelling catheters, prior hospitalization or intensive care unit (ICU) admission, and infection-related variables, including MASCC index score, shock, source of BSI, etc.
A mixed logistic regression model was used to estimate a predictive model for the development of multidrug resistance based on the patient’s medical history and clinical findings. The decision to use a mixed model was based on analysis of the variability in the rates of MDR infection between centers using funnel plots. Such plots allowed us to compare rates between centers/countries taking into account the number of patients in each.
First, we performed a descriptive analysis of the factors assessed for the development of MDR infections. Multiple imputation with chained equations (MICE) was then used to minimize the impact of missing data for those variables for which data were missing (34). Ten data sets were created, using the Gaussian normal regression method to impute continuous variables (MASCC risk index score) and the binomial logistic regression method to impute categorical variables (high-risk BSI, high-risk MASCC index score, comorbidities, the presence of a urinary catheter, hypotension, corticosteroid use, severe mucositis, prior hospital admission, prior fluoroquinolone prophylaxis, orotracheal intubation, ICU admission, a prior episode of BSI during hospitalization, the presence of any venous catheter, and septic shock). Each imputed data set was sampled by bootstrapping with replacement 100 times, totaling 1,000 samples. Models were fitted for each of the 1,000 samples using backwards elimination. Predictors retained in more than 80% of the 1,000 estimated models were considered for inclusion in the final model. A model including the predictors selected was then estimated using the 10 imputed samples and adjusting the coefficients and standard errors for the variability between imputations according to the Rubin rules (34, 35). Finally, discrimination was assessed by estimating the area under the ROC curve (AUC). This area indicates the probability that a patient with an infection due to an MDR strain had a higher predicted probability than a patient without one for random pairs of patients with and without such an infection. To assess calibration, observed versus expected episodes of MDR BSI were compared graphically by deciles of predicted risk. All validation analyses performed with the derived sample were also repeated with the reserved sample for validation (36). The TRIPOD checklist for development and validation of predictive models is provided in the supplemental material.
All analyses were performed with a two-sided significance level of 0.05 and R software, version 3.5 (37).
Supplementary Material
ACKNOWLEDGMENTS
The following are IRONIC study investigators: Guillermo Cuervo and Francesc Escrihuela-Vidal, Infectious Diseases Department, and Fe Tubau, Microbiology Department, Bellvitge University Hospital, IDIBELL, Barcelona, Spain; Cristian Tebé, Statistics Advisory Service, Institute of Biomedical Research of Bellvitge, Rovira i Virgili University; Marisol Rodríguez Arias, Oncology Department, Institut Català d’Oncologia (ICO)-Hospital Duran i Reynals, IDIBELL, Barcelona, Spain; Juan Aguilar-Company, Oncology Department, and Nieves Larrosa, Microbiology Department, Vall d’Hebron University Hospital, Barcelona, Spain; Celia Cardozo and Carolina Garcia-Vidal, Infectious Diseases Department, Hospital Clínic i Provincial, Barcelona, Spain; Ibrahim Karim-Yaqub, Instituto do Câncer do Estado de São Paulo da Faculdade de Medicina da Universidade de São Paulo, Brazil; Raffaella Greco, Haematology and Bone Marrow Transplantation Unit, and Paola Cichero, Microbiology and Virology Laboratory, IRCCS San Raffaele