Abstract
Objectives
The molecular mechanisms underlying the pathogenesis of human coronavirus OC43 (HCoV-OC43) infection are poorly understood. In this study, we investigated the ability of HCoV-OC43 to antagonize the transcriptional activation of antiviral response elements.
Methods
HCoV-OC43 structural (membrane M and nucleocapsid N) and accessory proteins (ns2a and ns5a) were expressed individually in human embryonic kidney 293 (HEK-293) cells. The transcriptional activation of antiviral response elements was assessed by measuring the levels of firefly luciferase expressed under the control of interferon (IFN)-stimulated response element (ISRE), IFN-β promoter, or nuclear factor kappa B response element (NF-κB-RE). The antiviral gene expression profile in HEK-293 cells was determined by PCR array.
Results
The transcriptional activity of ISRE, IFN-β promoter, and NF-κB-RE was significantly reduced in the presence of HCoV-OC43 ns2a, ns5a, M, or N protein, following the challenge of cells with Sendai virus, IFN-α or tumor necrosis factor-α. The expression of antiviral genes involved in the type I IFN and NF-κB signaling pathways was also downregulated in the presence of HCoV-OC43 structural or accessory proteins.
Conclusion
Both structural and accessory HCoV-OC43 proteins are able to inhibit antiviral response elements in HEK-293 cells, and to block the activation of different antiviral signaling pathways.
Keywords: Coronavirus OC43, Antiviral response elements, Luciferase reporter gene assay, Interferon antagonism
Introduction
Human coronavirus OC43 (HCoV-OC43) is an enveloped, positive-sense RNA virus classified as a Betacoronavirus, the same genus as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronaviruses. HCoV-OC43 infection has been associated mainly with upper respiratory tract symptoms and exacerbation of asthma and pneumonia in some groups and institutional settings [1, 2, 3, 4]. The virus has also been associated with severe neurological disorders like acute disseminated encephalomyelitis in children [5, 6].
While most studies have focused their attention on the immunopathology of SARS-CoV and MERS-CoV, there has not been the same interest for HCoV-OC43. Indeed, there are few studies concerned with the molecular mechanisms governing HCoV-OC43 infection and its effect on the intracellular host defenses. HCoV-OC43 is over 30 kb in length, and similarly to the other coronaviruses, it is composed of the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N) [7]. Uniquely, there are two accessory proteins interspaced between these structural proteins called ns2a and ns5a [8]. These proteins are not essential for replication; however, they might play a role in the pathogenesis of coronavirus infection [9].
The interferon (IFN) induction and signaling pathway is the major branch in the innate immune response against viruses. The induction of type I IFN is initiated by molecular sensing of viral RNA by pattern recognition receptors such as TLR, RIG-I, and MDA5 [10]. The adaptor protein MAVS mediates the signals from RIG-I and MDA5 to activate the kinases TBK1 and IKKε, which in turn phosphorylate IRF3 and IRF7 transcription factors [11]. IRF3 and IRF7 dimerize to undergo nuclear translocation and initiate the transcription of type I IFN. The adaptor proteins MYD88 and TRIF mediate signals from TLRs and activate IRAK1, TRAF6, and the aforementioned kinases [12]. The IRFs along with nuclear factor kappa B (NF-κB) can then be phosphorylated to translocate into the nucleus and establish the transcription of type I IFN by binding cognate sites on the IFN-β promoter [12]. NF-κB specifically binds to the NF-κB response element (NF-κB-RE) to regulate the expression of pro-inflammatory and cell survival genes [13].
IFN signaling via the JAK/STAT pathway is established when type I IFN binds to the IFN receptor. This leads to activation of the kinases JAK1 and TYK2. These kinases in turn phosphorylate the transcription factors STAT1 and STAT2 [14, 15]. Phosphorylated STAT1 and STAT2 then dimerize and undergo nuclear translocation, where they form a complex with IRF9 called the ISGF3. This complex will bind the IFN-stimulated response element (ISRE) and initiate the transcription of ISGs. These ISGs assist the cell in counteracting the viral infection by establishing an antiviral state [16].
