SUMMARY
In angiosperms, two sperm cells are transported and delivered by the pollen tube to the ovule to achieve double fertilization. Extensive communication takes place between the pollen tube and the female tissues until the sperm cell cargo is ultimately released. During this process, a pollen tube surface-located receptor complex composed of ANXUR1/2 (ANX1/2) and Buddha’s Paper Seal 1/2 (BUPS1/2) was reported to control the maintenance of pollen tube integrity by perceiving the autocrine peptide ligands rapid alkalinization factor 4 and 19 (RALF4/19). It was further hypothesized that pollen-tube rupture to release sperm is caused by the paracrine RALF34 peptide from the ovule interfering with this signaling pathway. In this study, we identified two Arabidopsis pollen-tube-expressed glycosylphosphatidylinositol-anchored proteins (GPI-APs), LORELEI-like-GPI-anchored protein 2 (LLG2) and LLG3, as co-receptors in the BUPS-ANX receptor complex. llg2 llg3 double mutants exhibit severe fertility defects. Mutant pollen tubes rupture early during the pollination process. Furthermore, LLG2 and LLG3 interact with ectodomains of both BUPSs and ANXURs, and this interaction is remarkably enhanced by the presence of RALF4/19 peptides. We further demonstrate that the N terminus (including a YISY motif) of the RALF4 peptide ligand interacts strongly with BUPS-ANX receptors but weakly with LLGs and is essential for its biological function, and its C-terminal region is sufficient for LLG binding. In conclusion, we propose that LLG2/3 serve as co-receptors during BUPS/ANX-RALF signaling and thereby further establish the importance of GPI-APs as key regulators in plant reproduction processes.
In Brief
GPI-anchored proteins (GPI-APs) are required for RALF ligand-CrRLK1L receptor-mediated signaling in regulating plant development, reproduction, and immunity. Ge et al. now report that two pollen-tube-specific GPI-APs, LLG2 and LLG3, control pollen tube integrity by serving as co-receptors of a BUPS-ANX receptor complex to perceive RALF4/19 signals.
INTRODUCTION
In flowering plants (angiosperms), the reproduction process depends on a unique double fertilization process involving the fusion of two sperm cells with two female gametes. One sperm cell fertilizes the egg cell and the other sperm the central cell to produce the zygote and endosperm, respectively, the major components of a future seed [1]. To achieve double fertilization, immotile sperm cells are delivered via pollen tubes, which interact during a tip-growing process with several cell types of the female pistil tissues to ultimately reach the deeply embedded female gametes inside the ovule [2–6]. During this long journey, proper maintenance of pollen tube integrity is critical for the success of double fertilization as tubes penetrate and interact with various female tissues and have to rupture timely to release their sperm cell cargo. Complex signaling mechanisms have evolved to ensure the precise control of each step in this crucial process. Recently, Catharanthus roseus RLK1-like (CrRLK1L) receptor-mediated signaling was discovered to play a key role for pollen tube cell wall integrity in Arabidopsis thaliana (hereafter named as Arabidopsis) [7–11]. The four CrRLK1L receptors ANX1/2 and BUPS1/2, all localized to the cell surface of Arabidopsis pollen tubes, were shown to work as a heteromeric receptor complex to coordinate the maintenance of pollen tube integrity and sperm cell discharge [8, 12–15]. Specifically, when pollen tubes grow, the self-generated autocrine signaling peptides RALF4/19 are perceived by the BUPS-ANX receptor complex to stabilize pollen tube cell integrity during the polarized cell growth process [14, 16]. However, when pollen tubes arrive inside ovules, ovule-generated paracrine signaling ligand RALF34 potentially competes with RALF4/19 for receptor binding and likely triggers pollen tube rupture and sperm release [11, 14]. Once pollen tubes enter the female gametophyte, they communicate with FERONIA (FER), another CrRLK1L receptor that was previously shown to possess important functions in pollen tube reception, root hair elongation, and plant immunity control [17–20]. Importantly, FER works together with a female-gametophyte-specific glycosylphosphatidylinositol-anchored protein (GPI-AP) named LORELEI (LRE) to control pollen tube reception [17, 21, 22]. Furthermore, a homolog of LRE, LORELEI-like-GPI-anchored protein 1 (LLG1), has been shown to facilitate efficient cell membrane localization of FER in root epidermal cells and participate in FER perception of the peptide ligand RALF1 to control root growth [23, 24]. It is unknown whether the four pollen-specific CrRLK1L receptors ANX1/2 and BUPS1/2 also require GPI-APs for their function. The receptors FER and FLS2 in plants [23–25] as well as receptors required for reproduction and development in animals interact with GPI-APs [23, 24, 26, 27], but it is mostly unclear how GPI-APs affect signaling processes.
Here, we report that two additional GPI-APs, LRE-like GPI-AP 2 (LLG2) and LLG3, are involved as co-receptors in BUPS/ANX-RALF4/19-mediated signaling and are both required to regulate pollen cell wall integrity. We further show how peptide ligands RALF4/19 affect the interactions among the receptor kinases and their interactions with LLG2/3. In conclusion, our findings further augment the understanding of the molecular mechanism on how BUPS/ANX-RALF-mediated signaling is regulated at the signal perception stage and underscore the multi-layered strategy used by flowering plants to ensure reproductive success.
RESULTS
LLG2 and LLG3 GPI-APs Are Strongly Expressed in Pollen and Pollen Tubes
We first conducted phylogenetic analysis of LRE-like GPI-AP proteins in the model plant Arabidopsis. We identified two so-far-uncharacterized members LLG2 and LLG3 sharing high sequence identity and thus clustering together (Figures 1A and S1A). High-throughput transcriptomic analysis [28, 29] showed that, unlike LRE that is predominately expressed in female tissues and seeds and LLG1 that is ubiquitously expressed in various tissues, LLG2 and LLG3 are highly expressed in pollen grains and pollen tubes (Figure 1A). Next, we generated transgenic plants expressing the GUS (β-glucoronidase) reporter gene driven by the LLG2 or LLG3 promoters, respectively. Strong GUS signals were detected in mature pollen grains in transgenic lines using both promoters, and very weak GUS signals were observed in young leaves of pLLG3::GUS transgenic plants (Figures 1B, 1E, and S1B–S1I). We also observed strong GUS activities of LLG2/LLG3 promoters in pollen tubes both in the in vitro germination assay and in vivo in the pistil (Figures 1C, 1D, 1F, and 1G). Subcellular localization studies of fusion proteins confirm the expression pattern. YFP-LLG3 was observed mainly in the pollen grains and pollen tubes accumulating predominately in vesicles-like structures. The same pattern was observed for mCitrine-LLG2 and mCitrine-LLG3 in transiently transformed tobacco pollen tubes (Figures S1J–S1M). Taken together, this suggests that the transcripts and proteins of both LLG2 and LLG3 especially accumulate in mature pollen and pollen tubes, implicating potential roles during pollen tube germination, growth, and/or fertilization.
Figure 1.
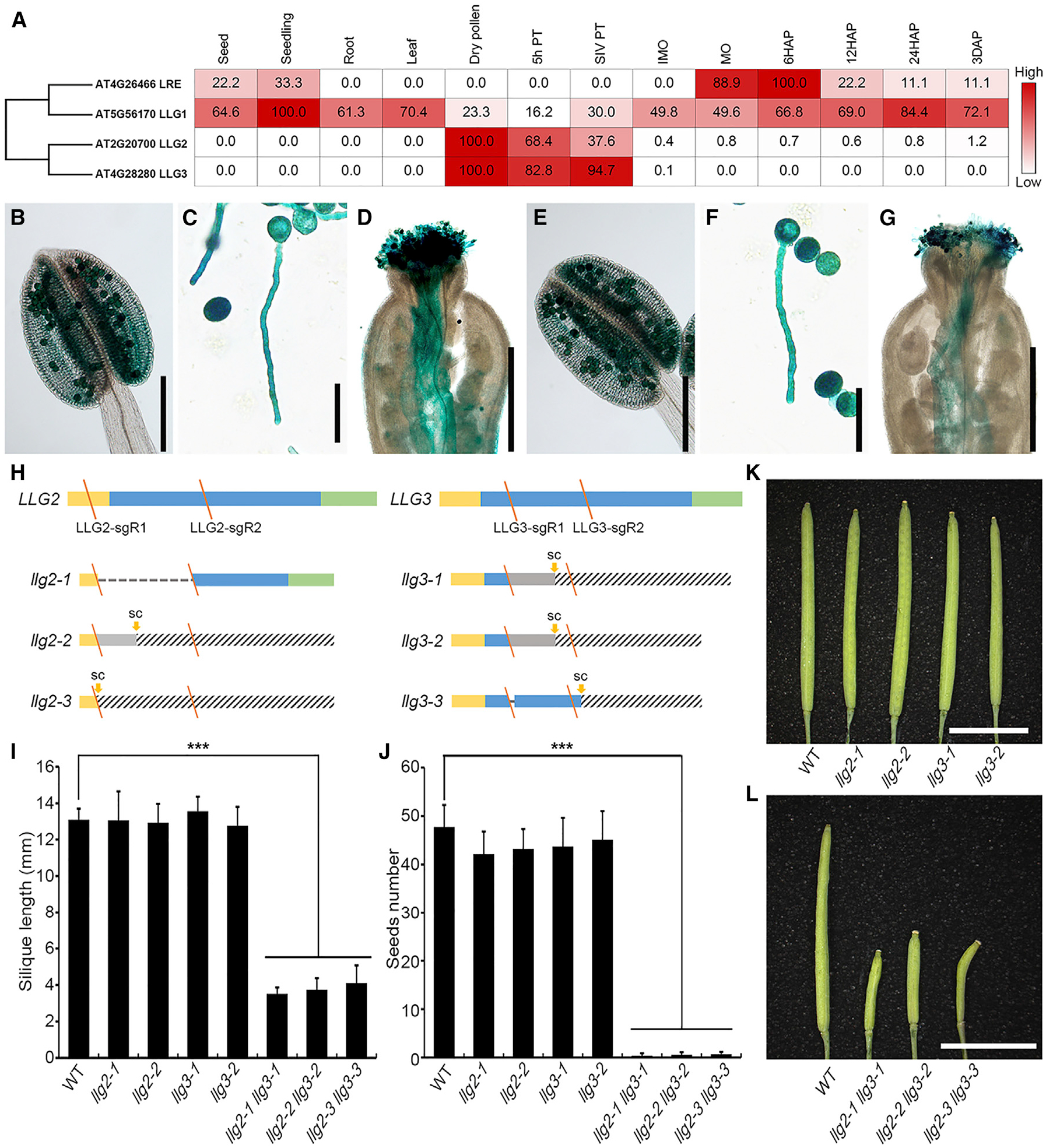
Pollen-Expressed LLG2/3 Function Redundantly in Controlling Fertility in Arabidopsis (A) Phylogenetic analysis of LLG proteins and heatmap showing LLG genes based on RNA sequencing (RNA-seq) data in Arabidopsis. Numbers in frames indicate relative expression levels of each gene in different tissues based on fragments per kilobase million (FPKM) values.
