SUMMARY
A fundamental question in microbial ecology is how microbes are spatially organized with respect to each other and their host. A test bed for examining this question is the tongue dorsum, which harbors a complex and important microbial community. Here, we use multiplexed fluorescence spectral imaging to investigate the organization of the tongue microbiome at micron to hundred-micron scales. We design oligonucleotide probes for taxa both abundant and prevalent, as determined by sequence analysis. Imaging reveals a highly structured spatial organization of microbial consortia, ranging in linear dimension from tens to hundreds of microns. The consortia appear to develop from a core of epithelial cells, with taxa clustering in domains suggestive of clonal expansion. Quantitative proximity analysis provides the basis for a model of tongue dorsum microbiome organization and dynamics. Our work illustrates how high-resolution analysis of micron-scale organization provides insights into physiological functions and microbiome-host interactions.
Graphical Abstract
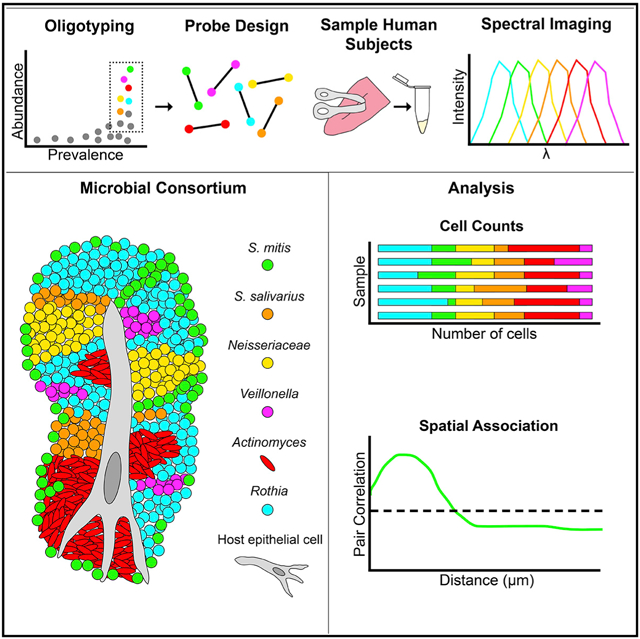
In Brief
How microbes are spatially organized is a fundamental question in microbial ecology about which little is known. Using fluorescence spectral imaging, Wilbert et al. discover dense, highly structured microbial consortia on the human tongue. Analysis of the organization of these consortia permits inferences about community dynamics and oral microbial ecology.
INTRODUCTION
The field of spatial ecology investigates the processes that give rise to spatial patterning in community structure. For macro-organisms, this field is generally referred to as landscape ecology, for which a rich conceptual framework, analytical procedures, and models have been developed (Turner and Gardner, 2015). Fundamental concepts in landscape ecology are those of “grain” and “extent,” namely the smallest unit of observation and the range over which the observations are made. The magnitude of grain and extent defines the scale of investigation. Choosing the appropriate scale of investigation is important for formulating hypotheses on the mechanisms underlying community organization (Levin, 1992). Although the concepts developed in macro-ecology apply, in principle, to the microbial world as well, scale and spatial patterning have received relatively little attention in microbial ecology (Ladau and Eloe-Fadrosh, 2019).
Spatial patterning in a microbial community is likely a result of multiple and complex processes. In the human mouth, landscape-scale factors patterning the community include temperature, moisture, salivary flow, pH, oxygen, and the frequency of disturbances, such as abrasion or oral hygiene (Kleinberg and Jenkins 1964; Marsh et al., 2016; Proctor and Relman 2017; Lamont et al., 2018; Mark Welch et al., 2019). Interacting with these landscape-scale factors are factors operating at the scale of individual microbial cells. Microbes influence their neighbors by acting as sources and sinks of metabolites, nutrients, and inhibitory molecules, such as hydrogen peroxide and antimicrobial peptides (Jakubovics et al., 2008; Zhu and Kreth 2012; Cotter et al., 2013; Goldford et al., 2018; Dal Co et al., 2019). By occupying space, microbes can physically exclude one another from desirable habitats, but their surfaces also present binding sites to which other microbes may adhere (Gibbons and Houte 1975; Kolenbrander and London 1993). Diversity and functional redundancy appear to be emergent properties of microbial communities (Louca et al., 2018), with networks of interactions among taxa influencing community stability (Levine et al., 2017), stabilization of diversity (Goldford et al., 2018), and metabolic efficiency (Yu et al., 2019). Thus, microbes themselves form an important part of the environment for neighboring microbes, and deciphering community spatial structure will be important for developing a predictive understanding of community ecology.
Key pieces of information for analyzing spatial patterning are the proximity relationships among microbes as well as the distance between a microbe and landmarks, such as the surface of the biofilm or the nearest host cell. Proximity relationships can be considered on a cell-by-cell basis but can also include measurement of the size, shape, and connectivity of single-taxon clusters or groups of interacting taxa. Imaging, as a tool to explore spatial patterning of microbial communities, can provide these types of information with a grain as small as a single bacterial cell and an extent of up to a millimeter in scale. The development of combinatorial labeling and spectral imaging-fluorescence in situ hybridization (CLASI-FISH) (Valm et al., 2011, 2016) enabled the localization and identification of many distinct microbial taxa simultaneously, thus opening a door to analysis of the micron-scale spatial organization of complex microbial communities at a system level.
In this study, we apply multi-spectral fluorescence imaging to the microbiota of the human tongue dorsum (TD) as a model system for microbial spatial ecology. The TD is home to a dense microbial community in direct contact with the human epithelium. Through saliva, it is connected with other habitats in the mouth, such as teeth and buccal mucosa, and thus, it provides an opportunity to investigate community structure in the absence of dispersal limitation. The functional significance for the human host of the tongue microbiota is only beginning to be explored but may include a significant role in the conversion of salivary nitrate to nitrite and, thus, to the regulation of host nitric oxide homeostasis (Lundberg et al., 2004; Hezel and Weitzberg 2015; Tribble et al., 2019).
Understanding the spatial ecology of the human tongue microbiota requires high-resolution taxonomic analysis as well as high-resolution spatial analysis. We analyzed sequence data to identify major bacterial taxa, and we characterized classes of microbial organization contained within samples from the tongue. Guided by sequence analysis, our imaging approach targeted major genera and selected species to obtain a comprehensive view of microbiome structure with spatial and taxonomic resolution sufficient to identify distinctive distributions of key microbial players. Our findings illustrate the use of spatial information to identify appropriate scales of analysis, analyze the proximity of taxa to host cells and other landmarks, and permit inferences about disturbance regimes and about the growth properties of individual taxa and of the community as a whole.
RESULTS
Identification of Bacterial Taxa Important in the Human TD
The human oral microbiome is complex, containing 756 species-level taxa (oral plus as yet unassigned), as documented in the expanded Human Oral Microbiome Database (eHOMD) (Dewhirst et al., 2010; Escapa et al., 2018; http://www.homd.org). For imaging the spatial organization of the microbiota, this high level of complexity poses formidable challenges and calls for a prioritization strategy for visualizing the taxa most likely to be present in the sample. Analysis of Human Microbiome Project (HMP) data for the 16S rRNA gene (Human Microbiome Project Consortium, 2012) showed that a limited number of high-resolution sequence groups termed oligotypes accounted for most of the microbiota of the TD (Eren et al., 2014). Building on that work, we updated the mapping of oral oligotypes to eHOMD taxa to connect the high-throughput sequencing data with the most up-to-date reservoir of knowledge about individual oral microbial species (Table S1).
By mapping oligotypes to genera, we identified 17 bacterial genera that were both abundant and prevalent on the tongue, with at least 0.5% mean abundance across individuals and detected in more than 80% of individuals (Figure 1A). Analysis of shotgun metagenomic sequencing data from the HMP (Kraal et al., 2014) identified a similar set of abundant genera, accounting for >95% of the bacterial sequences on the TD (Figure 1B). Because of their abundance and prevalence, these genera are likely to form both the spatial and the metabolic framework of the healthy TD microbiome.
Figure 1. Major Taxa Colonizing the TD: Site Specialists and Generalists.
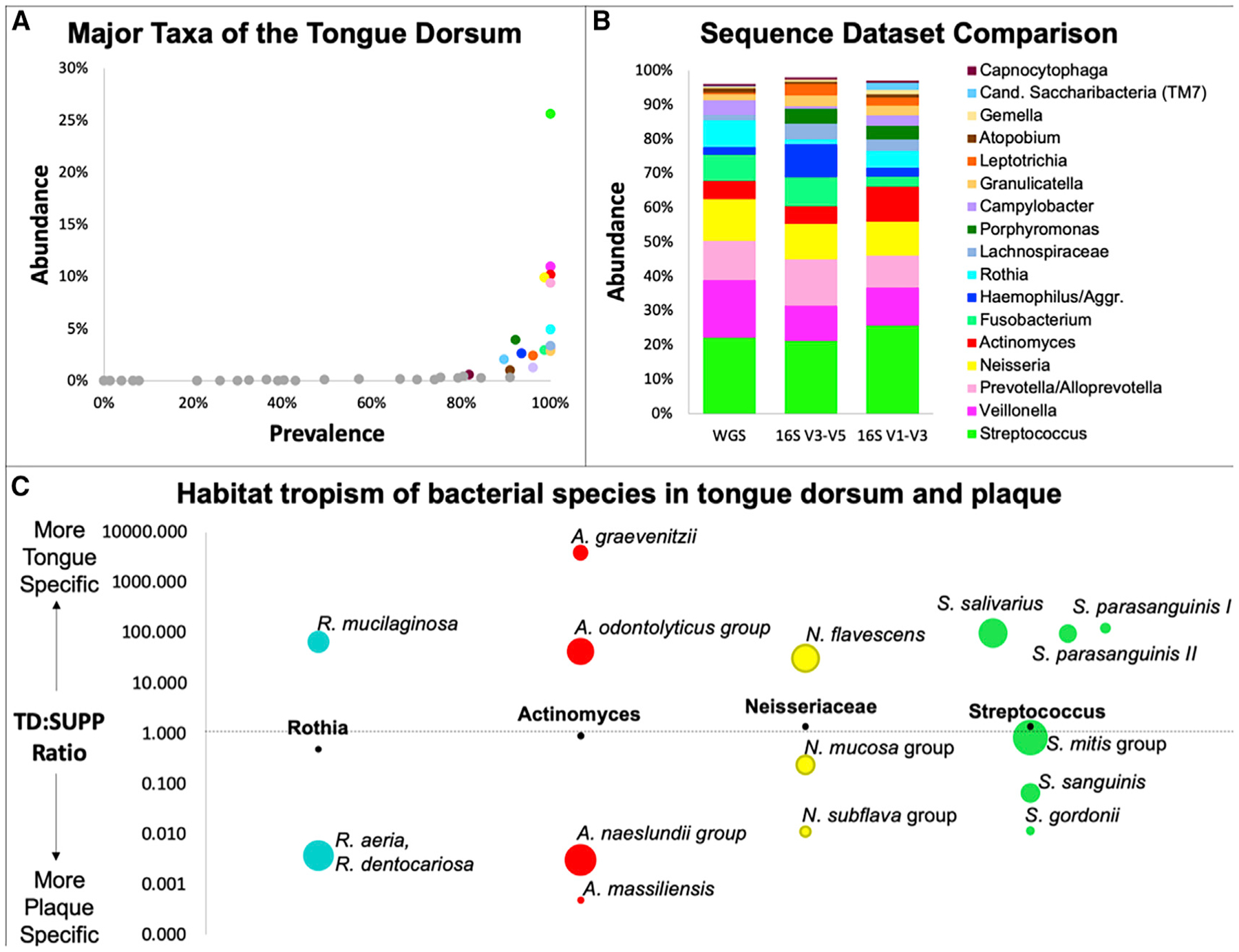
(A) A small number of genera are both prevalent and abundant on the tongue. Mean relative abundance from oligotyping reanalysis of HMP sequence data for the TD.