SARS-CoV and MERS-CoV were shown to inhibit the transcriptional activity of ISRE, IFN-β promoter or NF-κB-RE [17, 18, 19, 20, 21, 22, 23, 24]. However, the effect of different HCoV-OC43 proteins on the transcriptional activation of antiviral response elements has not yet been investigated. In this study, the ability of HCoV-OC43 structural (M and N) and accessory proteins (ns2a and ns5a) to antagonize the transcriptional activation of ISRE, IFN-β promoter, and NF-κB-RE was investigated.
Materials and Methods
Cell Culture
Human embryonic kidney 293 (HEK-293) cells (American Type Culture Collection ATCC, Manassas, VA, USA) were cultured in 25 cm2 tissue culture flasks in Dulbecco's minimal essential medium containing GlutaMAXTM (Life Technologies CorporationTM, Grand Island, NY, USA). The medium was supplemented with 10% fetal bovine serum, Fungizone® (250 μg/mL), penicillin G (10,000 U/mL), and streptomycin sulfate (10,000 pg/mL) (Life TechnologiesTM). Monolayers of HEK-293 cell culture flasks were incubated at 37°C in the presence of 5% carbon dioxide (CO2) and 90% humidity.
Amplification of HCoV-OC43 ns2a, ns5a, M, and N Genes by Reverse Transcription-Polymerase Chain Reaction
The QIAamp® Viral RNA Minikit (QiagenTM GmbH, Hilden, Germany) was used to isolate RNA from HCoV-OC43 (ATCC VR-1558) according to the manufacturer's instructions. The HCoV-OC43 ns2a, ns5a, M, and N genes were amplified by a two-step reverse transcription-polymerase chain reaction (RT-PCR) using the GeneAmp® RNA Core Kit (Applied BiosystemsTM, Foster City, CA, USA), and 10 pmol of previously described primers [8] on GeneAmp® PCR System 9700 (Applied BiosystemsTM). The RT reaction conditions were: annealing at 37°C for 60 min, denaturation at 90°C for 5 min, and cooling at 4°C. Thereafter, the cDNA was amplified by PCR using the following conditions: denaturation at 94°C for 10 min, then 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, followed by a final extension step at 72°C for 7 min, and cooling at 4°C. PCR products were run on agarose gel and the bands that were size-specific for the amplified gene of interest, were cut and purified by Wizard® SV Gel and PCR Clean Up System (PromegaTM, Madison, WI, USA) according to the manufacturer's instructions. RNA of Influenza A virus (H3N2, A/Hong Kong/8/68 strain; ATCC VR-544) was also isolated and used to amplify NS1 gene by RT-PCR using NS1-specific primers [25]. Influenza A NS1 protein was used in the following experiments as inhibitor of the innate immune host response [17, 26].
Cloning and Expression of HCoV-OC43 and Influenza A Genes in HEK-293 Cells
RT-PCR products were cloned using the pAcGFP1-N In-Fusion® Ready Vector (ClontechTM, Takara Bio Company, Mountain View, CA, USA) that encodes a green fluorescent protein (GFP) from Aequorea coerulescens allowing the expression of the fusion protein to the N-terminus of AcGFP1. The ligation reaction was set up by utilizing the In-Fusion® HD Cloning Kit (ClontechTM). Competent TOP10 Escherichia coli cells (InvitrogenTM, Carlsbad, CA, USA) were used for transformation. The PureYieldTM Plasmid Miniprep Kit (PromegaTM) was used to isolate the vector according to the manufacturer's instructions. Confirmation of successful cloning was achieved using restriction digestion and direct sequencing (data not shown). HEK-293 cells were seeded at 5 × 105 per well of 96-well plates, and transfected with ns2a-pAcGFP1, ns5a-pAcGFP1, M-pAcGFP1, N-pAcGFP1, or NS1-pAcGFP1 vector using Lipofectamine® 2000 (PromegaTM) according to the manufacturer's instructions. Expression of ns2a, ns5a, M, N and NS1 proteins was confirmed by indirect immunofluorescence assay using anti-GFP monoclonal antibody (10 μg/mL) (Clone JL-8, ClontechTM) (data not shown). Expression of N and NS1 proteins was further confirmed by immunofluorescence assay using monoclonal antibody against HCoV-OC43 N (EMD MilliporeTM, Billerica, MA, USA) and polyclonal antibody against influenza A NS1 protein (EMD MilliporeTM), respectively (data not shown).
