Abstract
SARS-CoV-2 is a betacoronavirus responsible for the COVID-19 pandemic. Although the SARS-CoV-2 genome was reported recently, its transcriptomic architecture is unknown. Utilizing two complementary sequencing techniques, we present a high-resolution map of the SARS-CoV-2 transcriptome and epitranscriptome. DNA nanoball sequencing shows that the transcriptome is highly complex owing to numerous discontinuous transcription events. In addition to the canonical genomic and 9 subgenomic RNAs, SARS-CoV-2 produces transcripts encoding unknown ORFs with fusion, deletion, and/or frameshift. Using nanopore direct RNA sequencing, we further find at least 41 RNA modification sites on viral transcripts, with the most frequent motif, AAGAA. Modified RNAs have shorter poly(A) tails than unmodified RNAs, suggesting a link between the modification and the 3′ tail. Functional investigation of the unknown transcripts and RNA modifications discovered in this study will open new directions to our understanding of the life cycle and pathogenicity of SARS-CoV-2.
Keywords: SARS-CoV-2, coronavirus, COVID-19, nanopore, direct RNA sequencing, RNA modification, discontinuous transcription, epitranscriptome, transcriptome, poly(A) tail
Graphical Abstract
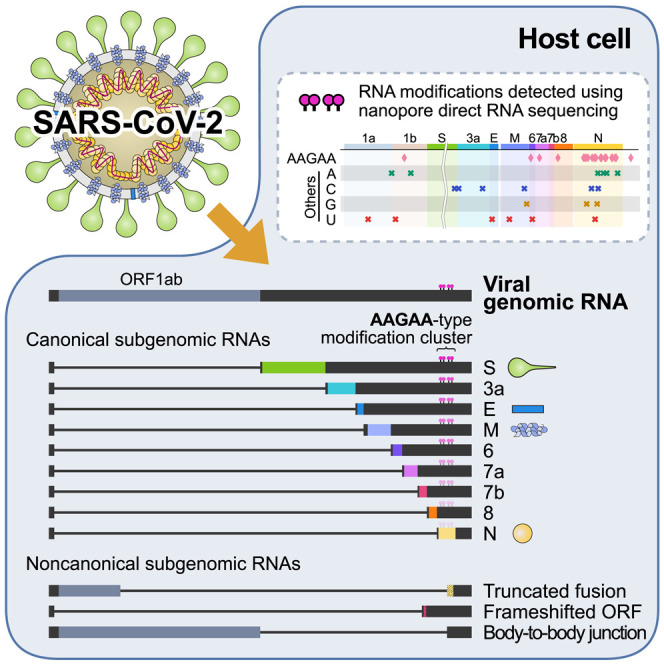
The SARS-CoV-2 transcriptome and epitranscriptome reveal a complex array of canonical and non-canonical viral transcripts with RNA modifications.
Introduction
Coronavirus disease 19 (COVID-19) is caused by a novel coronavirus designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Zhou et al., 2020, Zhu et al., 2020). Like other coronaviruses (order Nidovirales, family Coronaviridae, subfamily Coronavirinae), SARS-CoV-2 is an enveloped virus with a positive-sense, single-stranded RNA genome of ∼30 kb. SARS-CoV-2 belongs to the genus betacoronavirus, together with SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (with 80% and 50% homology, respectively) (Kim et al., 2020, Zhou et al., 2020). Coronaviruses (CoVs) were thought to primarily cause enzootic infections in birds and mammals. However, the recurring outbreaks of SARS, MERS, and now COVID-19 have clearly demonstrated the remarkable ability of CoVs to cross species barriers and transmit between humans (Menachery et al., 2017).
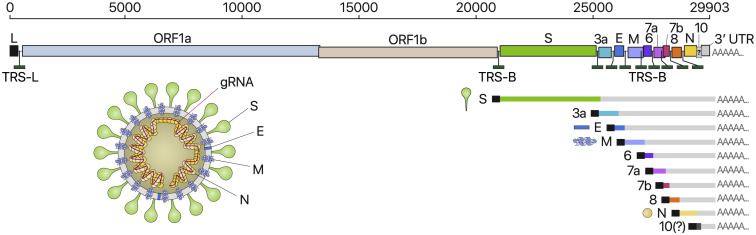
Among RNA viruses, CoVs have some of the largest genomes (26-32kb) (Figure 1 ). Each viral transcript has a 5′-cap structure and a 3′ poly(A) tail (Lai and Stohlman, 1981, Yogo et al., 1977). Upon cell entry, the genomic RNA is translated to produce nonstructural proteins (nsps) from two open reading frames (ORFs), ORF1a and ORF1b. The ORF1a produces polypeptide 1a (pp1a, 440–500 kDa) that is cleaved into 11 nsps. The −1 ribosome frameshift occurs immediately upstream of the ORF1a stop codon, which allows continued translation of ORF1b, yielding a large polypeptide (pp1ab, 740–810 kDa) which is cleaved into 15 nsps. The proteolytic cleavage is mediated by viral proteases nsp3 and nsp5 that harbor a papain-like protease domain and a 3C-like protease domain, respectively.
Figure 1.
Schematic Presentation of the SARS-CoV-2 Genome Organization, the Canonical Subgenomic mRNAs, and the Virion Structure
From the full-length genomic RNA (29,903 nt) that also serves as an mRNA, ORF1a and ORF1b are translated. In addition to the genomic RNA, nine major subgenomic RNAs are produced. The sizes of the boxes representing small accessory proteins are bigger than the actual size of the ORF for better visualization. The black box indicates the leader sequence. Note that our data show no evidence for ORF10 expression.
The viral genome is also used as the template for replication and transcription, which is mediated by nsp12 harboring RNA-dependent RNA polymerase (RdRP) activity (Snijder et al., 2016, Sola et al., 2015). Negative-sense RNA intermediates are generated to serve as the templates for the synthesis of positive-sense genomic RNA (gRNA) and subgenomic RNAs (sgRNAs). The gRNA is packaged by the structural proteins to assemble progeny virions. Shorter sgRNAs encode conserved structural proteins (spike protein [S], envelope protein [E], membrane protein [M], and nucleocapsid protein [N]), and several accessory proteins. SARS-CoV-2 is known to have at least six accessory proteins (3a, 6, 7a, 7b, 8, and 10) according to the current annotation (GenBank: NC_045512.2). However, the ORFs have not yet been experimentally verified for expression. Therefore, it is currently unclear which accessory genes are actually expressed from this compact genome.
Each coronaviral RNA contains the common 5′ “leader” sequence of ∼70 nt fused to the “body” sequence from the downstream part of the genome (Lai and Stohlman, 1981, Sola et al., 2015) (Figure 1). According to the prevailing model, leader-to-body fusion occurs during negative-strand synthesis at short motifs called transcription-regulatory sequences (TRSs) that are located immediately adjacent to ORFs (Figure 1). TRSs contain a conserved 6–7 nt core sequence (CS) surrounded by variable sequences. During negative-strand synthesis, RdRP pauses when it crosses a TRS in the body (TRS-B) and switches the template to the TRS in the leader (TRS-L), which results in discontinuous transcription leading to the leader-body fusion. From the fused negative-strand intermediates, positive-strand mRNAs are transcribed. The replication and transcription mechanism has been studied in other coronaviruses. However, it is unclear whether the general mechanism also applies to SARS-CoV-2 and if there are any unknown components in the SARS-CoV-2 transcriptome. For the development of diagnostic and therapeutic tools and the understanding of this new virus, it is critical to define the organization of the SARS-CoV-2 genome.
Deep sequencing technologies offer powerful means to investigate viral transcriptome. The “sequencing-by-synthesis (SBS)” methods such as the Illumina and MGI platforms confer high accuracy and coverage. However, they are limited by short read length (200–400 nt), so the fragmented sequences should be re-assembled computationally, during which the haplotype information is lost. More recently introduced is the nanopore-based direct RNA sequencing (DRS) approach. Although nanopore DRS is limited in sequencing accuracy, it enables long-read sequencing, which would be particularly useful for the analysis of long nested CoV transcripts. Moreover, because DRS detects RNA instead of cDNA, the RNA modification information can be obtained directly during sequencing (Garalde et al., 2018). Numerous RNA modifications have been found to control eukaryotic RNAs and viral RNAs (Williams et al., 2019). Terminal RNA modifications such as RNA tailing also play a critical role in cellular and viral RNA regulation (Warkocki et al., 2018).
In this study, we combined two complementary sequencing approaches, DRS and SBS. We unambiguously mapped the sgRNAs, ORFs, and TRSs of SARS-CoV-2. Additionally, we found numerous unconventional RNA joining events that are distinct from canonical TRS-mediated polymerase jumping. We further discovered RNA modification sites and measured the poly(A) tail length of gRNAs and sgRNAs.