(B) Three datasets show similar mean abundance of genera in TD: 16S rRNA V1–V3 and V3–V5 sequences (Eren et al., 2014) and whole-genome metagenomic sequencing (WGS) (Kraal et al., 2014).
(C) Oral site tropisms. Major genera are represented by different species in plaque and on tongue. Position of circles represents the ratio of TD abundance to supragingival plaque (SUPP) abundance in 16S rRNA V1–V3 data (Table S1). Black dots show TD:SUPP ratio of the genus; colored circles show TD:SUPP ratio of species within the genus with area proportional to mean abundance of the species in its preferred site. Within each genus, all species with >0.5% mean abundance in either TD or SUPP are shown.
See also Table S1.
High-resolution analysis of sequencing data enables one to examine habitat specificity below the genus level. A major habitat differentiation within the oral cavity is whether the host surface is mucosal, such as the TD, or non-mucosal, such as the teeth (Socransky and Manganiello, 1971; Mark Welch et al., 2019). A comparison of the relative abundance of a species on the tongue versus in a dental plaque showed that many genera consist of species found predominantly on either mucosal or dental surfaces but not both (Figure 1C; Table S1). For example, Rothia mucilaginosa is nearly 100-fold more abundant on the tongue than on the teeth and Rothia aeria and Rothia dentocariosa are more than 100-fold more abundant on the teeth than on the tongue. The genus Actinomyces is likewise represented by two groups of species on the tongue and two different groups of species in dental plaque. Other genera, such as Streptococcus contain not only habitat specialists but also habitat generalists. For example, Streptococcus mitis and its close relatives form a group that is abundant on both mucosal and non-mucosal surfaces (Figure 1C).
TD Microbial Landscape
The TD has a highly structured topography defined primarily by the filiform papillae that cover most of its surface (Kullaa-Mikkonen et al., 1985, 1987; Manabe et al., 1999; Just et al., 2005). Electron microscopy has documented the presence of bacteria on the papillae, in the valleys between them, and in high density on spines or hairs projecting from the papillae (Kullaa-Mikkonen and Sorvari 1985). Consequently, imaging the microbial landscape requires a sampling method that can capture the microbial populations from this topology. Our rationale for choosing a sampling method was based on minimizing perturbation of the collected biofilm while retaining non-invasiveness to the subject. After exploring a variety of sampling methods, including swabs, picks, and scrapers, we collected material with a ridged plastic tongue scraper, gently scraped over the tongue from back to front, and deposited the sample into ethanol. The community structures resistant to perturbation by this collection procedure were pieces of biofilm whose size and internal organization suggested that spatial structure in the size range of tens to hundreds of micrometers had been preserved (Figures 2 and 3). Operationally, the “extent” of our landscape analysis was, therefore, on the scale of hundreds of micrometers as opposed to the centimeter scale of the tongue as a whole.
Figure 2. Three Categories of Structures in TD Samples.
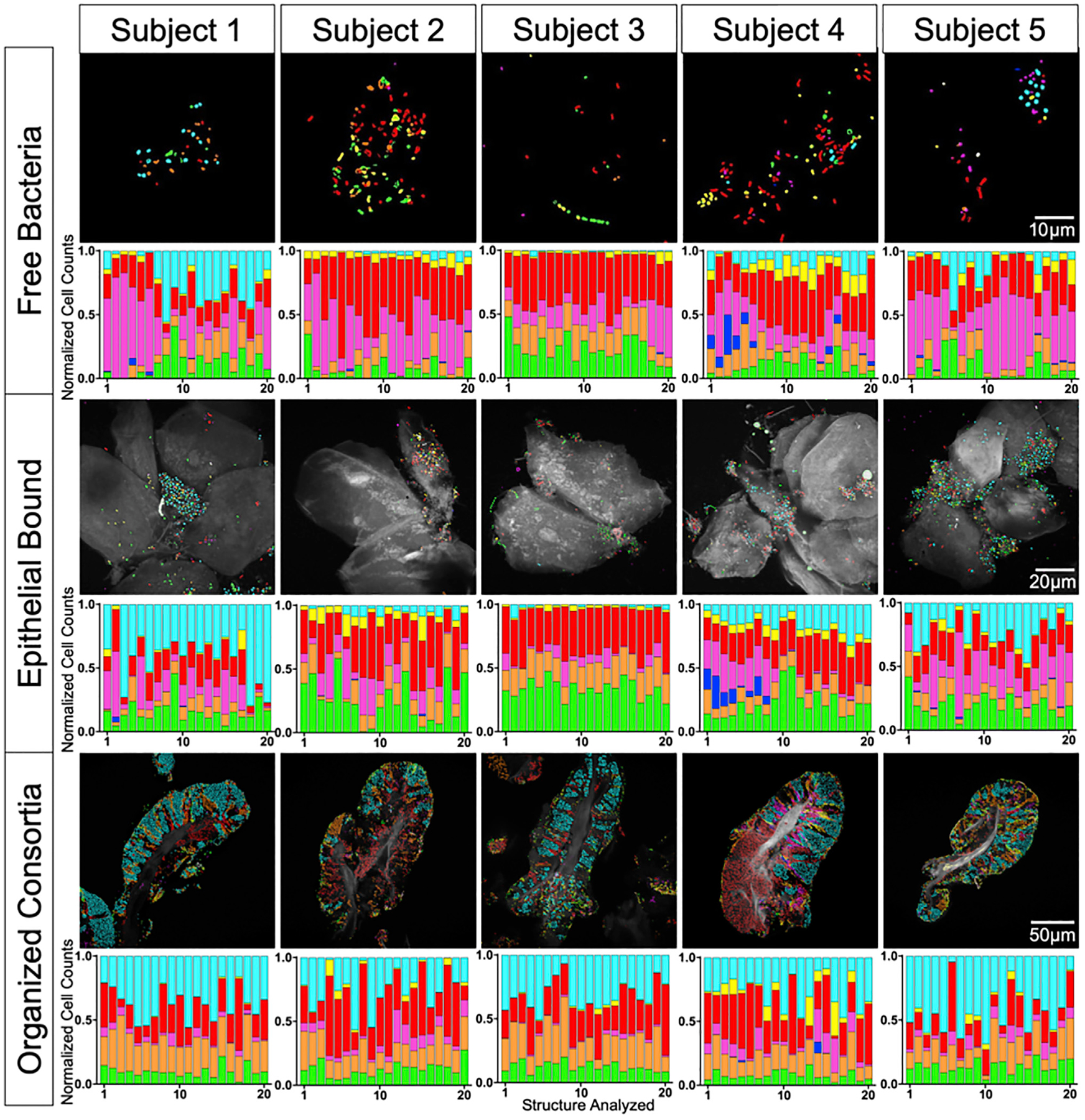
Spectral imaging shows microbes organized as free bacteria (no substrate visible; top), epithelial bound (epithelial cells visible and colonized; middle), or consortia (dense biofilm with clear perimeter and core of epithelial material; bottom). A total of 20 fields of view of each type were imaged from each of 5 subjects, for a total of 300 images. Representative images are displayed; normalized cell counts from each field of view are shown as bar charts. Rothia (cyan), Neisseriaceae (yellow), Actinomyces (red), Veillonella (magenta), Prevotella (blue), S. salivarius (orange), S. mitis (green), and autofluorescence (white). All 300 images are shown in Figure S1.
See also Figures S1 and S3 and Tables S2 and S3.
Figure 3. Tongue Biofilm Consortia Are Complex and Structurally Organized.
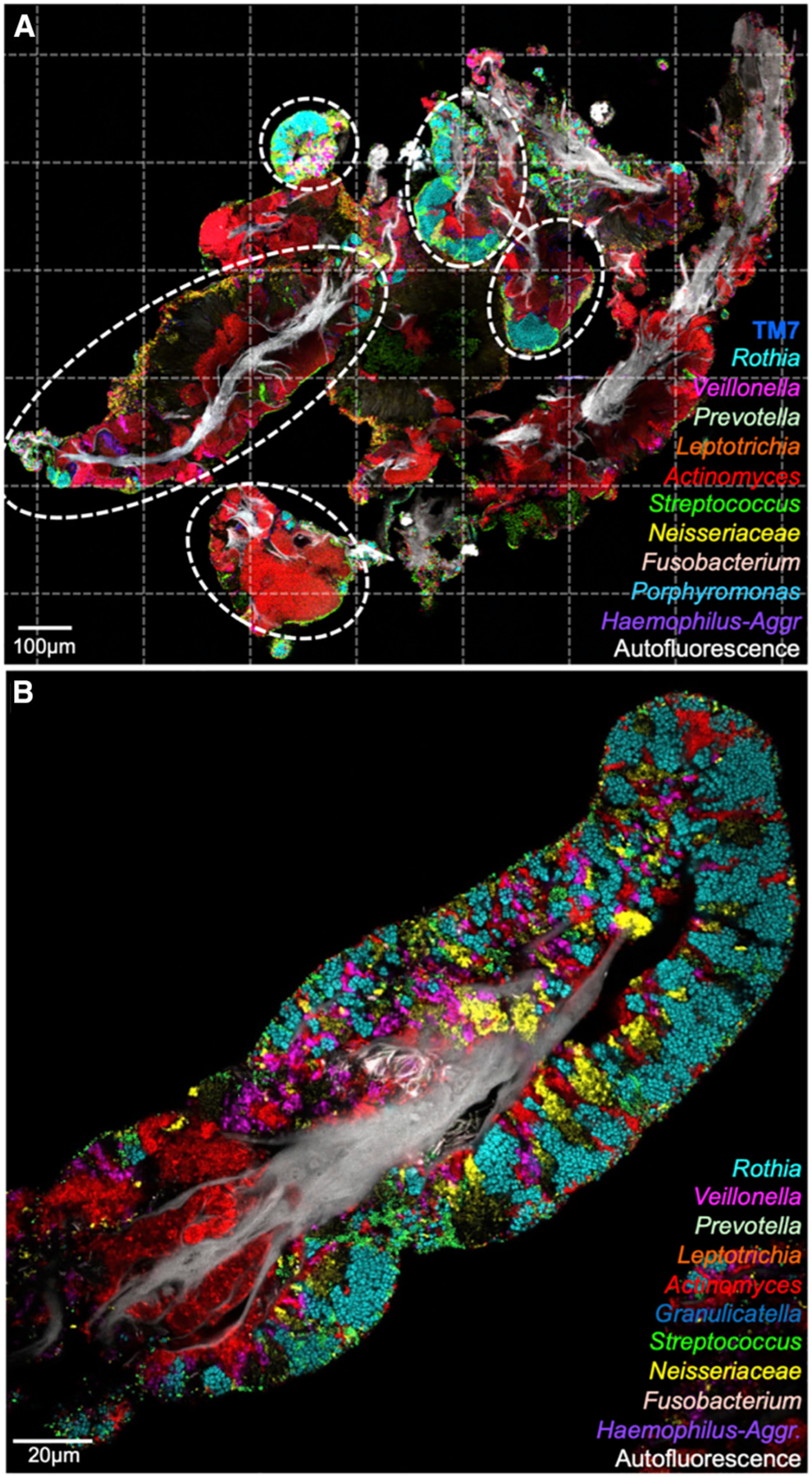
(A) Tile-scanned view (63 tiles; dotted lines) of material sampled from the TD. Host epithelial cells identified by autofluorescence are shown in white; colors indicate genera of bacteria. Bacterial consortia (circled) range in size from fifty to hundreds of microns.