Scientific Institute, Milan, Italy; Caglayan Merve Ayaz, Infectious Diseases Department, Hacettepe University School of Medicine, Ankara, Turkey; Roberto Céspedes, Haematology Department, and Leire López-Soria, Microbiology Department, Cruces University Hospital, Bilbao, Spain; Laura Magnasco, Division of Infectious Diseases, University of Genoa (DISSAL), and Ospedale Policlinico San Martino, Genoa, Italy; Jesús Fortún, Infectious Diseases Department, Ramón y Cajal Hospital, Madrid, Spain; Diego Torres, Infectious Diseases Section, Department of Medicine, Centro de Educación Médica e Investigaciones Clínicas (CEMIC), Buenos Aires, Argentina; Anna Boté, Infectious Diseases Department, and Mateu Espasa, Microbiology Department, Parc Taulí University Hospital, Sabadell, Barcelona, Spain; Mia Hold Montaguti, Infectious Diseases Department, Hospital Erasto Gaertner, Curitiba, Brazil; Pierre-Yves Bochud and Oriol Manuel, Infectious Diseases Department, Lausanne University Hospital (CHUV), Lausanne, Switzerland; Salvador Tabares Carrasco and Josefina Serrano López, Haematology Department, Reina Sofía University Hospital-IMIBIC-UCO, Córdoba, Spain; Hartmut Bertz, Haematology-Oncology Department, and Siegbert Rieg, Division of Infectious Diseases, Department of Medicine II, University of Freiburg Medical Center and Faculty of Medicine, Freiburg, Germany; Marina de Cueto and Jesús Rodríguez-Baño, Clinical Unit of Infectious Diseases, Microbiology and Preventive Medicine, University Hospitals Virgen Macarena and Virgen del Rocío-IBiS, Department of Medicine, University of Seville, Seville, Spain; Manuel Lizasoain and José María Aguado, Infectious Diseases Unit, 12 de Octubre University Hospital, Madrid, Spain; Juan Pablo Horcajada, Infectious Diseases Service, Hospital del Mar, Infectious Pathology and Antimicrobials Research Group (IPAR), Institut Hospital del Mar d’Investigations Mèdiques (IMIM), Universitat Autònoma de Barcelona (UAB), CEXS-Universitat Pompeu Fabra, Barcelona, Spain; Saeed El Zein and Jean-Francois Jabbour, Infectious Diseases Division, American University of Beirut Medical Center, Beirut, Lebanon; Ayse Uyan-Onal, Department of Infectious Diseases and Clinical Microbiology, Ege University Faculty of Medicine, Izmir, Turkey; Arzu Nazli-Zeka Department of Infectious Diseases and Clinical Microbiology, Dokuz Eylul University Faculty of Medicine, Izmir, Turkey; Begoña Palop, Microbiology Department, Hospital Regional de Málaga, Málaga, Spain; Lina Clemencia Correa, Clinica Maraya, Pereira, Colombia; Amanda Aparecida da Silva Machado and João Pedro Silva Tonhá, Instituto Oswaldo Cruz, Fundação Oswaldo Cruz, Rio de Janeiro, Brazil; Georg Maschmeyer, Department of Haematology, Oncology and Palliative Care, Klinikum Ernst von Bergmann, Academic Teaching Hospital of the Charité University Medical School, Berlin, Germany; Jose Munita, Instituto de Ciencias e Innovación en Medicina, Facultad de Medicina, Clínica Alemana Universidad del Desarrollo, Santiago de Chile, Chile, and Millennium Initiative for Collaborative Research on Bacterial Resistance (MICROB-R); Matteo Bassetti and Nadia Castaldo, Infectious Diseases Clinic, Department of Medicine, University of Udine and Azienda Sanitaria Universitaria Integrata, Udine, Italy; and Paloma Sangro del Alcázar, Internal Medicine Department, Navarra University Clinic, Pamplona, Spain.
We thank the ESGBIES and the ESGICH study groups for supporting the study.
This study was supported by the Spanish Plan Nacional de I+D+i 2013‐2016 and the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (grant REIPI RD16/0016/0001), cofinanced by the European Development Regional Fund A Way To Achieve Europe, Operative Program Intelligent Growth 2014‐2020.
