Abstract
Biochemical studies on human coronavirus, strain 229E, indicate that the RNA is present in the virion in association with protein as a ribonucleoprotein (RNP) complex. Morphological studies have revealed that this nucleocapsid is probably a continuous helical RNP.
Key Words: Coronavirus; Human coronavirus, strain 229E; Ribonucleoprotein; Helical or linear nucleocapsid of human coronavirus
KENNEDY/JOHNSON-LUSSKNBURG
The coronaviruses comprise a recently described virus group which shares many of the characteristics listed above [7, 8]. Although the external morphology of the virion is well defined, little is known about the internal organization of the particle. Apostolov et al. [9] reported evidence of a threadlike internal structure in thin sections of avian infectious bronchitis virus (IBV) but, as yet, there are no confirmatory data from negative staining or from isolation of internal structures. In fact, there is little agreement as to how the nucleic acid is packaged within the envelope; several interpretations of structure have been advanced based upon data from different laboratories [10].
Using a method based on that used by Davis and Rueckert [11] for the isolation of Rous sarcoma virus (RSV) nucleoprotein, we have isolated a ribonucleoprotein (RNP) complex from a human coronavirus, strain 229E (HCV/229E), one of the etiological agents of the common cold. Electron microscope examination of both spontaneously disrupting virions and isolated RNP has revealed structures which appear linear, suggesting a helical conformation.
Materials and Methods
Preparation of the virus. HCV/229E obtained through the courtesy of Dr. A.Z. Kapikian, National Institutes of Health, Bethesda, Md., was grown in monolayer cultures of L 132 cells [12], obtained from D. McLeod, Laboratory Centre for Disease Control, Ottawa, Canada. Virus inoculum was stored at-70° as infected cell cultures and subjected to three freeze-thaw cycles immediately prior to use. The cell monolayers were infected at a multiplicity of 3-5 PFU/cell and, following an adsorption period of 1 h at room temperature, were incubated at 33 for 40 h in medium 199 (M 199; Grand Island Biological Company, New York, N.Y.) with 0.2% bovine serum albumin (BSA).
Plaque assay. Virus was titrated by plaque assay in monolayers of L 132 cells in 75-cm2 disposable culture flasks (Falcon Plastics). 0.33-ml aliquots of appropriate virus dilutions were inoculated and allowed to adsorb for 1 h at room temperature, taking care to ensure even spreading. At the end of the adsorption period, the monolayers were overlaid with 30 ml per flask of M 199 containing 0.2% NaHCO3: 5-bromodeoxyuridinc (BUDR), 50 μg/ml; DEAE-dextran, 200 μg/ml; 2% fetal calf serum; antibiotics; and 0.6% Oxoid Agar No. 1. Following incubation at 33 for 6 days, the cells were fixed by the addition of formol-saline, the agar was removed and the cells were stained with crystal violet (approximately 1% in H2O) for plaque counting. DEAE-dextran [13) or both DEAE-dextran and BUDR [14] have been used in plaque assay overlay media for assay of HCV/229E, and using a different formulation we have confirmed that both contribute considerably to optimal plaque development [15].
Preparation of labelled virus. Cells were infected as above. At the end of the adsorption period, Eagle's MEM with 0.2% BSA was added to the monolayer and cither uridine-5-3H (10 μCi/ml; spec. act. = 28 Ci/mmol) alone or both uridine-5-3H (10 μCi/ml) and 14C-amino acid mixture (NEC 445, New England Nuclear), I μCi/ml, were included in the maintenance medium. Following concentration and purification, radioactivity was assayed by counting suitable aliquots (up to 500 μl) in a cocktail composed of BRS-3, 100 ml: butyl-PBD, 8 g (both from Beckman Instruments Inc.); water, 50 ml: and toluene to 1 liter using a Beckman LS-250 scintillation counter. Radiochemicals were obtained from New England Nuclear, Montreal, Canada.