(B) The spatial organization of an isolated consortium. Clusters of Rothia, Veillonella, Actinomyces, Neisseria, and Streptococcus cells comprise a large fraction of the consortium’s bacterial biomass. See also Figure S3 and Tables S2, S3, and S4.
Microbiota Organization within a TD Sample
Visualization of the microbiota in a TD sample, fixed in solution and spread onto slides in 50% ethanol, presents three major categories of microbial organization (Figure 2)—free bacteria, bacteria on squamous epithelial cells, and bacteria organized into structurally complex entities we term consortia. We define these categories operationally: free bacteria consisted of scattered bacterial cells attached to no identifiable substrate; epithelial-bound cells were similar to free bacteria but were associated with sheets of epithelial cells visible by their autofluorescence; and organized consortia were bacterial biofilms multiple layers thick, typically with a defined perimeter and a core of epithelial cells.
We analyzed bacterial communities in these different categories for overall composition as well as spatial organization. Taxa were quantified in each field of view by image segmentation and cell counting (Figures 2 and S1A) as well as by principal-component analysis (PCA). Ellipses of variation in the PCA plots (Figure S1B) showed that organized consortia were more homogeneous in composition than either free bacteria or epithelial-bound bacterial communities. In free bacteria and epithelial-bound bacteria, either Rothia, Veillonella, or Actinomyces could be dominant. In the epithelial-bound category, S. mitis was frequently at high levels. In contrast, within the organized consortia, the average proportions of each bacterial type were similar across samples. Structurally, the organized consortia displayed spatially localized domains dominated by a single taxon—equivalent to a patch structure as defined in the framework of landscape ecology (Turner and Gardner 2015; Proctor and Relman 2017). In contrast, free and epithelial-bound bacteria were organized as single cells or small clusters of cells. Because the consortia showed the densest, most highly structured and most consistent microbial colonization, we focused further analysis on the spatial organization of microbes in the organized consortia.
Spatial Organization of Consortia
Imaging at different spatial scales allowed visualization of both the complexity and the structural diversity of consortia. A composite image of a single biofilm, stitched together from 63 tiles (Figure 3A) and representing nearly a square millimeter of sample space, showed that the material was a mixture of microbes (colors) and host tissue (white). The ellipses in Figure 3A highlight several consortia, which varied in size, shape, and composition, but were identifiable by several common characteristics. Their size was typically in the range of tens to several hundred micrometers long; they had a well-defined perimeter, frequently displaying circular arc domains along their contour; and they were arranged around a core of autofluorescent material that we identify as epithelial cells derived from human mucosa, based on pan-cytokeratin immunostaining (Figure S2). Thus, the TD consortia occupied the space between a well-defined perimeter (the exterior of the biofilm exposed to saliva and oxygen) and a well-defined core composed of host epithelial cells. Similar microbial consortia were found among all subjects sampled and evaluated.
To assess the contribution of major TD microbes to the consortia, we performed multiplexed, spectral imaging FISH on hundreds of consortia by using 25 probes targeting the 17 abundant and prevalent genera as well as 7 abundant species within these genera, plus the phylum Saccharibacteria (TM7). Eight of the probes were newly designed for this study, and all probe sequences are shown in Table S2. We validated probe specificity and efficacy by hybridization to cultivated strains (Figures S3 and S4). We used probes in various combinations and conjugated with a variety of fluorophores (Table S3) in an effort to probe for all taxa and to optimize signal strength across taxa that differed significantly in brightness, perhaps due to differences in per-cell abundance of ribosomal RNA or cell wall permeability.
Among the 17 genera we targeted, 3 stood out as being present in 100% of individuals sampled and in ≥95% of images acquired (Table S4): Actinomyces, Rothia, and Streptococcus. Actinomyces frequently appeared in images as large uninterrupted domains near the epithelial cell core (Figures 2 and 3); it also was observed forming stripes located between patches of other taxa. Rothia was often observed in large patches toward the exterior of the consortium and forming a cortex around a core of epithelial material or other bacterial cells. This cortex of Rothia was often interrupted by veins or patches of other taxa. Streptococcus was observed forming a thin crust on the exterior of the consortia and also formed veins or patches in their interior. Other taxa that we observed in 100% of individuals sampled, but not in every consortium, included the genera Veillonella and Gemella, the family Neisseriaceae, and the phylum Saccharibacteria (TM7). Several taxa, notably Prevotella and the rod-shaped or filamentous Fusobacterium, Leptotrichia, and Lachnospiraceae, were less prominent in our images than would be expected based on the sequencing data. Images of consortia also contained individual cells and patches of cells that were not well labeled with taxon-specific probes. These non-labeled cells and patches generally comprised <20% of the cells that stained with the universal bacterial probe. Thus, our spectral imaging FISH assay was capable of identifying the majority of the cells in most consortia and provided significant information about their abundance and spatial organization relative to one another and to the epithelial core and the surface of the biofilm.
No single image can adequately represent all the detail of a three-dimensional microbial consortium. For efficiency, we imaged most structures by using a single image acquired at a plane in the center of the structure. At this plane, the well-defined perimeter and core of autofluorescent material were readily observed (Figure 3B). We carried out more complete analysis of the three-dimensional organization of selected consortia by acquiring z stacks of images throughout their depth (Figure S5). These three-dimensional images show the connectivity of single-taxon patches that in individual images might appear to be isolated (Videos S1, S2, and S3) and provide a more detailed view of the shape of single-taxon domains and the way in which the exterior contour remains relatively smooth even as the underlying taxa differ.
Species-Level Imaging Identifies Site Specialists
Sequence analysis led us to hypothesize that most bacterial genera are represented on the tongue by a small and well-defined set of species (Figure 1C; Table S1; Mark Welch et al., 2019). We tested this hypothesis by imaging consortia by using nested probe sets targeting the entire genus simultaneously with the species within the genus to evaluate what fraction of the cells of the genus were identified as the expected species. Probes for other abundant genera were also included in each set so as to visualize the target in context (see Table S3 for the full probe set composition). Prevalence within all donors and the frequency of identification across all images are reported in Table S4, and representative images are shown in Figure 4.
Figure 4. Species-Level Imaging Reveals Habitat Tropisms.
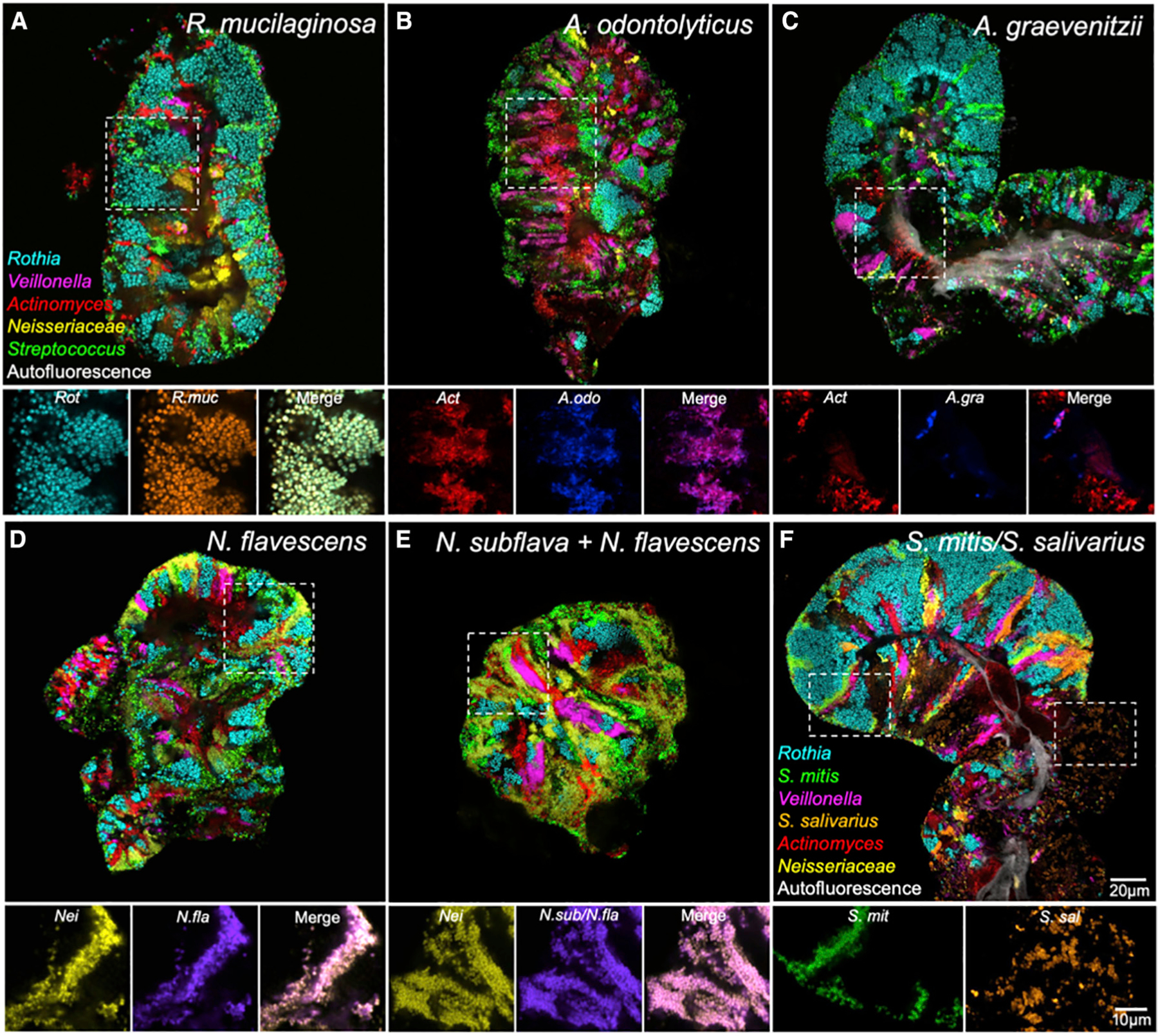
(A–E) Nested probe sets show cells hybridizing to both species-specific and genus-level probes.