A.-S.B. received a grant from Promex Stiftung fur die Forschung (via Carigest SA) and funding from Gilead to attend the ECCMID Congress (2018). O.R.S. received speaker honoraria from MSD, Astellas, Novartis, and Pfizer. S.S.K. received speaker honoraria from Pfizer, MSD, Astellas. F.H. received speaker honoraria from MSD, and Pfizer and a research and educational grant from Pfizer. The rest of the authors declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Collaborators: Guillermo Cuervo, Francesc Escrihuela-Vidal, Fe Tubau, Cristian Tebé, Marisol Rodríguez Arias, Juan Aguilar-Company, Nieves Larrosa, Celia Cardozo, Carolina Garcia-Vidal, Ibrahim Karim-Yaqub, Raffaella Greco, Paola Cichero, Caglayan Merve Ayaz, Roberto Céspedes, Leire López-Soria, Laura Magnasco, Jesús Fortún, Diego Torres, Anna Boté, Mateu Espasa, Mia Hold Montaguti, Pierre-Yves Bochud, Oriol Manuel, Salvador Tabares Carrasco, Josefina Serrano López, Hartmut Bertz, Siegbert Rieg, Marina de Cueto, Jesús Rodríguez-Baño, Manuel Lizasoain, José María Aguado, Juan Pablo Horcajada, Saeed El Zein, Jean-Francois Jabbour, Ayse Uyan-Onal, Arzu Nazli-Zeka, Begoña Palop, Lina Clemencia Correa, Amanda Aparecida da Silva Machado, João Pedro Silva Tonhá, Georg Maschmeyer, Jose Munita, Matteo Bassetti, Nadia Castaldo, and Paloma Sangro del Alcázar
REFERENCES
- 1.Gudiol C, Bodro M, Simonetti A, Tubau F, González-Barca E, Cisnal M, Domingo-Domenech E, Jiménez L, Carratalà J. 2013. Changing aetiology, clinical features, antimicrobial resistance, and outcomes of bloodstream infection in neutropenic cancer patients. Clin Microbiol Infect 19:474–479. doi: 10.1111/j.1469-0691.2012.03879.x. [DOI] [PubMed] [Google Scholar]
- 2.Gustinetti G, Mikulska M. 2016. Bloodstream infections in neutropenic cancer patients: a practical update. Virulence 7:280–297. doi: 10.1080/21505594.2016.1156821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikulska M, Viscoli C, Orasch C, Livermore DM, Averbuch D, Cordonnier C, Akova M, Fourth European Conference on Infections in Leukemia Group (ECIL-4), a joint venture of EBMT, EORTC, ICHS, ELN and ESGICH/ESCMID . 2014. Aetiology and resistance in bacteraemias among adult and paediatric haematology and cancer patients. J Infect 68:321–331. doi: 10.1016/j.jinf.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Trecarichi EM, Tumbarello M, Spanu T, Caira M, Fianchi L, Chiusolo P, Fadda G, Leone G, Cauda R, Pagano L. 2009. Incidence and clinical impact of extended-spectrum-beta-lactamase (ESBL) production and fluoroquinolone resistance in bloodstream infections caused by Escherichia coli in patients with hematological malignancies. J Infect 58:299–307. doi: 10.1016/j.jinf.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Satlin MJ, Cohen N, Ma KC, Gedrimaite Z, Soave R, Askin G, Chen L, Kreiswirth BN, Walsh TJ, Seo SK. 2016. Bacteremia due to carbapenem-resistant Enterobacteriaceae in neutropenic patients with hematologic malignancies. J Infect 73:336–345. doi: 10.1016/j.jinf.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spanik S, Kukuckova E, Pichna P, Grausova S, Krupova I, Rusnakova V, Kralovicova K, Krchnakova A, Mrazova M, Lacka J, Koren P, Stopkova K, Nogova J, Demitrovicova A, Helpianska L, Krcmery V Jr.. 1997. Analysis of 553 episodes of monomicrobial bacteraemia in cancer patients: any association between risk factors and outcome to particular pathogen? Support Care Cancer 5:330–333. doi: 10.1007/s005200050083. [DOI] [PubMed] [Google Scholar]
