Abstract
Objective
The specific effect of RNA interference on the replication of porcine hemagglutinating encephalomyelitis virus (PHE-CoV) was explored.
Methods
Four species of small interfering RNA (siRNA), targeting different regions of the PHE-CoV spike glycoprotein and replicase polyprotein genes, were prepared by in vitro transcription. After transfection of PK-15 cells with each of the siRNAs followed by infection with PHE-CoV, the cytopathic effect (CPE) was examined by phase-contrast microscope, and viral proliferation within cells was examined by indirect immunofluorescence microscopy, hemagglutination (HA) test, TCID50 assay and real-time RT-PCR.
Results
Examination of CPE demonstrated that the four siRNAs were capable of protecting cells against PHE-CoV invasion with very high specificity and efficiency. At 48 h post-infection, only a few siRNA-treated cells were positive for viral antigen staining, whereas most untreated virus-infected cells were positive. Transfection with siRNAs also suppressed the production of infectious virus by up to 18- to 32-fold as assessed by a HA test and 93- to 494-fold as assessed by TCID50 assay. Furthermore, treatment with siRNAs caused a 53-91% reduction in the viral genome copy number as assessed by real-time RT-PCR.
Conclusion
These results suggested that the four species of siRNAs can efficiently inhibit PHE-CoV genome replication and infectious virus production.
Key Words: Porcine hemagglutinating encephalomyelitis virus, Replication, Small interfering RNA
Introduction
RNA interference (RNAi) is a mechanism for the inhibition of expression of specific genes by double-stranded RNAs (dsRNAs) [1]. Transfection of 21-nt small interfering RNA (siRNA) molecules into mammalian cells results in degradation of the target gene mRNA and silencing of its expression [2]. The application of RNAi has had a significant impact in probing gene function and now promises to provide new strategies in the development of novel therapeutics and antivirals. The versatility of RNAi makes this technology particularly attractive in the pursuit of antiviral therapies directed against viral diseases, including the many viral diseases of livestock, and has been successfully applied to inhibit replication of some human and other animal viruses, including HIV-1 [3, 4, 5, 6], hepatitis C virus [7, 8, 9], hepatitis B virus [10], poliovirus [11], foot-and-mouth disease virus [12], transmissible gastroenteritis virus (TGEV) [13], bovine viral diarrhea virus [14], porcine reproductive and respiratory syndrome virus [15], and classical swine fever virus [16, 17], both in vitro and in vivo.
Porcine hemagglutinating encephalomyelitis virus (PHE-CoV) was first isolated in 1962 in Canada from suckling piglets with encephalomyelitis [18, 19] and can now be isolated from swine worldwide. Subclinical infections among swine are frequent [20, 21]. PHE-CoV causes encephalomyelitis, vomiting or wasting disease in suckling piglets. When piglets younger than 3 weeks are infected with PHE-CoV, their mortality rate ranges from 20 to 100%. Pigs older than 3 weeks show no obvious clinical signs [22]. PHE-CoV is a member of the antigenic group II of coronaviruses, along with bovine coronavirus (BCV), human coronavirus (HCV) strain OC43, turkey coronavirus (TCV), and mouse hepatitis virus (MHV). Members of this group possess five structural proteins: nucleocapsid (N), membrane (M), small membrane (sM), spike (S) and hemagglutinin-esterase (HE). HE is associated with granular projections located near the base of the typical large bulbous peplomers and displays hemagglutinating (HA) activities [23], Open reading frames (ORFs) potentially encoding non-structural (ns) proteins, including replicase polyprotein (R), have been identified. The number and size of R genes differ between coronavirus species, and they are important for RNA replication and transcription [24]. S possesses antigenic determinants that trigger the immune response for the production of antibodies that neutralize virus infectivity and inhibit HA activity [25]. S is a large, multifunctional protein that forms large petal-shaped spikes on the surface of the virions, and plays a central role in the biology and pathogenesis of coronaviruses [26], The availability of the complete genome sequence of PHE-CoV (GenBank accession No. NC_007732) enables the design of siRNA aimed at PHE-CoV. We designed specific siRNAs to target the S and R genes, in order to test whether RNAi could selectively target PHE-CoV viral RNAs. This study provides not only an experimental basis for the development of a new anti-PHE-CoV strategy, but also a new approach for studying PHE-CoV infection and replication.
Materials and Methods
Cell Culture, Virus Propagation, Titration and Sera
Prior to being infected with the HEV-67N strain (PHE-CoV) [18], the pig kidney cell line PK-15 was maintained in MEM supplemented with 10% fetal bovine serum and antibiotics (100 μg/ml streptomycin and 100 U/ml penicillin) in a 37°, 5% CO2 incubator overnight. When 70% of the virus-infected cells showed cytopathic effects (CPE), the cultures were collected, subjected to three freeze-thaw cycles, serially diluted 10-fold from 10−1 to 10−8, and added to PK-15 cells in 96-well plates. After 3 days of infection, the viral 50% tissue culture infectious dose (TCID50) was calculated using the Reed and Muench method [27, 28]. TCID50 assays were performed in triplicate 96-well plates with eight replicates per dilution. Positive anti-PHE-CoV serum and negative control serum were prepared as described previously [29].
