Abstract
Objective
To investigate the effect of the leucine zipper-like motif between HRA and HRB of the human parainfluenza virus 3 fusion protein on fusion activity.
Methods
Site-directed mutagenesis was utilized to substitute the heptadic residues at 257, 264, 271, 278, 285, 292, and 299 in this motif with alanine. Additionally, 3 middle heptadic leucine residues at 271, 278, and 285 were replaced with alanine singly or in combination. A vaccinia virus-T7 RNA polymerase transient expression system was employed to express the wild-type or mutated fusion (F) proteins. Three different types of membrane fusion assays were performed to analyze the fusogenic activity, fluorescence-activated cell sorting (FACS) analysis was executed to examine the cell surface expression level, and a coimmunoprecipitation assay was conducted to probe the hemagglutinin-neuraminidase (HN)-F interaction at the cell surface.
Results
All of the substitutions in this motif exhibited diminished or even lost fusion activity in all stages of fusion, although they all had no effect on cell surface expression. In the coimmunoprecipitation assay, all mutants resulted in decreased detection of the HN-F complexes compared with that of the wild-type F protein.
Conclusions
This motif has an important influence on fusion activity, and its integrality is indispensable for membrane fusion.
Key Words: Human parainfluenza virus 3, Fusion protein, Leucine zipper-like motif, Mutations, Membrane fusion
Introduction
Human parainfluenza virus 3 (hPIV-3) can cause serious respiratory illnesses including pediatric croup, bronchiolitis, and pneumonia in the human pediatric population worldwide [1, 2, 3]. At present, no licensed vaccines or antiviral agents are available for hPIV-3. The main hurdle for the development of safe and effective vaccines is that the mechanism of hPIV-3 infection of the host cells is unclear.
Penetration of this enveloped virus into host cells occurs via merging of viral envelopes with susceptible cell membranes, which results in delivery of the viral genome (a single-stranded, nonsegmented, negative-sense RNA) into the cytoplasm of the target cell [4]. Two types of virion-associated surface glycoproteins, i.e. the hemagglutinin-neuraminidase (HN) protein and the fusion (F) protein, cooperate to facilitate this process [5]. The HN glycoprotein is responsible for the initial binding between the virus and the target cells [6, 7], whereas the F glycoprotein is involved in disruption of the host-cell plasma membrane and direct induction of membrane fusion [8].
The hPIV-3 F protein, which is classified as a type I integral membrane protein [9], is synthesized as an inactive precursor, designated F0, that is then activated by proteolytic cleavage during transport through the Golgi membranes into a disulfide-linked heterodimer of F1and F2[10]. Several structural features of this protein are known to be important for its fusion activity. A stretch of hydrophobic amino acids at the amino terminus of the F1 subunit, termed the 'fusion peptide', is considered to be responsible for insertion into the host cell membrane to initiate the fusion process [5, 11, 12]. In addition, 2 heptad repeat regions (i.e. HRA and HRB) in the F1 polypeptide are believed to have important roles in membrane fusion. HRA is located at the carboxyl terminal of the fusion peptide, and HRB is located immediately upstream of the transmembrane region. Upon triggering, these 2 domains undergo conformational changes to form a stable 6- stranded coil that pulls the viral and target cell membranes together [9, 12, 13].
There is a large intervening region between HRA and HRB, and sequence analysis indicates that it contains a leucine zipper-like motif. In this highly conserved motif, characterized as a periodic repeat of leucine or isoleucine residues every 7 amino acids, nonpolar residues are found at all first and most fourth positions when displayed on an α-helical wheel [14, 15]. Previous studies of this motif in 3 different paramyxovirus systems have suggested that this region is important for fusion activity [16, 17, 18].
However, whether the leucine zipper-like motif located between HRA and HRB of the hPIV-3 F protein plays an important role in fusion activity has not yet been determined. To investigate the effect of this domain on fusion activity, point mutations were introduced by using site-directed mutagenesis to substitute the heptadic residues at amino acids 257, 264, 271, 278, 285, 292, and 299 in this motif with an alanine residue. Additionally, 3highly conserved middle heptadic leucine residues (i.e. L271, L278, and L285) were replaced with alanine in various combinations. The effects on the fusion activity, cell surface expression, and HN-F interactions of such mutated F proteins were examined. Our data showed that all of the mutants, whether individually or in combination, caused no reductions in the cell surface expression levels of F, but they led to a decreased fusogenic activity in all steps of membrane fusion. Thus, it seems that the defective fusogenic activity of these mutated F proteins is related to decreased HN-F interactions at the cell surface.
Materials and Methods
Cell and Viruses
BHK-21 cells, obtained from the American Type Culture Collection, were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, USA) supplemented with 10% fetal calf serum (FCS).
Fresh human erythrocytes (RBC) were obtained from the physical examination center of Qilu Hospital of Shandong University. This study was approved by the Institutional Review Board of Shandong University. Written informed consent was obtained from all patients, and the hospital ethics committee approved the experiments.
Wild-type (wt) vaccinia virus was used to quantify cell fusion, and recombinant vaccinia virus vTF7-3, a generous gift from Dr. Bernard Moss, was used to provide T7 RNA polymerase in the vaccinia-T7 RNA polymerase expression system [19]. Viruses were maintained in BHK-21 cells.
Recombinant Plasmid Vectors and the Transient-Expression System
The hPIV-3 HN and F genes were cloned into pBluescript SK(+) (pBSK+) at the BamH I site to generate pBSK-HN and pBSK-F, as previously described [20]. Recombinant plasmids were purified from Escherichia coli TG1 cells using an ENZA Plasmid Miniprep Kit (Omega Bio-Tek Inc., Norcross, Ga., USA).
The wt and all mutated F proteins were expressed in BHK-21 cells using the vaccinia virus-T7 (vTF7-3) RNA polymerase expression system, as previously described [19]. In all experiments, except in the spectrofluorometric analysis of lipid mixing, BHK-21 cells were seeded in 6-well plates for 1 day prior to use at 4 × 105 cells/well. Monolayer cells were infected with the recombinant vaccinia virus expressing vTF7-3 at a multiplicity of infection (MOI) of 10 for 1 h at 37°C in a 5% CO2 incubator and transfection was performed as recommended by Mirza et al. [21], using Lipofectamine™ 2000 (Invitrogen) at 1 μg for each plasmid.
Site-Directed Mutagenesis
Site-directed mutagenesis was performed according to protocols previously described [22]. The hPIV-3 F recombinant plasmid vector was used as the template in each PCR mutagenesis to generate alanine codon substitutions. Two pairs of reverse-complement mutagenesis primers (Sangon Biotech Co. Ltd., Shanghai, China) were designed to only mutate the target amino acids. The primers were as follows (sequences were started with the 5′ nucleotide, and the mutant sites are underlined in the primer sequences):
VP1 (vector): 5′-GTG ACT GGT GAG TAC TCA ACC AAG TC-3′
VP2 (vector): 5′-GA CTT GGT TGA GTA CTC ACC AGT CAC-3′
L257A-P1: 5′-T TAT GAT CTA GCA TTT ACA GAA T-3′
L257A-cP1: 5′-TGA TTC TGT AAA TGC TAG ATC ATA A-3′
V264A-P2: 5′-GAA TCA ATA AAG GCG AGA GTT ATA G-3′
V264A-cP2: 5′-C TAT AAC TCT CGC CTT TAT TGA TTC-3′
L271A-P3: 5′-ATA GAT GTT GAC GCG AAT GAT TAC T-3′
L271A-cP3: 5′-A GTA ATC ATT CGC GTC AAC ATC TAT-3′
L278A-P4: 5′-TAC TCA ATC ACC GCC CAA GTC AGA C-3′
L278A-cP4: 5′-G TCT GAC TTG GGC GGT GAT TGA GTA-3′
L285A-P5: 5′-AGA CTC CCT TTA GCA ACT AGA CTG C-3′
L285A-cP5: 5′-G CAG TCT AGT TGC TAA AGG GAG TCT-3′
Q292A-P6: 5′-G CTG AAC ACC GCG ATT TAC AGA GTA-3′
Q292A-cP6: 5′-TAC TCT GTA AAT CGC GGT GAA CAG C-3′
I299A-P7: 5′-A GTA GAT TCC GCA TCA TAT AAC ATC-3′
I299A-cP7: 5′-GAT GTT ATA TGA TGC GGA ATC TAC T-3′.