(B–G) LLG2 and LLG3 promoter-driven GUS expression pattern showing LLG2p::GUS activity in the anther (B), pollen tube in vitro (C), and pollen tube in vivo (D). LLG3p::GUS activity in the anther (E), pollen tube in vitro (F), and pollen tube in vivo (G) is shown.
(H) Mutant alleles of llg2 and llg3 generated by CRISPR/Cas9. Diagrams display coding sequences. Yellow rectangles indicate signal peptide (SP), and blue and green bars show predicted mature GPI-AP protein sequences and GPI-modified-dependent signal peptides (GPI-SPs), respectively. Slash (y) denotes localization of sgRNAs (sgRs), dashed lines indicate deletion of large fragments, gray rectangles indicate sequences of frameshift transcripts, and twill rectangles indicate sequences after a stop codon.
(I–L) Phenotypical analysis of single mutants and double mutants. Silique length (I) and seed number per silique (J) of WT and various mutants are shown. The data are mean ± SD; n = 9 (WT, llg2–1, llg2–2, llg3–1, llg2–1 llg3–1, and llg2–2 llg3–2); n = 8 (llg3–2); ***p < 0.001 (Student’s t test). (K) Siliques of WT and single mutants. (L) Siliques of WT and double mutants.
3 DAP, ovules 3 days after pollination; 5h PT, in vitro germinated pollen tubes after 5 h; 6/12/24 HAP, ovules after 6/12/24-h pollination; IMO/MO, immature/mature ovules; SC, stop codon; SIV PT, semi-in vivo germinated pollen tubes. Scale bars: 200 μm in (B and E); 50 μm in (C and F); 500 μm in (D and G); and 5 mm (K and L).
See also Figures S1, S2, and S3.
llg2 llg3 Double Mutants Generated by CRISPR/Cas9 Are Male Sterile
To analyze the biological function of LLG2/3, we next adopted the CRISPR/Cas9 technology to generate loss-of-function mutants for both genes [14, 30]. To ensure a high-efficiency genome-editing rate, we designed two single guide RNAs (sgRNAs) for each gene. The two sgRNAs for LLG2 targeted the coding sequence of the signal peptide and the predicted mature GPI-AP protein sequences, respectively, and those for LLG3 both targeted the GPI-AP region (Figure 1H). We successfully obtained multiple alleles of single and double mutants with different combination of llg2 and llg3 mutations (Figures 1H, S2A, and S2B). Compared to wild-type (WT) plants, single and double mutants showed normal vegetative growth and development, except that the llg2 llg3 double mutants exhibited severe defects in fertility (Figures 1I, 1J, and S3A). Statistical analysis of silique length and seed number per silique showed that both llg2 and llg3 single mutants were similar to those of the WT (silique length llg2 13.0 ± 1.3 mm and llg3 13.2 ± 1.0 mm, respectively; 42.7 ± 4.4 and 45.5 ± 5.2 seeds per silique, respectively; WT silique length 13.1 ± 0.6 mm and seed number 47.3 ± 3.6; Figures 1I–1K). However, llg2 llg3 produced much shorter siliques(3.8 ± 0.7 mm) with almost no seeds (0.6 ± 0.5/silique; Figures 1I, 1J, and 1L). In order to investigate whether this phenotype is caused by the loss of LLG2/3 functions, we transformed LLG2 into llg2 LLG3/llg3 mutant background and identified plants with llg2 llg3 homologous mutant background possessing normal siliques. This finding indicated that the fertility defects of llg2 llg3 were completely rescued by LLG2 (Figures S3B–S3D). Based on these results, we conclude that LLG2 and LLG3 act redundantly and play critical roles in fertility control.
Considering that LLG2/3 are primarily expressed in pollen grains and pollen tubes, we next investigated whether the fertility phenotypes in the llg2 llg3 double mutants were caused by male defects. Reciprocal crosses were performed between llg2 llg3 and WT or ms1 plants, a male-sterile mutant with normal functional pistils that are widely used as WT pistils in reciprocal crosses [31]. As shown in Figure 2A, silique length was comparable to that of WT ms1 pistils when WT pollen grains were pollinated to llg2 llg3 pistils. However, when WT pistils were pollinated with llg2 llg3 pollen grains, much shorter siliques with almost no seeds were obtained (Figures 2A, S3E, and S3F). This was consistent with severe male defects observed in llg2 llg3 mutant plants. To further determine whether it was a gametophytic and/or a sporophytic defect, we next performed crosses between llg2–1 llg3–1/LLG3 or llg2–1/LLG2 llg3–1 mutants with WT plants. We found that male transmission of llg2 llg3 double mutants was completely disrupted, and female transmission efficiency was comparable to WT controls (Table 1). In addition, although RNAi-mediated gene silencing of LLG2/3 did not fully abolish male fertility, male transmission deficiency and seed set were reduced by about 50% (Figures S4A–S4G), corroborating the findings from CRISPR/Cas9-generated mutants. In summary, the sterility phenotype of the double mutant llg2 llg3 was caused by a male gametophytic defect.
Figure 2. Loss of LLG2/3 Contributes to Defects in Pollen Tube Integrity.
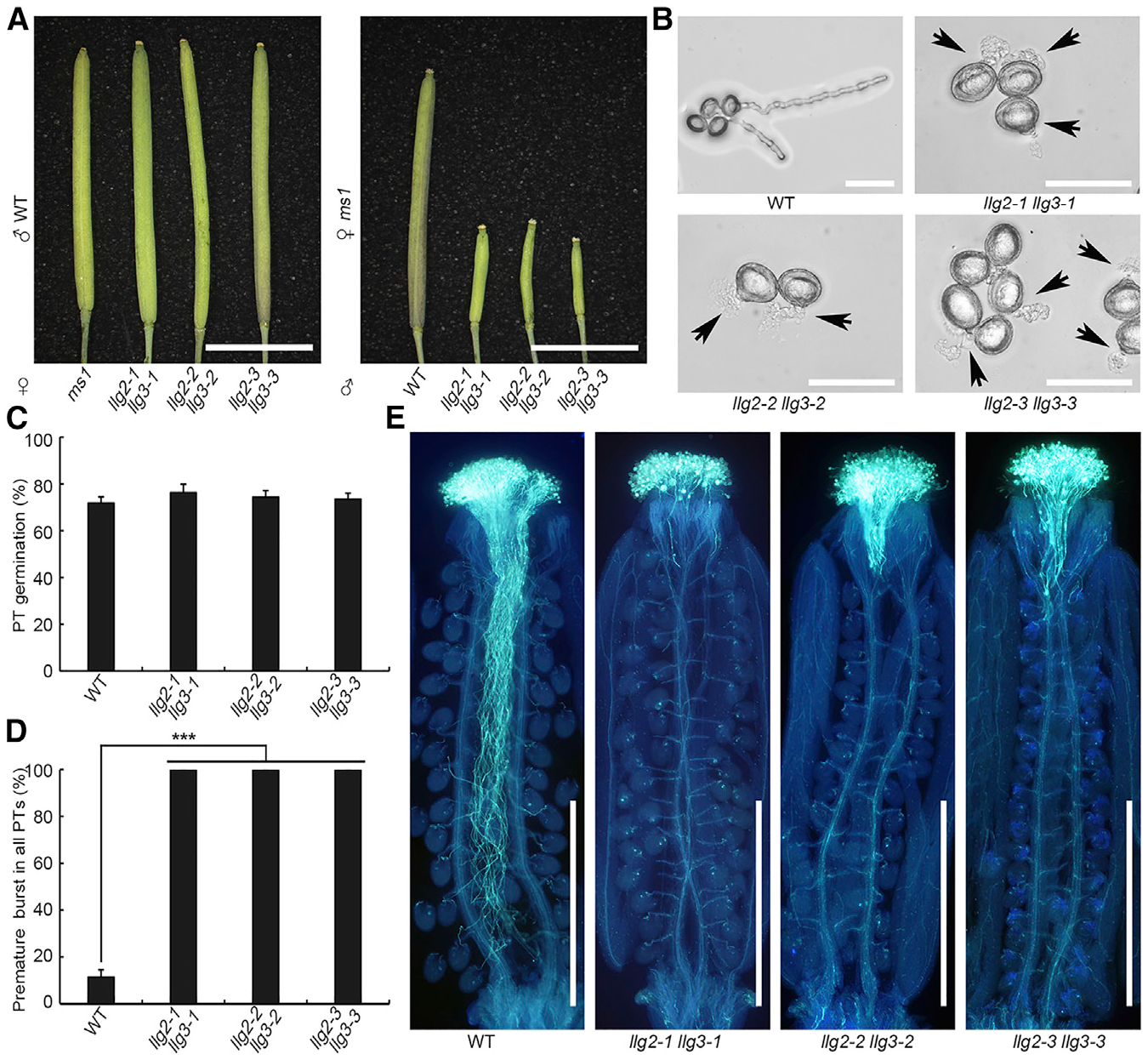
(A) Crosses between llg2 llg3 homozygous double mutant, WT, or ms1 plants. llg2 llg3 double mutant plants were crossed with WT pollen grains (left) or as pollen donors to the stigmas of ms1 mutant plants (right). More than 10 siliques in each group were observed, and pictures were taken 3 days after pollination (HAP).