Results and Discussion
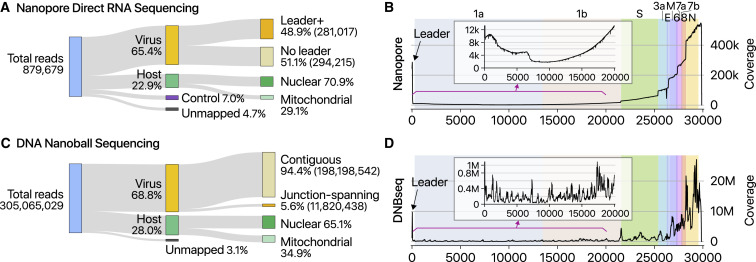
To delineate the SARS-CoV-2 transcriptome, we first performed DRS runs on a MinION nanopore sequencer with total RNA extracted from Vero cells infected with SARS-CoV-2 (BetaCoV/Korea/KCDC03/2020). The virus was isolated from a patient who was diagnosed with COVID-19 on January 26, 2020, after traveling from Wuhan, China (Kim et al., 2020). We obtained 879,679 reads from infected cells (corresponding to a throughput of 1.9 Gb) (Figure 2 A). The majority (65.4%) of the reads mapped to SARS-CoV-2, indicating that viral transcripts dominate the transcriptome while the host gene expression is strongly suppressed. Although nanopore DRS has the 3′ bias due to directional sequencing from the 3′ ends of RNAs, approximately half of the viral reads still contained the 5′ leader.
Figure 2.
Statistics of Sequencing Data
(A) Read counts from nanopore direct RNA sequencing of total RNA from Vero cells infected with SARS-CoV-2. “Leader+” indicates the viral reads that contain the 5′ end leader sequence. “No leader” denotes the viral reads lacking the leader sequence. “Nuclear” reads match mRNAs from the nuclear chromosome while “mitochondrial” reads are derived from the mitochondrial genome. “Control” indicates quality control RNA for nanopore sequencing.
(B) Genome coverage of the nanopore direct RNA sequencing data shown in (A). The stepwise reduction in coverage corresponds to the borders expected for the canonical sgRNAs. The smaller inner plot magnifies the 5′ part of the genome.
(C) Read counts from DNA nanoball sequencing using MGISEQ. Total RNA from Vero cells infected with SARS-CoV-2 was used for sequencing.
(D) Genome coverage of the DNA nanoball sequencing (DNB-seq) data shown in (C).
See also Figure S1.
The SARS-CoV-2 genome was almost fully covered, missing only 12 nt from the 5′ end due to the known inability of DRS to sequence the terminal ∼12 nt (Figure 2B). The longest tags (111 reads) correspond to the full-length gRNA (Figure 2B). The coverage of the 3′ side of the viral genome is substantially higher than that of the 5′ side, which reflects the nested sgRNAs. This is also partly due to the 3′ bias of the directional DRS technique. The common presence of the leader sequence (72 nt) in viral RNAs results in a prominent coverage peak at the 5′ end, as expected. We could also clearly detect vertical drops in the coverage, whose positions correspond to the leader-body junction in sgRNAs. All known sgRNAs are supported by DRS reads, with an exception of ORF10 (see below).
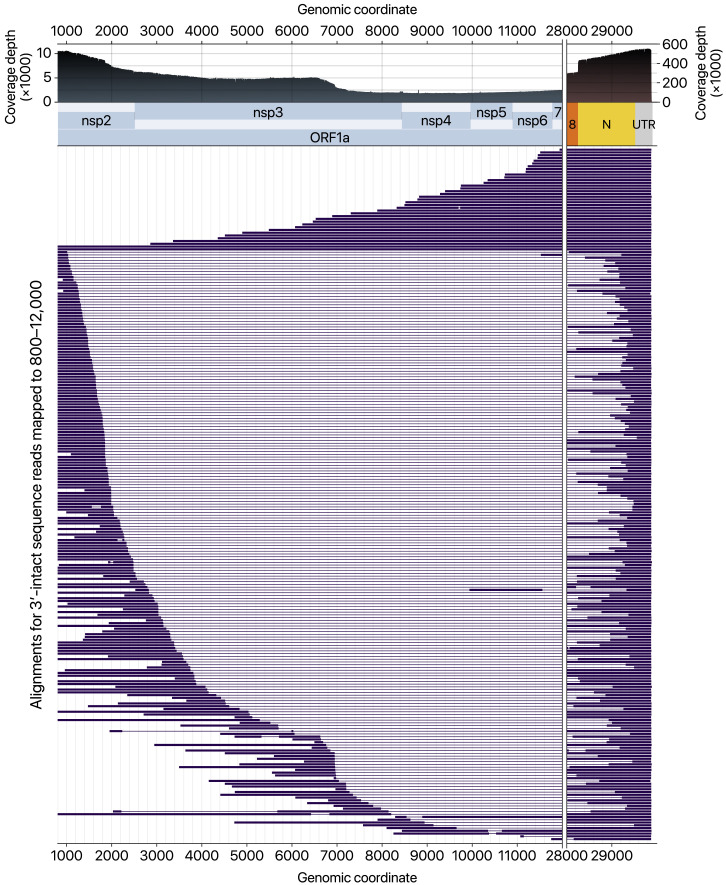
In addition, we observed unexpected reads reflecting noncanonical “splicing” events (Figure S1 ). Such fusion transcripts resulted in the increased coverage toward the 5′ end (Figure 2B, inset). Early studies on coronavirus mouse hepatitis virus reported that recombination frequently occurs (Furuya and Lai, 1993, Liao and Lai, 1992, Luytjes et al., 1996). Some viral RNAs contain the 5′ and 3′ proximal sequences resulting from “illegitimate” polymerase jumping.
Figure S1.
Subgenomic RNAs with Large Deletions between nsp2/3 and N Regions, Related to Figure 2
Sequence alignments of the 3′-intact DRS reads mapped to the genomic interval 800–12,000. The x axis highlights two separate ranges. The filled black curves on top show the read coverage. Single read alignment is shown as a set of thick bars and lines connected. Thick bars on the alignments indicate contiguous mappings consisting of matches, mismatches, insertions, and small deletions. The lines show the large gaps longer than 50 nt.
To further validate sgRNAs and their junction sites, we performed DNA nanoball sequencing (DNB-seq) based on the sequencing-by-synthesis principle and obtained 305,065,029 reads with an average insert length of 220 nt (Figure 2C). The results are overall consistent with the DRS data. The leader-body junctions are frequently sequenced, giving rise to a sharp peak at the 5′ end in the coverage plot (Figure 2D). The 3′ end exhibits a high coverage as expected for the nested transcripts.
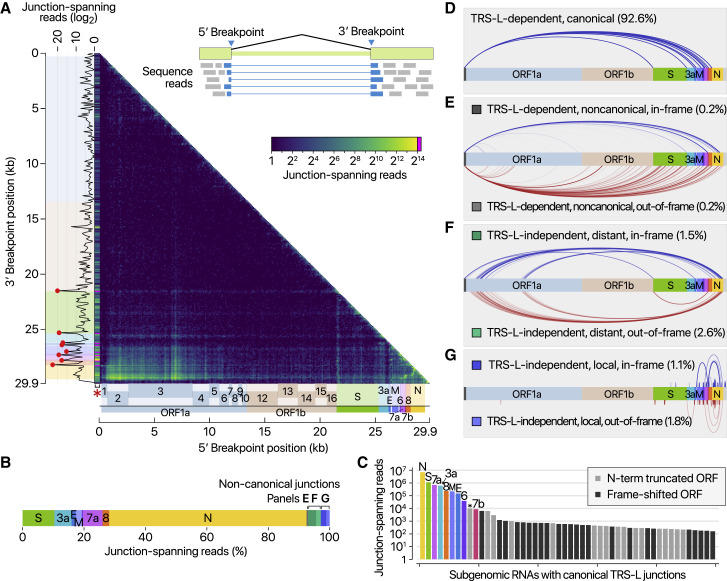
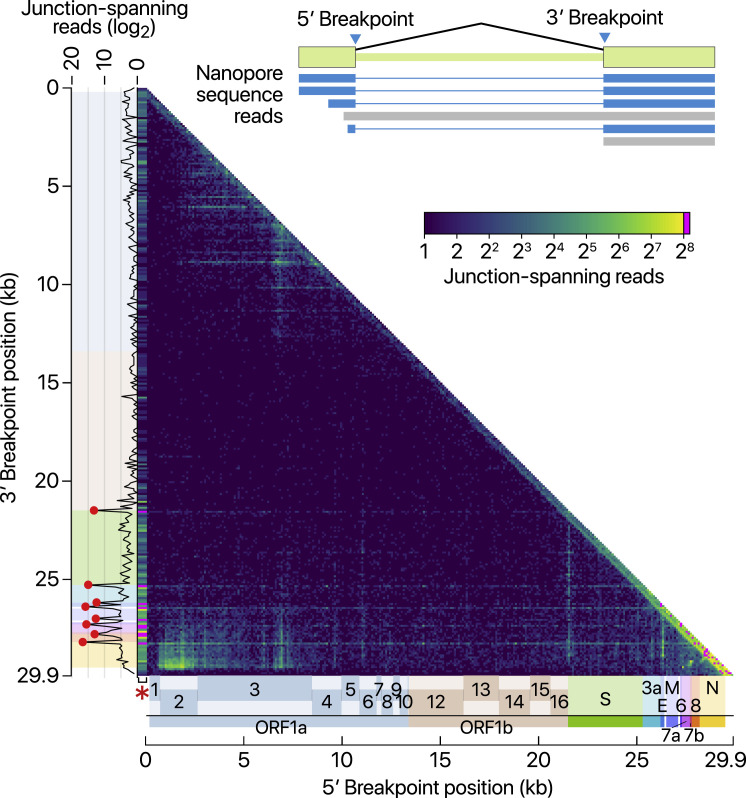
The depth of DNB-seq allowed us to confirm and examine the junctions on an unprecedented scale for a CoV genome. We mapped the 5′ and 3′ breakpoints at the junctions and estimated the fusion frequency by counting the reads spanning the junctions (Figure 3 A). The leader represents the most prominent 5′ site, as expected (Figure 3A, red asterisk on the x axis). The known TRS-Bs are detected as the top 3′ sites (Figure 3A, red dots on the y axis). These results confirm that SARS-CoV-2 uses the canonical TRS-mediated template-switching mechanism for discontinuous transcription to produce major sgRNAs (Figure 3B). Quantitative comparison of the junction-spanning reads shows that the N RNA is the most abundantly expressed transcript, followed by S, 7a, 3a, 8, M, E, 6, and 7b (Figure 3C).