The pBSK-F plasmid digested by Kpn I was used as the template to generate one PCR fragment with primers L257A-P1 (complemented with L257A-cP1) and VP2 (complemented with VP1). The pBSK-F plasmid digested by Not I was used as the template to generate the other PCR fragment with primers L257A-cP1 (complemented with L257A-P1) and VP1 (complemented with VP2). Two PCR products with homologous ends were transformed into E. coli TG1 cells that were then selected for ampicillin resistance. Identification of colonies carrying the mutated F genes was facilitated by screening for the presence of 2 unique restriction sites (i.e. the KPN I site and the Not I site). Finally, mutated F protein genes were sequenced in their entirety to verify the desired mutation(s) and that no other changes in the F protein were present. The other single mutants were obtained with the same procedures. Double and triple mutants were made by sequential mutagenesis on previously mutagenized templates. All mutations, as verified by DNA sequencing, were successfully introduced.
Syncytium Formation Assay
Monolayers of BHK-21 cells grown in 6-well plates were cotransfected with cDNA encoding wt or mutated F proteins along with hPIV-3 HN proteins or empty vector alone, with a total amount of 1 μg of DNA per well. At 36 h posttransfection, transfected monolayers were washed with phosphate-buffered saline (PBS) and fixed with methanol for 5 min. Then, the cells were stained with Giemsa Accustain (Sigma Chemical Co.) for syncytium observation [23]. In each field of view, all syncytia with 3 or more nuclei were counted. Representative fields were captured with an inverted microscope (Olympus IX71). Quantification of syncytia was accomplished by measuring the syncytia-covered area in 3 random fields and comparing this with the total area of the field. Areas were quantified using the analysis tool in Adobe Photoshop CS6 [24].
β-Galactosidase Assay for Content Mixing
The content-mixing assay, based on the cytoplasmic activation of the reporter gene β-galactosidase, was essentially performed as previously described [22], with slight modifications. One population of BHK-21 cells was infected with the recombinant vTF7-3 vaccinia virus as described above and cotransfected with the desired F and hPIV-3 HN genes. A second population of BHK-21 cells was infected with the wt vaccinia virus (MOI = 10) and transfected with 1 μg per well of plasmid pG1NT7β-gal, which carries the β-galactosidase gene under the control of a T7 promoter. At 22 h posttransfection, the cells were removed from the wells by trypsinization (0.05% trypsin, 0.53 mM EDTA; Gibco) and washed, pelleted, and resuspended in complete medium.
Equal numbers (1 × 105) of the 2 populations were mixed in triplicate in a 96-well microtiter plate. After incubation at 37°C for 5 h, the cells were lysed and the lysates were assayed for β-galactosidase activity according to the procedures of the High-Sensitivity β-Galactosidase Assay Kit (Stratagene, La Jolla, Calif., USA). The extent of fusion was quantified by measurement of the absorbance at 590 nm with a plate reader (ELX800; Bio-tek Instruments Inc.), with background fusion obtained from cells transfected with comparable amounts of the vector alone subtracted [23].
Dye Transfer Assay
The dye transfer assay was performed as previously described [22, 25]. Fresh human RBC were washed, resuspended in cold PBS (1% hematocrit), and incubated with 15 μl octadecyl rhodamine B (R18; Invitrogen) (1 mg/ml in ethanol) for 30 min at room temperature in the dark. Three volumes of complete medium were added, and incubation continued for an additional 30 min. The RBC were washed and then resuspended in PBS containing 0.1 mM CaCl2 and 1 mM MgCl2 (PBS-CM) (0.1% hematocrit) for downstream experiments. After 22 h of incubation, cell monolayers coexpressing wt or mutated F and HN proteins were washed and treated with 50 mU per ml of neuraminidase (Sigma Chemical Co.) in DMEM for 1 h at 37°C; then, the cells were washed again and overlaid with R18-labeled RBC at 4°C for 30 min with occasional gentle agitation. The cells were washed and then incubated for 60 min at 37°C. Finally, the cells were washed and immediately visualized, counted, and photographed using a Nikon fluorescence microscope.
Spectrofluorometric Analysis of Lipid Mixing
RBC were labeled with R18 in the same way as in the dye transfer assay. Monolayers of BHK-21 cells grown in 6-cm-diameter dishes were infected with vTF7-3 at an MOI of 10 per well for 1 h at 37°C. After being washed twice with DMEM (Gibco), cells were cotransfected with hPIV-3 wt or mutated F and HN genes. Plasmids encoding the HN and F proteins were kept at 2.5 μg, and the total transfected DNA was kept at 5 μg. At 22 h posttransfection, monolayers were washed with PBS and subjected to neuraminidase treatment as described above. Then, the cells were washed twice and incubated with 3 ml R18-labeled RBC (0.1% hematocrit) for 30 min at 4°C. Excess unbound RBC were removed, and the RBC-cell complexes were transferred from the dish into a new 2.0-ml Eppendorf tube for incubation for 30 min at 4°C in a solution of PBS containing 50 mM EDTA. The RBC-cell complexes were washed and placed on ice until further use. Fifty microliters of the R18-labeled RBC acceptor cell suspension was injected into a cuvette containing 3 ml PBS prewarmed to 37°C, and the fluorescence changes were continuously measured using an F-4500 spectrofluorometer (Hitachi, Tokyo, Japan) with 5 s of time resolution at 560- and 590-nm excitation and emission wavelengths, respectively. To normalize the data, the percent fluorescence dequenching (% FDQ) at each time point was calculated according to the following equation: % FDQ = 100(F - F0/Ft - F0), where F0 and F are the fluorescence intensities at time zero and at a given time point, respectively, and Ft is the fluorescence intensity in the presence of 0.1% Triton X-100 and taken as fluorescence at an infinite dilution of the probe [26, 27].
Flow Cytometry
The cell surface expression of hPIV-3 wt and mutated F proteins was examined by flow cytometry as previously described [23]. BHK-21 cells were transfected with plasmids encoding the hPIV-3 wt or mutated F proteins. At 22 h posttransfection, the monolayers were detached, pelleted, washed twice with PBS-FCS (PBS containing 5% FCS), and then incubated with an anti-F monoclonal antibody (CD9-4-3; Millipore) at a dilution of 1:100 in PBS containing 1% BSA at room temperature for 1 h. After 3 washes with PBS-FCS to remove unbound antibodies, the cells were incubated with 1:100 diluted Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G antibodies (Proteintech). Following 2 more washes, the cells were fixed in PBS containing 2% paraformaldehyde at 4°C for 10 min. After being washed 2 additional times, the cells were resuspended in 0.4 ml PBS and subjected to flow cytometry in a FACSCalibur flow cytometer (Becton Dickinson Biosciences). Cells transfected with vector alone were used as controls.