(B) In vitro pollen germination assays of WT and double mutants. Pictures were taken after 7 h incubation. Arrows show bursting pollen tubes.
(C and D) Pollen germination rate (C) and overall premature bursting rate of pollen tubes (D) of WT and llg2 llg3 double mutants in in vitro pollen germination assays. Phenotypes were analyzed after incubation for 7 h. Data are mean ± SD; at least 1,990 pollen grains and tubes were analyzed in each group; ***p < 0.001 (Student’s t test).
(E) Aniline blue staining of ms1 siliques pollinated with WT and llg2 llg3 pollen. Pictures were taken 20 HAP. More than 20 samples were observed in each group. PT, pollen tube.
Scale bars: 5 mm (A); 50 mm (B); and 1 mm (E).
See also Figures S3 and S4.
Table 1.
Transmission Efficiency Test based on Reciprocal Crosses
| Female × Male | LLG2 llg3/LLG3 | llg2/LLG2 llg3/LLG3 | TEM (%) | TEF (%) |
|---|---|---|---|---|
| WT × llg2-1/LLG2 llg3-1 | 102 | 0 | 0*** | NA |
| llg2-1/LLG2 llg3-1 × WT | 43 | 53 | NA | 123 |
| llg2/LLG2 LLG3 | llg2/LLG2 llg3/LLG3 | TEM (%) | TEF (%) | |
| WT × llg2-1 llg3-1/LLG3 | 97 | 0 | 0*** | NA |
| llg2-1 llg3-1/LLG3 × WT | 46 | 45 | NA | 98 |
NA, not applicable; TEF, female transmission efficiency; TEM, male transmission efficiency.
p < 0.001 (Chi-square test).
LLG2/3 Coordinate Pollen Tube Integrity In Vitro and In Vivo
To characterize the pollen defects and examine the cause of male transmission defect of llg2 llg3, we further conducted scanning electron microscopy (SEM) and Alexander staining. We found that llg2 llg3 pollen grains were morphologically normal and viable (Figures S4H and S4I). In vitro pollen germination assays showed that llg2 llg3 mutant pollen grains germinated normally (rates of 73.4%–76.2%, comparable to 71.7% of WT pollen grains; Figures 2B and 2C). However, almost all llg2 llg3 pollen tubes failed to maintain growth but instead burst immediately after germination (approximately 100%), and only about11.4% of WT pollen tubes ruptured within a 7-h incubation procedure on growth medium (Figures 2B and 2D). Pollen tube growth in the pistil was visualized by Aniline Blue staining. WT pollen tubes invaded the stigma and style, grew in the transmitting tract, and were guided to the ovules to penetrate the female gametophytes for fertilization (Figure 2E). However, the growth of llg2 llg3 pollen tubes was arrested before they reached the stylar-transmitting tract tissue (Figure 2E). Premature arrest of LLG2/3 RNAi pollen tube growth was also observed (Figures S4D–S4G). Thus, both in vitro and in vivo data indicate that LLG2/3 are essential for pollen tube integrity and growth control.
LLG2/3 Are Involved in RALF4/19-Mediated Signaling as Co-receptors
Because the pollen tube rupture phenotype of llg2 llg3 mutants highly resembled those of the receptor mutants anx1 anx2 and bups1 bups2 as well as that of the peptide ligand double mutant ralf4 ralf19 [12–14, 16], we hypothesized that LLG2/3 function in concert in the same signaling pathway and may physically interact with the ANX and BUPS receptors at the pollen tube cell surface. We first investigated whether LLG2/3, similar to ANXs and BUPSs, function in the signaling pathway of RALF4/19 peptide ligands. Previously, it was shown that RALF4 could inhibit pollen germination in vitro [16, 32]. We observed that the supply of 2 μM RALF4 in the growth medium could completely block WT pollen germination. However, bups1–1 bups2–1 mutant pollen grains were not affected by RALF4 treatment: they germinated normally and ruptured shortly after germination (Figure S5A). Similarly, llg2–1 llg3–1 mutant pollen grains were insensitive to RALF4 treatment, maintaining comparable germination rates with or without RALF4 treatment (Figure S5A). These findings supported the hypothesis that LLG2/3 may be directly involved in the perception of RALF4-mediated signaling to control the maintenance of pollen tube integrity.
LLG1 was previously suggested to function as a chaperone during root growth for the receptor FER. It was further proposed that the ligand RALF1 may interact with FER and LLG1 to form a trimeric complex and that FER depends on LLG1 for efficient cell membrane location [23, 24]. We therefore next tested membrane targeting of ANX1-GFP and BUPS2-GFP and observed that these receptors have normal membrane localization even without LLG2/3 (Figure S5B). A similar localization at the synergid plasma membrane was recently reported for the CrRLK1Ls HERK1 and ANJEA [33]. Next, we investigated the interactions among LLG2/3, the BUPS-ANX receptor complex, and the peptide ligands RALF4/19. By performing DUAL membrane yeast two-hybrid (Y2H) assays [34, 35], we found that yeast cells co-transformed with LLG2/3 and the ectodomains of BUPS1/2 or ANX1/2 could grow on selection media, indicating that ectodomains of BUPS1/2 and ANX1/2 interact with LLG2/3 (Figure 3A). To confirm these findings, we performed in vitro pull-down interaction assays and observed weak but detectable interaction between tobacco-expressed FLAG-tagged ectodomains of BUPS1/2 or ANX1/2 and E. coli-expressed His-tagged LLG2/3, respectively (Figures 3B and 3C).
Figure 3. Interactions of LLG2/3 with Ecto-domains of BUPS1/2 and ANX1/2.
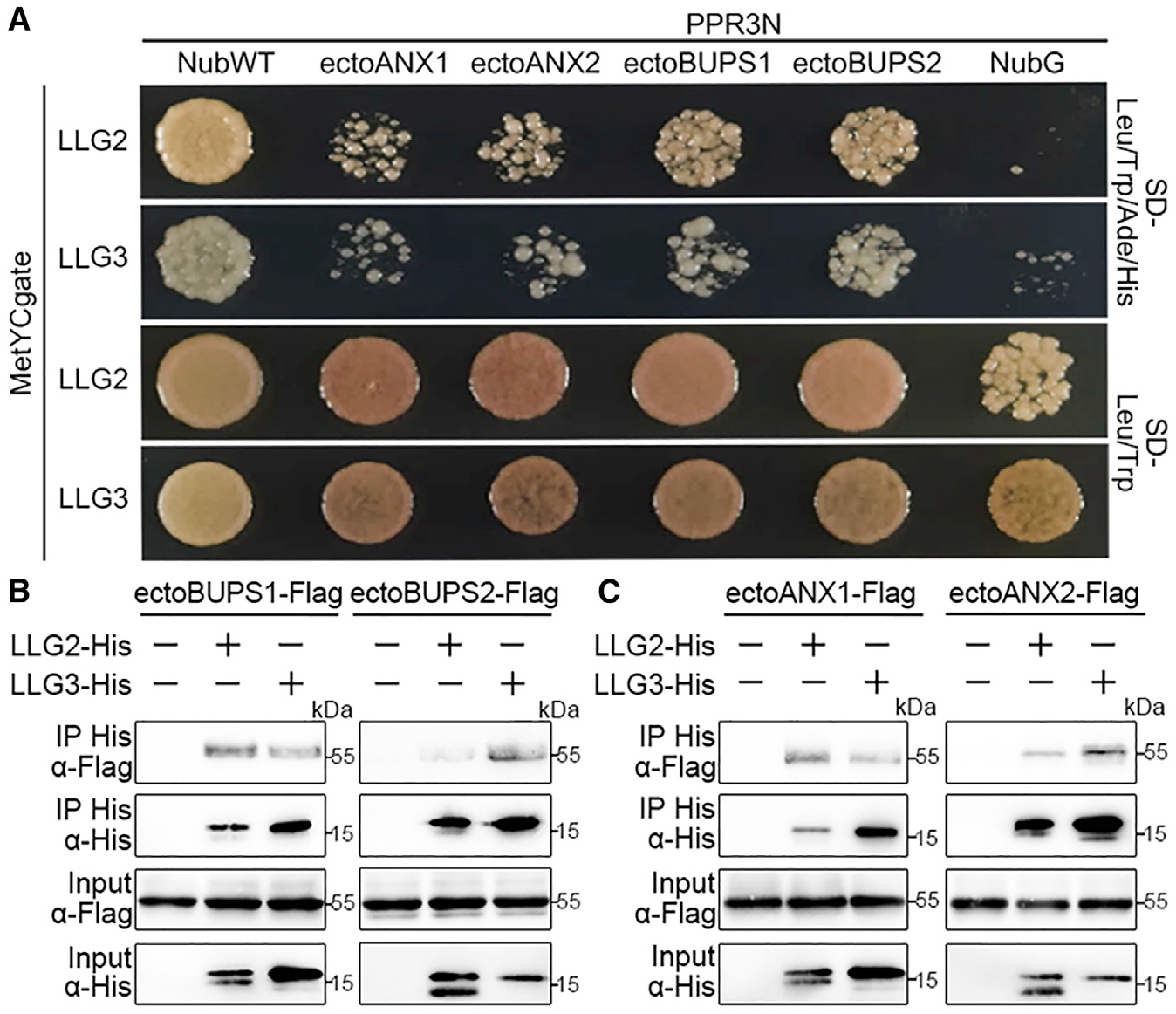
(A) Dual-membrane yeast two-hybrid assays showing interaction of LLG2/3 with ectodomains of BUPS1/2 and ANX1/2. Yeast cells containing indicated plasmids were plated on the growth medium (-Leu-Trp) or on selection medium (-Leu-Trp-His-Ade).
(B and C) Interactions between LLG2/3 with BUPS1/2 (B) and ANX1/2 (C) ectodomains by pull-down assays. LLG2/3 were expressed in E. coli, and ectodomains of BUPS1/2 and ANX1/2 were transiently generated in tobacco leaves. Pull-down assays were performed with Ni Sepharose, and western blots were probed with antibodies against FLAG and His (α-FLAG or α-His), respectively.
Similar results were obtained by three independent experiments.