Figure 3.
Viral Subgenomic RNAs and Their Recombination Sites
(A) Frequency of discontinuous mappings in the long reads from the DNB-seq data. The color indicates the number of reads with large gaps spanning between two genomic positions (starting from a coordinate in the x axis and ending in a coordinate in the y axis). The counts were aggregated into 100-nt bins for both axes. The red asterisk on the x axis indicates the column containing the leader TRS. Please note that the leftmost column was expanded horizontally on this heatmap to improve visualization. The red dots on the sub-plot alongside the y axis denote local peaks which coincide with the 5′ end of the body of each sgRNA.
(B) Transcript abundance was estimated by counting the DNBseq reads that span the junction of the corresponding RNA.
(C) Top 50 sgRNAs. The asterisk indicates an ORF beginning at 27,825 that may encode the 7b protein with an N-terminal truncation of 23 amino acids. The gray bars denote minor transcripts that encode proteins with an N-terminal truncation compared with the corresponding overlapping transcript. The black bars indicate minor transcripts that encode proteins in a different reading frame from the overlapping major mRNA.
(D) Canonical discontinuous transcription that is mediated by TRS-L and TRS-B.
(E) TRS-L-dependent noncanonical fusion between the leader TRS and a noncanonical 3′ site in the body.
(F) TRS-L-independent long-distance (>5,000 nt) fusion.
(G) TRS-L-independent local joining yielding a deletion between proximal sites (20–5,000 nt distance).
See also Figures S2 and S3 and Tables S2, S3, and S4.
It is important to note that ORF10 is represented by only one read in DNB data (0.000009% of viral junction-spanning reads) and that it was not supported at all by DRS data. ORF10 does not show significant homology to known proteins. Thus, ORF10 is unlikely to be expressed. The annotation of ORF10 should be reconsidered. Taken together, SARS-CoV-2 expresses nine canonical sgRNAs (S, 3a, E, M, 6, 7a, 7b, 8, and N) together with the gRNA (Figures 1 and 3C).
In addition to the canonical sgRNAs with expected structure and length (Figure 3D), our results show many minor junction sites (Figures 3E–3G; Table S2). There are three main types of such fusion events. The RNAs in the first group have the leader combined with the body at unexpected 3′ sites in the middle of ORFs or UTR (Figure 3E, TRS-L-dependent noncanonical; Table S3). The second group shows a long-distance fusion between sequences that do not have similarity to the leader (Figure 3F, TRS-L-independent distant). The last group undergoes local fusion, which leads to smaller deletions, mainly in structural and accessory genes, including the S ORF (Figure 3G, TRS-L-independent local recombination). These fusion transcripts were also found in DRS data (Figure S2 ). We verified the expression of some of these transcripts by RT-PCR (Figure S3 ).
Figure S2.
Map of Discontinuous Transcription Detected by Direct RNA Sequencing, Related to Figure 3
Frequency of discontinuous mappings in the long reads from the nanopore DRS data. The color indicates the number of reads with large gaps spanning between two genomic positions (starting from a coordinate in the x axis and ending in a coordinate in the y axis). The counts were aggregated into 100-nt bins for both axes. The red asterisk on the x axis indicates the column containing the leader TRS. Please note that the leftmost column containing the leader TRS was expanded horizontally on this heatmap to improve visualization. The red dots on the sub-plot alongside the y axis denote local peaks which coincide with the 5′ end of the body of each sgRNA.
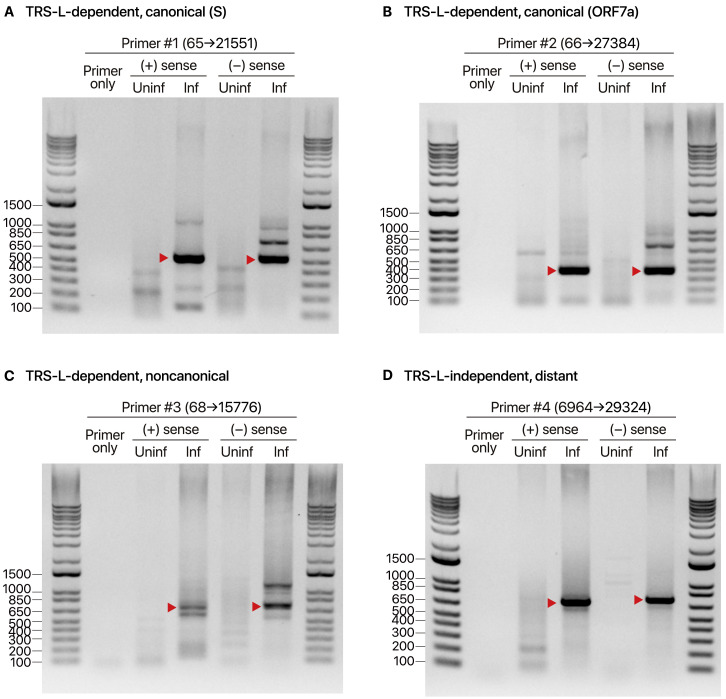
Figure S3.
Validation of Discontinuous Transcription Detected by RT-PCR, Related to Figure 3
To validate the sgRNAs found by sequencing, RT-PCR was performed to detect the sgRNAs and their negative-sense counterparts. (+) sense, cDNA from positive-strand specific reverse transcription; (−) sense, cDNA from negative-strand specific reverse transcription. ‘Primer only’ does not contain a cDNA template. cDNA from uninfected Vero cells (Uninf) were used as negative controls. Ladders are presented on the left (bp). A, RT-PCR spanning the canonical junction between TRS-L and the S ORF. B, RT-PCR spanning the canonical junction between TRS-L and the ORF7a. C, RT-PCR spanning the noncanonical junction between TRS-L and the middle of ORF1. D, RT-PCR spanning a noncanonical TRS-L-independent junction. The products were run on agarose gels. Red arrowheads denote the expected amplicons.
Of note, the junctions in these noncanonical transcripts are not derived from a known TRS-B. Some junctions show short sequences (3–4 nt) common between the 5′ and 3′ sites, suggesting a partial complementarity-guided template switching (“polymerase jumping”). However, the majority do not have any obvious sequences. Thus, we cannot exclude a possibility that at least some of these transcripts are generated through a different mechanism(s).
It was previously shown in other coronaviruses that transcripts with partial sequences are produced (Furuya and Lai, 1993, Liao and Lai, 1992, Luytjes et al., 1996). Recent sequencing analyses also revealed non-canonical sgRNAs from mouse hepatitis virus (genus betacoronavirus, subfamily Coronavirinae) (Irigoyen et al., 2016), HCoV-229E (genus alphacoronavirus, subfamily Coronavirinae) (Viehweger et al., 2019), and equine torovirus (genus Torovirus, subfamily Torovirinae, family Coronaviridae) (Stewart et al., 2018), suggesting this mechanism may be at least partially conserved in coronaviridae. Functionality of sgRNAs are not clear, and some of them have been considered as parasites that compete for viral proteins, hence referred to as “defective interfering RNAs” (DI-RNAs) (Pathak and Nagy, 2009).
Although the noncanonical transcripts may arise from erroneous replicase activity, it remains an open question if the fusion has an active role in viral life cycle and evolution. Although individual RNA species are not abundant, the combined read numbers are often comparable to the levels of accessory transcripts. Most of the RNAs have coding potential to yield proteins. Transcripts that belong to the “TRS-L-independent distant” group encode the upstream part of ORF1a, including nsp1, truncated nsp2, and/or truncated nsp3, whose summed abundance is ∼20% of gRNA. Depending on translation efficiency, the protein products may change the stoichiometry between nsps (Figure 3F; Table S4). Another notable example is the 7b protein with an N-terminal truncation that may be produced at a level similar to the annotated full-length 7b (Figure 3C, asterisk). Frameshifted or deleted ORFs may also generate shorter proteins that are distinct from known viral proteins (Figure 3C). It will be interesting in the future to examine if these unknown ORFs are actually translated and yield functional products.