Coimmunoprecipitation Assay
The abilities of hPIV-3 wt or mutated F proteins to interact with hPIV-3 HN proteins at the surface of transfected BHK-21 cells were assayed using a modification of a coimmunoprecipitation assay as previously described [22, 28, 29]. BHK-21 monolayers were grown in 6-well plates (~80% confluence) and transfected with cDNA encoding the HN protein as well as wt or mutated F proteins as described above. At 22 h posttransfection, the monolayers were washed and incubated with PBS-CM containing 1.0 mg/ml EZ-Link sulfo-NHS-SS [sulfosuccinimidyl-2-(biotinamido)-ethyl-1,3′-dithiopropionate]-biotin (Pierce, Rockford, Ill., USA) at 4°C for 40 min with gentle agitation. Unbound biotin was absorbed with 2 ml DMEM, and the cells were washed and lysed on ice using cold PBS containing 1% Triton X-100, 0.5% deoxycholate, and 1 mM phenylmethylsulfonyl fluoride. The cell lysates were clarified by centrifugation and the supernatants were collected into a 1.5-ml Eppendorf tube containing 50 μl anti-HN antibody at a 1:100 dilution that had been cross-linked to Dynabeads protein A (Invitrogen). The anti-HN antibody is a mixture of 2 monoclonal antibodies (i.e. M02122321 and M0301307) that were obtained from Abcam. The mixes were incubated with rotation at room temperature for 30 min to form the Dynabeads-HN Ab-HN Ag-F Ag complexes. After being washed with PBS using a magnet, the bound proteins were released by boiling for 10 min, resuspended in PBS, and then incubated with streptavidin beads (Pierce) overnight at 4°C. Following 2 washes, the beads were resuspended in gel sample buffer. The cells expressing vector, HN, or F alone were used as controls to ensure that the F protein coimmunoprecipitated by the HN antibody occurred only through its interaction with the HN protein.
Polyacrylamide Gel Electrophoresis and Western Blotting
Proteins diluted in gel sample buffer in the presence of 1 M β-mercaptoethanol, were separated in 10% polyacrylamide gels [30]. After electrophoresis, the gels were equilibrated in transfer buffer and transferred to Immobilon-P (Millipore Corp.) membranes. The membranes were blocked, washed, and incubated with the primary antibody (either a monoclonal antibody specific for the F protein or a mixture of 2 anti-HN monoclonal antibodies) diluted to 1:1,000 in PBS-Tween 20 and 0.5% nonfat milk overnight at 4°C. The anti-F antibody is a mouse monoclonal antibody and was purchased from Chemicon International Inc. (MAB10207). After being washed, the blots were incubated with the secondary antibody (goat anti-mouse immunoglobulin G coupled to horseradish peroxidase; ZSGB-BIO, Beijing, China) diluted to 1:2,000 in PBS-Tween 20 for 1 h at room temperature. Following extensive washes, the protein bands were detected using an enhanced chemiluminescence Western blotting detection reagent system (Amersham Biosciences). Quantification of the signal was accomplished using a Fluor-S imager (Bio-Rad).
Statistical Analysis
All results were expressed as means ± SD of at least 3 independent experiments. Statistical analysis was performed using Student's t test, and p < 0.05 was considered statistically significant.
Results
Mutagenesis of Heptadic Residues in the Leucine Zipper-Like Motif of the hPIV-3 F Protein by Alanine Substitution
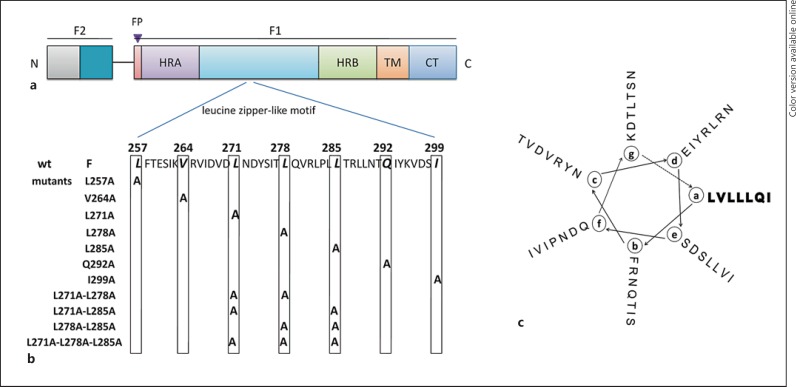
The leucine zipper-like motif of the hPIV-3 F protein is located between HRA and HRB at amino acids 257 to 299 (fig. 1a). The amino acid sequence in this region of the F protein is shown in figure 1b. To investigate the role of the leucine zipper-like motif in F-mediated membrane fusion, alanine substitution mutagenesis on the heptadic residues (LVLLLQI) was carried out. Mutations were introduced both individually and in combination as indicated in figure 1b. The reason we chose alanine rather than other amino acids as a substitute residue is that it has a short side chain that is believed to have a minimal disruptive impact on the potential protein structure. The locations of mutations on the α-helix are displayed in figure 1c.
Fig. 1.
Schematic representation of the locations of mutations. a Domain structure of the hPIV-3 F protein and relative location of the leucine zipper-like motif in relation to other domains in the F1 subunit. Structurally significant domains are shown as different colors of rectangles (colors refer to the online version only). The fusion peptide (FP), the heptad repeat (HR), the transmembrane domain (TM), and the cytoplasmic tail domain (CT) are shown. b Amino acid sequence of the leucine zipper-like motif in the hPIV-3 F protein from amino acids 257-299. Conserved residues forming the leucine zipper are indicated by vertical boxes. The mutant proteins are listed below the wt sequence and substitutions are indicated by the letter A. c Helical-wheel representation of the leucine zipper-like motif of the hPIV-3 F protein. The most amino-terminal residue is placed at position ‘a’ of the α-helix. The heptadic leucine and isoleucine residues are marked by bold letters.
All Mutants Reduced or Even Abrogated Fusion Activity in All Fusion Steps When Coexpressed with the hPIV-3 HN Protein
To investigate the abilities of different mutated F proteins to induce cell-cell fusion in the presence of HN coexpression, 3 different types of membrane fusion assays, i.e. a syncytium formation assay, a content-mixing assay, and a lipid-mixing assay, were performed.