Transcriptional Activation of Antiviral Response Elements in the Presence of HCoV-OC43 and Influenza A Proteins
The IFN-β promoter was amplified using previously described primers [27], and inserted into the reporter pGL4.19(luc2CP/Ne) vector (PromegaTM) between the restriction sites BglII and HindIII according to the manufacturer's instructions, creating the pIFNβ-luc plasmid. The reporter vectors coding for firefly luciferase under the control of ISRE, pISRE-luc (pGL4.45[luc2P/ISRE/Hygro]) or NF-κB-RE, pNFκB-RE-luc (pGL4.32[luc2P/NF-κB-RE/Hygro]) were purchased from PromegaTM. Following 48 h of transfection with one of the expression vectors cited above, HEK-293 cells were co-transfected with pIFNβ-luc, pISRE-luc or pNFκB-RE-luc vector at 0.1 μg/well using Lipofectamine® 2000 (PromegaTM). Renilla luciferase vector (pRL-TK) (PromegaTM) was used at 10 ng/well as an internal control for transfection efficiency. Following 24 h of incubation, the cells were stimulated for 6 or 24 h with IFN-α (PBL Assay ScienceTM, Piscataway, NJ, USA) at 5 × 104 units/well, Sendai virus (SeV) (ATCC VR-105) at multiple of infection 1, or tumor necrosis factor-α (TNF-α) (Life TechnologiesTM) at 0.1 μg/mL. Thereafter, promoter activity was assayed using the Dual-Luciferase® Reporter Assay System (PromegaTM) and the GloMax®-Multi Jr luminometer (PromegaTM) according to the manufacturer's instructions. Firefly luciferase levels were determined in relative light units and normalized against Renilla luciferase levels.
Antiviral Gene Expression Profile in Cells Expressing HCoV-OC43 ns2a, ns5a, M, and N Protein
Total RNA from transfected and mock-transfected HEK-293 cells was extracted using RNeasy® Kit (QiagenTM) with on-column DNase digestion according to the manufacturer's instructions. The RT2 First Strand Kit (QiagenTM) was used for cDNA synthesis. The RT reaction was then added to RT2 SYBR Green ROX qPCR Mastermix (QiagenTM), and the Human Antiviral Response PCR array (QiagenTM) was used to profile the antiviral gene expression in HEK-293 cells on ABI 7500 Fast Real-Time PCR system (Applied BiosystemsTM). The PCR array monitored the transcriptional activity of different genes involved in the activation of antiviral and pro-inflammatory proteins following amplification of RNA transcripts by real-time RT-PCR. However, we only profiled the expression of genes relevant to the antiviral response elements. Cycle threshold (CT) values were exported to an Excel file to create a table of CT values. This table was then uploaded on to the Qiagen data analysis web portal (http://www.qiagen.com/geneglobe). CT values were normalized to glyceraldehyde 3-phosphate dehydrogenase. Fold regulation comparison was used to profile antiviral gene expression. Fold change/regulation was calculated using the delta-delta CT method (ΔΔCT). The fold regulation threshold was ≥2 for upregulation and ≥-2 for downregulation.
