Abstract
Objective
This study was performed to further identify the previously uncharacterized human coronavirus 229E (hCoV-229E) and human coronavirus OC43 (hCoV-OC43) in Thailand by using the RT-PCR technique. In addition, we performed this study in order to delineate the prevalence, the potential clinical impacts and evaluation of the genetic characterization of this pathogen in young children who presented with acute lower respiratory tract infections (ALRI).
Methods
We obtained nasopharyngeal secretions (NPs) from 226 children <5 years of age who were either attending the outpatient department or hospitalized with ALRI from March 2002 to July 2003. All clinical, laboratory, RT-PCR, direct sequencing and phylogenetic analysis data were collected and analyzed.
Results
Of the 226 NPs samples from infants and young children presented with ALRI, 8 (3.54%) were positive for hCoV-229E, 2 (0.88%) were positive for hCoV-OC43, and 1 (0.44%) had co-infection. The following clinical presentations were noted: fever (100%), rhinitis (44%), acute bronchiolitis (44%), viral pneumonia (33%), viral pneumonia triggering asthma exacerbation (11%) as well as viral pneumonia causing BPD exacerbation (11%). All positive samples were subjected to direct sequencing. The amino acid sequences had 82-99% similarity to previous sequences stored in the GenBank database.
Conclusion
The molecular technique we applied to detect human coronavirus appears justified as a valuable diagnostic approach to elucidate the prevalence, cause and clinical implications of ALRI among infants and young children.
Key Words: Coronavirus, Human coronavirus, Respiratory tract infection
Introduction
The viruses most frequently associated with respiratory tract infections include respiratory syncytial virus (RSV), parainfluenza virus, adenovirus and influenza virus [1, 2]. RSV has been considered the most common respiratory virus affecting infants and young children worldwide. In contrast, coronavirus has been a well-known cause of upper respiratory tract infections, occasionally a more severe lower respiratory tract illness in this patient group [2, 3]. Due to the difficulty in distinguishing between respiratory pathogens clinically and by laboratory-based analysis, studies on the impact of respiratory infections are still quite limited. Despite improved sensitivity of diagnostic techniques, the causes of a significant part of lower respiratory tract infections have as yet eluded identification [4]. The development of new diagnostic techniques like RT-PCR has facilitated identification of coronavirus infection in recent years [5]. Hospitalized infants with acute bronchiolitis and pneumonia have previously been shown to harbor coronavirus as one of a minor contributory factor [6].
Coronavirus belongs to the Coronaviridae family and contains a single molecule of linear, single-stranded RNA of positive polarity, ranging from 27.6 to 31.3 kb (100–150 nm) in size, a nucleocapsid protein (N) and a lipid envelope with three major membrane proteins. Coronaviruses are subdivided into three groups, human corona viruses 229E (hCoV-229E) and human coronavirus OC43 (hCoV-OC43) have been recognized as causes of upper respiratory tract disease including common cold and also are group 1 and 2 coronaviruses, respectively [7]. Recently, another group (hCoV, hCoV-NL63) has been reported in the Netherlands [6]. Based on the structure and function of the polymerase gene, there is a valid region for phylogenetic comparisons as well as for developing a consensus polymerase chain reaction assay applicable to find the differentiation of novel coronaviruses such as SARS [8, 9]. This might prove very useful as these novel coronaviruses have been tentatively identified by electron microscopy in association with a variety of human and animal diseases, although further characterization and definite identification of these agents as coronaviruses has been extremely difficult and insensitive. RT-PCR is currently a rapid and sensitive method for the diagnosis of hCoV [5].
Coronaviruses are mostly associated with respiratory, enteric, hepatic and central nervous system diseases. Nevertheless, other organs such as kidney, heart, and eye can also be infected [7]. Coronavirus comprises a large family of viruses infecting a broad range of vertebrates, from mammalian to avian species. In humans and fowl, coronaviruses primarily lead to upper respiratory tract infections, whereas porcine and bovine coronaviruses cause enteric infections that result in high economic loss [7].