Virus purification and concentration. HCV/229E was harvested by subjecting the infected cultures to three freeze-thaw cycles to release virus and produce a cell lysate suspension. The lysate was clarified by centrifugation at low speed (IEC PRJ centrifuge, 2,000 rpm, for 20 min at 4°), after which the virus was spun down onto a 65% w/v sucrose cushion in a Spinco SW25.2 rotor at 20,000 rpm (48,000 g) for 60 min at 10°. 4 ml/tube of interface material was collected via tube bottom puncture, pooled, washed free of sucrose and concentrated in an Amicon ultrafiltration cell with an XM300 membrane using 0.001 m phosphate buffer, pH 7.2. The resulting virus concentrate, now in phosphate buffer, was partially purified by rate-zonal centrifugation through a preformed 10-35% w/w sucrose gradient in a Spinco SW25.2 rotor at 22,500 rpm (63,000 g) for 90 min at 10°. 2-ml samples were collected by tube bottom puncture, and virus-containing fractions located by assay of radioactivity were pooled, washed and concentrated by ultrafiltration as before.
Disruption of virus. Approximately 8 × 1010 PFU of 3H-labelled HCV/229E in 1 ml of 0.001 m phosphate buffer, pH 7.2, was mixed in an ice bath with an equal volume of a mixture containing 0.5% Nonidet P-40 (NP-40) (Shell Chemicals) and 2% dithiothreitol (DTT) in Tris-EDTA buffer (0.02 m Tris, 0.002 m EDTA, pH 7.2). The mixture was immediately layered (0.5 ml/tube) onto linear 25-75% w/w sucrose gradients containing 0.4% DTT and Tris-EDTA buffer, and centrifuged in a Spinco SW4I rotor at 37,000 rpm (170,000 g) for 16-22 h at 10°. Untreated labelled virus was mixed with an equal volume of 0.001 m phosphate buffer and run in phosphate-buffered sucrose gradients in parallel to serve as control. 0.5-ml fractions were then collected by lube bottom puncture and samples were assayed by liquid scintillation counting (LSC) using the cocktail described above.
Electron microscopy. All preparations were negatively stained following standard procedures [16]. Using finely drawn Pasteur pipets, virus preparations were placed directly onto carbon-stabilized formvar-coated grids and allowed to adsorb for 1 min. Excess sample was drained by touching filter paper to the edge of the grid, and the grid was usually then washed twice with distilled water, again draining excess with filter paper. The grid was stained with 2% sodium phosphotungstate, pH 6.5, for 30 or 60 sec, drained and allowed to air-dry. If grids were not examined immediately, they were stored in a desiccator until required. Grids were made directly from gradient fractions in sucrose or, more commonly, the fractions were diluted in 0.001 m phosphate buffer and centrifuged at 91,000 g for 60 min. Resulting pellets were resuspended in distilled water and grids immediately prepared by the above procedure. All grids were examined in a Philips EM 300 electron microscope.
Results
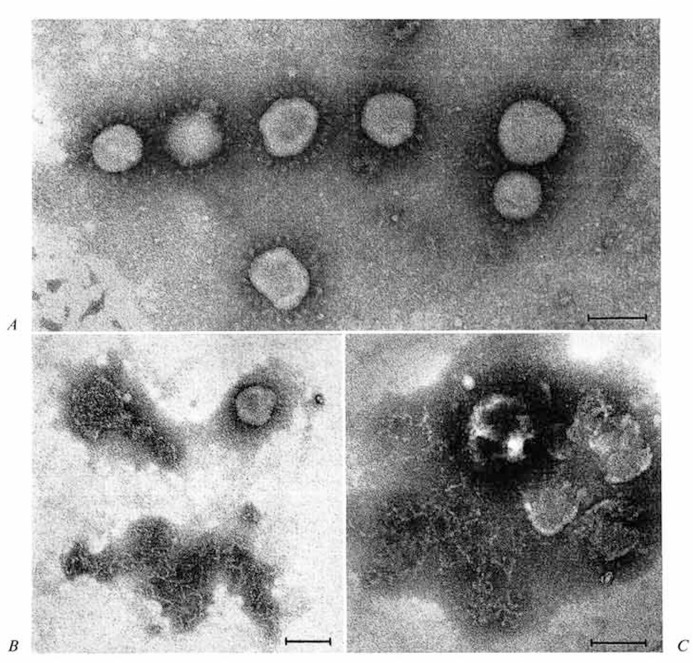
Morphology of spontaneously disrupting virions. Yields of human coronavirus strain 229E grown in L 132 cells were usually 200-300 PFU/cell, or 6 × 109 PFU per 75-cm2 flask. The virus was found to be quite stable when stored as a frozen infected monolayer, but loss of liter was inevitable during purification procedures. Batch preparations were routinely monitored by electron microscopy, and virions with typical coronavirus morphology were found in fresh preparations (fig. 1A). However, after the purified preparations in 0.001 m phosphate buffer had been stored overnight at 4°, fewer typical virions were evident, and disrupted or disintegrating forms were seen (fig. 1 B). Many particles appeared to have lost only their petal-like projections (peplomers). but in addition there were discrete tangles of threadlike strands which were 8-9 nm in width and appeared to be continuous. The possibility that these strands might be released internal component is supported by figure 1C, in which the same strands appear to be released from a collapsing virion. This was the first morphological evidence of a linear RNP-like internal structure of a coronavirus, and further studies were aimed at its isolation and characterization.