Statistical Analysis
Fold change in firefly luciferase from three different experiments was summarized as mean ± standard deviation. The difference in fold change mean between two groups was determined by independent-samples t test. p values < 0.05 were considered significant. All statistical analyses were performed using SPSS statistics software version 25.0 for windows (IBM Corporation, Armonk, NY, USA). GraphPad Prism 7 software (GraphPad Software Inc., La Jolla, CA, USA) was used to generate all charts.
Results
Effect of HCoV-OC43 proteins on the transcriptional activation of antiviral response elements.
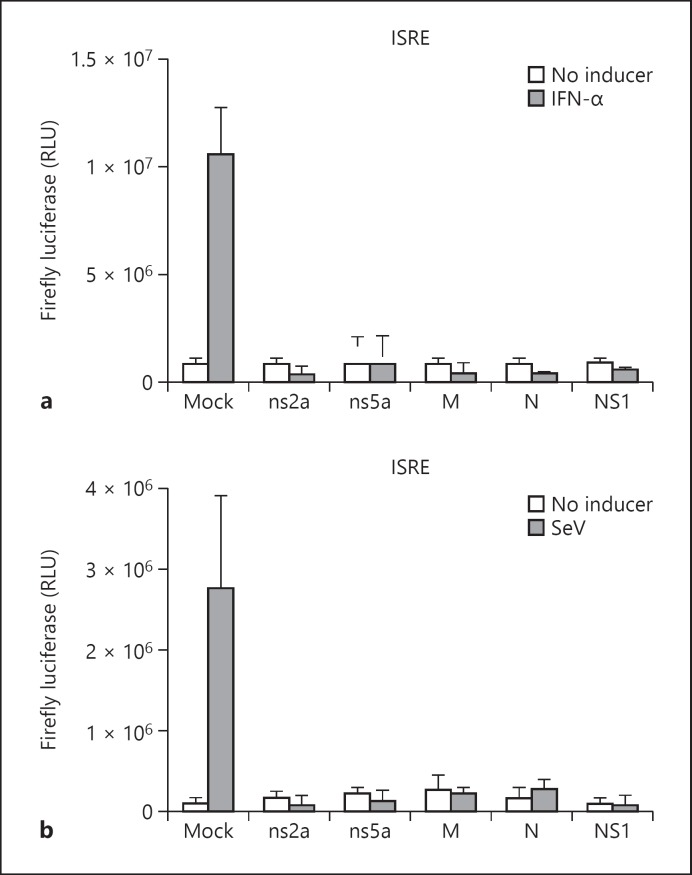
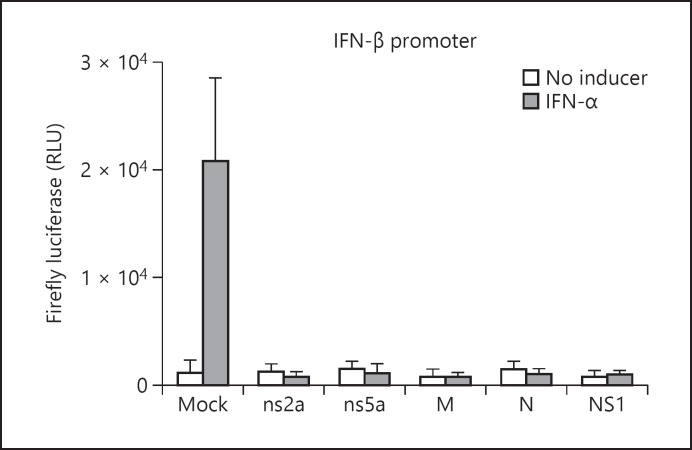
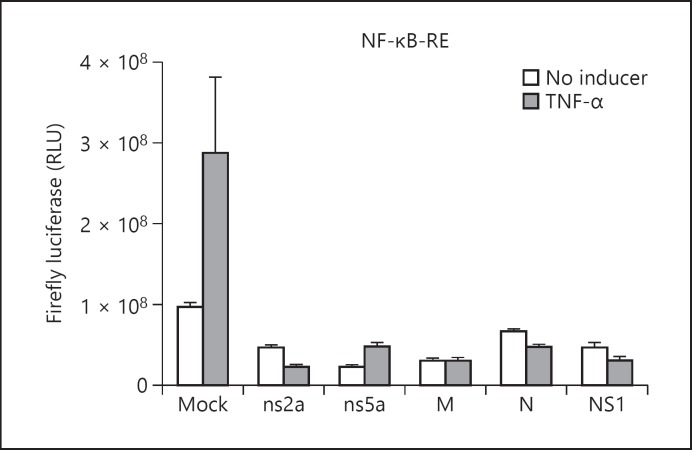
The transcriptional activity of ISRE, IFN-β promoter, and NF-κB-RE was assessed by measuring the firefly luciferase levels in HEK-293 cells transfected with ns2a-pAcGFP1, ns5a-pAcGFP1, M-pAcGFP1, N-pAcGFP1, or NS1-pAcGFP1 vector. In control cells nonexpressing viral protein, the firefly luciferase levels were highest following 6 h of stimulation with one of the inducers (data not shown). In the absence of an inducer, the firefly luciferase levels in HEK-293 cells expressing one HCoV-OC43 protein were similar to the background levels (Fig. 1a, b). In IFN-α-treated HEK-293 cells, the expression of firefly luciferase under the control of ISRE or IFN-β promoter was inhibited in the presence of one of the tested HCoV-OC43 proteins or in the presence of influenza A NS1 protein (Fig. 1a, 2). The transcriptional activity of ISRE was also inhibited in SeV-stimulated HEK-293 cells in the presence of one HCoV-OC43 