(A) All Rothia cells (Rot) imaged were identified as R. mucilaginosa (R.muc).
(B and C) Many Actinomyces cells (Act) were identified as members of the A. odontolyticus group (A.odo) (B), whereas some were A. graevenitzii (A.gra) (C). (D and E) Most Neisseriaceae (Nei) were identified as N. flavescens (N.fla) (D). A probe (N.sub/N.fla) that detected both N. subflava and N. flavescens also identified most cells (E).
(F) The Streptococcus mitis group (S.mit) occurred as a thin layer at the exterior of the structure and in stripes between domains of other taxa. In contrast, S. salivarius and S. vestibularis (S.sal) occurred as large clusters of cells within the consortium. See also Figures S4 and S6, Video S2, and Tables S2 and S3.
Imaging results were consistent with the predictions from sequence data. All visualized Rothia cells were identified as R. mucilaginosa (Figure 4A; Table S4), which is in agreement with the expectation based on HMP sequencing results (Figure 1C). In the genus Actinomyces, the probe targeting Actinomyces odontolyticus and its close relatives generally colocalized with the genus probe (Figure 4B), whereas the probe targeting Actinomyces graevenitzii highlighted a small fraction of the cells in the genus. Therefore, the A. odontolyticus species group appeared to be the dominant member and A. graevenitzii a lesser member of the genus on the tongue, which is, again, consistent with sequence data. This trend was also consistent across subjects (Table S4B, Figure S6).
For the genus Neisseria, we carried out separate hybridizations with two species probes, both of which were expected to hybridize with the major Neisseria species detected by sequencing, namely Neisseria flavescens. The Nfla469 probe was designed to hybridize only with N. flavescens and related species (N. flavescens group; Table S1), whereas the Nsubfla177 probe was designed to hybridize with both Neisseria subflava and N. flavescens (Figure S4). Both probes showed near-complete co-localization with the Neisseriaceae family probe (Figures 4D and 4E), suggesting that N. flavescens is dominant in these TD consortia as expected and that N. subflava is a minor player if present (Table S4).
Within the genus Streptococcus, imaging revealed differential patterns of spatial localization (Figure 4F). We used three subgenus-level probes: one targeting S. mitis and its close relatives Streptococcus oralis and Streptococcus infantis (S. mitis group in Table S1; hereafter referred to collectively as S. mitis); one targeting Streptococcus salivarius and the related Streptococcus vestibularis, hereafter referred to collectively as S. salivarius; and one targeting Streptococcus parasanguinis, including both biovars recognized in HOMD. In our imaging analysis, both S. mitis and S. salivarius showed 100% prevalence across subjects, with S. mitis showing 100% and S. salivarius showing 95% frequency within consortia. This indicates that both species play an important role in TD consortia. However, S. mitis was frequently located on the surface of structures as well as forming internal stripes, whereas S. salivarius occupied large patches with distinctive cellular morphology (Figure 4F). The third major Streptococcus on the tongue, S. parasanguinis, was detected primarily in stripes (Figure S6). These three subsets of Streptococcus all localized to the same oral site, namely the tongue, but demonstrated distinctive niche preferences at spatial scales of micrometers to tens of micrometers. Collectively, our species-level imaging results confirm and deepen our understanding of the habitat specificity of key players and show the value of investigating microbiomes at high imaging and identification resolution.
Quantification of Micron-Scale Spatial Organization of Consortia
Major, well-defined landmarks within tongue microbial consortia were the perimeter and the host epithelial core. To evaluate the spatial organization of consortia relative to these landmarks, we carried out imaging at a scale sufficient to reveal both core and perimeter and a magnification sufficient to resolve individual microbes. Because of the size of the consortia, these conditions generally required tile-scanning to embrace an entire consortium, as shown in Figure 5A with high magnification “call outs” (Figures 5B–5D) to examine detailed spatial relationships.
Figure 5. Linear Dipole Analysis Quantifies Spatial Organization of Taxa.
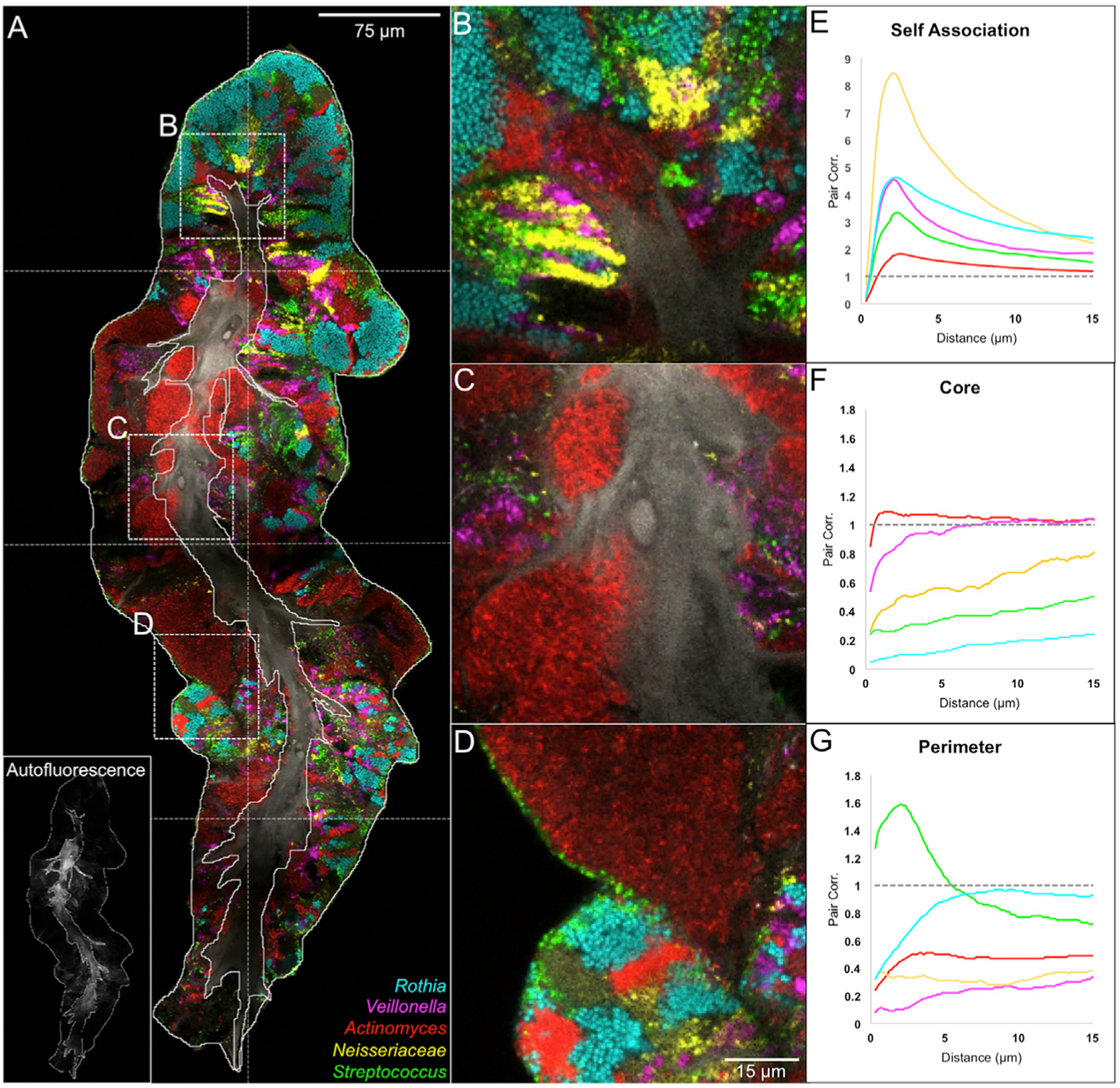
(A) A consortium imaged by tile scanning 8 fields of view. Outlines of perimeter and core were drawn by hand; the core outline was drawn based on autofluorescence of epithelial cells (inset in A). The presence of bacteria in the core represents coexistence of epithelial and microbial material in the same physical space.
(B–G) High-magnification images illustrate associations of taxa with themselves (B), with the core
(C), and with the perimeter (D). Linear dipole analysis of the image in (A) shows within-taxon autocorrelation (E); pair correlation between each taxon and the outline of the core (F); or pair correlation between each taxon and the perimeter (G). See also Figures S2, S3, S5, and S7; Videos S1 and S2; and Tables S2 and S3.
Spatial relationships were quantified by computing the pair cross-correlation function, g(r), of two features over a range of distances, r, by using linear dipole analysis. The values of g(r) indicate the probability of a randomly placed line segment (linear dipole) landing with one end touching each of the two features, normalized to the probability expected if the populations were randomly distributed so as to remove the dependence on density (Daims and Wagner 2011). Thus, g(r) indicates the degree to which the analyzed features are positively or negatively correlated as a function of distance: g(r) > 1 indicates an “attractive” interaction; g(r) < 1 indicates a “repulsive” interaction; and g(r) = 1 indicates a random, neutral relationship of the features.
All taxa evaluated showed a tendency toward self-association at short distances (1–3 μm) that weakened at longer distances (Figure 5E). The normalized self-association was stronger for taxa present only in a few patches and weaker for taxa that occupied a large fraction of the image. This finding is a logical result of the normalization because a taxon occupying a large fraction of the image will show a higher self-correlation in the randomized image to which the data are normalized. Self-association at short distances is also consistent with localized clonal expansion of individual taxa. The width of the correlation peak at half-maximum can be taken as a metric of the size of single-taxon clusters. For the consortium shown in Figure 5, this metric suggests that a typical cluster size is 7 to 10 μm for Neisseria and Veillonella and >15 μm for Rothia and Actinomyces.
Some pairs of taxa showed a significant tendency to associate with each other. Cross-correlation analysis of all possible pairs of taxa (Figure S7) revealed associations among S. mitis, S. salivarius, and Veillonella below 5-mm distance. Associations between Rothia and S. mitis and Actinomyces and S. mitis, evident in the images (Figures 3A, 4A, 4F, and 5D), did not show strongly in the cross-correlational analysis because of the strong tendency of Rothia and Actinomyces to form large single-taxon clusters.