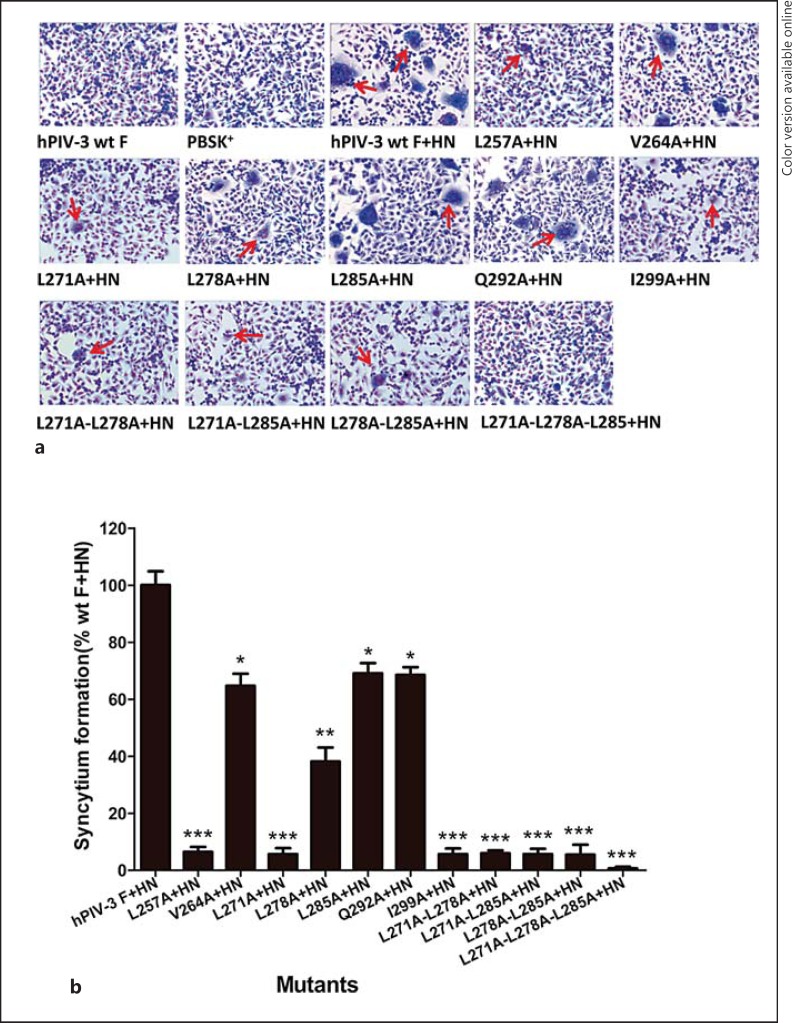
First, to test the mutant F proteins for an overall level of cell-cell fusion, recombinant vaccinia virus-infected BHK-21 cells were monitored for syncytium formation. Multinucleated giant cells were observed at 36 h posttransfection, and photomicrographs from a representative experiment exhibiting syncytia are shown in figure 2a. Cells expressing vector or hPIV-3 wt F protein alone, without coexpression of the hPIV-3 HN protein, were used as negative controls. In contrast to syncytia observed in cells coexpressing the hPIV-3 wt F and HN proteins, the syncytia formed in monolayer cells coexpressing mutated F and HN proteins were not only smaller but also fewer. Quantification of syncytia, expressed as a percentage of those detected in cells coexpressing wt F and HN proteins, is shown in figure 2b. Three single mutants (L257A, L271A, and I299A) and 3 double mutants (L271A-L278A, L271A-L285A, and L278A-L285A) were severely debilitated for syncytium formation (less than 6.0% of that of the wt F and HN proteins), whereas the V264A, L285A, and Q292A mutants displayed a slightly decreased extent of syncytium formation (64.7, 69.1, and 68.5% of the wt F and HN level, respectively). In addition, while the L278A mutant resulted in a decrease in syncytium formation, i.e. to 38.1% of the wt F and HN level, the triple mutant (L271A-L278A-L285A) lost its ability to form syncytia. These results suggest that in the presence of hPIV-3 HN coexpression all of these mutants mediated reduced levels of syncytium formation.
Fig. 2.
Syncytium formation in BHK-21 cells. Cell monolayers were infected with recombinant vaccinia virus and transfected with pBSK+, pBSK+-F, pBSK+-F and pBSK+-HN or pBSK+-HN and mutant F genes. At 36 h posttransfection, the monolayers were fixed with methanol and stained with Giemsa for syncytium observation. a Representative photomicrographs of syncytium formation. Syncytia are indicated by arrows. b Quantification of syncytia was accomplished by measuring the area covered by syncytia and comparing it with the total area of the field for 3 random fields. The data obtained for the mutated F proteins are expressed as percentages of the wt value. Values are shown as means ± SD of 3 separate experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
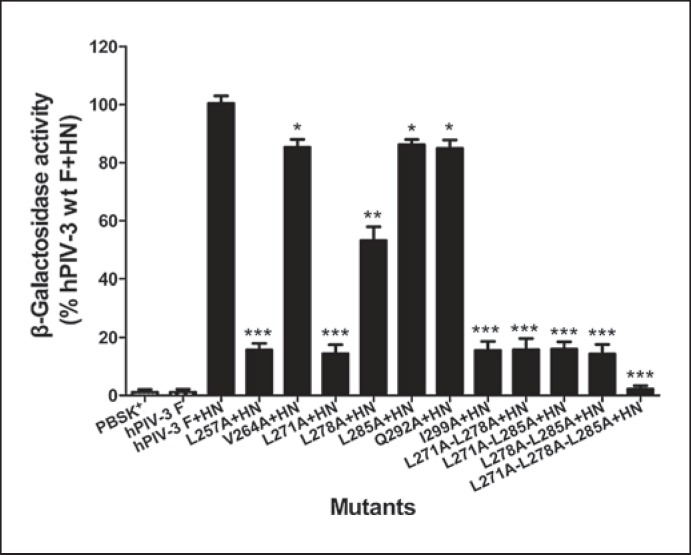
To more accurately quantify the membrane fusion activity of the mutants, we performed a β-galactosidase reporter gene assay to measure the levels of content mixing. For this assay, cells coexpressing the wt or mutated F along with hPIV-3 HN proteins and cells transfected with a plasmid containing the β-galactosidase gene were used to analyze the fusion activity. The results are reported in figure 3. Three single mutants (L257A, L271A, and I299A) and 3 double mutants (L271A-L278A, L271A-L285A, and L278A-L285A) had a markedly diminished fusion activity, corresponding to a percentage of less than 16.0% of the wt F and HN level. Whereas the V264A, L285A, and Q292A mutants retained about 85% of the wt level in content mixing, the L278A mutant reduced the fusion activity to approximately 50% of the wt F level. Additionally, only a background level of fusion activity (2.2% of wt F) was detected from the remaining triple mutant. These results recapitulate those obtained from the syncytium formation assay. These data demonstrate that alanine substitutions resulted in different levels of reductions in content mixing.
Fig. 3.