protein (Fig. 1b). In mock-transfected HEK-293 cells, SeV induced only low levels of firefly luciferase under the control of IFN-β promoter (data not shown), and therefore the effect of HCoV-OC43 proteins on the transcriptional activity of IFN-β promoter in SeV-stimulated HEK-293 cells could not be assessed. Following the challenge with TNF-α, the transcriptional activity of NF-κB-RE was inhibited in the presence of one of the tested HCoV-OC43 proteins or in the presence of influenza A NS1 protein (Fig. 3).
Fig. 1.
Mean fold change in firefly luciferase levels under the control of ISRE in IFN-α-treated (a) and SeV-treated (b) HEK-293 cells expressing HCoV-OC43 structural (M or N) or accessory (ns2a or ns5a) protein. Results are shown as the mean ± standard deviation (SD) of three independent experiments. Firefly luciferase activity measurements were normalized against Renilla luciferase levels.
Fig. 2.
Mean fold change in firefly luciferase levels under the control of IFN-β promoter in IFN-α-treated HEK-293 cells expressing HCoV-OC43 structural (M or N) or accessory (ns2a or ns5a) protein. Results are shown as the mean ± standard deviation (SD) of three independent experiments. Firefly luciferase activity measurements were normalized against Renilla luciferase levels.
Fig. 3.
Mean fold change in firefly luciferase under the control of NF-κB-RE in TNF-α-treated HEK-293 cells expressing HCoV-OC43 structural (M or N) or accessory (ns2a or ns5a) protein. Results are shown as the mean ± standard deviation (SD) of three independent experiments. Firefly luciferase activity measurements were normalized against Renilla luciferase levels.
