Abstract
Interferon (IFN)-α, -β and -γ have been shown to be only marginally effective against severe acute respiratory syndrome coronavirus (SARS-CoV) replication in Vero cell lines. We investigated the combination of type I IFNs (IFN-α or -β) and IFN-γ for antiviral activity and found that such combinations synergistically inhibited SARS-CoV replication in Vero cells, using yield reduction assay and the isobologram and combination index methods of Chou and Talalay for evaluation. The highly synergistic anti-SARS-CoV action of type I IFNs and IFN-γ parallels the marked increase in 2′-5′-oligoadenylate synthetase and p56 mRNAs following exposure in Vero cells to either IFN-α or -β and IFN-γ compared with the transcriptional levels obtained after stimulation with either IFN alone. These results demonstrate that SARS-CoV, although only moderately sensitive to the antiviral action of the individual types of IFN, is highly sensitive to a combination of type I and II IFNs, which suggests that such combinations may have potential in the treatment of SARS-CoV infections.
Key Words: Interferon, Severe acute respiratory syndrome coronavirus, 2′-5′-Oligoadenylate synthetase, p56
Severe acute respiratory syndrome (SARS) is a life-threatening disease caused by a new member of a diverse group of large, enveloped, positive-strand RNA viruses of the family Coronaviridae (SARS-CoV) [1]. Although in the 2 years since the discovery of SARS much has been learnt about its pathogenesis, epidemiology and laboratory diagnosis, progress has been less rapid in other areas, particularly in establishing an antiviral treatment for patients. Steroids with or without ribavirin have been widely employed in the treatment of SARS but have not been very effective [2, 3]. However, several studies have reported that type I interferons (IFNs) can inhibit SARS replication in vitro [4] and, specifically, that IFN-β exhibits the most potent anti-SARS-CoV activity in Vero cells, showing levels of inhibition at concentrations of 1,000 IU/ml or greater [5, 6, 7, 8]. IFN-α has been tested in non-human primates experimentally infected with SARS, but the actual therapeutic efficacy of post-exposure treatment with pegylated IFN-α has yet to be established [9]. With regard to the antiviral activity of IFNs, we have previously reported that simultaneous treatment with IFN-β, at various concentrations, and 100 IU/ml of IFN-γ acted synergistically against SARS-CoV replication in Vero cells [10]. Similar synergistic antiviral activity of IFN-β and IFN-γ against SARS-CoV has also been demonstrated by others [11] under different experimental conditions, namely using higher doses of IFN-γ in combination with IFN-β. However, recently, Paragas et al. [12] have reported that a combination of IFN-α consensus 1 and IFN-γ did not exhibit any synergistic antiviral activity against SARS-CoV.
Because of the importance of this issue and in support of the search for effective anti-SARS-CoV treatment, our previous study was extended to evaluate the in vitro antiviral activity of IFN-α or -β both alone and in combination with IFN-γ against SARS-CoV infection. Our specific goal was to determine whether combinations of both IFNs would produce enhanced antiviral effects that could, indirectly, suggest potential combination IFN therapy strategies for the treatment of SARS-CoV. The effect of combinations of both IFNs against SARS-CoV were analysed by yield reduction assay. Specifically, African green monkey kidney epithelial cells (Vero cells) were seeded into 96-well plates (2 × 104 cells per well) and cultured overnight. Triplicate cell cultures were treated with concentrations (1-100,000 IU/ml) of recombinant IFN-β (rIFN-β1a, specific activity 270 MIU/mg; Rebif®, Ares-Serono, Basel, Switzerland) or natural IFN-α (nIFN-α, leukocyte IFN, specific activity 200 MIU/mg; Alfaferone®, Alfa-Wasserman, Bologna, Italy), either alone or in combination with rIFN-γ (specific activity 20 MIU/mg; Imukin®, Boehringer Ingelheim, Reggello-Firenze, Italy) 24 h before infection with SARS-CoV [multiplicity of infection = 0.1 TCID50/cell (50% tissue culture infectious dose)]. Infection with SARS-CoV (HSR1 strain) [13] was allowed to develop for 72 h, depending on the time required for specific cytopathic effects to become clearly visible by optical microscopy [10, 13]. Culture supernatants were then collected and titration of SARS-CoV was performed in Vero cells by determination of the TCID50 using the method of Reed and Muench [14].