Fig. 1.
A Freshly prepared virus. Morphology is typical of the Coronaviridae. B Particles in three stages of disruption, seen after 24-hour storage at 4; upper right particle without petal-like projections is typical of many; upper left shows particle apparently losing internal component, but no structural detail is evident: bottom center shows typical discrete tangle of threadlike strands, 8-9 nm in diameter. C Preparation as in B, showing particle releasing filamentous material. Bars represent 100 nm in all micrographs.
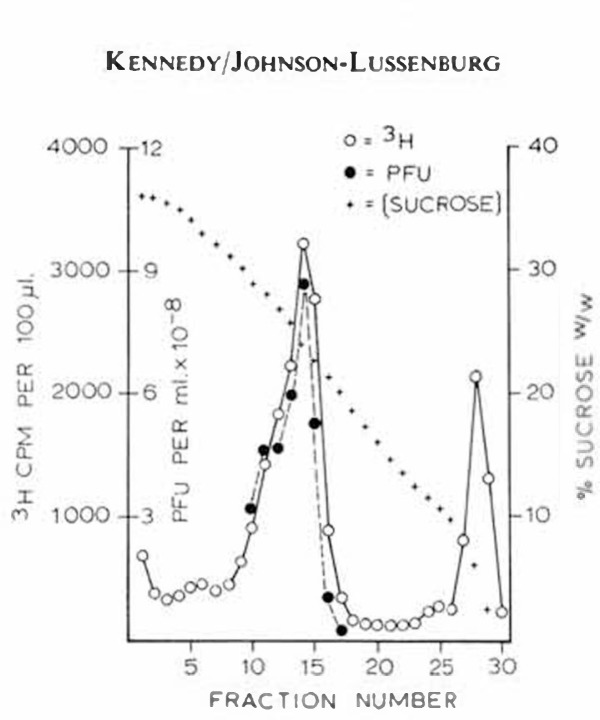
Correlation of virus infectivity with 3H-uridine incorporation. HCV/229E labelled with 3H-uridine was centrifuged in sucrose gradients at 63,000 g for 90 min and samples were collected and assayed by liquid scintillation counting. As can be seen in figure 2, the major peak of 3H-radioactivity was found in fraction 14, coinciding exactly with the peak of virus infectivity. Plaque assays of aliquots from the gradient fractions revealed that approximately 75% of the virus was recovered in five of the 3H-peak fractions. Since distribution of infectivity coincided with the uridine label, this indicates that 3H-uridine was incorporated into the RNA of the virion. Examination of these fractions by electron microscopy revealed some membranous debris in addition to typical coronavirions. Further purification by isopycnic centrifugation resulted in two visible bands, and in one (density 1.19), typical virions were again found to be associated with the peak of 3H-uridine activity [unpublished results].
Fig. 2.
Distribution of 3H-label and infectivity in sucrose gradient after rate-zonal centrifugation of HCV/229E.