As for the antigenic composition of coronaviruses, research of the human strains in particular has hardly advanced because the studies have been limited to cell culture or serology [10, 11, 12] and the apparently antigenic cross-reactivity between ASRA-CoV and hCoV-229E and hCoV-OC43 [13]. Thus, hCoVs are very difficult to detect and the prevalence and epidemiological data are scanty, especially in Asian countries. One recent report from Hong Kong showed that hCoV infections were also found in 4.4% (hCoV-NL63 1.5%, hCoV-OC43 0.3%, hCoV-229E 2.6%) of the children studied [14]. For isolation of suspected cases, contact tracing is only available for controlling the infection. A rapid and accurate diagnosis test is very important. Since the antibody response takes (such as SARS coronavirus) [14] 10–28 days after onset [19], detection of viral RNA by RT-PCR is likely to be the best option for early detection [6]. In this study we applied molecular biology techniques to identify hCoV in nasopharyngeal secretions (NPs) for the study on the prevalence of molecular characterization and clinical correlation of coronavirus infections in hospitalized infants and young children with acute lower respiratory tract infection (ALRI).
Materials and Methods
Samples
The study protocol was approved by the Ethics Committee of the Faculty of Medicine, Chulalongkorn University. Parents were informed as to the study objective and their written consents were obtained before specimen collection. We collected NP samples of 131 male and 95 female children at a mean age of 1.3 years (ranging from 24 days to 15 years) with ALRI attending either the outpatient clinic or admitted to King Chulalongkorn Memorial Hospital between March 2002 and July 2003. All samples were kept at −70°C until further examination. Patients' demographic data, clinical presentations, duration of hospitalization, basic laboratory data including complete blood count, chest radiographs and treatment were recorded for analysis.
Coronavirus Detection
NP specimens were centrifuged and the supernatant was analyzed for coronavirus RNA by RT-PCR. Briefly, RNA was extracted from 50 µl of NPs applying the guanidinium-isothiocyanate method as described elsewhere [15] and subsequently reverse transcribed into cDNA using primers MF3 for hCoV-OC43 and Pcon4.2 for hCoV-229E. The resulting cDNA was amplified by PCR using primer MF1 as a sense primer, and again MF3 as an antisense primer for hCoV-OC43 [16]. For a replicase gene, of hCoV-229E, Pcon3 was used as an outer sense primer, Pcon4.2 as an outer antisense primer, Pcon1 as an inner sense primer, and Pcon2.2 as an inner antisense primer base on the sequence accession No. X69721. The details of the primers are shown in table 1. The amplification reaction was performed in a thermocycler (9600 PerkinElmer Cetus, Norwalk, Conn., USA) applying the following conditions: initial denaturation at 95°C for 3 min, followed by 30 cycles each of 1 min at 95°C for denaturation, 1 min at 60°C for primer annealing, and 1 min at 72°C for extension and concluded by a final extension step at 72°C for 10 min. After electrophoresis in a 2% agarose gel stained with ethidium bromide on preparation, the expected 217-bp product for hCoV-229E and the 334-bp product for hCoV-OC43 were visualized on a UV transilluminator (Gel Doc 1000, Bio-Rad, Calif., USA). All the specimens were positive for β-actin which served as an internal control.
Table 1.
Location and sequences of the primers
| Primer | Gene | Position | Sequences 5′-3′ |
|---|---|---|---|
| hCoV-OC43 | |||
| MF1 | Matrix | 215-235 | GGCTTATGTGGCCCCTTACT |
| MF3 | Matrix | 549-530 | GGCAAATCTGCCCAAGAATA |
| hCoV-229E | |||
| Pcon2.2 | Replicase | 14245-14215 | TTGCATCACCAGAAGTTGTACCACCAGGTT |
| Pcon4.2 | Replicase | 14326-14305 | GACGAGTTAACGCTCAAAACGC |
| Pcon3 | Replicase | 13929-13948 | GCTACCAGAAATGCCACCGT |
| Pcon1 | Replicase | 14028-14056 | TGATGGGATGGGACTATCCTAAGTGTGA |
| Housekeeping gene | |||
| β-Actin F | β-Actin | 591-615 | ATGCCATCCTGCGTCTGGACCTGGC |
| β-Actin R | β-Actin | 1196-1172 | AGCATTTGCGGTGCACGATGGAGGG |
Coronavirus Sequencing
For further sequencing, the PCR product bands of interest were purified from residual agarose using the Perfect Prep Gel Cleanup Kit (Eppendorf, Westbury, N.Y., USA) according to the manufacturer's specifications and subsequently subjected to 2% agarose gel electrophoresis in order to ascertain their purity.