Taxa differed in their spatial relationship with the core (Figure 5F) or perimeter (Figure 5G). Streptococcus showed a strong affinity for the perimeter whereas Actinomyces, Neisseria, and Veillonella showed repulsion. Rothia showed intermediate behavior—repulsion from the core but not attraction to the perimeter. Streptococcus cells were frequently detected at the perimeter of the structure in a thin layer of 1–3 cells (Figure 5D), which accounts for the high cross correlation value with the perimeter at short distances. Although we cannot exclude the possibility that putative additional layers of Streptococcus were lost during sample collection, we think this possibility is unlikely because detached layers of Streptococcus were not observed. The affinity of Streptococcus for the perimeter was further investigated by high-magnification imaging and visualization in three dimensions (Figure S6; Video S2). Conversely, Actinomyces formed large solitary patches (Figure 5C), sometimes embedded between lobes of the epithelial core or making up almost the entire thickness of the consortium. Actinomyces was also present as clusters or stripes away from the perimeter, adjacent to Rothia and other taxa. Collectively, the Actinomyces distribution reports as negatively correlated with the perimeter but randomly distributed with respect to the core (Figures 5F and 5G). Neisseria and Veillonella showed similar cross-correlation trends as Actinomyces, but both typically occupied smaller patches located between domains of Rothia. These two-dimensional results were consistent with and reinforced by visual analysis of z stacks (Figures S5C–S5F). The specific and differing spatial relationships of the taxa suggest the existence of host and microbial physiological and landscape-ecological factors that influence their relative positions with respect to each other and to host features.
Consortial Development
Although it is not yet possible to observe growth of the TD consortia in situ, the thickness of the biofilm adhering to host epithelial cells can be used as a metric for organizing images of individual consortia in presumptive chronological order (Figure 6). Images from a single time point show a series of structures that have in common the presence of an epithelial core but that vary in the number of bacteria adherent to the core. In Figure 6A the biofilm is 5 to 8 cells thick; in Figure 6B it is 10 to 20 cells thick; and in Figure 6C it is more than 50 cells thick, with individual taxa clearly organized into distinct domains. The ordered sequence of images suggests a developmental pathway in which founder cells initially adhere to host epithelial cells and then proliferate and perhaps recruit additional taxa resulting in a progressive increase in size, complexity, and organization of the consortium.
Figure 6. Gradations in Biofilm Thickness and Shape of Clonal Domains Suggest Biofilm Growth and Selective Advantage.
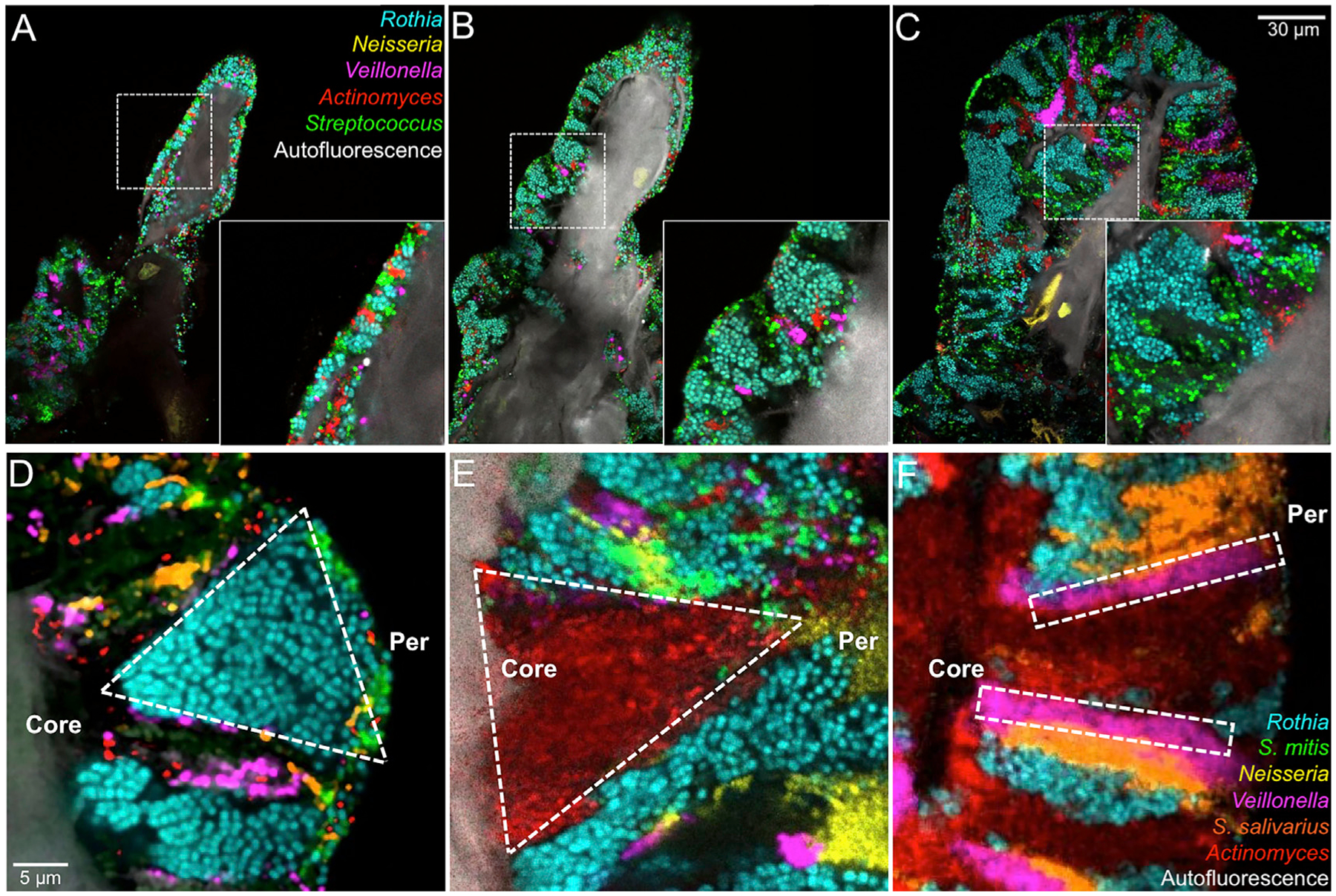
(A) A thin biofilm composed of small clusters of cells from each bacterial taxon.
(B) A thicker biofilm showing expansion of the facultative anaerobe Rothia and the beginnings of expansion of anaerobes Veillonella and Actinomyces.
(C) A mature structure showing well-defined domains.
(D) Increasing width of a clonal domain toward the perimeter suggests a selective advantage toward the periphery.
(E) Decreasing width toward the perimeter suggests a disadvantage at the periphery or selective advantage in the interior.
(F) Constant width suggests neither selective advantage nor disadvantage with respect to neighboring taxa. See also Figures S2 and S5, Video S3, and Tables S2 and S3.
The shape of single-taxon patches within the consortia may then be interpreted in terms of clonal dynamics. Experimental and theoretical analyses of clonal dynamics in model systems have been used to make inferences about selective advantage or disadvantage, based on whether the clonal domains show range expansion or contraction, respectively (Hallatschek et al., 2007; Blanchard et al., 2014). We qualitatively categorized taxon patch shapes and show examples of the more obvious patch types according to whether they apparently displayed range expansion toward the perimeter (Figure 6D) or range contraction (Figure 6E) or whether they developed linearly without widening in one direction or the other (Figure 6F). These observations provide clues to the dynamics of consortia and suggest complex community interactions.
DISCUSSION
TD consortia present a unique opportunity to observe spatial organization in a human-associated microbiome in a way that preserves key aspects of spatial information. Because of the distinct outer boundary and retention of the inner core, we deduce that important landmarks, such as the location of the substrate and the orientation of the community relative to sources of nutrients, are maintained by our sampling methods. Our data suggest a model for how the structured microbial communities harbored on our tongues are generated (Figure 7). We hypothesize that bacterial cells attach to the epithelium of the TD singly or in small clusters. During population growth, differing taxa push on one another and proliferate more rapidly in microenvironments that support their physiological needs. This differential growth results in the patch mosaic organization that we observe in larger, more mature structures. These snapshots in time of patch shapes, coupled with literature on the oral environment, enable formulation of hypotheses for the dynamics and micro-environmental interactions of community organization. We also note that because our study was on healthy subjects, different structures could occur as a result of differing microbiome diversity, host physiology, or disease state.
Figure 7. Inferred Development of TD Consortia.
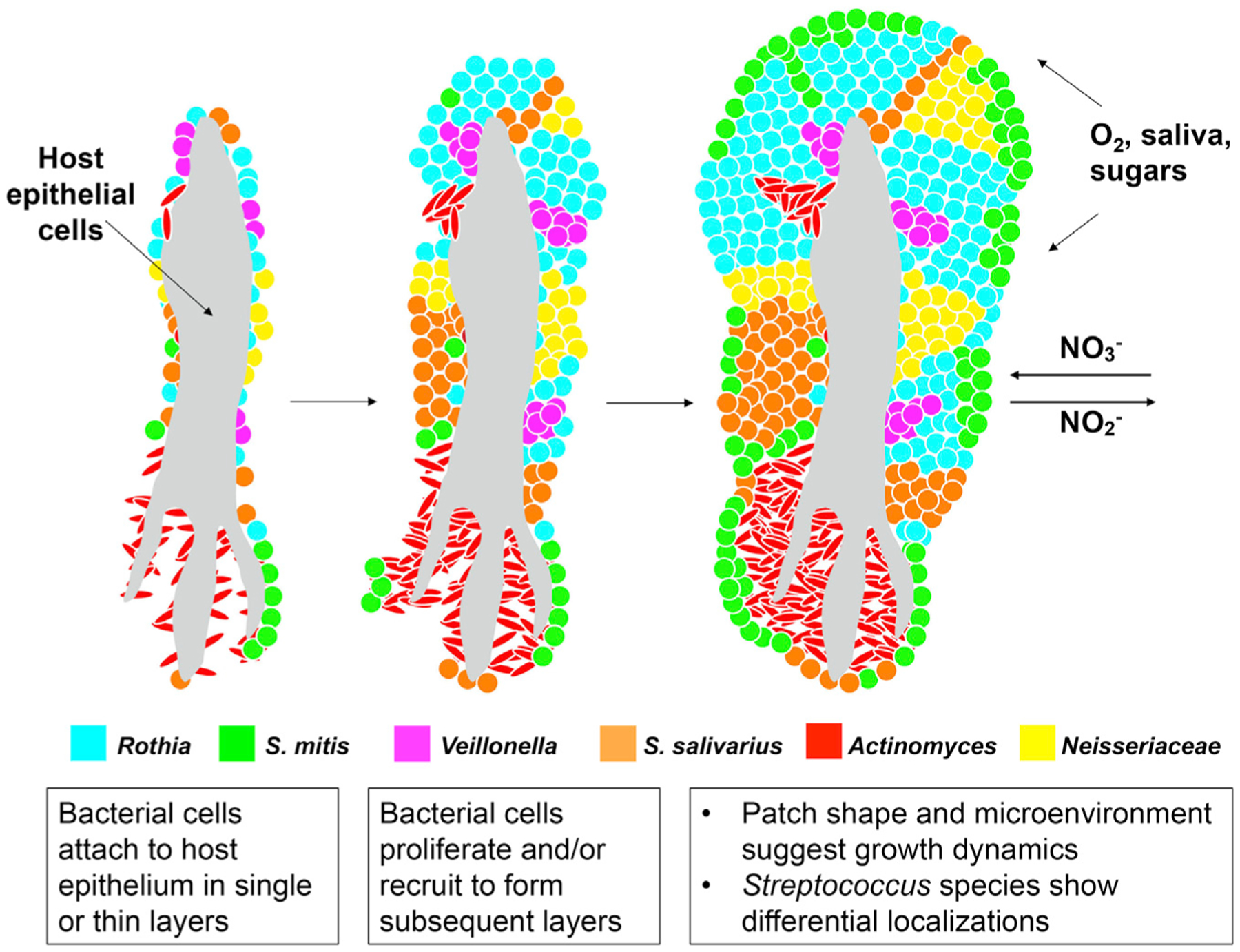
In this model, bacterial cells colonize host epithelial cells sparsely. As bacteria proliferate, layers of cells appear in a patch-like structure. Some S. mitis cells form a thin coat on the surface. Domain formation is dependent on neighbors and the microenvironment. Some nutrients may be gained from host epithelial material and other nutrients, O2, and NO3− from the oral cavity by saliva.