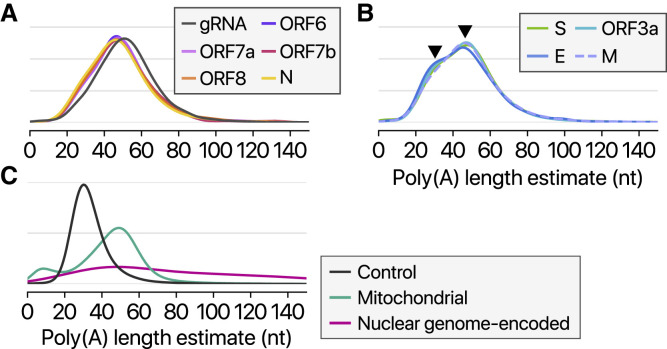
As nanopore DRS is based on single-molecule detection of RNA, it offers a unique opportunity to examine multiple epitranscriptomic features of individual RNA molecules. We recently developed software to measure the length of poly(A) tail from DRS data (Y. Choi and H.C., unpublished data). Using this software, we confirm that, like other CoVs, SARS-CoV-2 RNAs carry poly(A) tails (Figures 4 A–4B). The tail of viral RNAs is 47 nt in median length. The full-length gRNA has a relatively longer tail than sgRNAs. Notably, sgRNAs have two tail populations: a minor peak at ∼30 nt and a major peak at ∼45 nt (Figure 4B, arrowheads). Wu et al. (2013) previously observed that the poly(A) tail length of bovine CoV mRNAs changes during infection: from ∼45 nt immediately after virus entry to ∼65 nt at 6–9 hours post-infection and ∼30 nt at 120–144 hours post-infection. Thus, the short tails of ∼30 nt observed in this study may represent aged RNAs that are prone to decay. Viral RNAs exhibit a homogeneous length distribution, unlike host nuclear genome-encoded mRNAs (Figure 4C). The distribution is similar to that of mitochondrial chromosome-encoded RNAs whose tail is generated by MTPAP (Tomecki et al., 2004). It was recently shown that HCoV-229E nsp8 has an adenylyltransferase activity, which may extend poly(A) tail of viral RNA (Tvarogová et al., 2019). Because poly(A) tail should be constantly attacked by host deadenylases, the regulation of viral RNA tailing is likely to be important for the maintenance of genome integrity. Poly(A) tail of mRNA is also generally critical for stability control and translation through its interaction with poly(A) binding proteins (PABPs). Cytoplasmic PABPs facilitate deadenylation by the CCR4-NOT complex while blocking untimely decay by exosome and uridylation machinery. PABPs also interact with translation initiation factors to allow translation. Thus, the viral tail is likely to play multiple roles for translation, decay, and replication.
Figure 4.
Length of Poly(A) Tail
(A and B) Kernel density plots showing poly(A) tail length distribution of viral transcripts without (A) or with (B) a subpeak near 30 nt. Arrowheads indicate peaks at ~30 and ~45 nt.
(C) Kernel density plots showing poly(A) tail length distribution of quality control RNA that has a 30-nt poly(A) tail, host mRNAs from the nuclear chromosome, or host RNAs from the mitochondrial chromosome.
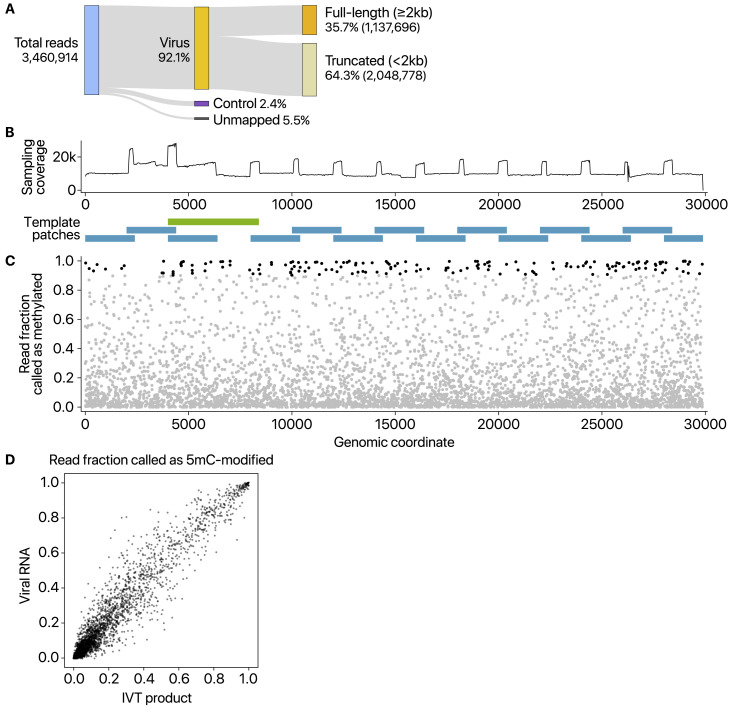
Next, we examined the epitranscriptomic landscape of SARS-CoV-2 by using the DRS data. Viral RNA modification was first described more than 40 years ago (Gokhale and Horner, 2017). N6-methyladenosine (m6A) is the most widely observed modification (Courtney et al., 2017, Gokhale et al., 2016, Krug et al., 1976, Lichinchi et al., 2016, Narayan et al., 1987), but other modifications have also been reported on viral RNAs, including 5-methylcytosine methylation (5mC), 2′-O-methylation (Nm), deamination, and terminal uridylation. In a recent analysis of HCoV-229E using DRS, modification calling suggested frequent 5mC signal across viral RNAs (Viehweger et al., 2019). However, because no direct control group was included in the analysis, the proposed modification needed validation. To unambiguously investigate the modifications, we generated negative control RNAs by in vitro transcription of the viral sequences and performed a DRS run on these unmodified controls (Figure S4 A). The partially overlapping control RNAs are ∼2.3 kb or ∼4.4 kb each and cover the entire length of the genome (Figure S4B). Detection using pre-trained models reported numerous signal level changes corresponding to 5mC modification, even with the unmodified controls (Figure S4C). We obtained highly comparable results from the viral RNAs from infected cells (Figure S4D). Thus, the 5mC sites detected without a control are likely to be false positives.
Figure S4.
False-Positive Calling of 5mC Modification Demonstrated by Using Unmodified Negative Control RNAs, Related to Figure 5
A, Read counts from nanopore direct RNA sequencing of in vitro transcribed (IVT) RNAs that have viral sequences. “Control” indicates quality control RNA for nanopore sequencing. B, The 15 partially overlapping patches cover the entire genome (blue rectangles at the bottom). Each RNA is ~2.3 kb in length. One fragment marked with a green rectangle is longer than others (~4.4 kb) to circumvent difficulties in the PCR amplification. The sequenced reads were downsampled so that every region is equally covered. The resulting balanced coverage is shown in the chart at the top. C, Detected 5mC modification from in vitro transcribed unmodified RNAs (IVT product) by the “alternative base detection” mode in Tombo. Black dots indicate the sites that satisfy the estimated false discovery rate cut-off calculated using unmodified yeast ENO2 mRNA (Viehweger et al., 2019). D, Comparison between the sites called from unmodified IVT products and those from viral RNAs expressed in Vero cells.
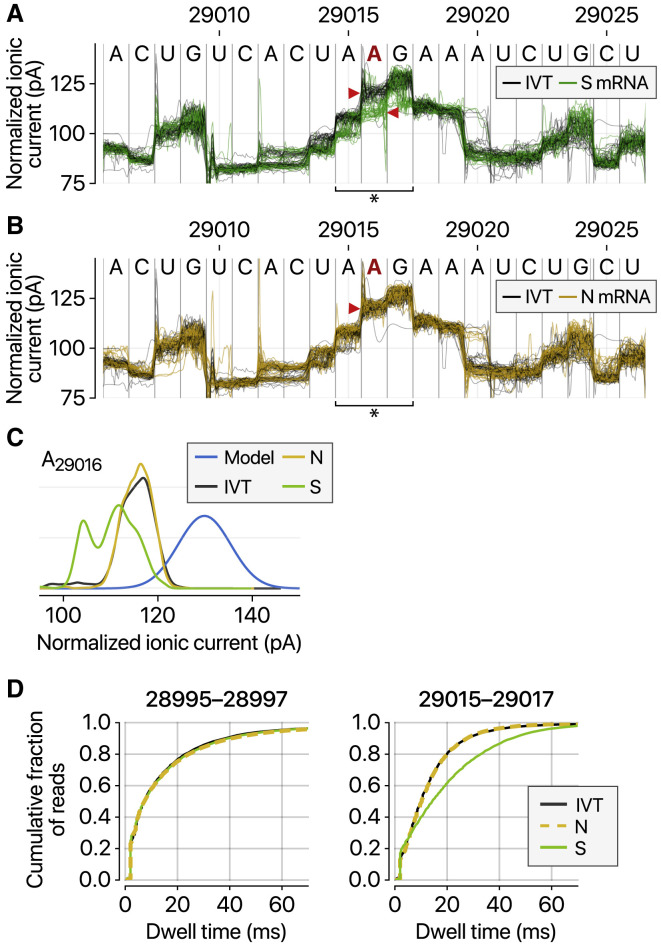
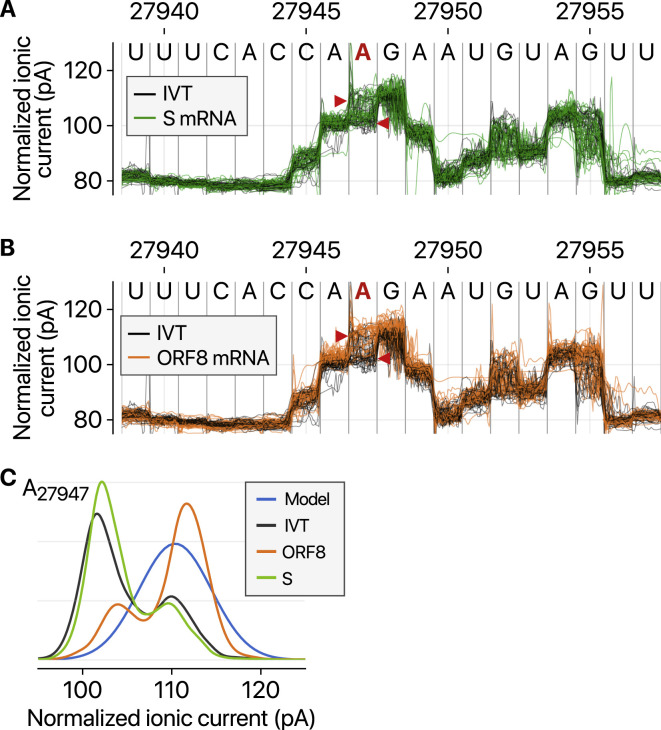
We, however, noticed intriguing differences in the ionic current (called “squiggles”) between negative control and viral transcripts (Figure 5 A). At least 41 sites displayed substantial differences (over 20% frequency), indicating potential RNA modifications (Table S5). Notably, some of the sites showed different frequencies depending on the sgRNA species. Figures 5A–5C show an example that is modified more heavily on the S RNA than the N RNA, while Figures S5 A–S5C present a site that is modified more frequently on the ORF8 RNA compared with the S RNA. Moreover, the dwell time of the modified base (Figure 5D, right) is longer than that of the unmodified base (Figure 5D, left), suggesting that the modification interferes with the passing of RNA molecules through the pore.