PHE-CoV siRNA Preparation
Based on recent research [2] and the successful experience of Ambion Corporation's researchers [4], using GenBank sequences (GenBank accession No. AY078417, NC_007732) for HEV-67N and VW527, the conservative areas were selected and Ambion's siRNA target design online tool was utilized to choose four sequences for targeting S and R mRNA. BLASTN searches were conducted on all sequences to ensure gene specificity. Two sequences, SR1 and SR2, were chosen as siRNA for R and two sequences, SS1 and SS2, were chosen for S. The reverse sequence of SR1 was used as a negative siRNA control (scramble siRNA). Five siRNA molecules were synthesized with the SilencerTM siRNA construction kit (Ambion) according to the manufacturer's protocol. For each sequence, oligo DNA was synthesized by Shanghai Sangon Biological Engineering Technology & Services Co. Ltd. The sequences are shown in table 1.
Table 1.
Sequences of template deoxynucleotides for siRNAs used for target genes
| Gene targets | Sequences | Position |
|---|---|---|
| T7 promoter | 5′-TAATACGACTCACTATAGGAGACAGG-3′ | |
| SRI Sense Antisense |
5′-GCAAATACTCTGACTATAA-3′ 5′-AATTATAGTCAGAGTATTTGCCCTGTCTC-3′ 5′-AAGCAAATACTCTGACTATAACCTGTCTC-3′ |
12361-12379 |
| SR2 Sense Antisense |
5′-GCACAAGTTCTTAGTGAAA-3′ 5′-AATTTCACTAAGAACTTGTGCCCTGTCTC-3′ 5′-AAGCACAAGTTCTTAGTGAAACCTGTCTC-3′ |
15282-15300 |
| SS1 Sense Antisense |
5′-TACACTATGTGTGAATACC-3′ 5′-AAGGTATTCACACATAGTGTACCTGTCTC-3′ 5′-AATACACTATGTGTGAATACCCCTGTCTC-3′ |
23910-23928 |
| SS2 Sense Antisense |
5′-TTTCTGTGGTAATGGTAAT-3′ 5′-AAATTACCATTACCACAGAAACCTGTCTC-3′ 5′-AATTTCTGTGGTAATGGTAATCCTGTCTC-3′ |
26789-26807 |
| Scramble siRNA Sense Antisense |
5′-ACATAAACCTATCAGTAGT-3′ 5′-AAACTACTGATAGGTTTATGTCCTGTCTC-3′ 5′-AAACATAAACCTATCAGTAGTCCTGTCTC-3′ |
|
The 8 nucleotide underlined sequences of template deoxynucleotides for each siRNA are complemented with those of the T7 promoter primer.
Cell Culture and Transfection
PK-15 cells were seeded in 24-well plates at 104 cells/well, and incubated for 24 h at 37° in a 5% CO2 atmosphere. When cells were 60-70% confluent, siRNA was introduced using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, Calif., USA) according to the manufacturer's protocol. Briefly, 1 μl of transfection reagent and 0.5 μg siRNA was added to each well and incubated for 6 h. The cells were then washed with MEM and further cultured for 16 h in MEM supplemented with 2% fetal bovine serum prior to viral infection. The siRNA-treated cells were infected with 400 TCID50 (104.37) of PHE-CoV, and the infection was allowed to proceed for the indicated time points. CPE were evaluated under an inverted fluorescence/phase-contrast microscopy (Nikon) at different time points post-infection and infected cells were visualized by indirect immunofluorescence assay (IFA) at 48 h post-infection. Culture supernatants and PHE-CoV-infected PK-15 cells were collected. Infectious virus production was determined by the hemagglutination (HA) test and the TCID50 assay. The copy number of PHE-CoV genomic RNA was determined by real-time RT-PCR.
Hemagglutination Assay
The HA titers of infectious virus production in 24-well plates were determined by the HA test, as described previously [30]. Chicken red blood cells, diluted 1:200 in physiological saline, were added in volumes of 0.5 ml to equal volumes of 2-fold antigen dilutions in plastic agglutination trays. Each sample was tested in duplicate. Results were read by the settling pattern method after 1 h at room temperature. The presence of HA was read as a positive test result for PHE-CoV, whereas the absence of HA was read as a negative test result.