In a previous study, we reported that RALF4/19 could bind to the BUPS/ANX receptor complex [14]. We therefore asked whether RALF4/19 could also directly interact with LLG2/3. As shown in Figures 4A and 4B, we found that E. coli-generated His-tagged LLG2 or LLG3 was pulled down together with biotinylated RALF4 and RALF19, respectively. This suggests that LLG2/3 directly interact with RALF peptides. Next, we investigated the effect of RALF ligands on the interaction between LLG2/3 and BUPS1/2 as well as between LLG2/3 and ANX1/2. When we applied 100 nM of RALF4 or RALF19 in pull-down assays, remarkably elevated levels of FLAG-tagged ectodomains of BUPS1/2 were pulled down by His-tagged LLG3 (Figure 4C). Similar results were obtained for RALF4 interaction with LLG3 and ANX1/2 ectodomains (Figure 4D). In conclusion, these results indicate that the interaction strength between LLG2/3 and the BUPS-ANX receptor complex is strongly enhanced in the presence of RALF4/19 peptides. In conclusion, this observation suggests a role of LLG2/3 as a co-receptor for ANX/BUPS receptors. Furthermore, we found that RALF34 also could induce the interaction between LLG2/3 and BUPS/ANX ectodomains (Figures 4C and 4D). Related RALF23 from leaves could promote the interaction of LLG2/3 with FER, and RALF4 is capable of inducing the interaction between LLG1 with BUPS/ANX (Figures 4E and 4F).
Figure 4. LRE-like GPI-APs Function as Co-receptors in RALF-CrRLK1L Complexes.
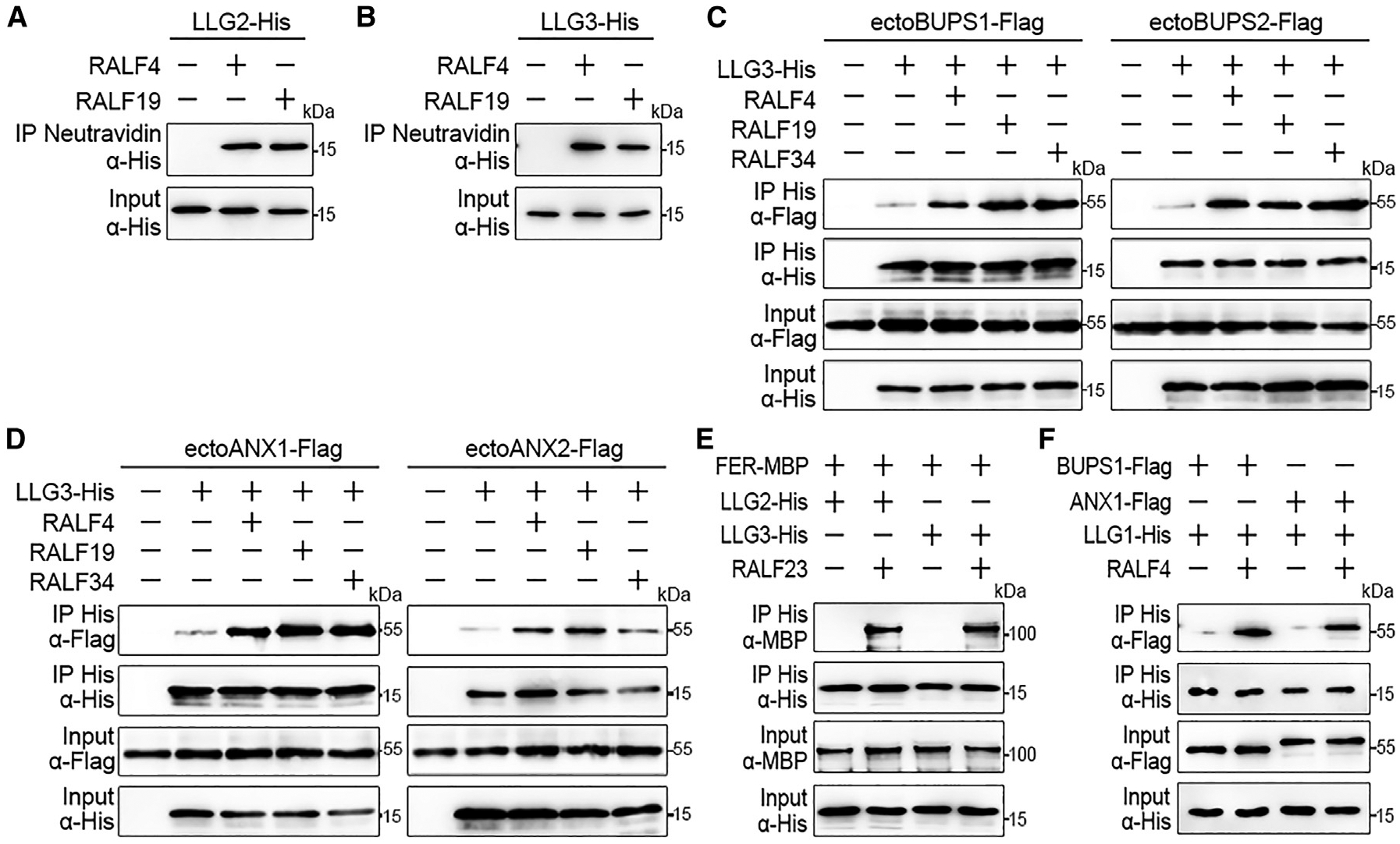
(A and B) In vitro binding assays with E. coli-expressed His-tagged LLG2 (A) and LLG3 (B) proteins. Both RALF4/19 were biotinylated, and pull-down assays were performed with streptavidin magnetic particles. Western blots were performed using an α-His antibody.
(C and D) Pull-down assays of His-tagged LLG3 and FLAG-tagged ectodomains of BUPS1/2 (C) and ANX1/2 (D) with or without the addition of RALF4/19/34 peptides (each 100 nM) as indicated. Pull-down assays were performed with Ni Sepharose, and western blots were probed with antibodies against FLAG and His (α-FLAG or α-His), respectively.
(E and F) Interactions between His-tagged LLG2/3 with ectoFER-MBP by pull-down assays in the presence or absence of RALF23 peptide (100 nM). (E) Pull-down assay using LLG1-His and FLAG-tagged BUPS1/ANX ectodomains with/without 100 nM RALF4 (F). LLG1/2/3-His and ectoFER-MBP were expressed in E. coli, and ectodomains of BUPS1/ANX1 were generated after transient expression in tobacco leaves. Pull-downs were performed with Ni Sepharose, and western blots were probed with antibodies against MBP, FLAG, and His (α-MBP, α-FLAG, or α-His), respectively.
All experiments were repeated at least three times with similar results.
Taken together, these results suggest that LLG2/3 most likely serve redundantly as co-receptors of the BUPS-ANX receptor complex in a RALF4/19-inducible manner to maintain pollen tube integrity. We further propose that LRE-related GPI-APs act in general in RALF/CrRLK1L-mediated signaling in multiple reproductive and developmental pathways.
N-Terminal Region of RALF4 Strongly Interacts with ANX/BUPS, Weakly with LLG2/3, and Is Essential for RALF4 Biological Function
In order to test how RALF4 peptide ligands bind to LLG2/3 proteins, we generated truncated variants of predicted mature RALF4 peptide for pull-down assays. The variants include RALF4-N19 (N-terminal 19 amino acids with one conserved cysteine residue), RALF4-N29 (N-terminal 29 amino acids with two conserved cysteines), RALF4-N48 (N-terminal 48 amino acids with four conserved cysteines), and their corresponding C-terminal truncated variants, i.e., RALF4-C34, RALF4-C24, RALF4-C11, and RALF4-C5 (Figures 5A and S5C). Biotinylated RALF4-N48, RALF4-C34, and RALF4-C24 interacted with His-tagged LLG3 in a similar strength compared to predicted mature RALF4 (Figures 5B and 5C), suggesting that the C-terminal region is sufficient for LLG binding. Meanwhile, RALF4-N19, RALF4-N29, and RALF4-C11 also possess a weak interaction with LLG3 (Figure 5B). This indicated that the region between the second and fourth cysteine residues is most important for RALF4 binding to LLG3. To identity the region that is responsible for RALF4 binding to BUPS/ANX receptors, we found that, although N-terminal 19 amino acid residues of RALF4 (i.e., RALF4-N19) was sufficient for RALF4 interaction with ectodomains of BUPS1 and ANX1 (Figure 5D), the C-terminal region between the second and third cysteine residues was also involved in RALF4 binding (Figure 5E). This finding suggests that RALF4 peptide binds to LLGs and BUPS1/ANX1 receptors through different regions. Using the same strategy, we found that N-terminal truncated RALF4 variants failed to enhance the interactions between LLG3 and BUPS1/ANX1 ectodomains, and RALF4-C34 and RALF4-C24 could weakly enhance the interactions (Figures S5D and S5E). This observation indicates that both N terminus and C terminus, particularly the N terminus of RALF4, are critical for interaction enhancement.
Figure 5. RALF4 Interacts with LLG3 and BUPS/ANX via Different Domains, with Its N-Terminal Region Including a YISY Motif Required for its Function.