Figure 5.
Frequent RNA Modification Sites
(A) Distinct ionic current signals (“squiggles”) from viral S transcript (green lines) and in vitro transcribed control (IVT, black lines) indicate RNA modification at the genomic position 29,016.
(B) The ionic current signals from viral N transcript at the genomic position 29,016 (yellow lines) are similar to those from IVT control (black lines), indicating that modification is rare on the N sgRNA.
(C) Kernel density estimations of ionic current distribution at A29016. Blue line shows the signal distribution in the standard model of tombo 1.5.
(D) Dwell time difference supports RNA modification. The dwell time of the region 29,015–29,017 of the S RNA (right) is significantly longer than those of IVT control and N RNAs. On the contrary, the neighboring region 28,995–28,997 of IVT, N, and S RNA is indistinguishable (left).
See also Figures S4 and S5.
Figure S5.
Detected Modified Sites in Viral RNAs, Related to Figure 5
A, Ionic current levels near the genomic position 27,947 in viral S RNA (green lines) and IVT control RNA (black lines). B, Ionic current levels for the identical region in the viral ORF8 RNA (orange lines) and IVT control RNA (black lines). C, Kernel density plots for signal distributions at the position 27,947 in the different RNAs. The blue line shows the standard model used for modification detections without controls (“alternative base detection” and “de novo” modes) in Tombo.
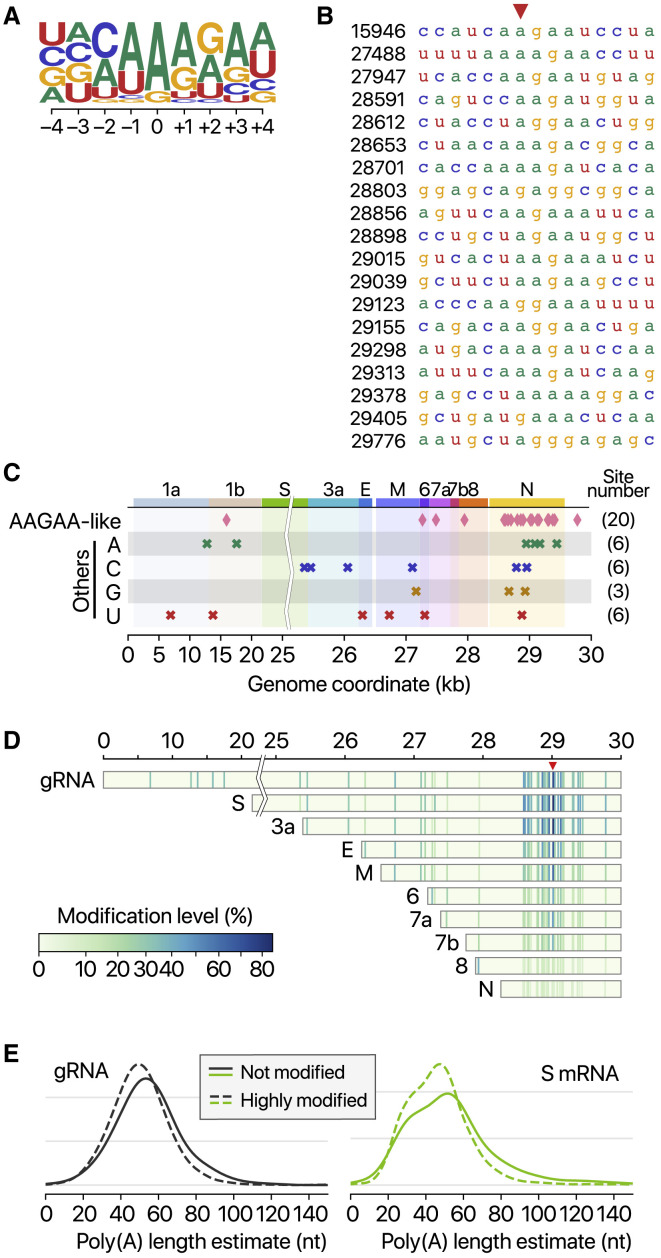
Among the 41 potential modification sites, the most frequently observed motif is AAGAA (Figures 6A and 6B). The modification sites on the “AAGAA-like” motif (including AAGAA and other A/G-rich sequences) are found throughout the viral genome but particularly enriched in genomic positions 28,500–29,500 (Figure 6C). Long viral transcripts (gRNA, S, 3a, E, and M) are more frequently modified than shorter RNAs (6, 7a, 7b, 8, and N) (Figure 6D), suggesting a modification mechanism that is specific for certain RNA species.
Figure 6.
Detected RNA Modifications Are Differentially Regulated
(A) Position-specific base frequency of a motif enriched in the frequently modified sites.
(B) Sequence alignment of the detected modification sites with “AAGAA”-like motif. Base positions on the left hand side correspond to the genomic coordinates denoted with red arrowhead.
(C) Genomic location of modification sites with the AAGAA-like motif (top row) and the others grouped by the detected nucleotide base.
(D) Location and modification levels in different RNA species.
(E) Kernel density plots showing poly(A) length distribution of gRNA (left) and S RNA (right). Modified viral RNAs carry shorter poly(A) tails.
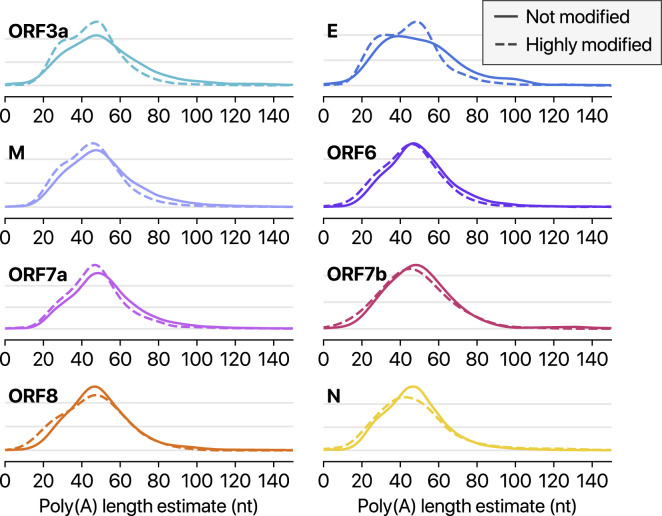
Because DRS allows simultaneous detection of multiple features on individual molecules, we cross-examined the poly(A) tail length and internal modification sites. Interestingly, modified RNA molecules have shorter poly(A) tails than unmodified ones (Figures 6E and S6 ; p < 9.8 × 10−5 and p < 7.3 × 10−12 for ORF1ab and S, respectively; Mann-Whitney U test). These results suggest a link between the internal modification and 3′ end tail. Because poly(A) tail plays an important role in RNA turnover, it is tempting to speculate that the observed internal modification is involved in viral RNA stability control. It is also plausible that RNA modification is a mechanism to evade host immune response. The type of modification(s) is yet to be identified, although we can exclude METTL3-mediated m6A (for lack of consensus motif RRACH), ADAR-mediated deamination (for lack of A-to-G sequence change in the DNBseq data), and m1A (for lack of the evidence for RT stop). Our finding implicates a hidden layer of CoV regulation. It will be interesting in the future to identify the chemical nature, enzymology, and biological functions of the modification(s).
Figure S6.
Highly Modified Viral RNAs Carry Shorter Poly(A) Tails, Related to Figure 6
Poly(A) tail length distribution of each viral transcript other than shown in Figure 6.