Indirect IFA
To study the inhibitory effects of RNAi on PHE-CoV replication, the level of viral antigen produced in PK-15 cell was examined by IFA using anti-PHE-CoV serum after siRNA transfection and viral infection at 48 h post-PHE-CoV infection. Infected cells were visualized by an indirect IFA as has been described previously [16]. Cells were washed 3 times with PBS and incubated with FITC-conjugated rabbit anti-pig IgG (Sigma; 1:60) for 1 h. After washing, cells were photographed and examined with a Zeiss Axioskop 40 fluorescence microscope.
Determination of Virus Genome Copy
Using GenBank sequences (GenBank accession No. AY078417, NC_007732) for HEV-67N and VW527, the conservative areas were selected and primers were designed for PHE-CoV using Primer Express 2.0 software. Three primers were synthesized (JiKang, Shanghai, China) for quantification of the PHE-CoV genome in real-time PCR: 5′-AGCGATGAGGCTATTCCGACTA-3′ (NFP primer), 5′-TTGCCAGAATTGGCTCTACTACG-3′ (NRP primer), and 5′-FTTC-CGCCAGGCACGGTACTCCCP-3′ (TaqMan probe). The target region of real-time RT-PCR was nucleotides 29344-29569 of the N protein gene of PHE-CoV. PHE-CoV-infected PK-15 cells, as well as culture supernatants, were collected 48 h after viral infection. Total RNA (100 ng) was extracted and purified using TRIzol LS Reagent (Invitrogen). PCR was used to amplify the N gene with NFP and NRP primers under conditions of 94° for 3 min, then 40 cycles of 94° for 30 s, 60° for 30 s, and 72° for 60 s, with a final extension at 72° for 7 min. The PCR product was gel-purified using the Agarose Gel DNA Extraction Kit (Roche) and then cloned into pGEM-18T vector (Promega Corp., Madison, Wisc., USA). The resulting plasmid, pT-N, with the correct sequence confirmed by direct sequencing, was selected as a quantitative standard for the determination of the viral RNA copy number. Real-time PCR was performed with the ABI Prism® 7000 Sequence Detection System using a QuantiTect™ Prob PCR Kit (Qiagen, Hilden, Germany) under the conditions of 95° for 3 min, followed by 40 cycles of denaturation at 95° for 30 s, annealing, and extension at 60° for 30 s. The quantitative standard curve for the determination of PHE-CoV genome copy number was created by real-time PCR of standard plasmid pT-N preparations at serial dilutions of 103, 104, 105, 106 and 107 copies/μl The specificity of the real-time PCR was confirmed by sequencing of the product.
Results
In vitro Transcription of siRNAs
For in vitro transcription of siRNAs, see figure 1.
Fig. 1.
Purity and integrity of the siRNAs transcribed in vitro: (1) SR1, (2) SR2, (3) SS1, (4) SS2, and (5) scramble siRNA.
CPE Analysis
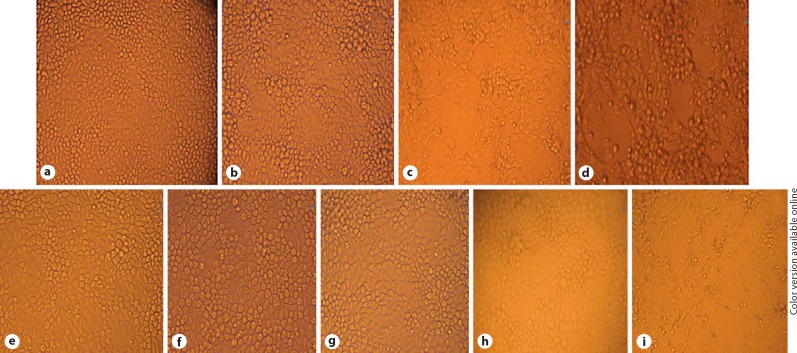
To study PHE-CoV-induced CPE (fig. 2), PK-15 cells were infected with PHE-CoV at 400 TCID50 (104.37) in 24-well plates. At 12 h after infection, cells were examined with by phase-contrast microscopy every 4 h. The virusinfected cells exhibited obvious morphological changes starting from the brim of wells at 56 h post-infection. About 72 h after viral infection, whole cells began to fall into pieces and lost their adhesion to the plate. PK-15 cells were transfected with four separate siRNA prior to being infected with PHE-CoV. At 56 h after viral infection, images of cells were captured. Analysis of cell morphology indicated that the negative control (scramble siRNA) had no inhibitory effect on PHE-CoV-induced CPE compared with the mock control. Non-infected cells stuck tightly to the plate and remained in good condition throughout the experiment. In contrast, the four species of siRNA, SR1, SR2, SS1, and SS2, effectively blocked CPE in the cell cultures until 96-120 h (data not shown) (fig. 2).
Fig. 2.