- 7.Viscoli C, Varnier O, Machetti M. 2005. Infections in patients with febrile neutropenia: epidemiology, microbiology, and risk stratification. Clin Infect Dis 40(Suppl 4):S240–S245. doi: 10.1086/427329. [DOI] [PubMed] [Google Scholar]
- 8.Gratwohl A, Baldomero H, Gratwohl M, Aljurf M, Bouzas LF, Horowitz M, Kodera Y, Lipton J, Iida M, Pasquini MC, Passweg J, Szer J, Madrigal A, Frauendorfer K, Niederwieser D, Worldwide Network of Blood and Marrow Transplantation (WBMT) . 2013. Quantitative and qualitative differences in use and trends of hematopoietic stem cell transplantation: a global observational study. Haematologica 98:1282–1290. doi: 10.3324/haematol.2012.076349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kern WV, Roth JA, Bertz H, Götting T, Dettenkofer M, Widmer AF, Theilacker C, Hospital Infection Surveillance System for Patients with Hematologic/Oncologic Malignancies Study Group (ONCO-KISS) . 2019. Contribution of specific pathogens to bloodstream infection mortality in neutropenic patients with hematologic malignancies: results from a multicentric surveillance cohort study. Transpl Infect Dis 21:e13186. doi: 10.1111/tid.13186. [DOI] [PubMed] [Google Scholar]
- 10.Caselli D, Cesaro S, Ziino O, Zanazzo G, Manicone R, Livadiotti S, Cellini M, Frenos S, Milano GM, Cappelli B, Licciardello M, Beretta C, Aricò M, Castagnola E, Infection Study Group of the Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP) . 2010. Multidrug resistant Pseudomonas aeruginosa infection in children undergoing chemotherapy and hematopoietic stem cell transplantation. Haematologica 95:1612–1615. doi: 10.3324/haematol.2009.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trecarichi EM, Tumbarello M, Caira M, Candoni A, Cattaneo C, Pastore D, Fanci R, Nosari A, Vianelli N, Busca A, Spadea A, Pagano L. 2011. Multidrug resistant Pseudomonas aeruginosa bloodstream infection in adult patients with hematologic malignancies. Haematologica 96:e1–e3. doi: 10.3324/haematol.2010.036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattaneo C, Antoniazzi F, Casari S, Ravizzola G, Gelmi M, Pagani C, D'Adda M, Morello E, Re A, Borlenghi E, Manca N, Rossi G. 2012. P. aeruginosa bloodstream infections among hematological patients: an old or new question? Ann Hematol 91:1299–1304. doi: 10.1007/s00277-012-1424-3. [DOI] [PubMed] [Google Scholar]
- 13.Trecarichi EM, Pagano L, Candoni A, Pastore D, Cattaneo C, Fanci R, Nosari A, Caira M, Spadea A, Busca A, Vianelli N, Tumbarello M, HeMABIS Registry—SEIFEM Group, Italy . 2015. Current epidemiology and antimicrobial resistance data for bacterial bloodstream infections in patients with hematologic malignancies: an Italian multicentre prospective survey. Clin Microbiol Infect 21:337–343. doi: 10.1016/j.cmi.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 14.Averbuch D, Tridello G, Hoek J, Mikulska M, Akan H, Yanez San Segundo L, Pabst T, Özçelik T, Klyasova G, Donnini I, Wu D, Gülbas Z, Zuckerman T, Botelho de Sousa A, Beguin Y, Xhaard A, Bachy E, Ljungman P, de la Camara R, Rascon J, Ruiz Camps I, Vitek A, Patriarca F, Cudillo L, Vrhovac R, Shaw PJ, Wolfs T, O’Brien T, Avni B, Silling G, Al Sabty F, Graphakos S, Sankelo M, Sengeloey H, Pillai S, Matthes S, Melanthiou F, Iacobelli S, Styczynski J, Engelhard D, Cesaro S. 2017. Antimicrobial resistance in Gram-negative rods causing bacteremia in hematopoietic stem cell transplant recipients: intercontinental prospective study of the Infectious Diseases Working Party of the European Bone Marrow Transplantation Group. Clin Infect Dis 65:1819–1828. doi: 10.1093/cid/cix646. [DOI] [PubMed] [Google Scholar]