As expected (table 1), exogenously added IFN-α, -β and -γ exhibited only a minor inhibitory effect on the yield of infectious SARS-CoV in Vero cells when used alone. Values of 50% inhibitory concentration (IC50) ranged from 400 IU/m (IFN-β) to 3,700 IU/ml (IFN-α). However, when different concentrations of IFN-α or -β were used in combination with different concentrations of IFN-γ, inhibition of the virus yield was more pronounced. Indeed, the IC50 values decreased markedly
Table 1.
Effect of the combination of type I IFNs (IFN-α or -β) and IFN-γ on infectious virus yield in Vero cells infected with HSR1 strain
| IC50 of type I IFNs obtained in combination with fixed concentrations of IFN-γ | IC50 of IFN-γ obtained in combination with fixed concentrations of type I IFNs (IFN-α or -β) | ||||
|---|---|---|---|---|---|
| IFN-γ | IFN-αa | IFN-βa | type I IFNs | IFN-γb | IFN-γc |
| IU/ml | IC50, IU/ml | IC50, IU/ml | IU/ml | IC50, IU/ml | IC50, IU/ml |
| 0 | 3,700 ± 650 | 400 ± 50 | 0 | 1,230 ± 300 | 1,230 ± 300 |
| 4 | 250 ± 80 | 74 ± 16 | 4 | 110 ± 21 | 100 ± 11 |
| 12 | 100 ±15 | 10 ± 2 | 12 | 80 ± 12 | 12 ± 3.4 |
| 37 | 40 ± 10 | 8.4 ± 1.3 | 37 | 50 ± 6.2 | 8.7 ± 2.0 |
| 111 | 8.3 ± 3.0 | 4.0 ± 1.3 | 111 | 12 ± 3.4 | 4.0 ± 1.1 |
| 333 | 2.1 ± 0.7 | 1.3 ± 0.8 | 333 | 4.1 ± 2.2 | 4.2 ± 1.3 |
| 1,000 | 1 ± 0.3 | <1 | 1,000 | 4.4 ± 3.4 | 2.5 ± 1.2 |
All data represent means ± standard deviations of three separate experiments.
p < 0.05 as determined by Student's t test, comparison of IC50 of type I IFN and IFN-γ combinations to IFN alone.
Vero cells were treated with a fixed concentration of IFN-7 and a different concentration of IFN-α or -β and infected with SARS-CoV HSR1 (multiplicity of infection 0.1).
when the different IFNs were used in combination. Specifically, the IC50 values for SARS-CoV yield for IFN-α in combination with IFN-γ were up to 1.2 and 3.6 log fold lower compared with the IC50 value for treatment with IFN-a alone. The IC50 values for IFN-γ showed a 1.0-2.5 log fold decrease when used in combination with IFN-α relative to the value recorded for IFN-γ alone. Interestingly, the synergistic antiviral effect of the combination of IFN-α or -β with IFN-γ was also observed with low concentrations of IFN-γ (<10 IU/ml).
As expected, these results (table 1) also confirmed our previous findings on the synergistic effects of IFN-β and IFN-γ [10]. Here, the extended experiments and statistical analysis clearly show that the concentrations of IFN-β or IFN-γ required to inhibit 50% of viral yield production were significantly decreased when used in combination compared with single IFN-β or IFN-γ treatments (p < 0.05).
A detailed analysis of the nature of antiviral interaction between type I and II IFNs on the replication of SARS-CoV was undertaken using the isobologram and combination index (CI) method of Chou and Talalay [15]. The general equation for the classic isobologram is given as follows:
CI = (D)1/(Dx)1 + (D)2/(Dx)2 + α(D)1(D)2/(Dx)1(Dx)2
where (Dx)1 and (Dx)2 are the concentrations for IFN-α or -β and IFN-γ alone that give x percent inhibition of SARS-CoV replication in Vero cells, whereas (D)1 and (D)2 are the concentrations of IFN-α or -β and IFN-γ that also inhibited x percent of SARS-CoV replication in the same cells. CI <1, CI = 1, and CI >1 indicate synergism, additive effect and antagonism, respectively. The CI values obtained from the mutually non-exclusive (α = 1) isobologram equation is presented in this study. Calcusyn software (Biosoft, Ferguson, Mo., USA) was used for data analysis.