Effect of siRNAs on PHE-CoV-induced CPE in PK-15 cells. PK-15 cells were transfected with different siRNAs and then infected with PHE-CoV at 400 TCID50. Images in the middle and brim of the same well, respectively, are shown. a-d Untreated cells infected with virus at 400 TCID50 in 24-well plates were observed at 24, 48, 56, and 72 h post-infection, respectively. e-i Cells were transfected with SR1, SS1, SR2, SS2 and scramble siRNA, respectively, and CPE was examined 56 h post-infection. Cell images were captured at the conclusion of the study. The experiment was performed in triplicate and repeated 3 times.
Examination of siRNA Effect by IFA
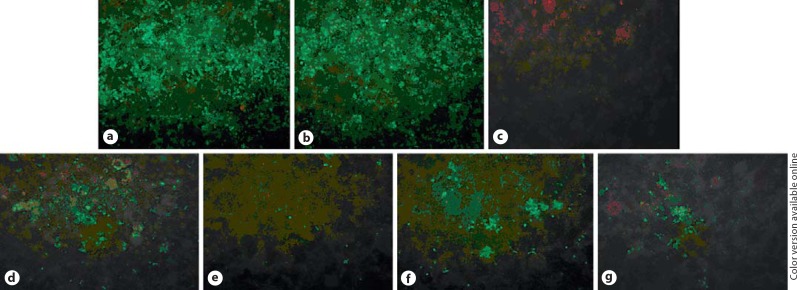
To study the inhibitory effects of RNAi on PHE-CoV replication, the level of viral antigen produced in PK-15 cell was examined by IFA using anti-PHE-CoV serum after siRNA transfection and viral infection. At 48 h post-PHE-CoV infection, most PK-15 cells that had been mock-transfected or that had received scramble siRNA exhibited bright green fluorescence in the cytoplasm, indicating that these cells were virus-producing (fig. 3a, b). By contrast, only a few cells in wells receiving SR1, SR2, SS1, or SS2 transfection displayed green fluorescence, indicating that most of these cells were effectively protected by the siRNA, and resisted viral infection (fig. 3d-g).
Fig. 3.
Protective effect of siRNA against PHE-CoV infection. The viral infection in PK-15 cells was examined by indirect immunofluorescence: a mock transfection (stained with PHE-CoV-positive serum); b scramble siRNA transfection; c mock transfection (stained with PHE-CoV-negative serum); d SR1 transfection; e SR2 transfection; f SS1 transfection, and g SS2 transfection.
Examination of siRNA Effect by HA Test and Infectious Virus Assay
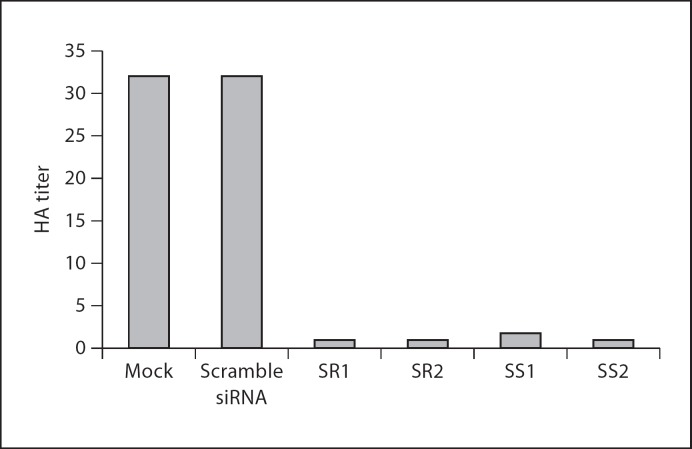
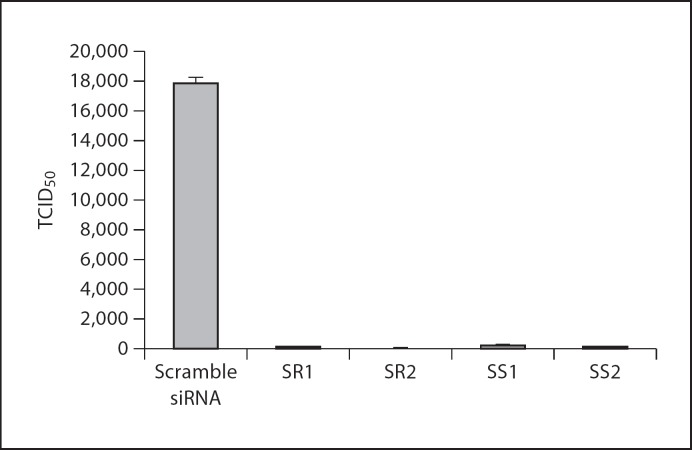
Culture supernatants and PHE-CoV-infected PK-15 cells were collected. The HA titer was determined to examine the effect of siRNA on production of viable virus in 24-well plates. The results (fig. 4) showed that in control cells transfected with scramble siRNA, the PHE-CoV HA titer reached 32 at 48 h post-infection, similar to that observed in mock-transfected cells. In contrast, titers at 48 h post-infection were 1, 1, 1.75, and 1 for cells transfected with SR1, SR2, SS1, and SS2 respectively, corresponding to 32-, 32-, 18-, and 32-fold reductions by comparison with scramble siRNA-transfected cells. The TCID50 assay was performed to examine the effect of siRNA on the production of viable virus, and the results (fig. 5) showed that in control cells transfected with scramble siRNA PHE-CoV titers reached 10425 TCID50/ml at 48 h post-infection, similar to that obtained from mock transfection. In contrast, titers at 48 h post-infection were 102.10, 101.56, 102.28, and 102.24 ml−1 for cells transfected with SR1, SR2, SS1, and SS2, respectively, corresponding to 141-, 494-, 93-, and 102-fold reductions in comparison to scramble siRNA-transfected cells.