To determine the concentration of the amplified DNA, we measured every sample's absorption at 260 nm in a UV spectrophotometer (Shimadzu UV 160 A, Tokyo, Japan). The concentration was calculated according to the formula 1 OD 260 = 50 µg double-stranded DNA. Between 10 and 30 ng/µl (3–6 µl) of each DNA sample were subjected to cycle sequencing using 4 µl of dye terminator, 2 µl of DNA sequencing buffer (Big Dye Terminator V.3.1 Cycle Sequencing Ready Reaction, Foster City, Calif., USA) and 3.2 pmol of specific primer in a final reaction volume of 20 µl in a thermocycler (9600 PerkinElmer Cetus). This amplification round was performed according to the manufacturer's specifications, using primers Pcon1 and Pcon2.2 for hCoV-229E and MF1 and MF3 for hCoV-OC43 to amplify the particular DNA strand of interest for further sequencing. The results were analyzed by the sequence navigator program and submitted to the GenBank database (AY751810–19, AY754762). The nucleotide sequences were also compared with the sequences published previously in GenBank, applying the BLAST program (www.ncbi.nlm.nih.gov/Blast).
Phylogenetic Analysis
In order to determine the relationship between various coronaviruses isolated from the patients, we generated unrooted phylogenetic trees of nucleotide sequences comprising the polymerase gene region for hCoV-229E and the M gene for hCoV-OC43. Ten nucleotide sequences from patients and references were multiple aligned to construct a phylogenetic tree using the ClustalW package running under Bioedit Version 5.0.9 (www.mbio.ncsu.edu/BioEdit). Phylogenetic trees were made with the DNAML software of the PHYLIP Version 3.6 (http://evolution.genetics.washington.edu/phylip.html) by using 1,000 bootstraps. The obtained coronavirus sequences were aligned with those of known species stored in GenBank and subjected to phylogenetic analysis. All isolates from this cluster grouped on the same node of the phylogenetic tree were assumed to be identical.
Statistical Analysis
The clinical data were analyzed and were expressed as percentage where applicable.
Results
In the present study, virus diagnosis was performed by virus identification from NPs obtained from the 226 infants and young children with ALRI attending our hospital. For HCoV RT-PCR analysis, we used primer sets specific for the known hCoV-OC43 and hCoV-229E (table 1). Eight of 226 (3.54%) proved positive for HCoV-229E and 2 of 226 (0.88%) for HCoV-OC43, 1 (0.44%) showed co-infection.
Out of the 226 specimens, we found hCoV infection in 9 patients. The clinical diagnoses of these patients with lower respiratory tract infection included acute bronchiolitis in 4 (44%), viral pneumonia in 3 (33%), viral pneumonia triggering asthma exacerbation in 2 (22%), and viral pneumonia causing BPD exacerbation in 1 (11%). Their common clinical presentations were fever (100%), upper respiratory tract infection (44%), and severe cough (33%). Wheezing was found in 4 (44%) and crepitations were more common in 6 (66%). Their mean age was 17 ± 14.9 months. The mean hospitalization required was at 6 ± 2.24 days, significantly more than that experienced by infants and children infected with RSV. There were 6 infants (66%) co-infected with RSV. All were treated with a bronchodilator yet only 4 (44%) responded to therapy. Nobody required intensive care unit admission. All were subjected to chest radiographs, which yielded abnormal results. Most chest radiographs showed perihilar infiltrates and hyperaeration. Three (33%) had patchy infiltrates simulating bacterial infection, although 5 (55%) had been given antibiotics and all required oxygen supplement for at least 1 day after admission. A summary of the clinical data is shown in table 2.