Content mixing of wt and mutated F proteins. Quantification of the extent of content mixing directed by wt or mutated F proteins with their homologous HN proteins as measured by β-galactosidase activity. Values are expressed as percentages of the fusion activity detected in cells coexpressing HN and wt F proteins. Results are expressed as means ± SD of 3 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
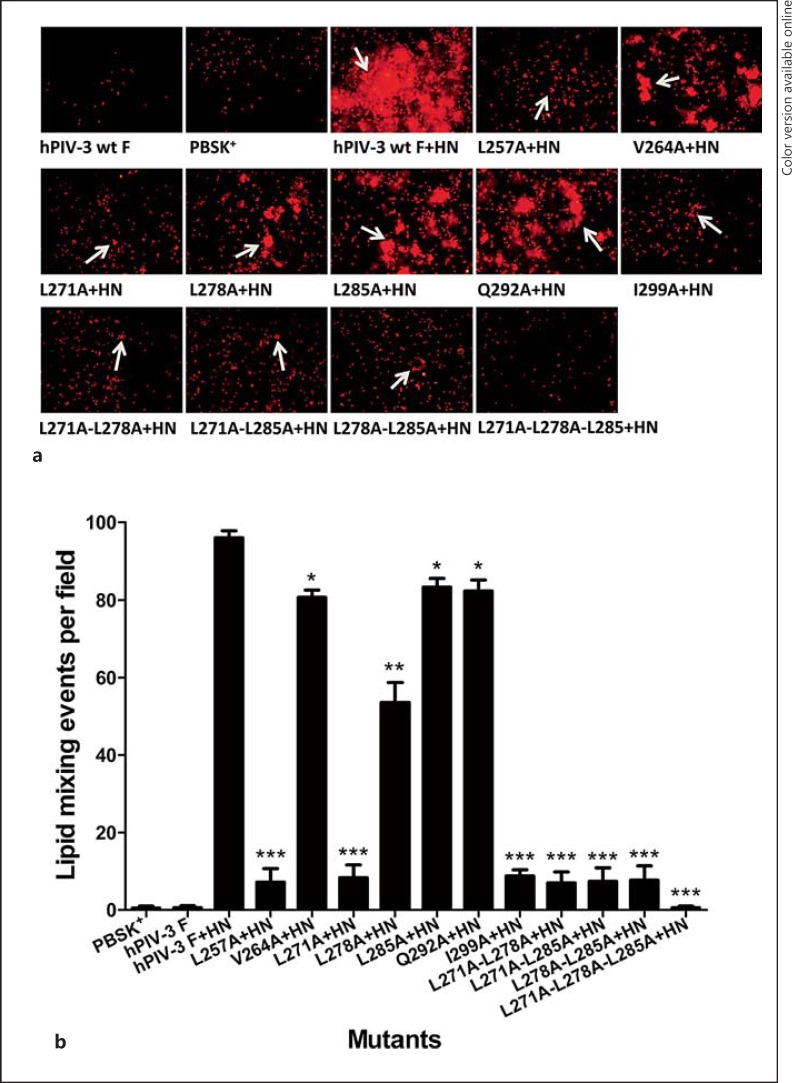
Membrane fusion is generally thought to include the following steps [31, 32]: hemifusion, pore formation, and pore expansion. Because alanine substitutions in the leucine zipper-like motif of the hPIV-3 F protein weakened these mutants' abilities to form syncytia and induce content mixing to various degrees, we decided to test whether these mutants were capable of mediating mixing of the outer leaflets of the plasma membrane, often referred to as hemifusion, which is assessed by transfer of the lipophilic R18 from the membrane of R18-labeled RBC to the membrane of cells coexpressing wt or mutated F and HN proteins. As demonstrated in figure 4a, there was dye transfer from the labeled RBC into underlying cells coexpressing the wt F and HN proteins, but not when labeled RBC were incubated with cells expressing pBSK+ or the wt F protein alone. The extent of dye transfer was expressed as the average number of dye transfer events in 6 microscopic fields. In the presence of hPIV-3 HN coexpression, the overall extent of the dye transfer promoted by the mutated F protein was lower than that of the wt F (fig. 4b). Three single mutants (L257A, L271A, and I299A) and all of the double mutants gave only a very small number of fusion events, almost similar to the negative control level, whereas the V264A, L285A, and Q292A mutants reduced the dye transfer events to 84.1, 86.8, and 85.7% of the wt F level, respectively. The L278A mutant caused approximately half of the dye transfer of wt F, and no dye transfer event was observed from the triple mutant. Therefore, the alanine substitutions resulted in the defective abilities of the mutated F proteins to mediate hemifusion.
Fig. 4.
Hemifusion directed by the wt or mutated F proteins when coexpressed with the hPIV-3 HN protein. Dye transfer was detected as described in Materials and Methods by the spread of fluorescence from R18-labeled red blood cell membranes to underlying transfected BHK-21 cell membranes. Images were monitored by fluorescent microscopy. a Representative photomicrographs of dye transfer caused by hPIV-3 wt or mutated F proteins in the presence of HN; dye transfer events are indicated by arrows. b Quantification of dye transfer caused by the mutated hPIV-3 F proteins. The extent of the dye transfer is expressed as the average number of dye transfer events in 6 microscopic fields. * p < 0.05, ** p < 0.01, *** p < 0.001.
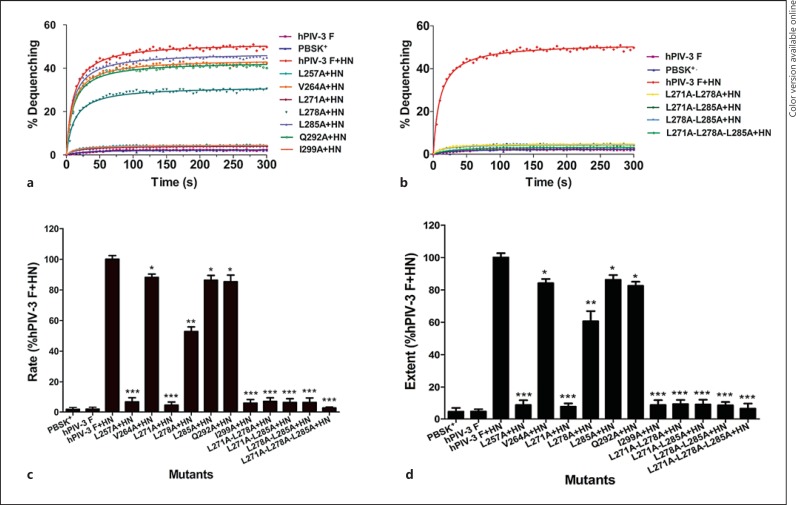
To obtain further quantitative information about the rate and extent of the initial fusion induced by hPIV-3 wt or mutated F along with HN proteins, the R18-labeled RBC were mixed with acceptor cells coexpressing hPIV-3 wt or mutated F and HN glycoproteins; the kinetics of fluorescence dequenching of the R18 probe were measured using a fluorometer. As shown in figure 5a, when cells individually expressing pBSK+ or the hPIV-3 F protein were mixed with R18-labeled RBC, no fluorescence dequenching was detected (blue curve or purple curve; colors refer to the online version only); however, with cells coexpressing wt F and its homologous HN proteins, fluorescence dequenching rapidly and significantly increased (red curve; color refers to the online version only). The rates of fluorescence dequenching were calculated from the maximum slopes of the curves, as shown in figure 5a and b, and normalized to the maximum rate of fusion induced by the wt F and HN proteins (fig. 4c). The maximum extent of fluorescence dequenching at 5 min was calculated from the kinetics curves, as shown in figure 5a and b, and are expressed as the percent activity relative to the level of the wt F and HN proteins (fig. 4d). When cells coexpressing 3 single mutant proteins (L257A, L271A, and I299A), 3 double mutant proteins (L271A-L278A, L271A-L285A, and L278A-L285A), or the triple mutant protein (L271A-L278A-L285A) along with hPIV-3 HN proteins were mixed with R18-labeled RBC, almost no fluorescence dequenching was observed; both the rate and the extent of hemifusion were less than 9.5% of the wt F and HN level. The initial rate of fusion for the V264A, L285A, and Q292A mutated F proteins coexpressed with hPIV-3 HN proteins was 88.8, 87.6, and 85.0%, respectively, and the extent of dequenching at 5 min was 85.5, 87.2, and 83.1%, respectively. Additionally, the initial rate and extent of fusion of cells cotransfected with L278A and hPIV-3 HN genes were reduced to 52.8 and 60.6% of the wt level, with both values being lower than those of cells cotransfected with wt F and HN genes. Consistent with the results obtained from the syncytium formation assay, the content-mixing assay, and the dye transfer assay, the results of spectrofluorometric analysis of lipid mixing showed that the mutated F proteins reduced the rate and extent of hemifusion. Therefore, mutations in the leucine zipper-like motif of the hPIV-3 F protein, when coexpressed with their homologous HN proteins, diminished the fusion activity to various degrees in all stages of membrane fusion.