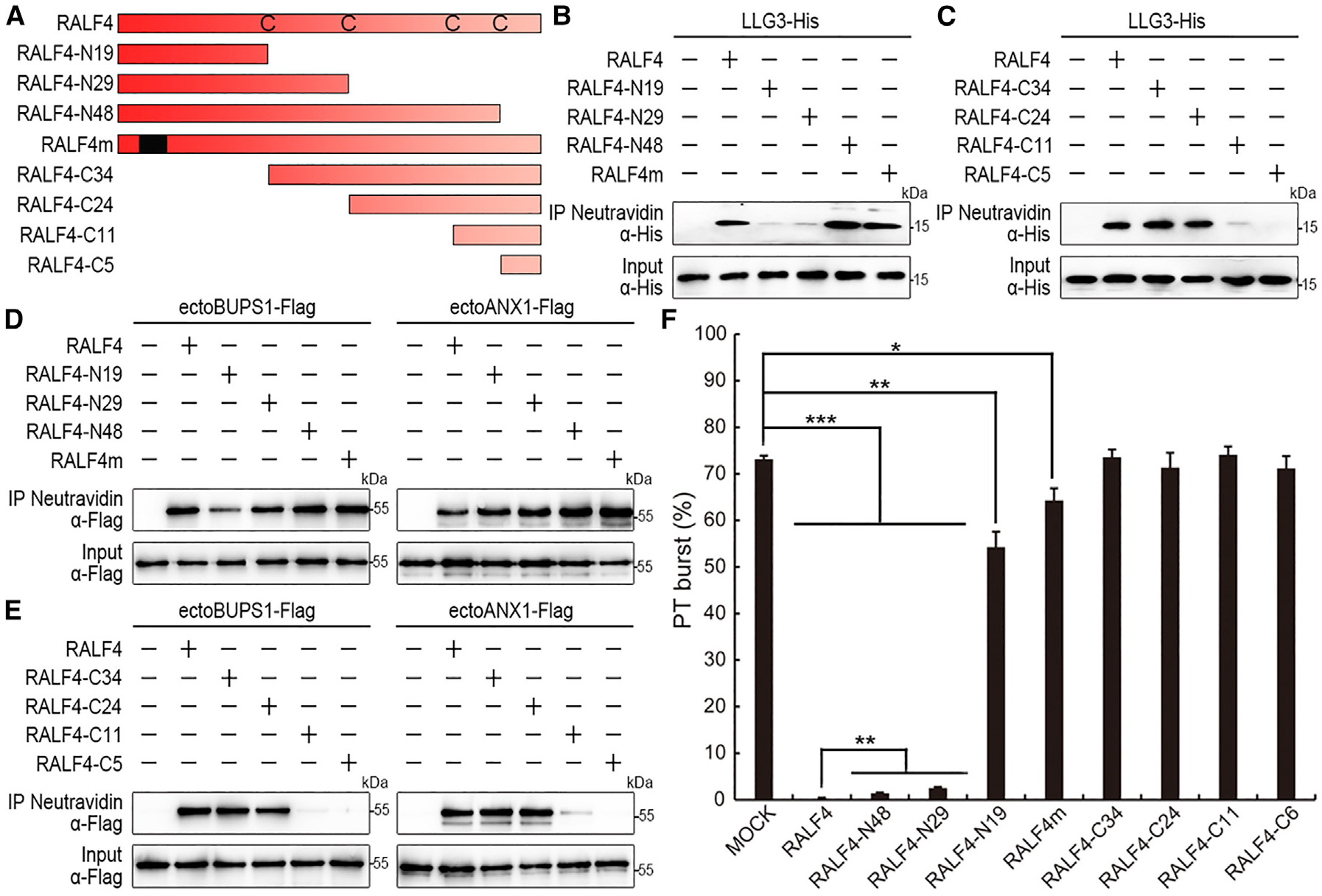
(A) Schematic diagram of RALF4 and RALF4 variants used in this study. Black frame in RALF4m indicates the mutated YIGY motif (mutated to AAAA).
(B and C) In vitro binding assays using His-tagged LLG3 proteins purified from E. coli. RALF4 N-terminal variants (B) and C-terminal variants (C) were biotinylated, and pull-down assays were performed with streptavidin magnetic particles. Western blots were performed using an α-His antibody. All experiments were repeated at least three times with similar results.
(D and E) Pull-down assays to study the interactions between biotinylated WT or mutated RALF4s and ectoBUPS1-FLAG and ectoANX1-FLAG proteins generated in tobacco leaves. N-terminal variants were tested in (D) and C-terminal in (E). Pull-down assays were performed using streptavidin magnetic particles and an α-FLAG antibody in western blots. Similar results were obtained by repeating experiments three times.
(F) Biological activities of RALF4 variants on the ralf4 ralf19 bursting phenotype. Pollen tube bursting (%) was analyzed 7 h after incubation of pollen on medium. The concentration of RALF4 variants were 2 μM in each treatment. Data are mean ± SD. More than 1,500 pollen grains and tubes were analyzed in each group.
*p < 0.05; **p < 0.01; ***p < 0.001 (Student’s t test).
See also Figure S5.
Next, we adopted in vitro pollen germination assays to test the biological activities of the truncated peptides on the pollen tube burst phenotype of the ralf4 ralf19 double mutant. When 2 μM of RALF4/19 was applied, less than 3% of the mutant pollen tubes ruptured, significantly lower compared to the 75% without RALF peptide treatments (Figure S5F). This indicates that RALF4/19 could rescue the pollen tube burst phenotypes at a concentration of 2 μM. Therefore, we used a concentration of 2 μM to test the activities of the RALF4 variants. We found that both RALF4-N48 and RALF4-N29, similar to RALF4, could significantly rescue ralf4 ralf19 pollen tube burst phenotype to 1.36% and 2.53%, respectively (Figure 5F). RALF4-N19 could partially complement the pollen tube burst ratio at 54.29%, which is also significantly lower compared to the control (Figure 5F). Similar results were observed when RALF4-N48, RALF4-N29, and RALF4-N19 were tested on the WT pollen germination (Figure S5G). Taken together, this indicates that the N terminus of RALF4, especially the 29 residues that interact strongly with BUPS/ANX and weakly with LLG3, is critical for RALF4 activity. Moreover, we noticed that neither RALF4-N48 nor RALF4-N29 could completely recapitulate the function of RALF4 in pollen tube rupture and pollen germination (Figure 5F). This finding suggests that the C terminus of RALF4 is also required for RALF4 signaling.
It was previously reported that the conserved YIXY motif of RALF peptides was essential for RALF activity and receptor binding [19, 36]. We therefore mutated YIGY (here X is G in RALF4) into AAAA to generate the RALF4m variant (Figures 5A and S5C). Afterward, we performed pull-down assays and biological function analysis. We found that RALF4m was still able to interact with BUPS1/ANX1 ectodomains (Figures 5D and 5E). This is consistent with the recent discovery showing that RALF1 containing a mutated YISY motif could still interact with the FER receptor [37]. Moreover, we found that RALF4m was able to interact with LLG3 and to induce LLG3-BUPS1/ANX1 association (Figures 5B, 5C, S5D, and S5E). However, in pollen germination assays using both WT and ralf4 ralf19 pollen, RALF4m significantly abolished its function, i.e., 49.24% of WT pollen and 64.11% of ralf4 ralf19 pollen tubes ruptured (Figures 5F and S5G). These results indicate that the YIGY motif is not essential for RALF4 interaction with LLG3 and ANX1/BUPS1 or for induction of the LLG-BUPS/ANX complex but significantly contribute to RALF4 activity.
In summary, our data demonstrate that the N terminus of RALF4, which is critical for RALF4 binding to BUPS/ANX receptors and partially for RALF4 interaction with LLG3, contribute significantly to the biological function of RALF4. The YIGY motif is essential for RALF4 activity but independent of peptide interaction with either receptors or LLG proteins. This observation suggests another binding partner of RALF4, which needs to be identified in future studies.
DISCUSSION
In comparison to animals, flowering plants have evolved a highly expanded family of receptor-like kinases (RLKs) to sense a diverse range of extrinsic and intrinsic signals and to trigger downstream signaling processes. For instance, the leucinrich repeat RLK (LRR-RLK) BRI1 senses brassinosteroids, the immune-related FLS2 perceives bacteria-derived flagellin 22 (flg22), and the TDIF receptor PXY/TDR regulates vasculature patterning [38–40]. These receptors functioning in plant growth, development, and immunity are usually facilitated by co-receptors, which are directly or indirectly involved in ligand recognition, by forming protein complexes to perceive endogenous peptides, plant hormones, or pathogen signatures [39, 41, 42]. Moreover, ligand-induced homodimerization or heterodimerization between receptors and co-receptors was reported as a common and essential step for signaling specificity in plant cells [42–45]. For example, the interaction between SERKs co-receptors with LRR-RLK HAE/HSL2 receptors depends on IDA peptides triggering organ abscission in plant development [45–48]. Meanwhile, GPI-APs were also reported to be involved in RLK-mediated signaling. In the chitin signaling pathway, for example, the GPI-AP OsCEBiP is able to directly bind to chitin and coordinates chitin perception in rice together with the receptor-like kinase OsCERK11 that is incapable of binding chitin itself [49–51]. In addition, the GPI-AP LLG1 was shown to serve as a chaperone for FER and FLS2 to regulate plant development and innate immunity [24, 25], whereas LRE and ENODLs were involved in FER-mediated pollen tube reception [21, 22, 52].
In this study, we provide genetic and cell biological evidence to demonstrate that two further GPI-APs, LLG2/3, are involved in RALF-mediated signaling for pollen tube integrity control. We demonstrate that LLG2/3 could interact with BUPS/ANX receptors, and this interaction is strongly enhanced by the presence of RALF peptides. This indicates that the ligand-induced heterodimerization between receptors BUPSs/ANXs and co-receptors LLG2/3 is a critical step for RALF signal perception, which is possibly conserved in BUPSs/ANXs-LLG2/3-RALF34 and other GPI-APs-CrRLK1L receptor complexes, e.g., within the LLG1-FER-RALF1 complex [24]. This hypothesis is supported by our findings, as we could also show that pollen-tube-expressed LLGs strongly enhance the interaction between FER and RALF23 that are not expressed in pollen tubes, and non-pollen-expressed LLG1 is capable to enhance RALF4-BUPS1 interaction. A recent report on the structure of the RALF23-LLG1/2-FER complex supports our findings and indicates that RALF23 serves as a molecular glue between FER and LLGs [53].