Spatial Ecology and Scale in a Microbial Community
The TD microbial community demonstrates the degree to which spatial organization can inform ideas about community structure and dynamics. Although the material we imaged was fixed and the images were, therefore, snapshots in time, the structures we observed—with an epithelial cell core, a smooth perimeter, and a mixed, patchy community—suggest a model for consortium growth as well as hypotheses about community dynamics. Differing thicknesses of the biofilm in different consortia suggest a growth series, and the shape of patches suggests a growth advantage of certain taxa as a function of their proximity to the host or the consortium perimeter.
As clearly explicated in two recent reviews (Proctor and Relman 2017; Ladau and Eloe-Fadrosh 2019), a critical question in the landscape ecology of microbiota is the appropriate spatial scale of analysis. If the scale is too coarse, the results become averages over heterogeneous features, whereas if the scale is too fine, questions arise as to the degree that any individual feature represents the population as a whole.
Our imaging results show that community composition on the tongue is effectively analyzed when the grain is small enough to identify individual cells, and the extent (the range over which observations are made) encompasses features that are hundreds of microns in size. Our results show patchiness in community structure at scales of tens of microns that would be invisible at a sampling scale of millimeters. The extent of hundreds of microns was sufficient to describe organized, spatially differentiated structures; the different types of material within a sample possessed different community structure that was evident with imaging but would have been missed by homogenizing and sequencing the sample.
Issues of the scale of analysis are related to estimates of community composition. Estimates of community composition, as measured by imaging and by sequencing, were consistent, in that the genera predicted to be prevalent by sequencing were also prevalent by imaging. However, some differences were apparent. In organized consortia, Rothia was more abundant and Prevotella was less abundant than might be expected from sequencing of whole samples. This result may stem from limitations in both sequencing and FISH for estimating taxon abundance. Sequencing is subject to amplification and other biases, whereas FISH is subject to limitations of target accessibility, target abundance, and signal intensity. However, the scale of sampling may also play a key role in this differential result. For sequencing, sampling is typically carried out on millimeter to centimeter scales, such as swabbing several cm2 of TD. The result of such sequencing will be an average over heterogeneous community types in the sample. In contrast, imaging provides sufficient resolution to differentiate planktonic, epithelial-bound, and mature consortium communities. Thus, micron-scale analysis enables definition of microbial communities at a scale commensurate with their biological organization.
Microbiome Structures: Compare and Contrast
The structures we visualize here on the tongue show similarities and differences compared with structures seen using the same methods in dental supragingival plaque (Mark Welch et al., 2016) or in the gnotobiotic mouse gut (Mark Welch et al., 2017). Like microbial structures on the tongue, dental plaque shows patchiness and distinctive consortial organization (Zijnge et al., 2010; Dige et al., 2014; Mark Welch et al., 2016) but with different morphology and composed of different microbes.
A fine-scale similarity between dental plaque and tongue biofilm communities is in the distribution of Streptococcus bacteria, which form a single-cell-thick layer in both tongue and plaque: in tongue, as a crust on the surface of Actinomyces as well as Rothia; and in plaque, as corncobs on the tips of Corynebacterium filaments. This spatial structure may reflect a strong competitive advantage of Streptococcus at the surface of a biofilm. Alternatively, it may signal a long-standing co-evolutionary relationship between Streptococcus and Actinobacteria, as Actinomyces, Corynebacterium, and Rothia are all genera within this phylum.
The mouth is colonized by bacterial biofilms, whereas the gut acts as a bioreactor in which peristalsis and a transient and rapidly renewing mucus layer are forces for mixing. In the mouse gut, the microbiota is extensively mixed at the single-cell level (Whitaker et al., 2017; Mark Welch et al., 2017). In the zebrafish gut, the microbiota are either mixed at the single-cell level or clustered into single-taxon aggregates (Schlomann et al., 2018). Although more investigation is clearly required, a general principle seems to be that microbial organization responds to environmental conditions created by host physiology.
Oral bacteria may play a role in host nitric oxide (NO) homeostasis (Lundberg et al., 2004, 2018) by the reduction of nitrate to nitrite (Doel et al., 2005; Hyde et al., 2014), a function that the human host genome does not have and that enables the conversion of dietary nitrate into NO. Our images demonstrate that some taxa capable of nitrate reduction—Actinomyces, Neisseria, Rothia, and Veillonella—are prominent in tongue consortia. Their prominence raises the possibility that human filiform papillae are structured to encourage growth of these bacteria on the tongue as an organ of nitrate reduction in the oral cavity. Further elucidation of the potential function of the tongue consortium in human physiology may have significant biomedical impact.
Species-Level Resolution Identifies Site Specialists
Much information can be extracted from imaging with probes directed at the genus level. Species-level identification, however, is valuable particularly when different species from the same genus show different spatial distribution suggestive of distinctive functional roles. The three major Streptococcus groups on the tongue, for example, are differently distributed among the surface crust, interior stripes, and large interior clusters in the observed consortia, features that would have been lost or lumped together without this level of identification.
Species-level imaging also tests hypotheses emerging from sequence data about the localization of species-level taxa at larger scales within the mouth. Evidence from DNA sequencing indicates that most oral microbes are site specialists, residing, for example, in dental plaque or on the TD but not abundantly in both (Aas et al., 2005; Eren et al., 2014; Lloyd-Price et al., 2017; Mark Welch et al., 2019). Our species-level imaging is consistent with sequencing data indicating dominance of certain species on the TD. An important remaining question is whether single-species patches are largely homogeneous or whether they contain sub-species-level mixtures of cells differentiated in genotype, metabolism, or gene expression.
Implications of Spatial Organization for Community Dynamics and Ecology
The tongue surface, like all mucosal epithelial surfaces, undergoes a constitutive process of renewal by progenitor cells pushing up from basal layers, whereas older, superficial cells complete differentiation and are shed into the oral cavity. Bacteria attached to the host epithelial cells will necessarily be shed as well, exposing nascent epithelial cells to colonization. One may predict that, in the steady state, microbial consortia should be present at varying stages of colonization, growth, and maturation. This steady-state model provides a conceptual framework for inferring community dynamics from image snapshots.
The size of the consortia we observed ranged from individual adherent cells to populations tens of layers in diameter. The observed differences in thickness suggest that microbial consortia are initiated by the adherence of founder microbes to the host material and then grow outward by clonal expansion. Consortia were rarely seen larger than a hundred microns in diameter. This upper size may reflect the maximal extent of proliferation that can occur before the underlying host cells, together with their adherent bacteria, are shed into the saliva.
Quantitative analysis showed that microbes were spatially structured with respect to themselves and with respect to the perimeter and the core. Measurements of auto-correlation showed that each taxon was positively correlated with itself, a quantitative confirmation of the visually apparent feature of single-taxon clusters microns to many tens of microns in size. The dynamics interpretation of this observation is that the clusters grew by clonal expansion.
Differences in selection or growth advantage of tongue biofilm taxa in different micro-environments were suggested by the shape of single-taxon patches—becoming wider toward the perimeter, wider toward the core, or in a stripe of constant thickness. Patches widening at the perimeter were predominantly the facultative aerobes Rothia and Neisseria, suggesting that they proliferated more rapidly than their neighbors in the surface layer. Anaerobes Veillonella and Actinomyces were more prominent near the core. One interpretation of this configuration is that they were disfavored in competition with Rothia and Neisseria as growth occurred near the surface. An alternate interpretation is that they persist in a relatively inactive state until the biofilm grows thick enough that a micro-aerobic environment develops near the core. Once oxygen levels drop, these cells begin to grow and either displace other microbes from binding sites on host material or expand into the host material itself. Measurements of growth rate and oxygenation state within the biofilm would differentiate among these alternatives. The complexity of the situation in vivo, however, is indicated by the complex and variable shape of patches and the difficulty in identifying many patches as clearly belonging to any of these categories. The shape of domains permits inference regarding the dynamics of selective growth, but these dynamics are likely to be complex and strongly dependent on details of the local environment.
In summary, we explored the spatial ecology of the TD and characterized the organization of its microbiome into free bacteria, epithelial-bound bacteria, and bacteria structured into consortia. We used spatial information to identify appropriate scales for analysis of its bacterial landscape ecology. By examining the shape of single-taxon domains and the proximity of taxa to host cells and other landmarks, we inferred properties of the dynamics of individual taxa and the overall community. These inferences allowed us to produce a model of the colonization and growth of bacteria as a consortium on tongue epithelial cells. The approaches taken in this study should be useful in the study of microbiome structural ecology at other human body sites, in the microbiomes of other macroscopic host organisms, and in environmental microbial ecology of complex biofilms and organized microbial communities.
However, our studies were limited by intrinsic properties of our methods and by barriers remaining in the microbiome field in general. What are some of these barriers and the prerequisites needed to overcome them? Our methods required removing small samples from the mouth. A barrier to be overcome in future work is examining microbial communities at high-resolution in situ, thus avoiding the perturbations of sampling. How can we expand the extent of sampling by several orders of magnitude while still maintaining the micron-scale grain? Such combination of extent and grain is necessary to be able to evaluate microbiome organization across the entire surface of the tongue. Another barrier to overcome is the necessity of fixing the samples. Fixed samples provide only snapshots in time, whereas studying live-cell dynamics is clearly needed to understand microbial assembly. Another barrier is how to create a manipulable model of the community so as to test well-formulated hypotheses. Can a suitable animal model system be developed? Current information on the oral microbiome of mice and other animals is far more limited than for the gut microbiome. Thus, validation of an animal model is a significant undertaking in itself. Can suitable reductionist experiments be designed with cultivars of selected organisms? The issues here revolve around the organisms themselves as well as their environment. To what extent do organisms under cultivation represent the genomic content of the organisms actually living in the mouth? Strain-level diversity of the microbial population may influence the functional interactions between the host and microbiota as well as between taxa, with the result that consortium structure and host-microbiota interaction may differ in detail from person to person within the population. ‘Omics technologies will be essential for sorting out the commonalities and the diversity of microbial interactions. Beyond the question of representativeness of cultured organisms is the question of simulating the environment in which they naturally occur. Bacteria growing on surfaces in the mouth experience localized gradients of molecules from the saliva and opposing gradients of molecules upwelling from host tissue secretions. Bacteria on the tongue grow on a complex papillary topography, which itself is differentiated at the micron scale. Neither the localized gradients nor the localized topography are captured by growing bacteria in broth, on plates, or on flat, inert substrates. Engineering suitable microenvironments that capture relevant biological community and host properties represents a challenge for future research. In short, many barriers and challenges remain in microbiome research. Although imaging is only one of several key technologies, it provides the unique benefit of showing us the target: the landscape and the structures that microbes build and that we need to explain and replicate in order to have achieved an understanding of microbial community organization.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILTY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jessica Mark Welch (jmarkwelch@mbl.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Human subjects were recruited and sampled according to protocol number 17–01r as approved by the Institutional Review Board of the Forsyth Institute, Cambridge, MA. Twenty-one healthy human subjects, 14 female, 7 male, aged 21 to 74 years, contributed to the study. Informed consent was obtained from all subjects prior to sampling.