Fig. 5.
Kinetics of hemifusion induced by hPIV-3 wt or mutated F and HN glycoproteins. BHK-21 cells coexpressing wt or mutated F and HN proteins were incubated with R18-labeled RBC to form the RBC-cell complexes, and the fluorescence measurements were performed as described in Materials and Methods. a, b Kinetics of fluorescence dequenching of the RBC-cell complexes. All data points represent the average of at least 3 independent experiments. Nonlinear regression on data points was performed with GraphPad Prism 5.0. c Quantification of the rates of hemifusion. The rates of fluorescence dequenching were calculated from the maximum slopes of fit curves as shown in a and b, and they were normalized to the maximum rate of hemifusion induced by the wt F and HN proteins. The results shown are the means ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001 of 3 independent experiments. d Measurement of the extent of R18 dequenching kinetics. The maximum extent of fluorescence dequenching at 5 min was calculated from kinetics curves analogous to those shown in figure a and b. Data are expressed as the percent activity with respect to the level of the wt F and HN proteins and are shown as means ± SD of 3 separate experiments. * p < 0.05, ** p < 0.01, *** p < 0.001.
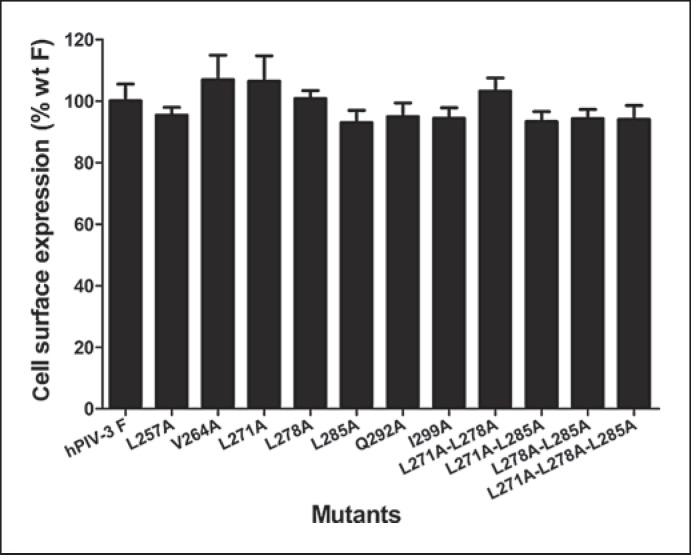
Substitutions in This Motif Cause No Reduction in the Cell Surface Expression of F
To inspect whether the alanine substitutions in the leucine zipper-like motif diminished or even abolished fusion activity by affecting the expression levels of each mutated F protein on the surface of transiently transfected BHK-21 cells, fluorescence-activated cell sorting (FACS) analysis was performed with an anti-F monoclonal antibody as described in Materials and Methods. The levels of mutated F proteins that were transported to the cell surface were indicated by the mean fluorescence intensity value for each mutant as a percentage of that of the wt F after subtracting the background. As shown in figure 6, all of the mutated F proteins were expressed at the cell surface at levels quite comparable to that of the wt F protein. These results indicate that each of the mutated F proteins retained its ability to be efficiently trafficked to the cell surface.
Fig. 6.
Cell surface expression of wt and mutated F proteins. The cell surface expression of the wt or mutated F proteins was detected via flow cytometry using an anti-F monoclonal antibody followed by anti-mouse Alexa Fluor 488-conjugated antibodies. The cell surface expression efficiency of each mutated F protein per cell was assessed using mean fluorescence intensity and is expressed as a percentage of wt F. The means and standard errors are from 3 experiments.
All Mutants Interfered with the Formation of HN-F Complexes at the Cell Surface
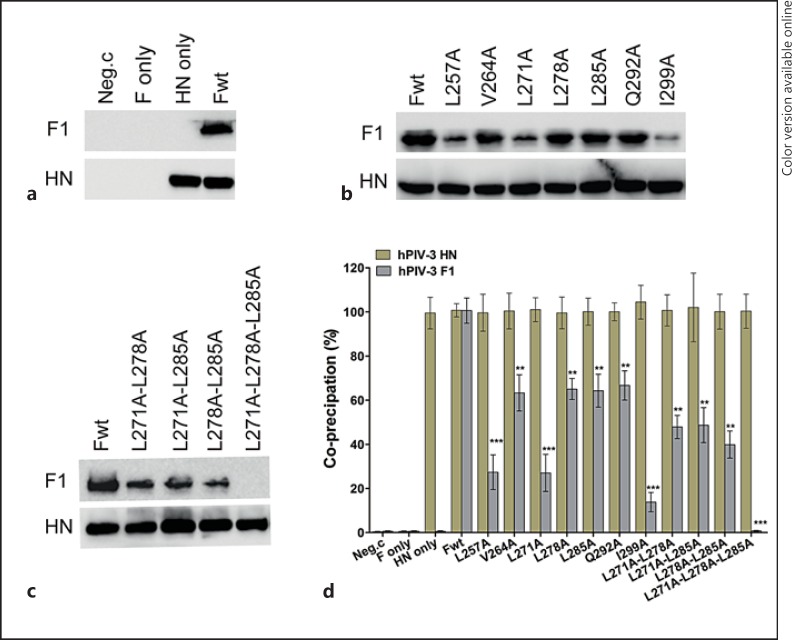
To probe whether the mutated F proteins in the leucine zipper-like motif weakened the membrane fusion activity by altering the interactions between the HN and F proteins at the cell surface, a modified coimmunoprecipitation assay was used to determine the amount of F brought down from extracts of HN-F-cotransfected cells through its interaction with HN by a mixture of 2 anti-HN monoclonal antibodies. The F protein existing in the immunoprecipitates was detected by Western blot analysis using an anti-F monoclonal antibody, while the HN protein was detected in a separate Western blot analysis using 2 anti-HN monoclonal antibodies. Critical controls for this assay are shown in figure 7a. The first lane shows that neither protein can be immunoprecipitated from control cells transfected with empty vector. The second lane demonstrates that the F protein is not immunoprecipitated with a mixture of 2 anti-HN monoclonal antibodies from the extracts of cells individually expressing the F protein, and the third lane demonstrates that the protein that coimmunoprecipitates with HN is not present in cells that have not been transfected with the hPIV-3 F gene. These controls established the specificity of the coimmunoprecipitation of F by the anti-HN antibodies. The relative levels of coimmunoprecipitated F proteins from the extracts of cells coexpressing mutated F and HN proteins were expressed as a percentage of that coimmunoprecipitated from extracts containing the hPIV-3 wt F and HN proteins.
Fig. 7.