In this study, we delineate the transcriptomic and epitranscriptomic architecture of SARS-CoV-2. Unambiguous mapping of the expressed sgRNAs and ORFs is a prerequisite for the functional investigation of viral proteins, replication mechanism, and host-viral interactions involved in pathogenicity. An in-depth analysis of the joint reads revealed a highly complex landscape of viral RNA synthesis. Like other RNA viruses, CoVs undergo frequent recombination, which may allow rapid evolution to change their host/tissue specificity and drug sensitivity. The frequent fusion detected in this study may provide a basis for variant generation and need to be investigated in detail. The new ORFs may serve as accessory proteins that modulate viral replication and host immune response. The RNA modifications may also contribute to viral survival and immune evasion in infected tissues as the innate immune system is known to be less sensitive to RNAs with nucleoside modification (Karikó et al., 2005). These new molecular features will need to be studied further in animal tissues and cell types that have an intact interferon system. It is also yet to be examined if the ORFs and RNA modifications are unique to SARS-CoV-2 or conserved in other coronaviruses. Comparative studies on their distribution and functional significance will help us to gain a deeper understanding of SARS-CoV-2 and coronaviruses in general. Our data provide a rich resource and open new directions to investigate the mechanisms underlying the pathogenicity of SARS-CoV-2.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| SARS-CoV-2 | National Culture Collection for Pathogens, Korea National Institute of Health, Korea | NCCP 43326 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Penicillin-streptomycin | GIBCO | Cat#15140163 |
| TRIzol | Invitrogen | Cat#15596018 |
| Actinomycin D | Sigma-Aldrich | Cat#A9415; CAS: 50-76-0 |
| Critical Commercial Assays | ||
| DMEM | GIBCO | Cat#11995065 |
| FBS | GIBCO | Cat#10082147 |
| Recombinant DNase I (RNase-free) | Takara | Cat#2270A |
| RNeasy MinElute Cleanup Kit | QIAGEN | Cat#74204 |
| SuperScript IV Reverse Transcriptase | Invitrogen | Cat#18090200 |
| Q5® High-Fidelity 2X Master Mix | NEB | Cat#M0492L |
| Gel Extraction Kit | Labopass | Cat#CMG0112 |
| MEGAscript T7 Transcription Kit | Invitrogen | Cat#AMB13345 |
| Oligo Clean & Concentrator | Zymo Research | Cat#D4061 |
| 1 Kb Plus DNA Ladder | Invitrogen | Cat#10787026 |
| Direct RNA sequencing kit | Oxford Nanopore Technologies | Cat#SQK-RNA002 |
| SUPERase•In RNase Inhibitor | Invitrogen | Cat#AM2696 |
| Flow Cell (R9.4.1) | Oxford Nanopore Technologies | Cat#FLO-MIN106D |
| MinION device | Oxford Nanopore Technologies | Cat#MinION; RRID: SCR_017985 |
| Dynabeads® mRNA Purification Kit | Invitrogen | Cat#61006 |
| MGIEasy RNA Directional Library Prep Set | MGI | Cat#1000006385 |
| MGISEQ-200RS High-throughput Sequencing Kit (PE100) | MGI | Cat#1000005233 |
| MGISEQ-200RS Sequencing Flow Cell | MGI | Cat#1000003794 |
| DNBSEQ-G50RS sequencer | MGI | Cat#DNBSEQ-G50RS |
| Deposited Data | ||
| RNA-seq using MGISEQ-200 | This study | https://doi.org/10.17605/OSF.IO/8F6N9 |
| Nanopore direct RNA sequencing | This study | https://doi.org/10.17605/OSF.IO/8F6N9 |
| Figure S3 original files | This study | https://doi.org/10.17632/bkhbpvtg7h.1 |
| Experimental Models: Cell Lines | ||
| Vero | ATCC | Cat#CCL-81 |
| Oligonucleotides | ||
| The oligonucleotides used in this study were listed in Table S1 | This study | N/A |
| Software and Algorithms | ||
| guppy 3.4.5 | Oxford Nanopore Technologies | https://community.nanoporetech.com/sso/login?next_url=%2Fdownloads |
| minimap2 2.17 | Li, 2018 | https://github.com/lh3/minimap2 |
| poreplex 0.5.0 | Hyeshik Chang, Seoul National University, Korea | https://github.com/hyeshik/poreplex |
| SciPy 1.4.1 | Virtanen et al., 2020 | https://www.scipy.org/; RRID: SCR_008058 |
| STAR 2.7.3a | Dobin et al., 2013 | https://github.com/alexdobin/STAR; RRID: SCR_015899 |
| tombo 1.5 | Oxford Nanopore Technologies, Stoiber et al., 2017 | https://github.com/nanoporetech/tombo |
| Other | ||
| Chlorosebus sabaeus genome and annotation | ENSEMBL release 99 | ChlSab1.1 (GCA_000409795.2) |
| yeast ENO2 cDNA | SGD | SGD: YHR174W |
| human ribosomal DNA complete repeat unit | GenBank | GenBank: U13369.1 |
| SARS-CoV-2 isolate Wuhan-Hu-1, complete genome | GenBank | GenBank: NC_045512.2 |
| SARS-CoV-2 isolate BetaCoV/Korea/KCDC03/2020, partial genome | GISAID | GISAID: EPI_ISL_407193 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, V. Narry Kim (narrykim@snu.ac.kr).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The source code for the data processing and analyses is available at https://github.com/hyeshik/sars-cov-2-transcriptome. The sequencing data were deposited into the Open Science Framework (OSF) with an accession number https://doi.org/10.17605/OSF.IO/8F6N9. The processed sequencing data can be accessed from the UCSC Genome Browser COVID-19 Pandemic Resources at https://genome.ucsc.edu/covid19.html. The original data for Figure S3 were deposited into Mendeley Data: https://doi.org/10.17632/bkhbpvtg7h.1.
Experimental Model and Subject Details
SARS-CoV-2 viral RNA was prepared by extracting total RNA from Vero cells (ATCC, CCL-81) infected with BetaCoV/Korea/KCDC03/2020 (Kim et al., 2020), at a multiplicity of infection (MOI) of 0.05, and cultured in DMEM (GIBCO) supplemented with 2% fetal bovine serum (GIBCO) and penicillin-streptomycin (GIBCO) at 37°C, 5% CO2. The virus is the fourth passage and not plaque-isolated. Cells were harvested at 24 hours post-infection. This study was carried out in accordance with the biosafety guideline by the KCDC. The Institutional Biosafety Committee of Seoul National University approved the protocol used in these studies (SNUIBC-200219-10).
Method Details
RNA purification
Cultured cells were washed once with PBS before adding TRIzol (Invitrogen). Purified total RNAs from non-infected and SARS-CoV-2-infected Vero cells were treated with DNaseI (Takara) followed by column purification (RNeasy MinElute Cleanup Kit [QIAGEN]) and used for the experiments.
In vitro transcription
With 0.5 μg of total RNA from SARS-CoV-2-infected Vero cell, reverse transcription (SuperScript IV Reverse Transcriptase [Invitrogen]) was done with each virus-specific RT primer (RTprimer1-8). Templates for in vitro transcription were prepared by PCR (Q5® High-Fidelity DNA Polymerase [NEB]) with each virus-specific PCR primer pair (i.e., IVT-frag1-F & IVT-frag1-R primers) followed by agarose gel purification (Gel Extraction Kit [Labopass]), in vitro transcription (MEGAscript T7 Transcription Kit [Invitrogen]) and RNA purification (Oligo Clean & Concentrator [Zymo Research]). The oligonucleotides used in this study are listed in Table S1.
Reverse transcription and PCR
With 0.25 μg of total RNA from SARS-CoV-2-infected and uninfected Vero cells, reverse transcription (SuperScript IV Reverse Transcriptase [Invitrogen]) was done with each forward and reverse PCR primer for negative-strand and positive-strand specific reverse transcription, respectively, with 6 ng/μl actinomycin D (Sigma-Aldrich). PCR (Q5® High-Fidelity DNA Polymerase [NEB]) was done with each PCR primer pair (i.e., Primer #1-F & Primer #1-R primers) followed by agarose gel running with ladder (1 Kb Plus DNA Ladder [Invitrogen]). The oligonucleotides used in this study are listed in Table S1.
Nanopore direct RNA sequencing
For nanopore sequencing on non-infected and SARS-CoV-2-infected Vero cells, each 4 μg of DNaseI (Takara)-treated total RNA in 8 μl was used for library preparation following the manufacturer’s instruction (the Oxford Nanopore DRS protocol, SQK-RNA002) with minor adaptations. 20 U of SUPERase-In RNase inhibitor (Ambion, 20 U/μl) was added to both adaptor ligation steps. SuperScript IV Reverse Transcriptase (Invitrogen) was adopted instead of SuperScript III, and the reaction time of reverse transcription was lengthened by 2 hours. The library was loaded on FLO-MIN106D flow cell followed by 42 hours sequencing run on MinION device (Oxford Nanopore Technologies).
For nanopore sequencing on SARS-CoV-2 RNA fragments produced by in vitro transcription, the same method was applied except for the RNA amount (a total 2 μg of in vitro transcribed RNAs) and reaction time for reverse transcription (30 minutes).