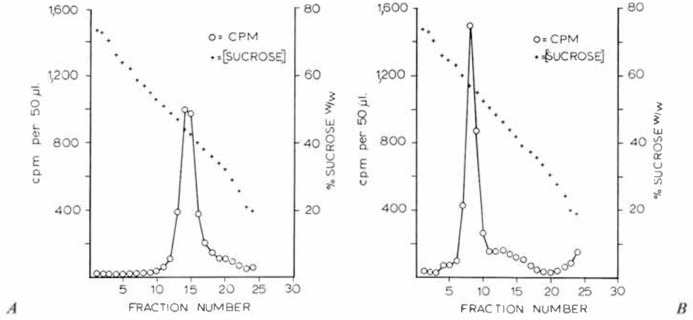
Isolation of virus ribonucleoprotein. 3H-uridine-labelled HCV/229E was purified by velocity gradient centrifugation, then treated with NP-40. In preliminary experiments, the resulting material was immediately layered onto 15-35% w/w sucrose gradients containing Tris/EDTA buffer and DTT above a 65% sucrose cushion and centrifuged at 110,000 g for 90 min. Untreated 3H-labelled HCV/229E was centrifuged in parallel. Analysis of the resulting fractions revealed no difference in the distribution of 3H-activity of either the untreated or the NP-40-trcated virus, and in both cases, only a single major peak was revealed which had sedimented through three-fifths of the gradients. Because we were confident that virus had been disrupted, judging by EM monitoring it was concluded that rate-zonal centrifugation under these conditions was not appropriate to resolve these components and that not only was a steeper gradient required, but centrifugation should be carried out to equilibrium. Consequently, NP-40-treated 3H-labelled and control, untreated 3H-labelled HCV/229E preparations were layered onto 25-75% w/w sucrose gradients and centrifuged at 170,000 g for 16-22 h at 10°. As shown in figure 3, untreated virus peaks were located at the sucrose concentration corresponding to a density of 1.19, whereas NP-40-trcatcd radioactive peaks occurred at a density of 1.27. The former density (1.19) corresponds to equilibrium densities reported for intact coronaviruses [8], while the latter (1.27) falls within the range of values reported for nucleoproteins isolated from a human coronavirus [8] as well as for several of the large RNA viruses [6]. Calculations revealed that 75% of the 3H-uridine label was regularly recovered in this peak.
Fig. 3.
Centrifugation of (A) control and (B) NP-40-trcated 3H-uridine-labelled HCV/229E in 25-75% w/w sucrose gradients at 170,000 g for 22 h.
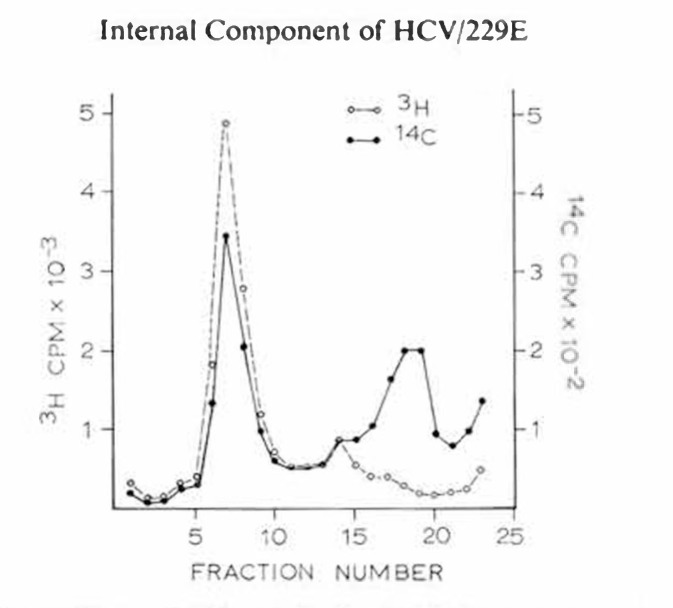
In a further series of experiments, HCV/229E was double-labelled with 3H-uridine and 14C-amino acids and once again treated with NP-40. The resulting preparations were centrifuged under reducing conditions as above. Analysis of these fractions revealed that both the 3H-uridine and 14C-amino acid activity occurred as major coincident peaks in the 1.3 density region of the gradient demonstrating the nucleoprotein nature of this complex (fig. 4). In addition, there was a minor peak of double-label activity in the 1.19 region, indicating the presence of a small proportion of nondisrupted virions, while a third peak at still lower density, showing only 14C-activity, probably consisted of detergent-released virus protein components.