Fig. 4.
Inhibition of virus production in siRNA-treated cells. HA titers are the means of three repeat titrations at the time points indicated.
Fig. 5.
Inhibition of virus production in siRNA-treated cells. TCID50 values are the means of three repeat titrations at the time points indicated.
Examination of siRNA Effect by RT-PCR
To quantify the effect of siRNA on viral replication at 48 h post-viral infection, the viral genome copy number was determined by real-time PCR, using the serially diluted plasmid pT-N as a standard. The R2 value of the standard curve was 0.99 and the average amplification efficiency E was 0.993. Sequencing showed that the fragment amplified was the expected part of PHE-CoV gene, thereby demonstrating the specificity and reliability of the analysis. As shown in figure 6, the copy number of the viral genome per nanogram of total RNA calculated from the standard curve was 7.62 × 104 copies/ng of viral genome in the total RNA from scramble siRNA-treated cells, whereas there were 2.11 × 104, 8.03 × 103, 3.65 × 104, and 3.11 × 104 copies/ng of the viral genome in total RNA from cells treated with SR1, SR2, SS1, and SS2, respectively. These values correspond to reductions of 73, 91, 53 and 61%, respectively.
Fig. 6.
Reduction of viral genome copy number by siRNA treatment in PK-15 cells 48 h post-infection. The PHE-CoV genome copy numbers are the means of three repeat experiments.
Discussion
Porcine hemagglutinating encephalomyelitis (PHE) is an infectious disease that mainly affects pigs younger than 3 weeks. Even if an infected pig survives the diseaseassociated immunopathology or the toxicity of the drug treatment, PHE can acutely affect its growth. Increasing research attention has been focused on PHE because infection rates have increased in some countries [31, 32, 33, 34]. PHE is caused by PHE-CoV, a virus that has only one known strain and is the only known CoV that is neurotropic in pigs [25, 35, 36]. The virus spreads via peripheral nerves to the CNS, its main propagation site [37]. Nerve cells are the primary target for viral replication. Because the regeneration capacity of nerve cells is weak, once these cells are damaged, pathology will remain even if the virus is eliminated. The study of PHE-CoV replication and the control or inhibition of this infection is of great significance because, currently, no effective preventative measures or cures for this disease exist. Recently, the replication of various viruses, including many coronaviruses, have been shown to be effectively inhibited in vitro or in vivo by knockdown with 21- to 25-base-pair (bp), sequence-homology-driven siRNAs [26, 38, 39, 40]. siRNA therapy can inhibit different levels of viral function, including transcription and translation [41]. Very effective and rapid silencing of a target gene can be achieved using in vitro transcribed siRNAs transfected into mammalian cells [16]. In the present study, we designed four 21-bp siRNA molecules against the PHE-CoV S and R genes, prepared these siRNAs by in vitro transcription, and tested the efficiency of RNAi on viral replication by assessing CPE, IFA, HA, TCID50 and RT- qPCR. Our results demonstrate that these four siRNAs were highly capable of inhibiting viral RNA genome replication.
PHE-CoV is a member of the Coronaviridae family, whose gene expression involves a series of complex transcriptional, translational, and post-translational regulatory mechanisms [42]. After receptor-mediated entry, the replicase polyprotein, encoded by ORFs, 1a and 1b, which comprise approximately two-thirds of the genome (beginning from the 5′ end), is the first viral protein to be translated. The translation of the replicase polyprotein involves ribosomal frameshifting to the −1 frame immediately upstream of the ORF 1a translation termination codon (GenBank accession No. NC_007732) [43]. Many studies have shown that siRNAs that target the R gene can efficiently inhibit viral replication [13]. In addition, similar to other coronaviruses spike proteins, the PHE-CoV spike glycoprotein contains two non-covalently associated functional subunits, S1 and S2 (GenBank accession No. AY078417, NC_007732). It is thought that the binding of the S1 subunit to its cellular receptor induces conformational changes in S2, which leads to the fusion between viral and cellular membranes and, thus, viral entry [44]. Variations in S proteins between strains of coronaviruses are responsible for differences in host range and tissue tropism [45]. Based on this knowledge, we chose the S and R genes as the RNAi targets, and our results demonstrate that these two key genes are important for PHE-CoV replication.