Table 2.
Summary of the clinical data of the infants with hCoV infection
| No. sex | Age months | Hospitalization days | Corona virus type accession No. | Diagnosis | Underlying disease | Signs/symptoms | CXR findings | Antibiotic | Response to bronchodilator | RSVAg detection |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (F) | 31 | 9 | 229 E/OC43 AY751815/17 |
Viral pneumonia | None | Fever for 1 week, dry cough crepitation both lungs | Perihilar infiltrates, bilaterally | Cefotaxime | No | Neg. |
| 2 (M) | 4 | 7 | 229E AY751814 |
Acute bronchiolitis | None | Low-grade fever 3 days, clear runny nose, medium rales, subcostal retraction | Perihilar infiltrates with bilateral hyperaeration | No | No | Pos. |
| 3 (F) | 48 | 2 | 229E AY754762 |
Pneumonia/asthma exacerbation | Asthma | Fever for 2 weeks, clear runny nose, generalized wheezing | Perihilar infiltrates with bilateral hyperaeration | No | Yes | Neg. |
| 4 (M) | 24 | 5 | 229E AY751812 |
Viral pneumonia | Neuroblastoma, asthma | Fever for 1 day, intractable cough, coarse crepitation, subcostal retraction | Perihilar infiltrates with bilateral hyperaeration | Ampicillin | Yes | Pos. |
| 5 (F) | 5 | 9 | 229E AY751816 |
Acute bronchiolitis | None | Fever for 3 days, runny nose expiratory rhonchi, wheezing | Perihilar infiltrates with bilateral hyperaeration | No | Yes | Pos. |
| 6 (M) | 4 | 5 | 229E AY751811 |
Acute bronchiolitis | None | Fever, productive cough, breathing difficulty, bilateral rales, expiratory wheezing | Minimal perihilar infiltrates, small RLL infiltrate | Cefotaxime | No | Pos. |
| 7 (F) | 16 | 5 | 229E AY751813 |
Viral pneumonia | BPD/G6PD deficiency | High fever for 6 days, severe productive cough, expiratory wheezing | Minimal perihilar infiltrates, patchy RML infiltrate | Cefotaxime | No | Pos. |
| 8 (F) | 11 | 7 | 229E AY751810 |
Acute bronchiolitis | None | Fever for 2 days, clear runny nose, medium rales | Minimal perihilar thickening, hyperaeration progress to RUL infiltrate | Ampicillin | No | Pos. |
| 9 (F) | 10 | 5 | OC43 AY751818 |
Viral pneumonia, asthma, acute exacerbation | Asthma | Low-grade fever for 4 days, breathing difficulty, generalized wheezing | Bilateral perihilar infiltrates, hyperaeration | No | Yes | Pos. |
BPD = Bronchopulmonary dysplasia; RLL = right lower lobe; RML = right middle lobe; RUL = right upper lobe.
We determined the cDNA sequences of hCoV-229E and hCoV-OC43 by direct sequencing of the RT-PCR products and submitted them to GenBank. The two samples positive for hCoV-OC43, CU60TH (AY751817) and CU82TH (AY751818), showed 99% relative similarity to the reference strain AY382775. The following 8 samples were positive for hCoV-229E: CU60TH (AY751815), CU65TH (AY751814), CU71TH (AY754762), CU109TH (AY751812), CU110TH (AY751816), CU122TH (AY751811), CU126TH (AY751813) and CU129TH (AY751810) exhibiting 82% similarity to the reference strains X69721 and AY518894 (hCoV-NL).