- 15.Kim HS, Park BK, Kim SK, Han SB, Lee JW, Lee DG, Chung NG, Cho B, Jeong DC, Kang JH. 2017. Clinical characteristics and outcomes of Pseudomonas aeruginosa bacteremia in febrile neutropenic children and adolescents with the impact of antibiotic resistance: a retrospective study. BMC Infect Dis 17:500. doi: 10.1186/s12879-017-2597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakki M, Humphries RM, Hemarajata P, Tallman GB, Shields RK, Mettus RT, Doi Y, Lewis JS. 2019. Fluoroquinolone prophylaxis selects for meropenem-nonsusceptible Pseudomonas aeruginosa in patients with hematologic malignancies and hematopoietic cell transplant recipients. Clin Infect Dis 68:2045–2052. doi: 10.1093/cid/ciy825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tofas P, Samarkos M, Piperaki E-T, Kosmidis C, Triantafyllopoulou I-D, Kotsopoulou M, Pantazatou A, Perlorentzou S, Poulli A, Vagia M, Daikos GL. 2017. Pseudomonas aeruginosa bacteraemia in patients with hematologic malignancies: risk factors, treatment and outcome. Diagn Microbiol Infect Dis 88:335–341. doi: 10.1016/j.diagmicrobio.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Viasus D, Puerta-Alcalde P, Cardozo C, Suárez-Lledó M, Rodríguez-Núñez O, Morata L, Fehér C, Marco F, Chumbita M, Moreno-García E, Fernández-Avilés F, Gutiérrez-Garcia G, Martínez JA, Mensa J, Rovira M, Esteve J, Soriano A, Garcia-Vidal C. 8 July 2019. Predictors of multidrug-resistant Pseudomonas aeruginosa in neutropenic patients with bloodstream infection. Clin Microbiol Infect doi: 10.1016/j.cmi.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Kappen TH, van Klei WA, van Wolfswinkel L, Kalkman CJ, Vergouwe Y, Moons K. 2018. Evaluating the impact of prediction models: lessons learned, challenges, and recommendations. Diagn Progn Res 2:11. doi: 10.1186/s41512-018-0033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Vidal C, Cardozo-Espinola C, Puerta-Alcalde P, Marco F, Tellez A, Agüero D, Romero-Santana F, Díaz-Beyá M, Giné E, Morata L, Rodríguez-Núñez O, Martinez JA, Mensa J, Esteve J, Soriano A. 2018. Risk factors for mortality in patients with acute leukemia and bloodstream infections in the era of multiresistance. PLoS One 13:e0199531. doi: 10.1371/journal.pone.0199531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilar-Guisado M, Espigado I, Martín-Peña A, Gudiol C, Royo-Cebrecos C, Falantes J, Vázquez-López L, Montero MI, Rosso-Fernández C, de la Luz Martino M, Parody R, González-Campos J, Garzón-López S, Calderón-Cabrera C, Barba P, Rodríguez N, Rovira M, Montero-Mateos E, Carratalá J, Pérez-Simón JA, Cisneros JM. 2017. Optimisation of empirical antimicrobial therapy in patients with haematological malignancies and febrile neutropenia (How Long study): an open-label, randomised, controlled phase 4 trial. Lancet Haematol 4:e573–e583. doi: 10.1016/S2352-3026(17)30211-9. [DOI] [PubMed] [Google Scholar]
- 22.Samonis G, Vardakas KZ, Kofteridis DP, Dimopoulou D, Andrianaki AM, Chatzinikolaou I, Katsanevaki E, Maraki S, Falagas ME. 2014. Characteristics, risk factors and outcomes of adult cancer patients with extensively drug-resistant Pseudomonas aeruginosa infections. Infection 42:721–728. doi: 10.1007/s15010-014-0635-z. [DOI] [PubMed] [Google Scholar]