In this study, knowledge of the kinetics of PHE-CoV replication in cultured cells was critical for determining the sampling points to estimate the siRNA effect in cells. Hence, the replication kinetics of PHE-CoV in PK-15 cells were studied by CPE, HA and TCID50 assays prior to siRNA transfection, and viral proliferation was shown to peak at 48-72 h post-infection (data not shown). Based on this information, the cell cultures for IFA, viral titration and RNA extraction were harvested at 48 h postinfection. The result of CPE analysis showed that virus-produced CPE was not observed at 56 h post-infection in cells treated with SR1, SR2, SS1, or SS2. To further examine the level of viral inhibition, we tested the viral titers in infected cells by TCID50, and showed that siRNAs caused an approximately 93- to 494-fold decrease in virus yield. Additionally, further analysis confirmed that the efficient inhibition of viral replication in this study was due to viral RNA degradation. We designed a pair of primers specific to the N gene of PHE-CoV and showed by real-time PCR that approximately 53-91% less viral RNA existed in the cells transfected with the siRNA compared to the control cells, which verified that the suppression of infection occurred at the RNA level. In fact, because the pair of primers designed for amplifying a region within the N gene could amplify the same regions within the PHE-CoV RNA genome and subgenomic mRNAs (except the shortest one) simultaneously, the reduction in viral RNAs suggests that not only the viral genome RNA itself but also all of the transcripts required for viral structural protein expression decreased [13].
Viruses are prone to errors during replication and continually produce mutated viral proteins that can escape attack by siRNAs [46]. In vitro transcribed SS1 and SS2 siRNAs were effective in reducing viral replication; however, the S1 subunit of the spike glycoprotein is not an essential structural protein and is prone to high mutation rates as the virus evolves in host populations [47]. To ensure that SS1 and SS2 could be used in a wide range of virus strains, we evaluated the cross-inhibitory capabilities of these siRNAs by conducting multiple alignments of S gene sequences from PHE-CoV strains based on their availability in GenBank. Our results showed that SS1 and SS2 could cover 67% (4/6) and 100% (6/6) of PHE-CoV strains, respectively (data not shown), indicating that additional studies should be conducted to search for siRNAs with the widest cross-inhibitory effects on a range of PHE-CoV strains.
The successful use of siRNAs as therapeutic or protective agents in the wild is limited, particularly for natural infection, because sequence data will not be available before successful isolation of a virus [17]. To circumvent this problem, the replicase genes of PHE-CoV, which are highly conserved among different strains, were targeted by SR1 and SR2 [13]. Our results demonstrated that both siRNAs could block viral replication with very high efficiency. Our study was performed utilizing infection with the HEV-67N strain, whose sequence is not available in GenBank; however, our study implies that the sequence identity of SR1, SR2 and the HEV-67N strain should be 100% because RNAi is highly sequence-specific and requires 100% identity for clearance of virus from cell culture between the target and targeting sequences [11, 17, 48]. Application of only one siRNA that is 100% identical to the sequence from the infecting virus or more than one siRNA sequence targeting different conserved regions of the gene are ways to overcome the lack of knowledge of the target sequence. However, additional studies should be performed to confirm this hypothesis both in vitro and in vivo.
In conclusion, our results indicate that PHE-CoV, like other ssRNA viruses, is susceptible to degradation via the RNAi pathway. This method may not completely inhibit viral growth but merits further animal studies to define its therapeutic potential. Additionally, siRNAs could potentially be delivered to animals via transgenes to eradicate animal viral infections [49]. Based on the present data and the advantages of siRNA technology, we propose that a combination of siRNAs targeting the S and R genes may be used as a tool to study PHE-CoV replication, antiviral therapy in transgenic animals in future studies.
Acknowledgement
This study was supported by the National Natural Science Foundation of China (Nos. 31072134, 30871849, 30671551), the Young Scientist Program of Science and Technology Development Plan in the Jilin Province (20090154), and the International Cooperation Program of Science and Technology Development Plan in the Jilin Province (20080722).