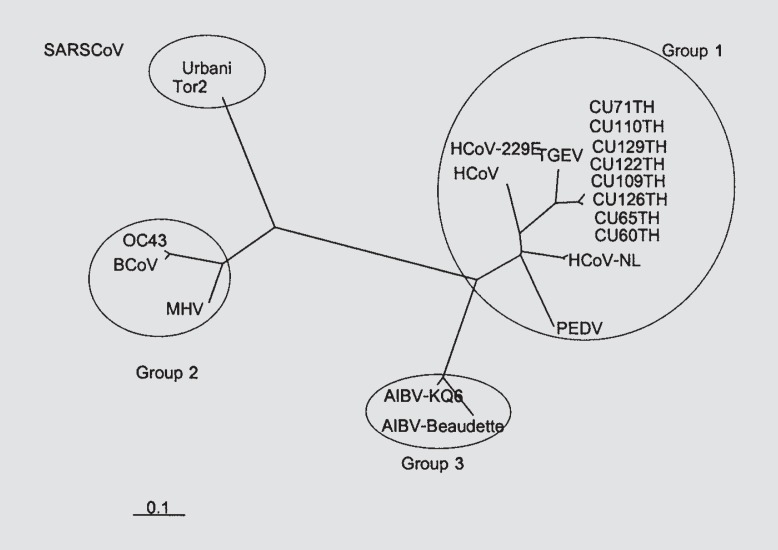
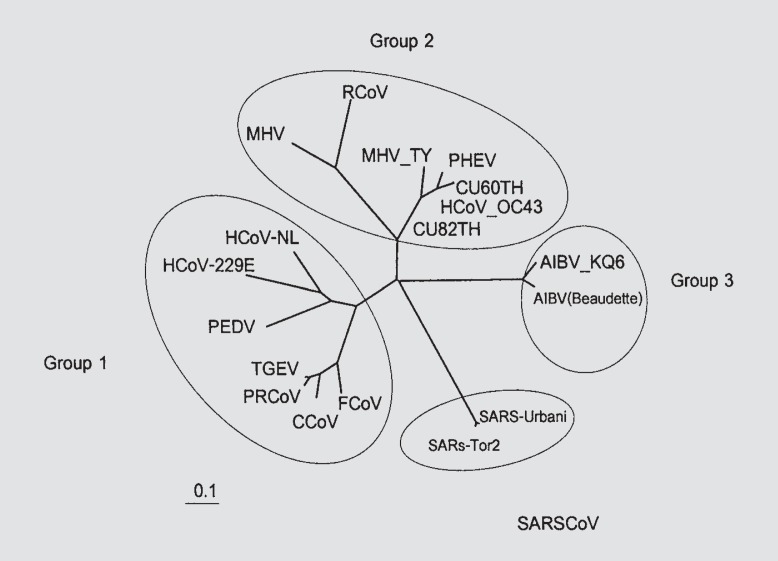
The accession numbers of references for both hCoV-229E and hCoV-OC43 to perform the phylogenetic analysis were: AY518894 (hCoV-NL), X69721 (hCoV) and AF304460 (hCoV-229E); AF353511 (porcine epidemic area virus, PEDV); AJ271965 (transmissible gastroenteritis virus, TGEV); AF220295 (bovine coronavirus, BCoV); NC_005147 (OC43) and AY382775 (hCoV-OC43); AF201929 (MHV) and AF190407 (MHV-TY); AF207551 (rat coronavirus, RCoV); AY078417 (porcine hemagglutinating encephalomyelitis virus, PHEV); D13096 (canine coronavirus, CCoV); AY204704 (feline coronavirus, FCoV); Z24675 (porcine respiratory coronavirus, PRCoV); M95169 (AIBV-Beaudette) and AY641576 (AIBV-KQ6); NC_004718 (SARS-Tor2) and AY278741 (SARS-Urbani). The hCoV-229E (AY751810–16, AY754762) and hCoV-OC43 (AY751817–18) sequences obtained in our study were aligned with those of known genotype stored in GenBank and subjected to phylogenetic analysis, shown along with the hCoV-229E and hCoV-OC43 in different geographical areas in figures 1 (hCoV-229E) and 2 (hCoV-OC43).
Fig. 1.
The phylogenetic analysis of HCoV-229E polymerase gene sequences in our study (initiated with CU) aligned with the known coronavirus sequences from GenBank.
Fig. 2.
The phylogenetic analysis of HCoV-OC43 matrix protein sequences in our study (initiated with CU) aligned with the known coronavirus sequences from GenBank.