Our data also provide further mechanistic and structural insight of the RALF-LLG-BUPS/ANX signaling complex. We could show, for example, that RALF4 is capable to interact with LLG3 and BUPS/ANX via different N-terminal domains, confirming findings in other tissues that CrRLK1L receptor complexes are composed at least of three different proteins. The finding that the N-terminal region of RALF4, including a YISY motif, is required for proper pollen tube integrity indicates that either an even higher order receptor complex is formed or interaction is required with other cell wall proteins, such as leucine-rich repeat extensions (LRX), which play a key role in cell wall formation and integrity during pollen tube growth [16, 54, 55]. It will now be exciting to elucidate the structure of the receptor complex in future studies as well as the cellular response of this interaction, especially the opposite readout of RALF4/19 and RALF34. Moreover, membrane targeting of BUPSs/ANXs seems to be independent of LLG2/3. The majority of LLG proteins were detected in punctuate structures likely representing vesicles and/or endosomes, indicating that LLG function in the pollen tube might also be involved, for example, in exocytosis of the receptor complex or endocytosis of the activated complex. In conclusion, this report adds to the understanding of the molecular mechanism on how RALF-CrRLK1L-mediated signaling is regulated and underscores the multi-layered strategy that flowering plants deploy to regulate pollen tube integrity and thus ensure reproductive success.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents or plasmids, and there are no restrictions to the availability of the plasmids/mutant plants reported. Requests for resources and reagents and further information should be directed to and will be fulfilled by the Lead Contact, Li-Jia Qu (qulj@pku.edu.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plant material and growth conditions
Arabidopsis thaliana (Columbia-0) was used as wild-type (WT) or transgenic material. Plants (both Arabidopsis and tobacco) were grown under long-day conditions (16 hr light/ 8 hr dark cycles) at 22 ± 2°C with a light intensity of ~170 mmol/m2/s using LED bulbs (Philips F17T8/TL841 17 W), as previously reported [14]. Wild-type plants were transformed by the floral dip method using Agrobacterium tumefaciens strain GV3101. Transient protein expression was conducted by agro-infiltration in Nicotiana benthamiana leaves and Nicotiana tabacum pollen tubes by particle bombardment [56], respectively. Pollination experiments were performed using llg2 llg3 mutant pollen on ms1 female plants [31].
METHOD DETAILS
Phylogenetic analyses and heat-map drawing
LRE, LLG1, LLG2, LLG3 and LYM1 protein sequences were obtained from the Arabidopsis Information Resource with the following gene accession numbers: LRE, At4G26466; LLG1, At5G56170; LLG2, At2G20700; LLG3, At4G28280; LYM1, At1G21880. Phylogenetic analysis was conducted using MEGA 6.0. Transcription profile heat-map was drawn based on RNA-seq data of different tissues as previously described [14] by using Morpheus.
Plasmid construction and plant transformation
2 kb of LLG2/3 promoters were cloned into pB7GUSWG0 [14] for GUS staining assays via TOPO and LR reactions according to protocols from Invitrogen. Gibson Assembly was adopted to generate a TOPO vector containing LLG2 promoter driving LLG2 genomic sequence with a GFP encoding sequence inserted in between the signal peptide and the mature protein, which was then cloned into pB7GUSWG0 [14] by LR reaction for complementary assays. Lat52p::SP-YFP-LLG3 was generated with the same procedure.
In order to generate LLG2 and LLG3 loss-of-function mutants, the CRISPR/Cas9 system based on the egg cell specific promoter EC1.2 was introduced [30, 57]. Two sgRNAs for LLG2 and LLG3, respectively, were cloned from pCBC-DT1T2 with primers consisting of sequences from sgRNAs, and then ligated into pENTR-MSR to obtain pENTR-LLG2-LLG3 as reported previously [14]. Later, these fragments containing four sgRNAs were cloned into the PHEE401E vector by using the Golden-Gate system [30]. Finally, these CRISPR/Cas9 constructs were transformed into Arabidopsis via flora dipping method. After generation of the transgenic plants, the genomic DNA fragments containing the target sites were amplified before they were subjected for Sanger sequencing. The sequencing results were analyzed by MEGA 6.0.
The sgRNA sequences for LLG2 and LLG3 mentioned above are as follows:
LLG2_sgRNA1: GTG TGT TGC TCC GCC TTC ANG G
LLG2_sgRNA2: GAA GAG AAA GCA GAC AGT ANG G
LLG3_sgRNA1: TCC ACA AGC CGA GCT CTT CNG G
LLG3_sgRNA2: GAC TTT GCT TGC CCA TTC GNG G.
Plasmids for transient expression in tobacco pollen tubes were generated as follows: to generate the Lat52:mCitrine-LLG2/3 vectors, mCitrine was inserted 69 base pairs after the start codon into the LLG2/3 coding sequence via DNA synthesis (GeneArt, Thermo Fisher Scientific). mCitrine-LLG2/3 was subcloned into pENTR/D-TOPO and the LR reaction performed between Lat52::GW and pENTR-mCitrine-LLG2/3 to obtain the final expression vectors. Coding sequence of ANX1 or BUPS2 without stop codon was separately cloned into pENTR vector, and LR reaction was then performed with Lat52p::GW-GFP to generate expression vectors Lat52p::ANX1-GFP and Lat52p::BUPS2-GFP, respectively.
Pollen analysis
Pollen grains of WT or mutant Arabidopsis plants were derived from freshly opened flowers and observed by a Scanning Electron Microscope (Hitachi S3000N, JOEL JSM-6610). For Alexander staining, mature pollen grains were collected and treated with Alexander solution directly [14].
Analysis of pollen germination was based on in vivo pollen germination assays. In brief, for in vitro pollen germination, mature pollen grains were dispersed on pollen germination medium (PGM) in Petri dishes containing 18% sucrose, 0.01% boric acid, 5 mM CaCl2, 5 mM KCl, 1 mM MgSO4, pH 7.5; 1.5% low-melting point agarose [58]. Petri dishes were then incubated in humid boxes at 22°C temperature for 7 hours. Pollen germination was observed under a microscope (Zeiss Axio Observer Z1). For RALF4 inhibition and complementation assays, 2 μM of peptide was pre-mixed in PGM and pollen germination assays performed with pollen of WT, ralf4 ralf19 and bups1 bups2 mutants, respectively. For subcellular localization analysis of LLG3 in Arabidopsis, pollen grains of LLG3-YFP transgenic plants were placed on the germination medium to germinate, and fluorescence signal was observed with a Spinning Disc Confocal Microscopy (Zeiss Cell Observer SD, Zeiss, Germany). For GUS staining, pollen tubes of LLGp::GUS transgenic plants were stained with GUS staining solution (2 mM X-Gluc, 50 mM NaHPO4) for 10 min before they were observed with a microscope (Olympus BX51, Japan).
For in vivo germination assays, pollen grains were applied on stigmata of ms1 plants [31]. In order to observe pollen tube growth, pollinated pistils were cut after 20 hours and stained by Aniline Blue as reported previously [59]. Specifically, pollinated samples were fixed in acetic acid/ethanol 1:3 for 2 hr and washed with 70% ethanol, 50% ethanol, 30% ethanol and ddH2O for 10 min each time. After being treated with 8 M NaOH for 10 hr, these samples were washed with ddH2O for three times and stained with aniline blue solution (0.1% aniline blue, 108 mM K3PO4) for more than 2 hr. A fluorescence microscope (Olympus BX51) was used to observe stained samples. Images shown are representative of more than 20 images captured in each group.
Transient expression in tobacco pollen tubes
Transient transformation of tobacco pollen tubes was performed as previously reported [56]. In short, gold particles (1 μm) were suspended in 1 mL 70% ethanol for 15 min, and after wash with water for three times, re-suspended in 1 mL 50% glycerol. For each transformation, 8 μg of a certain plasmid was mixed with 12.5 μL of gold particles and the final volume was adjusted to 75 μL with water. The mixture was vortexed for 5 s, then 50 μL of 2.5 M CaCl2 was added and vortexed for another 5 s. After adding 20 μL of 0.1 M spermidine, the mixture was vortexed for 90 s and centrifuged at 10 000 g for 10 s. The supernatant was removed and the mixture was washed twice with 200 μL ethanol and re-suspended in 30 μL ethanol. 10 μL of the mixture was loaded to a macro-carrier and air-dried at room temperature; 3 macro-carriers were used for each transformation.
For each bombardment, anthers from 5–10 flowers were collected and re-suspended in PGM by mild vortexing, and the medium (containing pollen) was transferred into a new tube. After a brief centrifuging, supernatant was removed and pollen grains at the bottom re-suspended in 180 μL PGM before they were loaded on 1.5 cm round filter paper, which was placed in a 9 cm Petri dish on top of a large filter paper soaked with PGM. The pollen was bombarded with the gold particles prepared as described above using rupture dicks with the strength of 1100 psi at a distance of 6 cm. After bombardment, pollen was incubated with 3 mL of PGM in a Petri dish on 100 rpm shaking platform at room temperature in dark. Confocal analysis was performed 4–8 hr after pollen germination.
DUAL membrane yeast two-hybrid assay
LLG2/3 used for protein-protein interaction studies in yeast were cloned into the pMetYCgate-GW destination vector. BUPS/ANX ectodomains were cloned into the pPR3N-GW vector by using the Gateway system [34]. Different combinations of bait and prey vectors, together with the positive and negative control plasmids provided, were individually co-transformed into yeast strain NMY51 using the lithium acetate (LiAc) transformation method according to the manufacturer’s instruction (Dualsystems Biotech AG, Switzerland). All clones successfully co-transformed could grow under moderate stringency on selective medium (-Trp/Leu). Only the positive control along with clones transformed with interacting proteins could grow on high-stringency medium (-His/Trp/Leu/Ade).