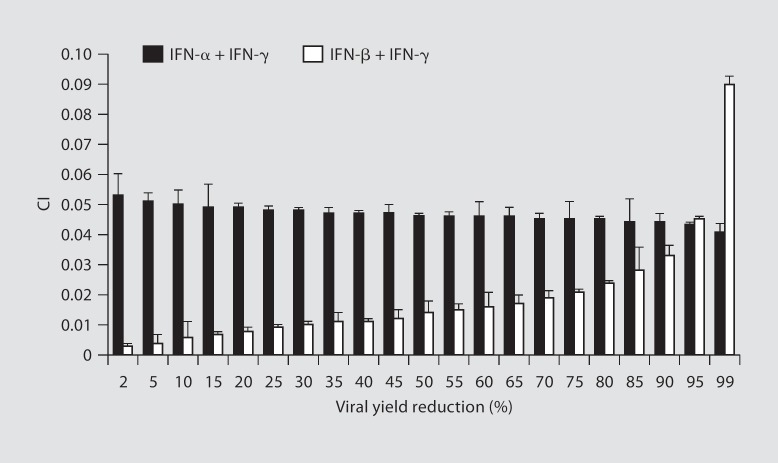
Isobolograms were constructed for viral yield reduction values ranging from 5 to 95%. Experimental combination therapy data points plotted well below the expected additive line at each viral yield reduction value for type I IFN (IFN-α or -β) plus IFN-γ combinations, indicating a strong synergism across a broad range of doses (data not shown). The CI values for the interaction between type I IFNs (IFN-α or -β) and IFN-γ were <1 over the entire range of viral yield reduction values tested (2-99%), indicating a strong synergism (fig. 1).
Fig. 1.
CI for type I IFN (IFN-α/IFN-β) and IFN-γ combinations as function of infectious virus yield in Vero cells infected with SARS-CoV HSR1 strain. CIs were calculated through the Calcusyn software (Biosoft) which uses the CI isobologram method of Chou and Talalay [15]. The constant ratio combination design was applied to asses the effect of IFN-α or -β in combination with IFN-γ against SARS-CoV replication in Vero cells. Interpretation of CI values in quantifying two drug antiviral interactions: CI = 1, additive; CI>1, antagonism; CI <1, synergism; CI<0.1, strong synergism. CI values are the means of three independent experiments.
The mechanism by which type I and II IFNs synergise to inhibit SARS-CoV is as yet unclear. Previous studies have suggested that MxA, a type-I-induced protein, is not the critical factor that mediates inhibition of SARS-CoV, despite its high-level induction by IFN-β as well as by a mixture of IFN-β and IFN-γ [10, 16]. Hence, in order to discover a possible mechanism responsible for the observed synergism between type I and II IFNs, we examined the expression of two other well-known IFN-inducible proteins: 2′-5′-oligoadenylate synthetase [17] and p56 (also known as IFIT1) [18]. The mRNA copy content of their genes was measured in untreated versus IFN-treated Vero cells by a real-time 5′-exonuclease reverse transcription polymerase chain reaction (TaqMan®) assay using the ABI 7700 sequence detector (Applied Biosystems, Monza, Italy). Briefly, total cellular RNA was extracted from 5 × 106 Vero cells using phenol and guanidine isothiocyanate reagent (Trizol®, Gibco BRL, Grand Island, N.Y., USA), following the manufacturer's instructions, and was transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems). Next, the following primer pair and probes were added to the Universal PCR Master Mix (Applied Biosystems) at 300 and 100 nm, respectively, in a final volume of 50 μl: 2′-5′-oligoadenylate synthetase [19] forward 5′-TCAG-CGAGGCCAGTAATCTTG-3′; reverse 5′-TCAGCCA-TTGCCAGCATATTT-3′; probe 6-carboxyfluorescein (FAM)-5′-TCCAGTTGACCCAACCAATAATGT-GAGTGG-3′-6-carboxy-tetramethyl rhodamine (TAMRA); p56 forward 5′-TGAAGAAGCTCTAGCCAACATGTC-3′; reverse 5′-GAGCTTTATCCACAGAGCCTTTTC-3′; probe 6′-FAM-5′-TATGTCTTTCGATATGCAGCCAAGTTTTACCG-3′-TAMRA). Co-amplification of the hypoxanthine phosphoribosyltransferase mRNA housekeeping gene (TaqMan endogenous controls, No. 4333768F, Applied Biosystems) was used to normalise the amount of total RNA present. Considering that the different IFN preparations have different specific activities (IU/mg protein), Vero cells were exposed to 5 ng/ml of either IFN-α (1,000 IU/ml), IFN-β (1,350 IU/ml), IFN-γ (100 IU/ml) or both type I IFNs (IFN-α/IFN-β) and IFN-γ (5 ng/ml each) for 18-20 h.