Discussion
ALRIs like acute bronchiolitis and viral pneumonia are major causes of morbidity in infants and young children <5 years of age. The majority of infections are caused by four groups of respiratory virus: RSV, parainfluenza viruses, influenza viruses and adenoviruses. In the past, virus detection was markedly limited due to technical obstacles and difficulties in virus isolation. However, owing to advances in molecular diagnostics in recent years, we are able to identify newly and deadly emerging respiratory pathogens such as SARS which is also caused by coronavirus infection. Thus, we realized the necessity to further explore these potential respiratory pathogens in more detail.
Coronaviruses have been divided into three groups or serotypes, based on their distinct genome organization and phylogenetic analyses [6, 7]. Group 1 comprises hCoV-229E, porcine enteric gastroenteritis virus, TGEV, PEDV and respiratory (PRCoV) coronavirus, canine coronavirus and feline coronavirus; group 2 consists of hCoV-OC43, bovine coronavirus, turkey coronavirus, murine coronaviruses, porcine hemagglutinating encephalomyelitis virus, rat coronavirus, and group 3 comprises avian coronavirus including infectious bronchitis virus, turkey coronavirus, rabbit coronavirus and the SARS coronavirus as a distinct entity located on a different node on the phylogenetic tree.
In this study, we have identified human coronavirus hCoV-OC43 and hCoV-229E infection in infants and young children presenting with ALRI. We found the human coronavirus-229E being predominant in our population. Only one sample contained both. hCoV-229E and hCoV-OC43 have previously been proven responsible for infecting people of all age groups and causing severe lower respiratory tract infection primarily in frail patients such as young children and elderly individuals [17, 18, 19].
Human coronavirus hCoV causes approximately 10–20% of common colds and 6% of influenza-like illnesses in all age groups [20]. The clinical features associated with coronavirus infection appear to be similar to those observed with other respiratory viruses, such as RSV, parainfluenza virus and human metapneumovirus. Symptoms of upper respiratory tract infection such as rhinitis, coryza, and of lower respiratory tract infection such as tachypnea, wheezing and chest wall retraction were frequently observed in our patients. Wheezing was observed in 44% compared to 9% reported from Hong Kong [14]. This may be explained by the difference in host response. Hypoxemia was present without exception. Thus, this may indicate yet another potentially severe effect of this virus infection and future impact on healthcare providers. Additionally, chest hyperinflation, which reflects small to medium airway obstruction caused by airway inflammation, has proven a relatively common finding in our population (>50°/o).
Co-infection with RSV and other pathogens has been previously described. Interestingly, we found 6 (66%) patients co-infected with RSV (table 1). Hence, RSV and human coronavirus might share a similar seasonal pattern. Nevertheless, confirmation of the underlying mechanism of co-infection will require further longitudinal studies conducted over several bronchiolitis seasons.
The role of viral respiratory tract infections in triggering reactive airway exacerbation, especially in asthma patients, has been the subject of much research interest. RSV, parainfluenza and rhinoviruses have previously been recognized as a primary trigger of reactive airway exacerbation. Our data indicate that human coronavirus may also be important in this regard, although its role in the pathogenesis of wheezing still warrants further study. More importantly, the protection from this virus may reduce the cost of hospitalization and reduce the morbidity of these children. The hCoV-229E sequences obtained from our samples showed 82% similarity to previously published data. Hence, the availability of the hCoV, hCoV-NL and our genome sequences might contribute to the development of a variety of diagnostic assays to determine both prevalence and clinical impact of hCoV infections in humans. Moreover, further studies conducted on the remaining genes would be required in order to arrive at more specific diagnoses and elucidate the related clinical implications.
Acknowledgement
We are grateful to the Thailand Research Fund, Senior Research Scholar and Center of Excellence in Viral Hepatitis Fund, Chulalongkorn University. Also, we would like to express our gratitude to the entire staff of the Center of Excellence in Viral Hepatitis, Chulalongkorn University, for their efforts in contributing to the present study. We would also like to thank Venerable Dr. Mettanando Bhikkhu of The Foundation of King Rama IX the Great and Ms. Petra Hirsch for reviewing the manuscript.