Protein expression in E. coli and tobacco leaves
His-tagged LLG2 and LLG3 proteins were expressed in E. coli. Sequences of the domain between signal peptides and GPI-modification sites were cloned from LLG2/3 CDS and inserted into pET28GW by using the Gateway system from Invitrogen. Constructs were then transformed into the BL21 E. coli strain obtained from TransGen Biotech (China). Bacteria were grown at 37°C to an OD600 of 0.5 and thereafter induced with 1/1000 1M IPTG at 18°C overnight. Induced bacteria were collected at room temperature and centrifuged at a speed of 4,000 rpm for 20 minutes. Pellets were re-suspended with His lysis buffer (50 mM NaH2PO4, pH8.0, 300 mM NaCl, 10 mM imidazole, 1 mM PMSF, 1 tablet of Protease Inhibitor Cocktail per 50 mL lysis buffer), and sonicated at regular intervals: 10 s-work and 20 s-rest as one cycle, for up to 30 minutes. After centrifugation, insoluble pellets were removed and suspensions transferred into a new tube. Protein extracts were incubated with His-beads (Ni Sepharose 6 FF, GE Healthcare) at 4°C overnight. His washing buffer (50 mM NaH2PO4, pH8.0, 300 mM NaCl, 50 mM imidazole) was used to clean beads for 5 times and His elution buffer (50 mM NaH2PO4, pH8.0, 300 mM NaCl, 500 mM imidazole) was added to elute His-tagged proteins from beads. pMAL-tagged ectodomains in pMALGW vectors [60] were expressed and purified in the same system with the same procedures but different buffer. MBP lysis buffer (pH 7.5) consists of 50 mM Tris, 100 mM NaCl, 10% Glycerol, 0.2% Triton X-100, 1 tablet of Protease Inhibitor Cocktail per 50 mL lysis buffer; MBP wash buffer is made by Tris, NaCl and Glycerol at the same concentration as in MBP lysis buffer; MBP elution buffer consists of 0.01 M PBS buffer and 30 mM maltose. Protein extracts were incubated with MBP-beads (Amylose resin, NEB).
Flag-tagged ectodomains were transiently expressed in N. benthamiana leaves and extracted as previously described [14]. More specifically, ectodomains of BUPS1/BUPS2/ANX1/ANX2 were cloned into pB7FLAGWG2, respectively and each transformed into Agrobacterium tumefaciens GV3101, and infiltrated into tobacco leaves. After 60-hr incubation (24 hr in dark and 36 hr under 16 hr light/8 hr dark growth condition), infiltrated leaves were collected and ground in the lysis buffer containing 150 mM NaCl, 10% Glycerol, 1 mM PMSF, 5 mM DTT, 50 mM Tris-HCl (pH8.0), 0.5% NP-40, 1xC-complete protease inhibitor (Roche, Switzerland). Supernatants were collected from the extracts afert being centrifuged for 15 min and three times. After that, the extracts were incubated with anti-Flag gel (Bimake, USA, B23102) in 4°C for 3 hr, washed with the lysis buffer for 5 times in 4°C, eluted with 3xFlag Peptide (Sigma, F4799–4 MG) for 3 times at room temperature, and collected for further use.
RALF4/19/23/34 peptides were synthesized by Scilight Biotechnology LLC (Beijing, China) as described previously [14].
in vitro protein binding assays
Pull-down assays between LLG2/3-His proteins and Flag-tagged ectodomains of above receptor-like kinases were conducted as described previously [14, 20]. Samples were mixed in 500 μL binding buffer (20 mM Tris-HCl, pH7.5, 1% IGEPAL) and kept at 4°C for 2 hours before His-beads (Ni Sepharose 6 FF, GE Healthcare) were added. Two hours after binding, beads were pelleted and samples washed for 5 times. Afterward, samples were boiled with SDS-loading buffer and subjected to SDS-PAGE and western blot performed by using anti-Flag-HRP (Sigma-Aldrich, A8592–2MG), ProteinFind Anti-His Mouse Monoclonal Antibody (TransGen Biotech, HT501–02) and Goat Anti-Mouse IgG-HRP (CWBIO, CW0102S). For interaction studies between LLG2/3-His proteins and Flag-tagged ectodomains with RALF peptides, 100 nM of biotin-RALF peptide was each added into the pre-mixed samples, kept at 4°C for 3 hours and tested by the steps mentioned above. Pull-down assays in LLG1-BUPS/ANX-RALF4 and LLG2/3-FER-RALF23 were conducted under the same condition, except that FER-MBP was tested by an Anti-MBP Monoclonal Antibody (NEB, E8038S). The binding assays for RALF4s to LLG2/3 or BUPS/ANX were performed as previously described [14, 20] using Streptavidin Magnetic Particles (Spherotech, SVMS-30–10).
QUANTIFICATION AND STATISTICAL ANALYSIS
Siliques length measurement and Plot assays were carried out using ImageJ. In each related experiment, number of repeats (n), sample sizes and P values were indicated in the figure legends or the results. Statistical significance was calculated using Student’s t test and Chi-square test with Microsoft Excel and indicated in the legends.
DATA AND CODE AVAILABILITY
This study did not generate/analyze datasets/code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ProteinFind Anti-His Mouse Monoclonal Antibody | TransGen Biotech | HT501-02 |
| anti-Flag-HRP | Sigma-Aldrich | A8592-2MG |
| Goat Anti-Mouse IgG-HRP | CWBIO | CW0102S |
| Anti-MBP Monoclonal Antibody | NEB | E8038S |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Biotin-RALF4 | Scilight Biotechnology | N/A |
| RALF4 | Scilight Biotechnology | N/A |
| Biotin-RALF4-N19 | Scilight Biotechnology | N/A |
| RALF4-N19 | Scilight Biotechnology | N/A |
| Biotin-RALF4-N29 | Scilight Biotechnology | N/A |
| RALF4-N29 | Scilight Biotechnology | N/A |
| Biotin-RALF4-N48 | Scilight Biotechnology | N/A |
| RALF4-N48 | Scilight Biotechnology | N/A |
| Biotin-RALF4m | Scilight Biotechnology | N/A |
| RALF4m | Scilight Biotechnology | N/A |
| Biotin-RALF4-C34 | Scilight Biotechnology | N/A |
| RALF4-C34 | Scilight Biotechnology | N/A |
| Biotin-RALF4-C24 | Scilight Biotechnology | N/A |
| RALF4-C24 | Scilight Biotechnology | N/A |
| Biotin-RALF4-C11 | Scilight Biotechnology | N/A |
| RALF4-C11 | Scilight Biotechnology | N/A |
| Biotin-RALF4-C5 | Scilight Biotechnology | N/A |
| RALF4-C5 | Scilight Biotechnology | N/A |
| Biotin-RALF19 | Scilight Biotechnology | N/A |
| RALF19 | Scilight Biotechnology | N/A |
| Biotin-RALF23 | Scilight Biotechnology | N/A |
| RALF23 | Scilight Biotechnology | N/A |
| Biotin-RALF34 | Scilight Biotechnology | N/A |
| RALF34 | Scilight Biotechnology | N/A |
| 3xFlag Peptide | Sigma | F4799-4 MG |
| SuperSignal@West Femto Maximum Sensitivity Substrate | Thermo Fisher | 34095 |
| Copmplete Protease Inhib. Cocktail | Roche | 11836145001 |
| Ni Sepharose 6 FF | GE Healthcare | 17531801 |
| Anti-Flag Affinity Gel | Bimake | B23101 |
| Streptavidin Magnetic Particles | Spherotech | SVMS-30-10 |
| Amylose Resin | NEB | E8021L |
| Experimental Models: Organisms/Strains | ||
| Arabidopsis thaliana: Col0 | ABRC | N/A |
| Arabidopsis thaliana: llg2-1 | This study | N/A |
| Arabidopsis thaliana: llg2-2 | This study | N/A |
| Arabidopsis thaliana: llg2-3 | This study | N/A |
| Arabidopsis thaliana: llg3-1 | This study | N/A |
| Arabidopsis thaliana: llg3-2 | This study | N/A |
| Arabidopsis thaliana: llg3-3 | This study | N/A |
| Arabidopsis thaliana: bups1-2 bups2-1 | [14] | N/A |
| Arabidopsis thaliana: LLG2p::GUS | This study | N/A |
| Arabidopsis thaliana: LLG3p::GUS | This study | N/A |
| Arabidopsis thaliana: ms1 | [31] | N/A |
| Arabidopsis thaliana: LLG2p::SP-GFP-LLG2 (llg2 llg3) | This study | N/A |
| Arabidopsis thaliana: Lat52:SP-YFP-LLG3 | This study | N/A |
| Nicotiana tabacum cultivar SR 1 | This study | N/A |
| Oligonucleotides | ||
| Primers used in this study, see Table S1 | This study | N/A |
| Recombinant DNA | ||
| Plasmid LLG2p::GUS | This study | N/A |
| Plasmid LLG3p::GUS | This study | N/A |
| Plasmid pHEE401E-LLG2-LLG3 | This study | N/A |
| Plasmid pENTR-LLG2-LLG3 | This study | N/A |
| Plasmid pET28GW-LLG1 | This study | N/A |
| Plasmid pET28GW-LLG2 | This study | N/A |
| Plasmid pET28GW-LLG3 | This study | N/A |
| Plasmid pMALGW-ectoFER | This study | N/A |
| Plasmid 35Sp::ectoBUPS1-Flag | [14] | N/A |
| Plasmid 35Sp::ectoBUPS2-Flag | [14] | N/A |
| Plasmid 35Sp::ectoANX1-Flag | [14] | N/A |
| Plasmid 35Sp::ectoANX2-Flag | [14] | N/A |
| Plasmid LLG2p::SP-GFP-LLG2 | This study | N/A |
| Plasmid Lat52:SP-YFP-LLG3 | This study | N/A |
| Plasmid Lat52:SP-mCitrine-LLG2 | This study | N/A |
| Plasmid Lat52:SP-mCitrine-LLG3 | This study | N/A |
| Software and Algorithms | ||
| Arabidopsis Information Resource | http://www.arabidopsis.org/ | N/A |
| Morpheus | https://software.broadinstitute.org/morpheus | N/A |
| UniProt | https://www.uniprot.org/ | N/A |
| MEGA 6.0 | http://www.megasoftware.net/ | N/A |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
Highlights.