The results show that all IFNs tested induced detectable amounts of 2′-5′-oligoadenylate synthetase and p56 mRNAs (fig. 2). Interestingly, the 2′-5′-oligoadenylate synthetase and p56 transcriptional levels increased significantly after stimulation of the Vero cells by both IFN-α or -β and IFN-γ, compared with the transcriptional levels obtained after stimulation with either IFN alone.
Fig. 2.
2′-5′-Oligoadenylate synthetase and p56 are up-regulated in Vero cells following treatment with type I IFN and IFN-γ. Vero cells were exposed to either IFN-α, IFN-β, IFN-γ or both type I IFNs (IFN-α/IFN-β) and IFN-γ for 18–20 h. Cells were harvested and RNA isolated 18–20 h after exposure. TaqMan reverse transcriptase polymerase chain reaction was used to quantify 2′-5′-oligoadenylate synthetase (OAS) and p56 mRNA expression. Data, normalised to hypoxanthine phosphoribosyltransferase mRNA, were calculated by using the arithmetic formula 2 – ΔΔCt according to the supplier's guidelines. * p < 0.05 compared with fold change of 2′-5′-oligoadenylate synthetase and p56 after exposure of Vero cells to IFN-α, IFN-β or IFN-γ separately using Student's t test.
It is tempting to speculate that the increased expression of 2′-5′-oligoadenylate synthetase and p56, when a combination of both types of IFN is used, could be due to a synergistic interaction between type I and II IFNs through the induction of the IFN-stimulated gene factor 3 and type I receptors [20, 21]. Such a possibility should be addressed by performing further and more focused studies; at present, no definite conclusions can be drawn on the mechanism underlying the strong synergism recorded here.
In summary, our data extending and accomplishing previous results demonstrate that the combination of IFN-α or -β with IFN-γ produced significantly enhanced antiviral activity against SARS-CoV infection compared with any individual IFN, and this synergism parallels the increase in expression of 2′-5′-oligoadenylate synthetase and p56 mRNAs. It should be emphasised that our in vitro study has thus far been performed only in Vero cells. We were unable to obtain viral progeny from other human cell lines, including Caco and Huh7, employed by others [5, 22, 23]; thus, we were unable to assess the activity of the various IFNs in a proper human system. Further, no firm conclusions or extrapolations can be drawn from the IFN inhibitory concentration values obtained with regard to potential therapeutic concentrations of IFNs that might be expected to be required in a clinical situation. Nevertheless, the findings suggest that further investigation may be warranted to determine the role of IFN-α or -β plus IFN-γ as a therapeutic strategy in the treatment of SARS-CoV infections.
Acknowledgement
This work was supported in part by a grant to G.A. from University La Sapienza (Fondi Ateneo quota 60%).