- 23.Satlin MJ, Chavda KD, Baker TM, Chen L, Shashkina E, Soave R, Small CB, Jacobs SE, Shore TB, van Besien K, Westblade LF, Schuetz AN, Fowler VG, Jenkins SG, Walsh TJ, Kreiswirth BN. 2018. Colonization with levofloxacin-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae and risk of bacteremia in hematopoietic stem cell transplant recipients. Clin Infect Dis 67:1720–1728. doi: 10.1093/cid/ciy363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown KA, Khanafer N, Daneman N, Fisman DN. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 57:2326–2332. doi: 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couderc C, Jolivet S, Thiébaut AC, Ligier C, Remy L, Alvarez AS, Lawrence C, Salomon J, Herrmann JL, Guillemot D, Antibiotic Use and Staphylococcus aureus Resistant to Antibiotics (ASAR) Study Group . 2014. Fluoroquinolone use is a risk factor for methicillin-resistant Staphylococcus aureus acquisition in long-term care facilities: a nested case-case-control study. Clin Infect Dis 59:206–215. doi: 10.1093/cid/ciu236. [DOI] [PubMed] [Google Scholar]
- 26.Mikulska M, Cordonnier C. 2018. Fluoroquinolone prophylaxis during neutropenia: what can we expect nowadays? Clin Microbiol Infect 24:678–679. doi: 10.1016/j.cmi.2018.02.031. [DOI] [PubMed] [Google Scholar]
- 27.Gudiol C, Tubau F, Calatayud L, Garcia-Vidal C, Cisnal M, Sánchez-Ortega I, Duarte R, Calvo M, Carratalà J. 2011. Bacteraemia due to multidrug-resistant Gram-negative bacilli in cancer patients: risk factors, antibiotic therapy and outcomes. J Antimicrob Chemother 66:657–663. doi: 10.1093/jac/dkq494. [DOI] [PubMed] [Google Scholar]
- 28.Albasanz-Puig A, Gudiol C, Parody R, Tebe C, Akova M, Araos R, Bote A, Brunel AS, Calik S, Drgona L, García E, Hemmati P, Herrera F, Ibrahim KY, Isler B, Kanj S, Kern W, Maestro de la Calle G, Manzur A, Marin JI, Márquez-Gómez I, Martín-Dávila P, Mikulska M, Montejo JM, Montero M, Morales HMP, Morales I, Novo A, Oltolini C, Peghin M, Del Pozo JL, Puerta-Alcalde P, Ruiz-Camps I, Sipahi OR, Tilley R, Yáñez L, Gomes MZR, Carratalà J, for the IRONIC Study Group . 2019. Impact of antibiotic resistance on outcomes of neutropenic cancer patients with Pseudomonas aeruginosa bacteraemia (IRONIC study): study protocol of a retrospective multicentre international study. BMJ Open 9:e025744. doi: 10.1136/bmjopen-2018-025744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klastersky J, Paesmans M, Rubenstein EB, Boyer M, Elting L, Feld R, Gallagher J, Herrstedt J, Rapoport B, Rolston K, Talcott J. 2000. The Multinational Association for Supportive Care in Cancer Risk index: a multinational scoring system for identifying low-risk febrile neutropenic cancer patients. J Clin Oncol 18:3038–3051. doi: 10.1200/JCO.2000.18.16.3038. [DOI] [PubMed] [Google Scholar]
- 30.Elting LS, Rubenstein EB, Rolston K, Bodey GP. 1997. Outcomes of bacteremia in patients with cancer and neutropenia: observations from two decades of epidemiological and clinical trials. Clin Infect Dis 25:247–259. doi: 10.1086/514550. [DOI] [PubMed] [Google Scholar]
- 31.European Committee on Antimicrobial Susceptibility Testing. 2018. Breakpoint tables for interpretation of MICs and zone diameters, version 8.0.
- 32.BSAC. 2015. BSAC methods for antimicrobial susceptibility testing, version 14. BSAC, Birmingham, United Kingdom. [Google Scholar]
- 33.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 34.Heymans MW, van Buuren S, Knol DL, van Mechelen W, de Vet H. 2007. Variable selection under multiple imputation using the bootstrap in a prognostic study. BMC Med Res Methodol 7:33. doi: 10.1186/1471-2288-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Little RJA, Rubin DB. 2002. Statistical analysis with missing data. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 36.Collins GS, de Groot JA, Dutton S, Omar O, Shanyinde M, Tajar A, Voysey M, Wharton R, Yu LM, Moons KG, Altman DG. 2014. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Med Res Methodol 14:40. doi: 10.1186/1471-2288-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.