LLG2/3 are pollen tube GPI-anchored proteins required for pollen tube integrity
LLG2/3 interact with BUPS/ANX receptors
RALF4/19 peptide ligands enhance the strength of LLG2/3-BUPS/ANX interaction
RALF4 interacts with LLG3 and BUPS/ANX via different domains
ACKNOWLEDGMENTS
This work is supported by National Natural Science Foundation of China (31830004, 31620103903, and 31621001), China Postdoctoral Science Foundation (2018M640020 and 2019T120012), US Natural Science Foundation (IOS-1645854 and MCB-1715764), and National Institute of Food and Agriculture, US Department of Agriculture (MAS00525). Financial support of the Qu laboratory by the Peking-Tsinghua Joint Center for Life Sciences and the Dresselhaus lab by the German Research Council DFG via the Collaborative Research Center SFB924 is gratefully acknowledged. L.W. is recipient of a fellowship from the China Scholarship Council. We also thank the Research Coordination Network on Integrative Pollen Biology (US-NSF MCB0955910) for supporting activities that forged this collaboration.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.08.032.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Liu J, and Qu L-J (2008). Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol. Plant 1, 564–574. [DOI] [PubMed] [Google Scholar]
- 2.Palanivelu R, and Tsukamoto T (2012). Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdiscip. Rev. Dev. Biol 1, 96–113. [DOI] [PubMed] [Google Scholar]
- 3.Higashiyama T, and Yang WC (2017). Gametophytic pollen tube guidance: attractant peptides, gametic controls, and receptors. Plant Physiol. 173, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Huang Q, Zhong S, Bleckmann A, Huang J, Guo X, Lin Q, Gu H, Dong J, Dresselhaus T, and Qu LJ (2017). Sperm cells are passive cargo of the pollen tube in plant fertilization. Nat. Plants 3, 17079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong S, and Qu L-J (2019). Peptide/receptor-like kinase-mediated signaling involved in male-female interactions. Curr. Opin. Plant Biol 51, 7–14. [DOI] [PubMed] [Google Scholar]
- 6.Zhong S, Liu M, Wang Z, Huang Q, Hou S, Xu Y-C, Ge Z, Song Z, Huang J, Qiu X, et al. (2019). Cysteine-rich peptides promote inter-specific genetic isolation in Arabidopsis. Science 364, eaau9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson MA, Harper JF, and Palanivelu R (2019). A fruitful journey: pollen tube navigation from germination to fertilization. Annu. Rev. Plant Biol 70, 809–837. [DOI] [PubMed] [Google Scholar]
- 8.Franck CM, Westermann J, and Boisson-Dernier A (2018). Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol 69, 301–328. [DOI] [PubMed] [Google Scholar]
- 9.Stegmann M, and Zipfel C (2017). Complex regulation of plant sex by peptides. Science 358, 1544–1545. [DOI] [PubMed] [Google Scholar]
- 10.Higashiyama T (2018). Plant reproduction: autocrine machinery for the long journey of the pollen tube. Curr. Biol 28, R266–R269. [DOI] [PubMed] [Google Scholar]
- 11.Liang X, and Zhou J-M (2018). The secret of fertilization in flowering plants unveiled. Sci. Bull. (Beijing) 63, 408–410. [DOI] [PubMed] [Google Scholar]
- 12.Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, and Grossniklaus U (2009). Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyazaki S, Murata T, Sakurai-Ozato N, Kubo M, Demura T, Fukuda H, and Hasebe M (2009). ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol 19, 1327–1331. [DOI] [PubMed] [Google Scholar]
- 14.Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, García-Valencia LE, Zhong S, et al. (2017). Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge Z, Cheung AY, and Qu L-J (2019). Pollen tube integrity regulation in flowering plants: insights from molecular assemblies on the pollen tube surface. New Phytol. 222, 687–693. [DOI] [PubMed] [Google Scholar]
- 16.Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, and Grossniklaus U (2017). RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358, 1600–1603. [DOI] [PubMed] [Google Scholar]
- 17.Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, and Grossniklaus U (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317, 656–660. [DOI] [PubMed] [Google Scholar]
- 18.Duan Q, Kita D, Li C, Cheung AY, and Wu H-M (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 107, 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haruta M, Sabat G, Stecker K, Minkoff BB, and Sussman MR (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, and Zipfel C (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. [DOI] [PubMed] [Google Scholar]
- 21.Tsukamoto T, Qin Y, Huang Y, Dunatunga D, and Palanivelu R (2010). A role for LORELEI, a putative glycosylphosphatidylinositol-anchored protein, in Arabidopsis thaliana double fertilization and early seed development. Plant J. 62, 571–588. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Castro C, Wang Y, Noble J, Ponvert N, Bundy M, Hoel C, Shpak E, and Palanivelu R (2016). The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell 28, 1035–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Wu H-M, and Cheung AY (2016). FERONIA and her pals: functions and mechanisms. Plant Physiol. 171, 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Yeh FL, Cheung AY, Duan Q, Kita D, Liu MC, Maman J, Luu EJ, Wu BW, Gates L, et al. (2015). Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4, e06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Q, Bourdais G, Pan H, Robatzek S, and Tang D (2017). Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl. Acad. Sci. USA 114, 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianchi E, Doe B, Goulding D, and Wright GJ (2014). Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, Hamada S, Bianco C, Mancino M, Nagaoka T, Gonzales M, Bailly V, Strizzi L, and Salomon DS (2007). Requirement of glycosylphosphatidylinositol anchor of Cripto-1 for trans activity as a Nodal co-receptor. J. Biol. Chem 282, 35772–35786. [DOI] [PubMed] [Google Scholar]
- 28.Huang Q, Dresselhaus T, Gu H, and Qu L-J (2015). Active role of small peptides in Arabidopsis reproduction: Expression evidence. J. Integr. Plant Biol 57, 518–521. [DOI] [PubMed] [Google Scholar]
- 29.Lalanne E, Honys D, Johnson A, Borner GH, Lilley KS, Dupree P, Grossniklaus U, and Twell D (2004). SETH1 and SETH2, two compnents of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, and Chen QJ (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, and Shinozaki K (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19, 3549–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morato do Canto A, Ceciliato PHO, Ribeiro B, Ortiz Morea FA, Franco Garcia AA, Silva-Filho MC, and Moura DS (2014). Biological activity of nine recombinant AtRALF peptides: implications for their perception and function in Arabidopsis. Plant Physiol. Biochem 75, 45–54. [DOI] [PubMed] [Google Scholar]
- 33.Galindo-Trigo S, Blanco-Touriñán N, DeFalco TA, Zipfel C, Gray JE, and Smith LM (2018). CrRLK1L receptor-like kinases HERK1 and ANJEA are female determinants of pollen tube reception. bioRxiv. 10.1101/428854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao L, Liu J, Zhong S, Gu H, and Qu L-J (2016). AtVPS41-mediated endocytic pathway is essential for pollen tube-stigma interaction in Arabidopsis. Proc. Natl. Acad. Sci. USA 113, 6307–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stagljar I, Korostensky C, Johnsson N, and te Heesen S (1998). A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95, 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearce G, Yamaguchi Y, Munske G, and Ryan CA (2010). Structure-activity studies of RALF, rapid alkalinization factor, reveal an essential–YISY–motif. Peptides 31, 1973–1977. [DOI] [PubMed] [Google Scholar]
- 37.Liu P, Haruta M, Minkoff BB, and Sussman MR (2018). Probing a plant plasma membrane receptor kinase’s three-dimensional structure using mass spectrometry-based protein footprinting. Biochemistry 57, 5159–5168. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Lin X, Han Z, Qu L-J, and Chai J (2016). Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 26, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zipfel C, and Oldroyd GE (2017). Plant signalling in symbiosis and immunity. Nature 543, 328–336. [DOI] [PubMed] [Google Scholar]
- 40.Liang X, and Zhou J-M (2018). Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol 69, 267–299. [DOI] [PubMed] [Google Scholar]
- 41.Ma X, Xu G, He P, and Shan L (2016). SERKing coreceptors for receptors. Trends Plant Sci. 21, 1017–1033. [DOI] [PubMed] [Google Scholar]
- 42.Song W, Han Z, Wang J, Lin G, and Chai J (2017). Structural insights into ligand recognition and activation of plant receptor kinases. Curr. Opin. Struct. Biol 43, 18–27. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Lin X, Han Z, Wang J, Qu L-J, and Chai J (2016). SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol. Plant 9, 1406–1414. [DOI] [PubMed] [Google Scholar]
- 44.Meng X, Chen X, Mang H, Liu C, Yu X, Gao X, Torii KU, He P, and Shan L (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol 25, 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng X, Zhou J, Tang J, Li B, de Oliveira MVV, Chai J, He P, and Shan L (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14, 1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chinchilla D, Shan L, He P, de Vries S, and Kemmerling B (2009). One for all: the receptor-associated kinase BAK1. Trends Plant Sci. 14, 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Zhou J, Shan L, and Meng X (2018). Plant cell surface receptor-mediated signaling - a common theme amid diversity. J. Cell Sci 131, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somssich M, Ma Q, Weidtkamp-Peters S, Stahl Y, Felekyan S, Bleckmann A, Seidel CA, and Simon R (2015). Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci. Signal 8, ra76. [DOI] [PubMed] [Google Scholar]
- 49.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, and Shibuya N (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, and Shibuya N (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K, et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 111, E404–E413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hou Y, Guo X, Cyprys P, Zhang Y, Bleckmann A, Cai L, Huang Q, Luo Y, Gu H, Dresselhaus T, et al. (2016). Maternal ENODLs are required for pollen tube reception in Arabidopsis. Curr. Biol 26, 2343–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, and Chai J (2019). Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Wang K, Yin G, Liu X, Liu M, Cao N, Duan Y, Gao H, Wang W, Ge W, et al. (2018). Pollen-expressed leucine-rich repeat extensins are essential for pollen germination and growth. Plant Physiol. 176, 1993–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sede AR, Borassi C, Wengier DL, Mecchia MA, Estevez JM, and Muschietti JP (2018). Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett. 592, 233–243. [DOI] [PubMed] [Google Scholar]
- 56.Müller AO, Blersch KF, Gippert AL, and Ischebeck T (2017). Tobacco pollen tubes - a fast and easy tool for studying lipid droplet association of plant proteins. Plant J. 89, 1055–1064. [DOI] [PubMed] [Google Scholar]
- 57.Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, and Dresselhaus T (2012). Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 58.Boavida LC, and McCormick S (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52, 570–582. [DOI] [PubMed] [Google Scholar]
- 59.Mori T, Kuroiwa H, Higashiyama T, and Kuroiwa T (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol 8, 64–71. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Wei B, Yuan R, Wang J, Ding M, Chen Z, Yu H, and Qin G (2017). The Arabidopsis RING-type E3 ligase TEAR1 controls leaf development by targeting the TIE1 transcriptional repressor for degradation. Plant Cell 29, 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code.