References
- 1.Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:88–97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsang K, Seto WH. Severe acute respiratory syndrome: scientific and anecdotal evidence for drug treatment. Curr Opin Investig Drugs. 2004;5:179–185. [PubMed] [Google Scholar]
- 3.Cinatl J, Jr, Michaelis M, Hoever G, Preiser W, Doerr HW. Development of antiviral therapy for severe acute respiratory syndrome. Antiviral Res. 2005;66:81–97. doi: 10.1016/j.antiviral.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cinatl J, Jr, Michaelis M, Scholz M, Doerr HW. Role of interferons in the treatment of severe acute respiratory syndrome. Expert Opin Biol Ther. 2004;4:827–836. doi: 10.1517/14712598.4.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362:748. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahl H, Linde A, Strannegard O. In vitro inhibition of SARS virus replication by human interferons. Scand J Infect Dis. 2004;36:829–831. doi: 10.1080/00365540410021144. [DOI] [PubMed] [Google Scholar]
- 7.Hensley LE, Fritz LE, Jahrling PB, Karp CL, Huggins JW, Geisbert TW. Interferon-β1a and SARS coronavirus replication. Emerg Infect Dis. 2004;10:317–319. doi: 10.3201/eid1002.030482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng B, He ML, Wong KL, Lum CT, Poon LL, Peng Y, Guan Y, Lin MC, Kung HF. Potent inhibition of SARS-associated coronavirus (SCoV) infection and replication by type I interferons (IFN-α/β) but not by type II interferon (IFN-γ) J Interferon Cytokine Res. 2004;24:388–390. doi: 10.1089/1079990041535610. [DOI] [PubMed] [Google Scholar]
- 9.Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, van Amerongen G, van Riel D, de Jong T, Itamura S, Chan KH, Tashiro M, Osterhaus AD. Pegylated interferon-α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scagnolari C, Vicenzi E, Bellomi F, Stillitano MG, Pinna D, Poli G, Clementi M, Dianzani F, Antonelli G. Increased sensitivity of SARS-coronavirus to a combination of human type I and type II interferons. Antivir Ther. 2004;9:1003–1011. [PubMed] [Google Scholar]
- 11.Sainz B, Jr, Mossel EC, Peters CJ, Garry RF. Interferon-β and interferon-γ synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paragas J, Blatt LM, Hartmann C, Huggins JW, Endy TP. Interferon-αcon1 is an inhibitor of SARS-corona virus in cell-based models. Antiviral Res. 2005;66:99–102. doi: 10.1016/j.antiviral.2005.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicenzi E, Canducci F, Pinna D, Mancini N, Carletti S, Lazzarin A, Bordignon C, Poli G, Clementi M. Coronaviridae and SARS-associated coronavirus strain HSR1. Emerg Infect Dis. 2004;10:413–418. doi: 10.3201/eid1003.030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed LJ, Muench H. A simple method of estimating fifty percent end-points. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- 15.Chou TC, Talalay YP. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 16.Spiegel M, Pichlmair A, Muhlberger E, Haller O, Weber F. The antiviral effect of interferon-β against SARS-coronavirus is not mediated by MxA protein. J Clin Virol. 2004;30:211–213. doi: 10.1016/j.jcv.2003.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebouillat D, Hovanessian AG. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J Interferon Cytokine Res. 1999;19:295–308. doi: 10.1089/107999099313992. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar SN, Sen GC. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Luo S, Cassidy W, Jeffers L, Reddy KR, Bruno C, Howell CD. Interferon-stimulated gene expression in black and white hepatitis C patients during peginterferon α-2a combination therapy. Clin Gastroenterol Hepatol. 2005;3:499–506. doi: 10.1016/s1542-3565(04)00615-9. [DOI] [PubMed] [Google Scholar]
- 20.Levy DE, Lew DJ, Decker T, Kessler DS, Darnell JE., Jr Synergistic interaction between interferon-α and interferon-γ through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizukoshi E, Kaneko S, Yanagi M, Ohno H, Matsushita E, Kobayashi K. Upregulation of type I interferon receptor by IFN-γ. J Interferon Cytokine Res. 1999;19:1019–1023. doi: 10.1089/107999099313235. [DOI] [PubMed] [Google Scholar]
- 22.Tang BS, Chan KH, Cheng VC, Woo PC, Lau SK, Lam CC, Chan TL, Wu AK, Hung IF, Leung SY, Yuen KY. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J Virol. 2005;79:6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgenstern B, Michaelis M, Baer PC, Doerr HW, Cinatl J., Jr Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem Biophys Res Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]