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz RJ. RNA interference meets HIV-1: will silence be golden? Nat Med. 2002;8:659–660. doi: 10.1038/nm0702-659. [DOI] [PubMed] [Google Scholar]
- 4.Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kok KH, Lei T, Jin DY. siRNA and shRNA screens advance key understanding of host factors required for HIV-1 replication. Retrovirology. 2009;6:78. doi: 10.1186/1742-4690-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber N, Ortega P, Clemente MI, Shcharbin D, Bryszewska M, de la Mata FJ, Gomez R, Munoz-Fernandez MA. Characterization of carbosilane dendrimers as effective carriers of siRNA to HIV-infected lymphocytes. J Control Release. 2008;132:55–64. doi: 10.1016/j.jconrel.2008.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783–2788. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YL, Guo YJ, Bin L, Sun SH. Hepatitis C virus single-stranded RNA induces innate immunity via toll-like receptor 7. J Hepatol. 2009;51:29–38. doi: 10.1016/j.jhep.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Kretschmer-Kazemi Far R, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian ZQ, Cui ZL, Chen WZ, Liu MQ, Yan WY, Zheng ZX. Transfection of siRNA expressing plasmids targeting S gene inhibits replication and expression of hepatitis B virus in hepatic cancer cells (in Chinese) Zhonghua Yi Xue Za Zhi. 2009;89:347–351. [PubMed] [Google Scholar]
- 11.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Yan W, Du Q, Fei L, Liu M, Ni Z, Sheng Z, Zheng Z. RNA interference targeting VP1 inhibits foot-and-mouth disease virus replication in BHK-21 cells and suckling mice. J Virol. 2004;78:6900–6907. doi: 10.1128/JVI.78.13.6900-6907.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou JF, Hua XG, Cui L, Zhu JG, Miao DN, Zou Y, He XZ, Su WG. Effective inhibition of porcine transmissible gastroenteritis virus replication in ST cells by shRNAs targeting RNA-dependent RNA polymerase gene. Antiviral Res. 2007;74:36–42. doi: 10.1016/j.antiviral.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambeth LS, Moore RJ, Muralitharan MS, Doran TJ. Suppression of bovine viral diarrhea virus replication by small interfering RNA and short hairpin RNA-mediated RNA interference. Vet Microbiol. 2007;119:132–143. doi: 10.1016/j.vetmic.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Li G, Jiang P, Li Y, Wang X, Huang J, Bai J, Cao J, Wu B, Chen N, Zeshan B. Inhibition of porcine reproductive and respiratory syndrome virus replication by adenovirus-mediated RNA interference both in porcine alveolar macrophages and swine. Antiviral Res. 2009;82:157–165. doi: 10.1016/j.antiviral.2009.02.202. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Guo H, Xiao C, Zha Y, Shi Z, Xia X, Tu C. In vitro inhibition of classical swine fever virus replication by siRNAs targeting Npro and NS5B genes. Antiviral Res. 2008;78:188–193. doi: 10.1016/j.antiviral.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Porntrakulpipat S, Supankong S, Chatchawanchonteera A, Pakdee P. RNA interference targeting nucleocapsid protein (C) inhibits classical swine fever virus replication in SK-6 cells. Vet Microbiol. 2010;142:41–44. doi: 10.1016/j.vetmic.2009.09.041. [DOI] [PubMed] [Google Scholar]
- 18.Mengeling WL, Boothe AD, Ritchie AE. Characteristics of a coronavirus (strain 67N) of pigs. Am J Vet Res. 1972;33:297–308. [PubMed] [Google Scholar]
- 19.Greig AS, Mitchell D, Corner AH, Bannister GL, Meads EB, Julian RJ. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can J Comp Med Vet Sci. 1962;26:49–56. [PMC free article] [PubMed] [Google Scholar]
- 20.Andries K, Pensaert MB. Immunofluorescence studies on the pathogenesis of hemagglutinating encephalomyelitis virus infection in pigs after oronasal inoculation. Am J Vet Res. 1980;41:1372–1378. [PubMed] [Google Scholar]
- 21.Vijgen L, Keyaerts E, Lemey P, Maes P, Van Reeth K, Nauwynck H, Pensaert M, Van Ranst M. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pensaert MB, Hemagglutinating encephalomyelitis virus . In: Diseases of Swine, ed 9. Straw BL, Zimmerman JJ, D' Allaire S, Taylor DJ, editors. Ames, Blackwell; 2006. pp. pp 353–358. [Google Scholar]
- 23.Vlasak R, Luytjes W, Leider J, Spaan W, Palese P. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J Virol. 1988;62:4686–4690. doi: 10.1128/jvi.62.12.4686-4690.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spaan W, Cavanagh D, Horzinek MC. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 25.Sasseville AM, Boutin M, Gelinas AM, Dea S. Sequence of the 3′-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J Gen Virol. 2002;83:2411–2416. doi: 10.1099/0022-1317-83-10-2411. [DOI] [PubMed] [Google Scholar]