Fig. 4.
Distribution of 3H and 14C activity in 25-75% sucrose gradient centrifuged at 170,000 g for 22 h. HCV/229E was labelled with 3H-uridine and 14C-amino acids and treated with NP-40 before centrifugation.
We therefore conclude that an RNP component with a density of 1.27 has been released by NP-40 disruption of HCV/229E and isolated by equilibrium centrifugation on a sucrose gradient.
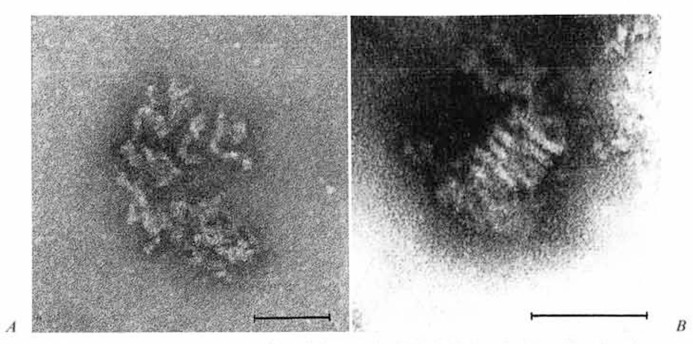
Morphology of the RNP of NP-40 disrupted HCV/229E. EM examination of fractions from the nucleoprotein peak region of NP-40-treated HCV/229E revealed a new type of particle, the majority showing the characteristic morphology illustrated in figure 5A. In addition, there were a few more expanded forms which resembled the internal componbent described above in association with spontaneously disrupting virions (fig. 1B, C). These structures could not be associated with the other fractions from the same gradient, nor were they found in any fractions of the control virus gradient. They appear to consist of a loosely twisted, helical, continuous strand which has an average width of 9 nm. The more tightly coiled structure shown in figure 5B was seen in one of the NP-40-treated HCV/229E preparations. At each end the strands of this coil seem to have dissolved into amorphous, unresolved material: however, over the space of five turns, the average strand diameter is 9 nm.
Fig. 5.
A Typical appearance of particles seen in 3H-labelled peak of gradient fractions of NP-40-trcated HCV/229E preparations. B Tightly coiled particle, also from NP-40-trcated HCV/229E samples. Bar represents 100 nm.
It is tempting to speculate that these structures (fig. 1B, C, 5A, B) depict various stages in the uncoiling of an originally tightly coiled or ‘doughnut’ type of packing arrangement of the internal component within the virion and that the resulting conformations seen arc strictly dependent on physico-chemical parameters as yet undefined. However, further work is required to confirm such speculations. Nevertheless, it would seem justified to conclude that the nucleocapsid of HCV/229E is a continuous helical RNP complex.
Discussion
Until recently the coronaviruses were believed to be RNA viruses on the basis of indirect evidence such as fluorescent and metabolic inhibition studies [10]. Direct confirmation of the RNA content of infectious bronchitis virus (IBV) has now been provided by Tannock [17] and Watkins el at. [18] and of HCV by J.C. Hierholzer (reported by Tyrrell el al. [8]). However, attempts to resolve the structure of the internal component of these viruses have, until now, proven unsuccessful.
Our studies reported here, both biochemical and morphological, leave little doubt that the RNA is associated with protein in an RNP complex. The conformation of the RNP complex is quite similar to that reported for the other large RNA viruses in that it appears to be a continuous, linear strand; however, the packing arrangement within the virus has not been established. We do not regard the lightly coiled appearance seen in one of our preparations as conclusive: however, suggestions of coiling have been seen in other preparations and it is possible that the nucleocapsid exists as a coiled structure within the virion. The diameter of the strand we have seen is similar to that reported by Apostolov el al. [9] on the basis of thin-section studies and the arrangement suggested is not incompatible with their observations.