PCR Array Profiling of Antiviral Genes in HEK-293 Cells Expressing HCoV-OC43 Structural and Accessory Proteins
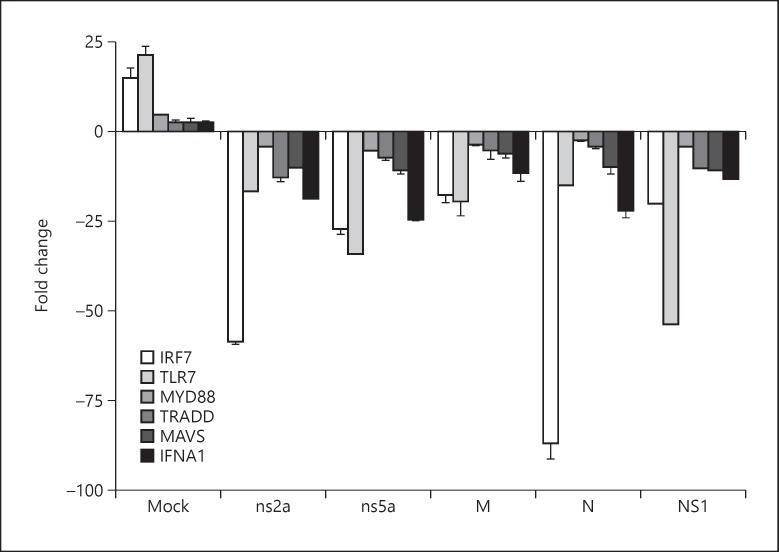
To investigate whether the inhibition of the transcriptional activity of antiviral response elements in the presence of HCoV-OC43 proteins can be associated with a downregulation of the expression of antiviral genes, PCR array profiling of antiviral genes was carried out in transfected and mock-transfected HEK-293 cells. Following SeV challenge of HEK-293 cells, the expression of genes involved in the type I IFN and NF-κB signaling pathways was downregulated in the presence of HCoV-OC43 structural or accessory proteins (Fig. 4). Similar results were obtained in the presence of the influenza A NS1 protein. The mean negative fold change for TRADD gene in the presence of accessory protein ns2a was greater than in the presence of other HCoV-OC43 proteins (p < 0.05). The expression of IRF7 gene was more downregulated in the presence of ns2a or N protein than in the presence of other HCoV-OC43 proteins (p < 0.001), whereas the expression of TLR7 gene was more downregulated in the presence of ns5a protein than in the presence of other HCoV-OC43 proteins (p < 0.01). Remarkably, IRF7 and IFNA1 gene expression was more downregulated in the presence of ns2a, ns5a or N protein than in the presence of influenza A NS1 protein.
Fig. 4.
Mean fold change in antiviral gene expression in SeV-stimulated HEK-293 cells expressing HCoV-OC43 structural (M or N) or accessory (ns2a or ns5a) protein. Results are shown as the mean ± standard deviation (SD) of two independent experiments. Mean ± SD was not available for influenza A NS1 because only one experiment was performed.
Discussion
Similar to influenza A NS1 protein, HCoV-OC43 structural (M and N) and accessory (ns2a and ns5a) proteins were able to inhibit the transcriptional activity of antiviral response elements, ISRE, IFN-β promoter, and NF-κB-RE, and to downregulate the expression of several genes involved in the activation of an antiviral response.
The inhibition of the ISRE, IFN-β promoter, and NF-κB-RE activity, and the downregulation of the expression of IFNA1, IRF7, TLR7, MYD88, TRADD, and MAVS in the presence of each HCoV-OC43 accessory protein suggest that HCoV-OC43 ns2a and ns5a have the potential to block the type I IFN and NF-κB pathways. Previous studies have shown that SARS-CoV and MERS-CoV accessory proteins have a role in innate immune evasion [9, 17, 20, 21, 22]. However, HCoV-OC43 accessory proteins are not homologous to SARS-CoV and MERS-CoV accessory proteins. Also of interest, the murine coronavirus accessory protein 5a, which is homologous to HCoV-OC43 ns5a protein, was shown to antagonize IFN induction [28].
SARS-CoV and MERS-CoV M proteins can suppress the activity of IFN-β promoter and ISRE [18, 19, 22]. NF-κB activation is also inhibited when SARS-CoV M protein is expressed in Vero or HeLa cells [23]. Our data showed that the transcriptional activity of ISRE, IFN-β promoter, and NF-κB-RE was inhibited in the presence of HCoV-OC43 M protein. In addition, like the accessory proteins, the HCoV-OC43 M protein was able to downregulate the expression of IFNA1, IRF7, TLR7, MYD88, TRADD, and MAVS. These findings suggest that in addition to its function in the assembly of virions [29], HCoV-OC43 M protein is an antagonist of the host antiviral defenses.