The nanopore direct sequencing data were basecalled by guppy 3.4.5 (Oxford Nanopore Technologies) using the high-accuracy model. The sequence reads were aligned to the reference sequence database composed of the C. sabaeus genome (ENSEMBL release 99), a SARS-CoV-2 genome, yeast ENO2 cDNA (SGD: YHR174W), and human ribosomal DNA complete repeat unit (GenBank: U13369.1) using minimap2 2.17 (Li, 2018) with options “-k 13 -x splice -N 32 -un.” We used the sequence of the Wuhan-Hu-1 strain (GenBank: NC_045512.2) as a backbone for the viral reference genome, then corrected the four single nucleotide variants found in BetaCoV/Korea/KCDC03/2020; T4402C, G5062T, C8782T, and T28143C (GISAID: EPI_ISL_407193). The sequence alignments were further improved by re-mapping the identified viral reads to the viral genome using minimap2 options “-k 8 -w 1–splice -g 30000 -G 30000 -A1 -B2 -O2,24 -E1,0 -C0 -z 400,200–no-end-flt–junc-bonus=100 -F 40000 -N 32–splice-flank=no–max-chain-skip=40 -un–junc-bed=FILE -p 0.7.” Chimeric reads were filtered out according to the flag from minimap2.
DNBseq RNA sequencing
With 1 μg of total RNA from SARS-CoV-2-infected Vero cell, Dynabeads mRNA Purification Kit (Invitrogen) was applied to deplete rRNA and enrich poly(A)+ RNA by using oligo d(T). RNA-seq library for 250 bp insert size was constructed following the manufacturer’s instruction (MGIEasy RNA Directional Library Prep Set). The library was loaded on MGISEQ-200RS Sequencing flow cell with MGISEQ-200RS High-throughput Sequencing Kit (PE 100), and the library was run on DNBSEQ-G50RS (paired-end run, 100 × 100 cycles).
The sequences from DNBseq were aligned to the reference sequences used in nanopore DRS. We used STAR 2.7.3a (Dobin et al., 2013) with many switches to completely turn off the penalties of non-canonical eukaryotic splicing: “–outFilterType BySJout–outFilterMultimapNmax 20–alignSJoverhangMin 8–outSJfilterOverhangMin 12 12 12 12–outSJfilterCountUniqueMin 1 1 1 1–outSJfilterCountTotalMin 1 1 1 1–outSJfilterDistToOtherSJmin 0 0 0 0–outFilterMismatchNmax 999–outFilterMismatchNoverReadLmax 0.04–scoreGapNoncan −4–scoreGapATAC −4–chimOutType WithinBAM HardClip–chimScoreJunctionNonGTAG 0–alignSJstitchMismatchNmax −1 −1 −1 −1–alignIntronMin 20–alignIntronMax 1000000–alignMatesGapMax 1000000.”
Quantification and Statistical Analysis
RNA-seq coverage depth plots (Figures 2B and 2D)
Sequencing read coverage was calculated using bedtools genomecov of version 2.27.1. The coverage depths were binned to 30-nt (wide views) or 15-nt (insets) bins and plotted by using medians in the plots.
Heatmaps showing discontinuous mappings (Figures 3A and S2)
Start and end positions of large gaps (≥20nt) were collected from the CIGAR strings of all high-quality (≥100 in the STAR mapping quality) alignments to the viral genome. The positions were counted into 100-nt bins in the zero-based coordinate. The read counts were mapped to a colormap “viridis” in matplotlib 3.1.3 after log-transformation with a pseudocount of 1. The detected most-frequent canonical sites (red dots in the line plots on the left-hand sides) were detected by using signals.find_peaks in the SciPy 1.4.1 (prominence = 4 and height = 8 for the DRS data; prominence = 8 and height = 13 for the DNBseq data) (Virtanen et al., 2020).
Counting and classifying reads from subgenomic RNAs (Figures 3B and 3C)
The junction-spanning reads (JSRs) were categorized by the position of 5′ and 3′ site positions. A JSR was marked as a leader-to-body junction when the 5′ site of the deletion is mapped to a genomic position between 55 and 85. In the cases where the 5′ site is in the 5′ UTR region, the sgRNA identity and the frame matching were determined by the first appearance of AUG in the downstream of the 3′ site. In the cases where the 5′ site is in a known ORF or an AUG is introduced by fusion, we checked if the concatenated sequence generates a protein product with the same reading frame as a canonical ORF after the 3′ site.
For the analyses of sgRNA reads using the nanopore DRS data, the mapped reads from canonical sgRNAs were identified using the start and end positions of large deletions ≥ 10000 nt. For a valid assignment to a species of sgRNA, we required that the start position is between 55 and 85 in the genomic coordinate. The first AUG in the downstream of the end position of a large deletion was used for identification of the “spliced” product.
Poly(A) length distribution analysis (Figures 4, 6E, and S6)
The dwell time of poly(A) tails were measured using poreplex 0.5.0 (https://github.com/hyeshik/poreplex). For the conversion from a dwell time to a nucleotide length, we divided a poly(A) dwell measurement by 1/30 of the mode of the poly(A) dwell time of the ONT sequencing calibration control which has a 30 nt-long poly(A) tail.
Balancing IVT product reads and modified base detection by sample level comparison (Figures 5A, 5B, S5A, and S5B)
The DRS reads of the IVT RNAs were downsampled to balance the coverage between different fragments that were split into equal-sized patches. Sampling frequency of a fragment was controlled by the read counts within a 100-nt bin with the lowest coverage in each fragment. We sampled the reads so that the result contains roughly 10,000 reads from every IVT fragment. The viral RNA reads and the downsampled IVT reads were processed for squiggle analyses by ONT tombo 1.5 (Stoiber et al., 2017) with a minor tune to improve the sensitivity of sequence alignments (-k8 -w1). The modified base detection was done by using the “model_sample_compare” mode with an option “–sample-only-estimates” unless otherwise specified.
Sequence contexts of detected modified positions (Figures 6A–6D)
The classification of sequence context near the modified sites was first done by the existence of four consecutive purine bases within 5-nt from the position with the highest modification fraction reported by tombo. Then, the rest were further divided into four groups according to the nucleotide base with the highest modification fraction.
Statistical analysis of modified bases by alternative model (Figures S4C and S4D)
The candidate sites of 5-methylcytidine were detected using a bundled “alternative model” of tombo 1.5. Figure S4C shows all positions with at least 500 supporting reads. Significantly methylated sites (black dots on top) were selected by applying the 5% false-discovery rate cut-off estimated by Viehweger et al. (2019). Figure S4D shows all positions with enough coverage depth (≥100 reads) in both IVT products and viral RNAs
Poly(A) length analysis depending on modification rates (Figures 6E and S6)
“Highly modified” sgRNA reads were detected by referring to eight modification sites which were at least 40% modified in any species of sgRNAs: 28591, 28612, 28653, 28860, 28958, 29016, 29088, 29127. We used the reads that were reported as modified at three or more sites with a statistic < 0.01 as “highly modified” reads. “Not modified” reads were reported with the statistic ≥ 0.01 in all eight sites. Statistical tests for shorter poly(A) length of highly modified sgRNAs were carried out using wilcox.test() function in R 3.6.1.
Visualization of sequence alignment (Figure S1)
To visualize the sequences mapped near CDS regions of nsp2–8, the alignments were first selected by the “intersect” command of bedtools 2.29.2 for the region 800–12000 (zero-based coordinates). The survived alignments filtered again intersecting with the region 29850–29950 to enrich the 3′-intact reads. The resulting alignments further filtered so that we only keep alignments with (1) minimum alignment length of 1000 nt excluding insertions or deletions, (2) minimum contiguously mapped length of 50 nt in the 5′-most block to suppress noisy short alignments. 250 Randomly chosen alignments passing the criteria were sorted by the 5′ site position of the largest deletion within each alignment. Alignments without a large gap (≥100nt) were ordered by the first mapped coordinate.
Acknowledgments
We thank members of our institutions for discussion and help, particularly Eun-jin Chang, Inhye Park, and Young-suk Lee at IBS. We are grateful to Drs. Jung-Hye Roe, Nam-Hyuk Cho, Kwangseog Ahn, and Jae-Hwan Nam for their advice and comments. We thank Kyung-Chang Kim and Sung Soon Kim at Korea National Institute of Health for their support. The pathogen resource (NCCP43326) for this study was provided by the National Culture Collection for Pathogens, Korea National Institute of Health. This work was supported by the Institute for Basic Science from the Ministry of Science and ICT of Korea (IBS-R008-D1 to D.K., H.C., and V.N.K.); a BK21 Research Fellowship from the Ministry of Education of Korea (to D.K.); the New Faculty Startup Fund from Seoul National University (to H.C.); and funding from Korea Centers for Disease Control and Prevention (4845-300[19-NG044]) to J.-Y.L., J.-S.Y., J.W.K., and H.C.).
Author Contributions
Conceptualization, H.C., J.-Y.L., and V.N.K.; Methodology, H.C. and D.K.; Software, H.C.; Investigation, D.K., J.-S.Y., and J.W.K.; Writing – Original Draft, V.N.K.; Writing – Review & Editing, H.C., J.-Y.L., and V.N.K.; Formal Analysis, H.C.; Visualization, H.C.; Supervision, V.N.K., H.C., and J.-Y.L.
Declaration of Interests
The authors declare no competing interests.
Published: April 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.04.011.
Supplemental Information
Refer to STAR Methods for the experimental procedure.
The genomic coordinates in the “5′ site” and “3′ site” point to the 3′-most and the 5′-most nucleotides that survive the recombination event, respectively. “Count” is the number of the junction-spanning reads that support the recombination event. “Skip” is the size of the deletion. “5p_before” is the 3′-most sequence from the 5′-side of the recombination event. “5p_after” is the bases immediately following the “5′ site.” The sequence is the first nucleotides of the region that is excluded by the recombination event. “3p_before” is the last nucleotide sequence of the region that is excluded by the recombination. “3p_after” is the 5′-most nucleotides of the 3′-side of the recombination product.