Investigations are in progress to further characterize this nucleoprotein. In particular, preliminary evidence of RNA-dependent RNA polymerase activity associated with purified virions has been obtained [unpublished results] which may prove to be associated with this nucleoprotein complex. Attempts to demonstrate infectivity of the complex have so far given equivocal results. The techniques reported here for the isolation of intact RNP from the virus offer a most encouraging basis for future progress.
References
- 1.Apostolov K., Flewett T.H. Internal structure of influenza virus. Virology. 1965;26:506–508. doi: 10.1016/0042-6822(65)90014-0. [DOI] [PubMed] [Google Scholar]
- 2.Horse R.W., Waterson A.P. A helical structure in mumps, Newcastle disease and Sendai viruses. J. molec. Biol. 1960;2:75–77. [Google Scholar]
- 3.Howatson A.F., Whitmore G.F. The development and structure of vesicular stomatitis virus. Virology. 1962;16:466–478. doi: 10.1016/0042-6822(62)90228-3. [DOI] [PubMed] [Google Scholar]
- 4.Simpson R.W., Hauser R.E. Structural components of vesicular stomatitis virus. Virology. 1966;29:654–667. doi: 10.1016/0042-6822(66)90289-3. [DOI] [PubMed] [Google Scholar]
- 5.Nowinski R.C., Lloyd J.O., Sarkar N.H., Moore D.H. Common properties of the oncogenic RNA viruses (oncornaviruses) Virology. 1970;42:1152–1157. doi: 10.1016/0042-6822(70)90367-3. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinskaya A.G. Nucleocapsids of large RNA viruses as functionally active units in transcription. Adv. Virus Res. 1973;18:195–255. [Google Scholar]
- 7.Almeida J.D., Berry D.M., Cunningham C.H., Hamre D., Hofstad M.S., Malluci L., McIntosh K., Tyrrell D.A.J. Coronaviruses. Nature, Lond. 1968;220:650. [Google Scholar]
- 8.Tyrrell D.A.J., Almeida J.D., Cunningham C.H., Dowdle W.R., Hofstad M.S., McIntosh K., Tajima M., Zakstelskaya L.Ya., Easterday B.C., Kapikian A., Bingham R.W. Coronaviridae. Intervirology. 1975;5:76–82. doi: 10.1159/000149883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apostolov K., Flewett T.H., Kendal A.P. Symp. Cambridge. (Academic Press, London 1970): 1969. Morphology of influenza A, B, C and infectious bronchitis virus (IBV) virions and their replication; in Barry and Mahy The biology of large RNA viruses; pp. pp.3–26. [Google Scholar]
- 10.McIntosh K. Coronaviruses: a comparative review. Curr. Top. Microbiol. Immunol. 1974;63:85–129. [Google Scholar]
- 11.Davis N.L., Rueckert R.R. Properties of a ribonucleoprotein particle isolated from Nonidet P-40 treated Rous sarcoma virus. J. Virol. 1972;10:1010–1020. doi: 10.1128/jvi.10.5.1010-1020.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis E.V., Bolin V.S. Continuous cultivation of isogenous cell lines from the human embryo. Fed. Proc. 1960;19:386. [Google Scholar]
- 13.Hamre D., Kindig D.A., Mann J. Growth and intracellular development of a new respiratory virus. J. Virol. 1967;1:810–816. doi: 10.1128/jvi.1.4.810-816.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradburne A.F., Tyrrell D.A.J. The propagation of coronaviruses in tissue culture. Arch. ges. Virusforsch. 1969;28:133–150. doi: 10.1007/BF01249379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy D.A. PhD thesis. Ottawa, Ontario: University of Ottawa; 1976. Studies on the biology of a human coronavirus. [Google Scholar]
- 16.Howatson A.F. Electron microscopic procedures; in Habel and Salzman Fundamental techniques in virology. (Academic Press, New York 1969); pp. pp.505–524. [Google Scholar]
- 17.Tannock G.A. The nucleic acid of infectious bronchitis virus. Arch. ges. Virusforsch. 1973;43:259–271. doi: 10.1007/BF01250421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watkins H., Reeve P., Alexander D.J. The ribonucleic acid of infectious bronchitis virus. Archs Virol. 1975;47:279–286. doi: 10.1007/BF01317815. [DOI] [PMC free article] [PubMed] [Google Scholar]