Coimmunoprecipitation of hPIV-3 wt or mutated F proteins with hPIV-3 attachment proteins. Surface proteins of cells transfected with the desired plasmids were biotinylated, and then the cells were lysed. Dynabeads-HN Ab-HN Ag-F Ag immune complexes were formed with the Dynabeads-cross-linked anti-HN antibody; after being washed sufficiently, they were resuspended in PBS and then incubated with streptavidin beads as described in Materials and Methods to precipitate the cell surface complexes. The coimmunoprecipitated F proteins in the complexes were detected by Western blot analysis with the anti-F monoclonal antibody (top), while the HN proteins were detected using a mixture of anti-HN monoclonal antibodies (bottom). All proteins were electrophoresed in the presence of a reducing agent. a Critical controls in this coimmunoprecipitation assay: Neg.c = Empty vector; HN only and F only = these plasmids alone; Fwt = wt F protein and HN protein. b, c The results of a coimmunoprecipitation assay for each of the proteins carrying alanine substitutions in the leucine zipper-like motif. wt F protein or the mutants indicated were cotransfected with hPIV-3 HN and are indicated above the gels. d Quantification analysis for b and c using the densitometry method from 3 independent experiments. Data are shown as means ± SD. ** p < 0.01, *** p < 0.001.
As shown in figure 7b and c, almost all mutated F proteins could be coimmunoprecipitated with the hPIV-3 HN protein, except for the triple mutated F protein (L271A-L278A-L285A). The quantification analysis, using the densitometry method, is presented in figure 7d, and the results are from 3 independent experiments. There is no detectable coimmunoprecipitation of the F protein for the triple mutant (L271A-L278A-L285A), consistent with its lost fusogenic activity. Three single mutant proteins (L257A, L271A, and I299A), which had a fusion activity below 16.0% of the wt level, were coimmunoprecipitated with anti-HN monoclonal antibodies below 30% of the wt F amount. Three double mutant proteins (L271A-L278A, L271A-L285A, and L278A-L285A), which exhibited a fusion activity similar to that of L257A, coimmunoprecipitated less than half of the wt F protein. The V264A, L285A, and Q292A mutant F proteins, which had no significant effect on fusion activity (retaining about 85% of the wt F level), could be coimmunoprecipitated at 63.3, 64.3, and 66.7% of the wt level, respectively. In addition, the L278A mutant fused at 53.1% of the wt level and coimmunoprecipitated at 65.1% of the wt F level. Taken together, these results imply that the reduction in fusion activity correlates with the defective abilities of these mutated F proteins to interact with hPIV-3 HN proteins at the cell surface.
Discussion
The leucine zipper motif which contains a periodic repeat of leucine or isoleucine residues every 7 amino acids [33] was found to have an important effect on membrane fusion in paramyxoviruses [16, 17, 34], and there is more than one leucine zipper motif in paramyxovirus fusion proteins. To analyze the role of this motif located between HRA and HRB of the hPIV-3 F protein in fusogenic activity, we performed site-directed mutagenesis to substitute the heptadic leucine residues with the alanine residue, either singly or in combination. Cell surface expression and the interaction between F and HN proteins, which may affect the fusion activity of the F protein, were also analyzed. Interestingly, all of the mutants in this motif exhibited a decreased fusion activity.
In this study, we used 3 different types of membrane fusion assays to quantitatively analyze the effect of mutations in this motif on the fusogenic activity of the F protein. Our results indicated that amino acid residues L257 and I299, which are positioned at either end of the leucine zipper-like domain, displayed an extremely exhausted fusion activity in all stages of the fusion process when they were replaced by alanine. One nonexclusive possibility is that L257 and I299 are not only key residues in stabilizing the structure of this motif but they are also involved in fusion. While V264A and Q292A mutants both retained about 85% of the fusion activity compared with the wt F protein, it appears that the 2 residues were much more tolerant of alanine substitution than the ones located at either end of this motif without significantly affecting its fusogenicity. As shown in figure 8, comparison of the primary amino acid sequence of the leucine zipper-like motifs of different paramyxovirus F proteins suggested that the 3 middle heptadic leucine residues were highly conserved. Analysis of these 3 heptadic leucines, which were replaced individually or in combination by alanine, revealed that single substitution was able to diminish the fusion activity of the protein, while replacement in combination could further reduce or even abrogate fusion activity. These data are in agreement with the results from the mutational analysis of the leucine zipper domain of the murine coronavirus spike (S) protein; substitution of at least 2 heptadic leucine residues is necessary to abrogate fusion activity [35].
Fig. 8.
Comparisons of the primary amino acid sequence of the leucine zipper-like domains of different paramyxovirus F proteins. The heptadic leucine and isoleucine residues are indicated by vertical boxes and are shown in italics.
Since these mutants exhibited similar levels of cell surface expression with respect to the wt F protein, the lack of their fusion activity could not be accounted for by an insufficient amount of F proteins available on the cell surface.
Traditionally, membrane fusion can be divided into 3 sequential stages [30, 31]. Hemifusion, the first step, is the merger of the outer leaflets of the target and effector lipid bilayers and can be measured by the R18 assay [36, 37]. Pore formation follows hemifusion and results in mixing of both outer and inner leaflet membrane lipids with concomitant mixing of aqueous contents; it is measured by a reporter gene assay [26, 27]. Pore expansion is the final stage, it leads to the formation of multinucleated giant cells, and it is measured by syncytium formation assay [24, 38]. Six of the mutated F proteins (L257A, L271A, I299A, L271A-L278A, L271A-L285A, and L278A-L285A), which nearly lost their ability to form syncytia, directed less than 16.0% of the levels of pore formation of the wt F protein and exhibited a hemifusion activity almost similar to the negative control level, with the rate and extent of initial fusion below 9.5% of the wt F level. The minimal levels of hemifusion, the low levels of pore formation, and the almost negative control levels of syncytium formation detected with these mutants suggest that the low levels of syncytium formation were likely due to slow fusion kinetics being unable to efficiently support pore expansion.
In most paramyxovirus systems, including hPIV-3, membrane fusion requires HN and F proteins derived from homologous viruses [39], which suggests that there is a virus-specific interaction between the 2 membrane glycoproteins that is crucial for the induction of cell fusion [5]. Upon attachment of the HN protein to its sialic acid receptor, the HN protein undergoes a conformational change that stimulates the associated F protein refolding into its postfusion conformation. Our study shows that all of the mutations in the leucine zipper-like motif of the hPIV-3 F protein not only diminished or even abrogated fusion activity but also reduced the amount of HN-F complexes. There seems to be a correlation between fusion deficiency and the decreased ability of F to interact with HN. This result seems to be in line with the findings of Melanson and Iorio [40]. Thus, the most reasonable explanation for the defective fusion activity is that these mutant F proteins failed to form a sufficient amount of functional complexes containing the hPIV-3 HN protein, thereby blocking signal transmission between the HN and F proteins. These signals are presumably involved in modulation of the membrane fusion process. It has been reported that the domains M1, M2, and B of the PIV5 F protein and the M1 region of the PIV2 F protein are involved in the HN-F interaction [41, 42]; the corresponding region of this motif of the hPIV-3 F protein in the PIV5 and PIV2 F proteins was identified as being included in these domains by amino acid sequence alignment. Our present findings thus do not exclude the possibility that this motif per se was the constituent of its specific interaction domains with HN.