This table extends the information for the recombination events using the canonical leader TRS. “5′ site” and “3′ site” are the end positions of the most frequently observed gap among the ones that are expected to produce the identical ORF. “Startpos” is the genomic coordinate where the first AUG appears in the resulting recombination product. “Resultingsequence” shows the sequence near the recombination site with the nucleotides from the 5′-side in lowercase, and those from the 3′-side in uppercase. “Annot_cds” is the name of an ORF that shares the start codon position with the recombination product by the “first AUG” rule. “Annot_cds_start” and “annot_cds_end” indicate the interval of the annotated canonical ORF that matches to the start codon in the recombination product. “Startmatch_name” and “startmatch_offset” show the canonical ORF that overlaps with the position of the first appearing AUG in the recombination product, and the distance between the AUG and the start codon position of the ORF. “Stopmatch_name” and “stopmatch_offset” show the canonical ORF that overlaps with the expected stop codon position in case the translation starts at the first appearing AUG, and the relative coordinate of the “expected” stop codon position from the stop codon of the canonical ORF. “Translation” shows the peptide sequence that is expected to be produced by the first ORF in the recombination product.
Many distant recombination events do not involve the leader sequence. Instead, the 5′ sites are often found in “hot spots” near ORFs or nsp2 and nsp3. “Count” indicates the number of reads that share the same 5′ and 3′ sites. “Skip” is the size of the deletion in the given combination of 5′ and 3′ sites. “Truncated_prod” is the name of a mature peptide that is truncated by recombination. “Lost_aa” is the number of amino acids lost from the mature peptide by recombination. “Added_aa” is the number of amino acids introduced to the potentially truncated product by recombination. By the recombination event, the sequence indicated in “ctx_orig” turns into the sequence in “ctx_new.”
The “pos” column shows all genomic positions reported to be modified at least 20% in any species of sgRNA. “Location” indicates the name of the ORF or mature peptide that the position codes. The percentage numbers below the names of sgRNAs are the estimated fraction of modified reads at the position. “Context” shows the sequence context around the modified bases.
References
- Courtney D.G., Kennedy E.M., Dumm R.E., Bogerd H.P., Tsai K., Heaton N.S., Cullen B.R. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe. 2017;22:377–386. doi: 10.1016/j.chom.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T., Lai M.M. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J. Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- Gokhale N.S., Horner S.M. RNA modifications go viral. PLoS Pathog. 2017;13:e1006188. doi: 10.1371/journal.ppat.1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N.S., McIntyre A.B.R., McFadden M.J., Roder A.E., Kennedy E.M., Gandara J.A., Hopcraft S.E., Quicke K.M., Vazquez C., Willer J. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N., Firth A.E., Jones J.D., Chung B.Y., Siddell S.G., Brierley I. High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling. PLoS Pathog. 2016;12:e1005473. doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Chung Y.S., Jo H.J., Lee N.J., Kim M.S., Woo S.H., Park S., Kim J.W., Kim H.M., Han M.G. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res. Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M., Morgan M.A., Shatkin A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.M., Stohlman S.A. Comparative analysis of RNA genomes of mouse hepatitis viruses. J. Virol. 1981;38:661–670. doi: 10.1128/jvi.38.2.661-670.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.L., Lai M.M. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J. Virol. 1992;66:6117–6124. doi: 10.1128/jvi.66.10.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G., Zhao B.S., Wu Y., Lu Z., Qin Y., He C., Rana T.M. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Gerritsma H., Spaan W.J. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology. 1996;216:174–183. doi: 10.1006/viro.1996.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Graham R.L., Baric R.S. Jumping species-a mechanism for coronavirus persistence and survival. Curr. Opin. Virol. 2017;23:1–7. doi: 10.1016/j.coviro.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P., Ayers D.F., Rottman F.M., Maroney P.A., Nilsen T.W. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol. Cell. Biol. 1987;7:1572–1575. doi: 10.1128/mcb.7.4.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak K.B., Nagy P.D. Defective Interfering RNAs: Foes of Viruses and Friends of Virologists. Viruses. 2009;1:895–919. doi: 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart H., Brown K., Dinan A.M., Irigoyen N., Snijder E.J., Firth A.E. Transcriptional and Translational Landscape of Equine Torovirus. J. Virol. 2018;92 doi: 10.1128/JVI.00589-18. e00589-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber M., Quick J., Egan R., Eun Lee J., Celniker S., Neely R.K., Loman N., Pennacchio L.A., Brown J. De novo Identification of DNA Modifications Enabled by Genome-Guided Nanopore Signal Processing. bioRxiv. 2017 doi: 10.1101/094672. [DOI] [Google Scholar]
- Tomecki R., Dmochowska A., Gewartowski K., Dziembowski A., Stepien P.P. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvarogová J., Madhugiri R., Bylapudi G., Ferguson L.J., Karl N., Ziebuhr J. Identification and Characterization of a Human Coronavirus 229E Nonstructural Protein 8-Associated RNA 3′-Terminal Adenylyltransferase Activity. J. Virol. 2019;93:e00291-19. doi: 10.1128/JVI.00291-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viehweger A., Krautwurst S., Lamkiewicz K., Madhugiri R., Ziebuhr J., Hölzer M., Marz M. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29:1545–1554. doi: 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., SciPy 1.0 Contributors SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z., Liudkovska V., Gewartowska O., Mroczek S., Dziembowski A. Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20180162. doi: 10.1098/rstb.2018.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D., Gokhale N.S., Horner S.M. Regulation of Viral Infection by the RNA Modification N6-Methyladenosine. Annu. Rev. Virol. 2019;6:235–253. doi: 10.1146/annurev-virology-092818-015559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Ke T.Y., Liao W.Y., Chang N.Y. Regulation of coronaviral poly(A) tail length during infection. PLoS ONE. 2013;8:e70548. doi: 10.1371/journal.pone.0070548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y., Hirano N., Hino S., Shibuta H., Matumoto M. Polyadenylate in the virion RNA of mouse hepatitis virus. J. Biochem. 1977;82:1103–1108. doi: 10.1093/oxfordjournals.jbchem.a131782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Refer to STAR Methods for the experimental procedure.
The genomic coordinates in the “5′ site” and “3′ site” point to the 3′-most and the 5′-most nucleotides that survive the recombination event, respectively. “Count” is the number of the junction-spanning reads that support the recombination event. “Skip” is the size of the deletion. “5p_before” is the 3′-most sequence from the 5′-side of the recombination event. “5p_after” is the bases immediately following the “5′ site.” The sequence is the first nucleotides of the region that is excluded by the recombination event. “3p_before” is the last nucleotide sequence of the region that is excluded by the recombination. “3p_after” is the 5′-most nucleotides of the 3′-side of the recombination product.
This table extends the information for the recombination events using the canonical leader TRS. “5′ site” and “3′ site” are the end positions of the most frequently observed gap among the ones that are expected to produce the identical ORF. “Startpos” is the genomic coordinate where the first AUG appears in the resulting recombination product. “Resultingsequence” shows the sequence near the recombination site with the nucleotides from the 5′-side in lowercase, and those from the 3′-side in uppercase. “Annot_cds” is the name of an ORF that shares the start codon position with the recombination product by the “first AUG” rule. “Annot_cds_start” and “annot_cds_end” indicate the interval of the annotated canonical ORF that matches to the start codon in the recombination product. “Startmatch_name” and “startmatch_offset” show the canonical ORF that overlaps with the position of the first appearing AUG in the recombination product, and the distance between the AUG and the start codon position of the ORF. “Stopmatch_name” and “stopmatch_offset” show the canonical ORF that overlaps with the expected stop codon position in case the translation starts at the first appearing AUG, and the relative coordinate of the “expected” stop codon position from the stop codon of the canonical ORF. “Translation” shows the peptide sequence that is expected to be produced by the first ORF in the recombination product.
Many distant recombination events do not involve the leader sequence. Instead, the 5′ sites are often found in “hot spots” near ORFs or nsp2 and nsp3. “Count” indicates the number of reads that share the same 5′ and 3′ sites. “Skip” is the size of the deletion in the given combination of 5′ and 3′ sites. “Truncated_prod” is the name of a mature peptide that is truncated by recombination. “Lost_aa” is the number of amino acids lost from the mature peptide by recombination. “Added_aa” is the number of amino acids introduced to the potentially truncated product by recombination. By the recombination event, the sequence indicated in “ctx_orig” turns into the sequence in “ctx_new.”
The “pos” column shows all genomic positions reported to be modified at least 20% in any species of sgRNA. “Location” indicates the name of the ORF or mature peptide that the position codes. The percentage numbers below the names of sgRNAs are the estimated fraction of modified reads at the position. “Context” shows the sequence context around the modified bases.
Data Availability Statement
The source code for the data processing and analyses is available at https://github.com/hyeshik/sars-cov-2-transcriptome. The sequencing data were deposited into the Open Science Framework (OSF) with an accession number https://doi.org/10.17605/OSF.IO/8F6N9. The processed sequencing data can be accessed from the UCSC Genome Browser COVID-19 Pandemic Resources at https://genome.ucsc.edu/covid19.html. The original data for Figure S3 were deposited into Mendeley Data: https://doi.org/10.17632/bkhbpvtg7h.1.