The coronavirus N protein binds genomic RNA to form the helical capsid, and has a critical role in coronavirus replication [30, 31]. The SARS-CoV N protein can activate NF-κB-RE in Vero E6 cells [32]. However, SARS-CoV N protein inhibits the promoter activity of ISRE, IFN-β promoter, and NF-κB-RE in 293T cells [17]. Our study showed that the transcriptional activity of ISRE, IFN-β promoter, and NF-κB-RE was inhibited in the presence of HCoV-OC43 N protein. Lai et al. [33] showed that after 24 h of TNF-α treatment, HCoV-OC43 N protein potentiates NF-κB expression in 293T cells by binding its inhibitor, miRNA 9. miRNA 9 expression is induced by stimuli which activate NF-κB, such as TNF-α and lipopolysaccharide [34]. Unfortunately, the authors did not show results for NF-κB-RE activation after 6 h of TNF-α treatment, the optimal time for NF-κB-RE activation in our experiments. The influenza A virus that is known to inhibit IRF3, AP-1, and NF-κB activation [35, 36] was shown to upregulate the miRNA 9 expression [37]. We speculate that the expression of N protein results in transient downregulation of NF-κB expression through downregulation of its mediators (e.g., TLR7, MYD88, TRADD), leading to early inhibition of NF-KB-RE activation, and that the accumulation of miRNA9 in cells with time will lead to the formation of N protein/miRNA 9 complex and the removal of inhibition on NF-KB and NF-KB-RE.
In conclusion, HCoV-OC43 has the ability to inhibit the transcriptional activation of antiviral response elements, and the expression of antiviral genes. This would lead to the disruption of several antiviral signaling pathways such as the type I IFN and NF-κB pathways. Further studies are needed to confirm the role of HCoV-OC43 proteins in antagonizing the host innate immunity.
Disclosure Statement
The authors report no conflicts of interest.
Acknowledgement
This work was in part supported by Kuwait University Research Administration Grant No. YM 04/15.
References
- 1.Pitkaranta A, Jero J, Arruda E, Virolainen A, Hayden FG. Polymerase chain reaction-based detection of rhinovirus, respiratory syncytial virus, and coronavirus in otitis media with effusion. J Pediatr. 1998;133:390–394. doi: 10.1016/S0022-3476(98)70276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lepiller Q, Barth H, Lefebvre F, Herbrecht R, Lutz P, Kessler R, et al. High incidence but low burden of coronaviruses and preferential associations between respiratory viruses. J Clin Microbiol. 2013;51:3039–3046. doi: 10.1128/JCM.01078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razuri H, Malecki M, Tinoco Y, Ortiz E, Guezala MC, Romero C, et al. Human coronavirus-associated influenza-like illness in the community setting in Peru. Am J Trop Med Hyg. 2015;93:1038–1040. doi: 10.4269/ajtmh.15-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talbot HK, Shepherd BE, Crowe JEJ, Griffin MR, Edwards KM, Podsiad AB, et al. The pediatric burden of human coronaviruses evaluated for twenty years. Pediatr Infect Dis J. 2009;28:682–687. doi: 10.1097/INF.0b013e31819d0d27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeh EA, Collins A, Cohen ME, Duffner PK, Faden H. Detection of coronavirus in the central nervous system of a child with acute disseminated encephalomyelitis. Pediatrics. 2004;113:e73–6. doi: 10.1542/peds.113.1.e73. [DOI] [PubMed] [Google Scholar]
- 6.Morfopoulou S, Brown JR, Davies EG, Anderson G, Virasami A, Qasim W, et al. Human coronavirus OC43 associated with fatal encephalitis. N Engl J Med. 2016;375:497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 7.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 8.Vijgen L, Keyaerts E, Lemey P, Moës E, Li S, Vandamme A-M, et al. Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology. 2005;337:85–92. doi: 10.1016/j.virol.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McBride R, Fielding BC. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses. 2012;4:2902–2923. doi: 10.3390/v4112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber G. The molecular basis for differential type I interferon signaling. J Biol Chem. 2017;292:7285–7294. doi: 10.1074/jbc.R116.774562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 12.Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15:407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy DE, Darnell JEJ. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 15.Stark GR, Darnell JEJ. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siu K-L, Chan C-P, Kok K-H, Chiu-Yat Woo P, Jin D-Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell Mol Immunol. 2014;11:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lui P-Y, Wong L-YR, Fung C-L, Siu K-L, Yeung M-L, Yuen K-S, et al. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg Microbes Infect. 2016;5:e39. doi: 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, et al. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Ye F, Zhu N, Wang W, Deng Y, Zhao Z, et al. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, et al. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4:951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang X, Gao J, Zheng H, Li B, Kong L, Zhang Y, et al. The membrane protein of SARS-CoV suppresses NF-kappaB activation. J Med Virol. 2007;79:1431–1439. doi: 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siu K-L, Kok K-H, Ng M-HJ, Poon VKM, Yuen K-Y, Zheng B-J, et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HK, Tang JWT, Kong DHL, Koay ESC. Simplified large-scale sanger genome sequencing for influenza A/H3N2 virus. PLoS One. 2013;8:e64785. doi: 10.1371/journal.pone.0064785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayman A, Comely S, Lackenby A, Murphy S, McCauley J, Goodbourn S, et al. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology. 2006;347:52–64. doi: 10.1016/j.virol.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Wang G, Zhang H, Xin G, Zhang D, Zeng J, et al. Effects of NS1 variants of H5N1 influenza virus on interferon induction, TNF a response and p53 activity. Cell Mol Immunol. 2010;7:235–242. doi: 10.1038/cmi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koetzner CA, Kuo L, Goebel SJ, Dean AB, Parker MM, Masters PS. Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus. J Virol. 2010;84:8262–8274. doi: 10.1128/JVI.00385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locker JK, Rose JK, Horzinek MC, Rottier PJ. Membrane assembly of the triple-spanning coronavirus M protein. Individual transmembrane domains show preferred orientation. J Biol Chem. 1992;267:21911–21918. doi: 10.1016/S0021-9258(19)36699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker MM, Masters PS. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 1990;179:463–468. doi: 10.1016/0042-6822(90)90316-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo L, Masters PS. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Q-J, Ye L-B, Timani KA, Zeng Y-C, She Y-L, Ye L, et al. Activation of NF-kappaB by the full-length nucleocapsid protein of the SARS coronavirus. Acta Biochim Biophys Sin (Shanghai) 2005;37:607–612. doi: 10.1111/j.1745-7270.2005.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai FW, Stephenson KB, Mahony J, Lichty BD. Human coronavirus OC43 nucleocapsid protein binds microRNA 9 and potentiates NF-κB activation. J Virol. 2014;88:54–65. doi: 10.1128/JVI.02678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao S, Song L, Li J, Zhang Z, Peng H, Jiang W, et al. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol. 2012;14:1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 36.Ruckle A, Haasbach E, Julkunen I, Planz O, Ehrhardt C, Ludwig S. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-kappaB pathway and p52/RelB-dependent gene expression in lung epithelial cells. J Virol. 2012;86:10211–10217. doi: 10.1128/JVI.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong C, Sun X, Guan Z, Zhang M, Duan M. Modulation of influenza A virus replication by microRNA-9 through targeting MCPIP1. J Med Virol. 2017;89:41–48. doi: 10.1002/jmv.24604. [DOI] [PubMed] [Google Scholar]