- 26.Qin ZL, Zhao P, Zhang XL, Yu JG, Cao MM, Zhao LJ, Luan J, Qi ZT. Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293t cells. Biochem Biophys Res Commun. 2004;324:1186–1193. doi: 10.1016/j.bbrc.2004.09.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed LJ, Muench HA. A simple method of estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 28.Zhen Y, Jinghua L. Animal Virology. Peking. Science Press; 1997. pp. pp 690–692. [Google Scholar]
- 29.Yu X, Tu C, Li H, Hu R, Chen C, Li Z, Zhang M, Yin Z. DNA-mediated protection against classical swine fever virus. Vaccine. 2001;19:1520–1525. doi: 10.1016/s0264-410x(00)00334-0. [DOI] [PubMed] [Google Scholar]
- 30.Girard AA, Mitchell DS. Encephalomyelitis of swine caused by a haemagglutinating virus. III. Serological studies. Res Vet. 1964;5:294–302. [Google Scholar]
- 31.Hara Y, Hasebe R, Sunden Y, Ochiai K, Honda E, Sakoda Y, Umemura T. Propagation of swine hemagglutinating encephalomyelitis virus and pseudorabies virus in dorsal root ganglia cells. J Vet Med Sci. 2009;71:595–601. doi: 10.1292/jvms.71.595. [DOI] [PubMed] [Google Scholar]
- 32.Cartwright SF, Lucas M, Cavill JP, Gush AF, Blandford TB. Vomiting and wasting disease of piglets. Vet Rec. 1969;84:175–176. doi: 10.1136/vr.84.7.175. [DOI] [PubMed] [Google Scholar]
- 33.Mengeling WL, Cutlip RC. Pathogenicity of field isolants of hemagglutinating encephalomyelitis virus for neonatal pigs. J Am Vet Med Assoc. 1976;168:236–239. [PubMed] [Google Scholar]
- 34.Quiroga MA, Cappuccio J, Pineyro P, Basso W, More G, Kienast M, Schonfeld S, Cancer JL, Arauz S, Pintos ME, Nanni M, Machuca M, Hirano N, Perfumo CJ. Hemagglutinating encephalomyelitis coronavirus infection in pigs, Argentina. Emerg Infect Dis. 2008;14:484–486. doi: 10.3201/eid1403.070825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greig AS, Girard A. Serological comparison of hemagglutinating encephalomyelitis viruses isolated from different outbreaks. Can J Comp Med. 1969;33:25–28. [PMC free article] [PubMed] [Google Scholar]
- 36.Greig AS, Girard A. Encephalomyelitis of swine caused by a haemagglutinating virus. V. Response to metabolic inhibitors and other chemical compounds. Res Vet Sci. 1969;10:509–513. [PubMed] [Google Scholar]
- 37.Yagami K, Hirai K, Hirano N. Pathogenesis of haemagglutinating encephalomyelitis virus (HEV) in mice experimentally infected by different routes. J Comp Pathol. 1986;96:645–657. doi: 10.1016/0021-9975(86)90061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pushparaj PN, Aarthi JJ, Manikandan J, Kumar SD. siRNA, miRNA, and shRNA: in vivo applications. J Dent Res. 2008;87:992–1003. doi: 10.1177/154405910808701109. [DOI] [PubMed] [Google Scholar]
- 39.Wu CJ, Huang HW, Liu CY, Hong CF, Chan YL. Inhibition of SARS-CoV replication by siRNA. Antiviral Res. 2005;65:45–48. doi: 10.1016/j.antiviral.2004.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J, Huang F, Hua X, Cui L, Zhang W, Shen Y, Yan Y, Chen P, Ding D, Mou J, Chen Q, Lan D, Yang Z. Inhibition of porcine transmissible gastroenteritis virus replication in mini-pigs by shRNA. Virus Res. 2010;149:51–55. doi: 10.1016/j.virusres.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou XM, Lin JS, Shi Y, Tian DA, Huang HJ, Zhou HJ, Jin YX. Establishment of a screening system for selection of siRNA target sites directed against hepatitis B virus surface gene. Acta Biochim Biophys Sin (Shanghai) 2005;37:310–316. doi: 10.1111/j.1745-7270.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- 42.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 43.Brierley I, Digard P, Inglis SC. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zelus BD, Schickli JH, Blau DM, Weiss SR, Holmes KV. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J Virol. 2003;77:830–840. doi: 10.1128/JVI.77.2.830-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carmichael GG. Medicine: silencing viruses with RNA. Nature. 2002;418:379–380. doi: 10.1038/418379a. [DOI] [PubMed] [Google Scholar]
- 47.Ruan YJ, Wei CL, Ee AL, Vega VB, Thoreau H, Su ST, Chia JM, Ng P, Chiu KP, Lim L, Zhang T, Peng CK, Lin EO, Lee NM, Yee SL, Ng LF, Chee RE, Stanton LW, Long PM, Liu ET. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGrew MJ, Sherman A, Ellard FM, Lillico SG, Gilhooley HJ, Kingsman AJ, Mitrophanous KA, Sang H. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 2004;5:728–733. doi: 10.1038/sj.embor.7400171. [DOI] [PMC free article] [PubMed] [Google Scholar]