In summary, the substitutions we have introduced into the leucine zipper-like motif located between HRA and HRB of the hPIV-3 F protein resulted in these mutants having a defective fusion activity in all stages of the fusion process. The mutants formed by replacing the 3highly conserved middle heptadic leucine residues (L271, L278, and L285) in combination, and 2 single mutants (L257A and I299A) at the end of the motif drastically diminished or even abrogated fusogenic activity. Based on these findings, we propose that the leucine zipper-like motif of the hPIV-3 F glycoprotein has an important effect on its fusion activity, and the integrality of this motif is indispensable for membrane fusion. Studying the influence of this motif of the hPIV-3 F protein on fusion activity could provide clues to develop antiviral vaccines and therapeutic drugs to prevent and cure paramyxovirus infection.
Acknowledgements
The authors thank Dr. Ronald Iorio for providing the hPIV-3 F and HN genes, and Dr. Bernard Moss for the recombinant vaccinia virus vTF7-3. Thanks to Dr. Edward C. Mignot (Shandong University) for linguistic advice.
This research was supported by grants from the National Natural Science Foundation of China (No. 81271806) and the Natural Science Foundation of Shandong Province (No. 2009ZRB019VW).
References
- 1.Liu WK, Liu Q, Chen DH, Liang HX, Chen XK, Huang WB, Qin S, Yang ZF, Zhou R. Epidemiology and clinical presentation of the four human parainfluenza virus types. BMC Infect Dis. 2013;13:28. doi: 10.1186/1471-2334-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinberg GA, Hall CB, Iwane MK, Poehling KA, Edwards KM, Griffin MR, Staat MA, Curns AT, Erdman DD, Szilagyi PG. Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. J Pediatr. 2009;154:694–699. doi: 10.1016/j.jpeds.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 3.Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC, Anderson LJ. Seasonal trends of human parainfluenza viral infections: United States, 1990-2004. Clin Infect Dis. 2006;43:1016–1022. doi: 10.1086/507638. [DOI] [PubMed] [Google Scholar]
- 4.Stegmann T, Doms RW, Helenius A. Protein-mediated membrane fusion. Annu Rev Biophys Biophys Chem. 1989;18:187–211. doi: 10.1146/annurev.bb.18.060189.001155. [DOI] [PubMed] [Google Scholar]
- 5.Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 6.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, Colman PM. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Choppin PW, Scheid A. The role of viral glycoproteins in adsorption, penetration, and pathogenicity of viruses. Rev Infect Dis. 1980;2:40–61. doi: 10.1093/clinids/2.1.40. [DOI] [PubMed] [Google Scholar]
- 9.Lamb RA, Paterson RG, Jardetzky TS. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheid A, Choppin PW. Identification of biological activities of paramyxovirus glycoproteins: activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- 11.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 12.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 13.Yu M, Wang E, Liu Y, Cao D, Jin N, Zhang CW, Bartlam M, Rao Z, Tien P, Gao GF. Six-helix bundle assembly and characterization of heptad repeat regions from the F protein of Newcastle disease virus. J Gen Virol. 2002;83:623–629. doi: 10.1099/0022-1317-83-3-623. [DOI] [PubMed] [Google Scholar]
- 14.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 15.O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243:538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 16.Buckland R, Malvoisin E, Beauverger P, Wild F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 17.Reitter JN, Sergel T, Morrison TG. Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J Virol. 1995;69:5995–6004. doi: 10.1128/jvi.69.10.5995-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh JK, Ovadia M, Shai Y. A leucine zipper motif in the ectodomain of Sendai virus fusion protein assembles in solution and in membranes and specifically binds biologically-active peptides and the virus. Biochemistry. 1997;36:15451–15462. doi: 10.1021/bi971152i. [DOI] [PubMed] [Google Scholar]
- 19.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng R, Wang Z, Mirza AM, Iorio RM. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209:457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 21.Mirza AM, Deng R, Iorio RM. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J Virol. 1994;68:5093–5099. doi: 10.1128/jvi.68.8.5093-5099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu FL, Wen HL, Hou GH, Lin B, Zhang WQ, Song YY, Ren GJ, Sun CX, Li ZM, Wang Z. Role of N-linked glycosylation of the human parainfluenza virus type 3 hemagglutinin-neuraminidase protein. Virus Res. 2013;174:137–147. doi: 10.1016/j.virusres.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Ren G, Wang Z, Wang G, Song Y, Yao P, Xu H, Wen H, Zhang W. Effects of heptad repeat regions of F protein on the specific membrane fusion in paramyxoviruses. Intervirology. 2006;49:299–306. doi: 10.1159/000094246. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Felipe L, Villar E, Munoz-Barroso I. Entry of Newcastle disease virus into the host cell: role of acidic pH and endocytosis. Biochim Biophys Acta. 2014;1838:300–309. doi: 10.1016/j.bbamem.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris SJ, Sarkar DP, White JM, Blumenthal R. Kinetics of pH-dependent fusion between 3T3 fibroblasts expressing influenza hemagglutinin and red blood cells: measurement by dequenching of fluorescence. J Biol Chem. 1989;264:3972–3978. [PubMed] [Google Scholar]
- 26.Dutch RE, Joshi SB, Lamb RA. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J Virol. 1998;72:7745–7753. doi: 10.1128/jvi.72.10.7745-7753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagai S, Lamb RA. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apte-Sengupta S, Negi S, Leonard VH, Oezguen N, Navaratnarajah CK, Braun W, Cattaneo R. Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J Biol Chem. 2012;287:33026–33035. doi: 10.1074/jbc.M112.373308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinnes LW, Morrison TG. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J Virol. 2006;80:2894–2903. doi: 10.1128/JVI.80.6.2894-2903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravel KA, McGinnes LW, Reitter J, Morrison TG. The transmembrane domain sequence affects the structure and function of the Newcastle disease virus fusion protein. J Virol. 2011;85:3486–3497. doi: 10.1128/JVI.02308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 33.Buckland R, Wild F. Leucine zipper motif extends. Nature. 1989;338:547. doi: 10.1038/338547a0. [DOI] [PubMed] [Google Scholar]
- 34.Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR., Jr Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo Z, Matthews AM, Weiss SR. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J Virol. 1999;73:8152–8159. doi: 10.1128/jvi.73.10.8152-8159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain S, McGinnes LW, Morrison TG. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J Virol. 2007;81:2328–2339. doi: 10.1128/JVI.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 38.West DS, Sheehan MS, Segeleon PK, Dutch RE. Role of the simian virus 5 fusion protein N-terminal coiled-coil domain in folding and promotion of membrane fusion. J Virol. 2005;79:1543–1551. doi: 10.1128/JVI.79.3.1543-1551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu XL, Ray R, Compans RW. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melanson VR, Iorio RM. Amino acid substitutions in the F-specific domain in the stalk of the Newcastle disease virus HN protein modulate fusion and interfere with its interaction with the F protein. J Virol. 2004;78:13053–13061. doi: 10.1128/JVI.78.23.13053-13061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsurudome M, Ito M, Nishio M, Nakahashi M, Kawano M, Komada H, Nosaka T, Ito Y. Identification of domains on the fusion (F) protein trimer that influence the hemagglutinin-neuraminidase specificity of the F protein in mediating cell-cell fusion. J Virol. 2011;85:3153–3161. doi: 10.1128/JVI.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsurudome M, Ito M, Nishio M, Kawano M, Okamoto K, Kusagawa S, Komada H, Ito Y. Identification of regions on the fusion protein of human parainfluenza virus type 2 which are required for haemagglutinin-neuraminidase proteins to promote cell fusion. J Gen Virol. 1998;79:279–289. doi: 10.1099/0022-1317-79-2-279. [DOI] [PubMed] [Google Scholar]