References
- 1.Bulla A, Hitze KL. Acute respiratory infections: a review. Bull World Health Organ. 1978;56:481–498. [PMC free article] [PubMed] [Google Scholar]
- 2.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis. 2003;15:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gagneur A, Sizun J, Vallet S, Legr MC, Picard B, Talbot PJ. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J Hosp Infect. 2002;51:59–64. doi: 10.1053/jhin.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA. 2000;283:499–505. doi: 10.1001/jama.283.4.499. [DOI] [PubMed] [Google Scholar]
- 5.Myint S, Johnston S, Sanderson G, Simpson H. Evaluation of nested polymerase chain methods for the detection of human coronaviruses 229E and OC43. Mol Cell Probes. 1994;8:357–364. doi: 10.1006/mcpr.1994.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, Jong JC, Simon JH, Osterhaus ADME. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enjuanes L, Sola I, Almazan F, Ortego J, Izeta A, Gonzalex JM, Alonso S, Sanchez JM, Escors D, Calvo E, Riquelme C, Sanchez C. Coronavirus derived expression systems. J Biotechnol. 2001;88:183–204. doi: 10.1016/S0168-1656(01)00281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drosten C, Gunther S, Preiser W, Werf SVD, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM, Berger A, Burguiere AM, Chinatl J, Eickmann M, Escrio N, Grywna K, Kramme S, Manguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus ADME, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 9.Ksiazek TG, Erdman D, Goldsmith CH, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey C, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ; SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 10.Gill EP, Dominguez EA, Greenberg SB, Atmar RL, Hogue BG, Baxter BD, Couch RB. Development and application of an enzyme immunoassay for coronavirus OC43 antibody in acute respiratory illness. J Clin Microbiol. 1994;32:2372–2376. doi: 10.1128/jcm.32.10.2372-2376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradburne AF. Antigenic relationships amongst coronaviruses. Arch Gesamte Virusforsch. 1970;31:352–364. doi: 10.1007/BF01253769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hierholzer JC. Purification and biophysical properties of human coronavirus 229E. Virology. 1976;75:155–165. doi: 10.1016/0042-6822(76)90014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Che XY, Qiu LW, Liao ZY, Wang YD, Wen K, Pan YX, Hao W, Mei YB, Cheng VC, Yuen KY. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu SS, Chan KH, Chu KW, Kwan SW, Guan Y, Poon LL, Peiris JS. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thanasugarn W, Samransamruajkit R, Vanapongtipagorn P, Prapphal N, Van den Hoogen B, Osterhaus AD, Poovorawan Y. Human metapneumovirus infection in Thai children. Scand J Infect Dis. 2003;35:754–756. doi: 10.1080/00365540310000094. [DOI] [PubMed] [Google Scholar]
- 16.Vabret A, Mouthon F, Mourez T, Gouarin S, Petitjean J, Freymuth F. Direct diagnosis of human respiratory coronaviruses 229E and OC43 by the polymerase chain reaction. J Virol Methods. 2001;97:59–66. doi: 10.1016/S0166-0934(01)00343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falsey AR, Walsh EE, Hayden FG. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J Infect Dis. 2002;185:1338–1341. doi: 10.1086/339881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIntosh K, Chao RK, Krause HE, Wasil R, Mocega HE, Mufson MA. Coronavirus infection in acute lower respiratory tract disease of infants. J Infect Dis. 1974;130:502–507. doi: 10.1093/infdis/130.5.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treanor J, Falsey A. Respiratory viral infections in the elderly. Antiviral Res. 1999;44:79–102. doi: 10.1016/S0166-3542(99)00062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lina B, Valette M, Foray S, Lucian J, Stagnara J, See DM, Aymard M. Surveillance of community-acquired viral infections due to respiratory viruses in Rhône-Alpes (France) during winter 1994 to 1995. J Clin Microbiol. 1996;34:3007–3011. doi: 10.1128/jcm.34.12.3007-3011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]