Microbe strains and culture
Microbe strains used for probe validation are listed in the Key Resources Table. Microbes were cultured in standard media under standard aerobic or anaerobic conditions as appropriate for the microbe in question. Cultures were typically grown in liquid culture to exponential phase and then fixed in 2% paraformaldehyde for 1.5 h at 4°C. Fixed cells were then washed and stored in 50% ethanol at −20°C until use.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse IgG1 anti-pan-cytokeratin (AE1/AE3) | Santa Cruz Biotechnology, Inc. | Cat# sc-81714; RRID: AB_2191222 |
| Goat anti-mouse IgG1 AlexaFluor 647-conjugate | Molecular Probes | Cat# A-21240; RRID: AB_141658 |
| Bacterial and Virus Strains | ||
| Actinomyces graevenitzii | Forsyth Institute | F0582 |
| Actinomyces naeslundii | Forsyth Institute | F0404a |
| Actinomyces odontolyticus | Forsyth Institute | XH001 |
| Aggregatibacter aphrophilus | Forsyth Institute | F0387 |
| Atopobium sp. HMT 810 | Forsyth Institute | F0209 |
| Campylobacter gracilis | Forsyth Institute | F0073 |
| Capnocytophaga ochracea | ATCC | ATCC 33596 |
| Fusobacterium nucleatum subsp. vincentii | ATCC | ATCC 49256 |
| Gemella haemolysans | Forsyth Institute | F0145 |
| Granulicatella adiacens | ATCC | ATCC 49175 |
| Leptotrichia buccalis | ATCC | ATCC 14201 |
| Neisseria mucosa | ATCC | ATCC 19696 |
| Neisseria subflava | ATCC | ATCC 49275 |
| Oribacterium asaccharolyticum | Forsyth Institute | F0425 |
| Porphyromonas gingivalis | ATCC | ATCC 33277 |
| Prevotella melaninogenica | Forsyth Institute | F0114 |
| Rothia aeria | Forsyth Institute | F0184 |
| Rothia dentocariosa | ATCC | ATCC 17931 |
| Rothia mucilaginosa | ATCC | ATCC 25296 |
| Streptococcus mitis | ATCC | ATCC 49456 |
| Streptococcus parasanguinis | Forsyth Institute | F0613 |
| Streptococcus salivarius | ATCC | ATCC 7073a |
| TM7 + Actinomyces odontolyticus (helper strain) | Forsyth Institute | 1906 |
| Veillonella dispar | Forsyth Institute | F0600 |
| Biological Samples | ||
| Adult human tongue dorsum scrapings | Forsyth Institute | N/A |
| Oligonucleotides | ||
| See Table S2 for list of oligos used in this study | https://www.Biomers.net | N/A |
| Software and Algorithms | ||
| DAIME | Daims et al., 2006 | https://dome.csb.univie.ac.at/daime |
| FIJI | Schindelin et al., 2012 | https://fiji.sc/ |
| Imaris Version 8.0 | Bitplane Inc | https://imaris.oxinst.com/ |
METHOD DETAILS
Sampling of the tongue dorsum
Subjects were instructed not to eat, drink (except water), or brush their tongue during the 12 h before sampling and not to brush their teeth during the 2 h before sampling. The subjects sampled themselves under supervision by gently scraping the dorsal surface of the tongue from back to front using a ridged plastic tongue scraper (BreathRx Gentle Tongue Scraper, Discus Dental, Culver City, CA).
Sample fixation and storage
Collected samples adherent to the tongue scraper were fixed by bending the scraper into a “U” and immersing the bent region (containing the sample) into 30 mL of 50% ethanol in a 50 mL conical centrifuge tube. Samples detached from the scraper upon immersion and became dispersed in the fixative as flocculent material. The scraper was removed, and the samples were put at −20°C for storage. Some samples were processed as above but fixed with 2% paraformaldehyde in 1x phosphate buffered saline (PBS) or 1× TE (10 mM Tris pH 7.5, 1 mM EDTA) for 1.5 h at 4°C, then washed in PBS or TE, then washed with 50% ethanol and stored in 50% ethanol at −20°C. Finally, some samples were transferred from the tongue scraper directly to slides and fixed on the slides by addition of drops of 2% paraformaldehyde in PBS.
Sequence analysis
Sequence analysis was carried out with previously published oligotypes (Eren et al., 2014) of the V1–V3 region of the 16S ribosomal RNA gene defined by re-analysis of publicly available data from the Human Microbiome Project (Human Microbiome Project Consortium, 2012). To map oligotypes to human oral microbiome taxa, each oligotype was compared by BLAST to the extended Human Oral Microbiome Database, eHOMD version 15.1 (http://www.homd.org). Oligotypes were assigned to the most closely matching eHOMD reference sequence. Of the 494 oral oligotypes, 479 had at least 98% identity to the closest-match reference sequence in eHOMD and the remaining 15 were low-abundance, collectively accounting for 0.3% of the oligotype data from tongue dorsum. Oligotypes mapping to the same species were combined; abundance was calculated as the mean relative abundance over the 77 individuals analyzed (4) and prevalence was calculated as the fraction of these 77 individuals in which the taxon was detected at any non-zero abundance. The mapping information is shown in Table S1.
Probe design and validation
For design of new probes, near-full-length aligned 16S rRNA sequences of bacteria identified by sequence analysis as abundant in the healthy human oral microbiome were downloaded from the Human Oral Microbiome Database (http://HOMD.org) and inspected by eye for candidate target sites 18 to 22 nucleotides long at which target taxa were identical and differed from other oral taxa by at least 2 central mismatches. Candidate oligonucleotides were evaluated in silico using MathFISH (Yilmaz et al., 2011) and probeCheck (Loy et al., 2008) to estimate binding affinity to target and non-target taxa. Fluorophore-labeled oligonucleotide probes were purchased from ThermoFisher (Waltham, MA) or Biomers (Ulm, Germany). To test specificity and efficacy, probes were hybridized with pure cultures of bacterial cells of target and non-target taxa and imaging and linear unmixing were carried out with the same settings as those used for tongue dorsum samples.
Multiplexed FISH
Tongue dorsum samples
Typically, 100 μL of fixed samples in 50% ethanol were applied to silanized slides (Gold Seal UltraStick slides, Electron Microscopy Sciences, Hatfield, PA) and air-dried. The dried samples were then incubated with hybridization solution (900mM NaCl, 20mM Tris pH 7.5, 0.01% SDS, 20% formamide, 2 uM each FISH probe) for 2 to 4 h at 46°C in a humid chamber, washed in wash buffer (215mM NaCl, 20mM Tris pH7.5, 5mM EDTA) for 15 min at 48°C in a humid chamber, dipped into ice cold water and then immediately into 100% ethanol, drained, and air-dried. Slides were mounted with Prolong Gold antifade mounting medium (Thermo Scientific) with a glass coverslip of #1.5 thickness and stored flat to cure overnight in the dark.
Reference spectra
Reference spectra were acquired from cultured Leptotrichia buccalis cells, which were cultured as described in Experimental Model and Subject Details. L. buccalis cultures were concentrated by centrifugation and fixed with 2% paraformaldehyde in 1x phosphate buffered saline (PBS) for 1.5 h at 4°C, washed in PBS and stored in 50% ethanol at −20°C. Typically, 10–30 μL of dense fixed cells were hybridized in suspension with the Eub338 oligonucleotide probe conjugated to the fluorophore of interest in solution. Hybridized cells were then collected by centrifugation, washed as above, resuspended in 100 μL of wash buffer and 10–30 μL was applied to silanized slides either for 2 h at room temperature or overnight at 4°C in a humid chamber to allow cells to settle and adhere to the slide. Once settled, slides were dipped into ice-cold water, then immediately in 100% ethanol, drained, air-dried and mounted for microscopy as above.
Probe validation
Cultures of pure bacterial strains representing distinct taxa examined in this study were cultured as described in Experimental Model and Subject Details, then concentrated, fixed, and stored as described above for L. buccalis. To test the reactivity of probe sets with these known reference cells, 10–30 μl of fixed cultures were adhered to silanized slides by air drying, then processed for FISH as described above for tongue dorsum samples, using probe sets as described in the text.
Immunohistochemistry
10–30 μl of fixed tongue dorsum scrapings were adhered to silanized slides as above, washed in 1× phosphate buffered saline with 0.1% Tween20 (PBST), then blocked with 10% goat serum in PBST for 1 h at room temperature in a humid chamber. Block was removed and anti-pan-cytokeratin (1:100, Santa Cruz Biotechnology #sc-81714) in 10% goat serum in PBST was applied to samples. A coverslip was applied to prevent evaporation and the slide was incubated in a humid chamber overnight at 4°C. After three PBST washes, samples were then incubated in AlexaFluor 647-conjugated goat anti-mouse IgG1 secondary antibody (1:500, Invitrogen #A-21240) in 10% goat serum in PBST for 1 h at room temperature in a humid chamber, washed with PBST, and cured overnight in the dark in Prolong Gold antifade mounting medium (Thermo Scientific).
Image acquisition and spectral unmixing
Spectral confocal imaging was performed using either a Zeiss LSM 780 or Zeiss LSM 880 (Carl Zeiss) with a 40× or 63× 1.4 numerical aperture Plan-Apochromat objective. Spectral datasets were collected either by sequential excitation with single laser lines (633nm followed by 594nm, 561nm, 514nm, 488nm, and 405nm) or by simultaneous excitation with 405nm, 488nm, 561nm, and 633nm laser lines using a triple dichroic beam splitter. Reference spectra were collected using the same laser lines and dichroic filters as the experimental acquisitions. Images were acquired at a pixel size of 0.11 × 0.11 μm. This size was found to be sufficient for segmentation of individual bacterial cells. Some consortia were imaged as three-dimensional image stacks. Other consortia were imaged in a single representative plane chosen by focusing along the z axis to estimate the thickness of the consortium and then selecting the medial plane for image acquisition. Linear unmixing, to express the observed data as a weighted sum of the reference spectra while minimizing residuals, was performed using Zeiss ZEN software or using the nonlinear least-squares function in MATLAB.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
For presentation of individual 2D images and Z stacks, unmixed fluorophore channels were pseudo-colored and adjusted for visual presentation using FIJI (Schindelin et al., 2012). Three-dimensional renderings were produced using Imaris version 8.0 (Bitplane Inc.). Segmentation of images was performed in FIJI by applying a 3×3 median filter followed by thresholding using the “Auto Local Threshold” function with the Bernsen method, followed by a watershed function “Find Maxima” with output “Segmented Particles”; the output of “Segmented Particles” was copied onto the thresholded image using the Boolean operator “AND” to separate adjacent cells. Objects smaller than bacterial cells (noise, dust, artifactual segmentation) were removed from the segmented image using a size filter with a 0.5 μm diameter threshold in the “Analyze Particles” function. Auto-correlation and cross-correlation analysis was carried out by 2D linear dipole analysis using the DAIME software package (Daims et al., 2006) to generate the output presented in Figures 5 and S5. The analysis was carried out using manually drawn biomass masks to minimize the possibility of spurious correlations resulting from unoccupied area surrounding the biomass. The mask included biomass from both microbial cells and epithelial cell core regions.
Principal component analysis for cell count data were carried out using Clustvis (Metsalu and Vilo, 2015).
DATA AND CODE AVAILABILITY
The datasets supporting the current study have not been deposited in a public repository because of large image file sizes, but they are available from the corresponding author on request.
Supplementary Material
Highlights.
Bacteria on the tongue form large organized consortia with a patch mosaic structure
Consortia are organized around a core of keratinized epithelial cells
Spatial organization can be quantified and permits inferences on dynamics
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant DE022586 (to G.G.B.). We thank Floyd Dewhirst for helpful discussions and a critical reading of the manuscript; Alex Valm for design of the Gra65 and TM7550 probes; Blair Rossetti for help with MATLAB spectral unmixing; and Jordan Briscoe, Andrew Kempchinsky, and Janina Schumann for technical assistance. Some of the imaging was performed in the Biological Imaging Facility of the California Institute of Technology, with the support of the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.097.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Amann et al., 1990; Chalmers et al., 2008; Diaz et al., 2006; Kong et al., 2010; Paster et al., 1998.
REFERENCES
- Aas JA, Paster BJ, Stokes LN, Olsen I, and Dewhirst FE (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, and Stahl DA (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol 56, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard AE, Celik V, and Lu T (2014). Extinction, coexistence, and localized patterns of a bacterial population with contact-dependent inhibition. BMC Syst. Biol 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers NI, Palmer RJ Jr., Cisar JO, and Kolenbrander PE (2008). Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol 190, 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Ross RP, and Hill C (2013). Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol 11, 95–105. [DOI] [PubMed] [Google Scholar]
- Daims H, and Wagner M (2011). In situ techniques and digital image analysis methods for quantifying spatial localization patterns of nitrifiers and other microorganisms in biofilm and flocs. Methods Enzymol. 496, 185–215. [DOI] [PubMed] [Google Scholar]
- Daims H, Lücker S, and Wagner M (2006). daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol 8, 200–213. [DOI] [PubMed] [Google Scholar]
- Dal Co A, van Vliet S, Kiviet DJ, Schlegel S, and Ackermann M (2019). Short-range interactions govern cellular dynamics in microbial multi-genotype systems. bioRxiv. 10.1101/530584. [DOI] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, and Wade WG (2010). The human oral microbiome. J. Bacteriol 192, 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ Jr., and Kolenbrander PE (2006). Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol 72, 2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dige I, Grønkjær L, and Nyvad B (2014). Molecular studies of the structural ecology of natural occlusal caries. Caries Res. 48, 451–460. [DOI] [PubMed] [Google Scholar]
- Doel JJ, Benjamin N, Hector MP, Rogers M, and Allaker RP (2005). Evaluation of bacterial nitrate reduction in the human oral cavity. Eur. J. Oral Sci 113, 14–19. [DOI] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, and Mark Welch JL (2014). Oligotyping analysis of the human oral microbiome. Proc. Natl. Acad. Sci. USA 111, E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, and Lemon KP (2018). New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): a Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 3, e00187–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, and Houte JV (1975). Bacterial adherence in oral microbial ecology. Annu. Rev. Microbiol 29, 19–44. [DOI] [PubMed] [Google Scholar]
- Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segrè D, Mehta P, and Sanchez A (2018). Emergent simplicity in microbial community assembly. Science 361, 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, and Nelson DR (2007). Genetic drift at expanding frontiers promotes gene segregation. Proc. Natl. Acad. Sci. USA 104, 19926–19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel MP, and Weitzberg E (2015). The oral microbiome and nitric oxide homoeostasis. Oral Dis. 21, 7–16. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde ER, Andrade F, Vaksman Z, Parthasarathy K, Jiang H, Parthasarathy DK, Torregrossa AC, Tribble G, Kaplan HB, Petrosino JF, and Bryan NS (2014). Metagenomic analysis of nitrate-reducing bacteria in the oral cavity: implications for nitric oxide homeostasis. PLoS One 9, e88645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Gill SR, Vickerman MM, and Kolenbrander PE (2008). Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol. Ecol 66, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just T, Stave J, Pau HW, and Guthoff R (2005). In vivo observation of papillae of the human tongue using confocal laser scanning microscopy. ORL J. Otorhinolaryngol. Relat. Spec 67, 207–212. [DOI] [PubMed] [Google Scholar]
- Kleinberg I, and Jenkins GN (1964). The pH of dental plaques in the different areas of the mouth before and after meals and their relationship to the pH and rate of flow of resting saliva. Arch. Oral Biol 9, 493–516. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, and London J (1993). Adhere today, here tomorrow: oral bacterial adherence. J. Bacteriol 175, 3247–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, He M, McAlister T, Seviour R, and Forster R (2010). Quantitative fluorescence in situ hybridization of microbial communities in the rumens of cattle fed different diets. Appl. Environ. Microbiol 76, 6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraal L, Abubucker S, Kota K, Fischbach MA, and Mitreva M (2014). The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS One 9, e97279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullaa-Mikkonen A, and Sorvari TE (1985). A scanning electron microscopic study of the dorsal surface of the human tongue. Acta Anat. (Basel) 123, 114–120. [DOI] [PubMed] [Google Scholar]
- Kullaa-Mikkonen A, Sorvari T, and Kotilainen R (1985). Morphological variations on the dorsal surface of the human tongue. Proc. Finn. Dent. Soc 81, 104–110. [PubMed] [Google Scholar]
- Kullaa-Mikkonen A, Hynynen M, and Hyvönen P (1987). Filiform papillae of human, rat and swine tongue. Acta Anat. (Basel) 130, 280–284. [DOI] [PubMed] [Google Scholar]
- Ladau J, and Eloe-Fadrosh EA (2019). Spatial, Temporal, and Phylogenetic Scales of Microbial Ecology. Trends Microbiol. 27, 662–669. [DOI] [PubMed] [Google Scholar]
- Lamont RJ, Koo H, and Hajishengallis G (2018). The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol 16, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin SA (1992). The problem of pattern and scale in ecology. Ecology 73, 1943–1967. [Google Scholar]
- Levine JM, Bascompte J, Adler PB, and Allesina S (2017). Beyond pair-wise mechanisms of species coexistence in complex communities. Nature 546, 56–64. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, et al. (2017). Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Polz MF, Mazel F, Albright MBN, Huber JA, O’Connor MI, Ackermann M, Hahn AS, Srivastava DS, Crowe SA, et al. (2018). Function and functional redundancy in microbial systems. Nat. Ecol. Evol 2, 936–943. [DOI] [PubMed] [Google Scholar]
- Loy A, Arnold R, Tischler P, Rattei T, Wagner M, and Horn M (2008). probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ. Microbiol 10, 2894–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg JO, Weitzberg E, Cole JA, and Benjamin N (2004). Nitrate, bacteria and human health. Nat. Rev. Microbiol 2, 593–602. [DOI] [PubMed] [Google Scholar]
- Lundberg JO, Carlström M, and Weitzberg E (2018). Metabolic Effects of Dietary Nitrate in Health and Disease. Cell Metab. 28, 9–22. [DOI] [PubMed] [Google Scholar]
- Manabe M, Lim HW, Winzer M, and Loomis CA (1999). Architectural organization of filiform papillae in normal and black hairy tongue epithelium: dissection of differentiation pathways in a complex human epithelium according to their patterns of keratin expression. Arch. Dermatol 135, 177–181. [DOI] [PubMed] [Google Scholar]
- Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, and Borisy GG (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 113, E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, and Borisy GG (2017). Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 114, E9105–E9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark Welch JL, Dewhirst F, and Borisy GG (2019). Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis. Annu. Rev. Microbiol 73, 335–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P, Lewis M, Rogers H, Williams D, and Wilson M (2016). Marsh and Martin’s Oral Microbiology, Sixth Edition (Churchill Livingstone; ). [Google Scholar]
- Metsalu T, and Vilo J (2015). ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43, W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Bartosyk IM, and Dewhirst FE (1998). Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization In Methods for Studying the Genetics, Molecular Biology, Physiology, and Pathogenesis of the Streptococci, Fives-Taylor PM and LeBlanc DJ, eds. (Springer; ), pp. 223–231. [Google Scholar]
- Proctor DM, and Relman DA (2017). The Landscape Ecology and Microbiota of the Human Nose, Mouth, and Throat. Cell Host Microbe 21, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomann BH, Wiles TJ, Wall ES, Guillemin K, and Parthasarathy R (2018). Bacterial Cohesion Predicts Spatial Distribution in the Larval Zebrafish Intestine. Biophys. J 115, 2271–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, and Manganiello SD (1971). The oral microbiota of man from birth to senility. J. Periodontol 42, 485–496. [DOI] [PubMed] [Google Scholar]
- Tribble GD, Angelov N, Weltman R, Wang B-Y, Eswaran SV, Gay IC, Parthasarathy K, Dao DV, Richardson KN, Ismail NM, et al. (2019). Frequency of Tongue Cleaning Impacts the Human Tongue Microbiome Composition and Enterosalivary Circulation of Nitrate. Front. Cell. Infect. Microbiol 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MG, and Gardner RH (2015). Landscape Ecology in Theory and Practice, Second Edition (Springer-Verlag; ). [Google Scholar]
- Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, and Borisy GG (2011). Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. USA 108, 4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valm AM, Oldenbourg R, and Borisy GG (2016). Multiplexed spectral imaging of 120 different fluorescent labels. PLoS One 11, e0158495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker WR, Shepherd ES, and Sonnenburg JL (2017). Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell 169, 538–546.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz LS, Parnerkar S, and Noguera DR (2011). mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol 77, 1118–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Polz MF, and Alm EJ (2019). Interactions in self-assembled microbial communities saturate with diversity. ISME J. 13, 1602–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, and Kreth J (2012). The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell. Longev 2012, 717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmür R, and Harmsen HJ (2010). Oral biofilm architecture on natural teeth. PLoS One 5, e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository because of large image file sizes, but they are available from the corresponding author on request.


