Tigecycline serves as one of the antibiotics of last resort to treat multidrug-resistant (including carbapenem-resistant) pathogens. However, the recently emerged plasmid-mediated tigecycline resistance mechanism, Tet(X), challenges the clinical efficacy of this class of antibiotics. In this study, we detected 180 tet(X)-harboring Acinetobacter isolates (8.9%, n = 180) from 2,018 samples collected from avian farms and adjacent environments in China.
KEYWORDS: Acinetobacter spp., blaNDM-1, coharboring plasmid, tet(X), tigecycline resistance, waterfowls
ABSTRACT
Tigecycline serves as one of the antibiotics of last resort to treat multidrug-resistant (including carbapenem-resistant) pathogens. However, the recently emerged plasmid-mediated tigecycline resistance mechanism, Tet(X), challenges the clinical efficacy of this class of antibiotics. In this study, we detected 180 tet(X)-harboring Acinetobacter isolates (8.9%, n = 180) from 2,018 samples collected from avian farms and adjacent environments in China. Eighteen tet(X)-harboring isolates (10.0%) were found to cocarry the carbapenemase gene blaNDM-1, mostly from waterfowl samples (94.4%, 17/18). Interestingly, among six Acinetobacter strains, tet(X) and blaNDM-1 were found to colocalize on the same plasmids. Moreover, whole-genome sequencing (WGS) revealed a novel orthologue of tet(X) in the six isolates coharboring tet(X) and blaNDM-1. Inverse PCR suggested that the two tet(X) genes form a single transposable unit and may be cotransferred. Sequence comparison between six tet(X)- and blaNDM-1-coharboring plasmids showed that they shared a highly homologous plasmid backbone even though they were isolated from different Acinetobacter species (three from Acinetobacter indicus, two from Acinetobacter schindleri, and one from Acinetobacter lwoffii) from various sources and from different geological regions, suggesting the horizontal genetic transfer of a common tet(X)- and blaNDM-1-coharboring plasmid among Acinetobacter species in China. Emergence and spread of such plasmids and strains are of great clinical concern, and measures must be implemented to avoid their dissemination.
INTRODUCTION
Antibiotic resistance poses a significant threat to public health. Multidrug-resistant (MDR) pathogens cause nearly 700,000 death worldwide each year (1). Carbapenems have been used as last-line antibiotics for the treatment of infections caused by MDR bacteria (2). Unfortunately, carbapenem-resistant Gram-negative bacteria (CRGN), especially carbapenem-resistant Enterobacteriaceae (CRE), Pseudomonas aeruginosa (CRPA) and Acinetobacter baumannii (CRAB), have emerged in recent years, seriously threatening this class of lifesaving drugs (3–5). Effective antibiotics against CRGN remain limited, with polymyxin and tigecycline being the two choices of last resort. However, the recent discovery of a plasmid-mediated colistin resistance gene, mcr-1, challenges the clinical utility of colistin to treat these infections.
Although colistin and ceftazidime-avibactam have recently been approved for clinical usage in China, tigecycline remains one of the most common antibiotics used to treat complicated CRGN infections in the country. Tigecycline resistance was sporadically reported in clinical isolates, primarily due to chromosome-mediated mechanisms (6, 7). However, two very recent studies described the emergence of a plasmid encoding tigecycline enzymatic inactivation mechanism, Tet(X), in Acinetobacter spp. and Enterobacteriaceae bacteria, including isolates coharboring mcr-1, from animal and human samples in China (8, 9), threatening the clinical efficiency of tigecycline as a last-line antibiotic.
In addition to being common nosocomial pathogens (10, 11), Acinetobacter species strains are widely distributed in a variety of environmental sources, including water, soil, foods, arthropods, and livestock. Thus, antimicrobial resistance genes (ARGs), including tet(X), should be closely monitored in Acinetobacter species isolates from humans, animals (especially livestock), and environment microbiomes. Interestingly, Acinetobacter spp. appear to be common bacterial hosts for the mobile tigecycline resistance gene tet(X) (8), and a further concern is the convergence of carbapenem and tigecycline resistance in Acinetobacter spp., which will simultaneously deactivate the two most important families of antibiotics. Indeed, sporadic cases of tet(X)- and blaNDM-1-positive Acinetobacter isolates have been reported from livestock samples in Jiangsu province in China (8).
In addition to livestock, poultry and other avian species constitute an important reservoir for the transmission of antimicrobial resistance genes. In this study, we described a high prevalence of Acinetobacter isolates coharboring tet(X) and blaNDM-1 from avian and especially waterfowl samples in different regions in China. Interestingly, genomic sequencing analysis revealed the occurrence of two tet(X) genes, tet(X) [formerly named tet(X3), GenBank accession number MK134375] and a novel tet(X) orthologue, in the same Acinetobacter isolates cocarrying blaNDM-1. More importantly, we were the first to describe six novel plasmids concomitantly carrying tet(X) and blaNDM-1 with highly similar backbone structures from various Acinetobacter species and different sources. These findings greatly deepened our understanding of the landscape of tet(X)-harboring plasmids in Acinetobacter spp.
It should be noted that according to the standards of the nomenclature center (http://faculty.washington.edu/marilynr/), all tet(X) genes [formerly named tet(X1) to tet(X5)] at present can only be called tet(X). Therefore, the tet(X3) (GenBank accession no. MK134375) we detected in this study has been replaced by the name tet(X) in the following text.
RESULTS
Molecular epidemiology of tet(X) from avian samples and their adjacent environments in China.
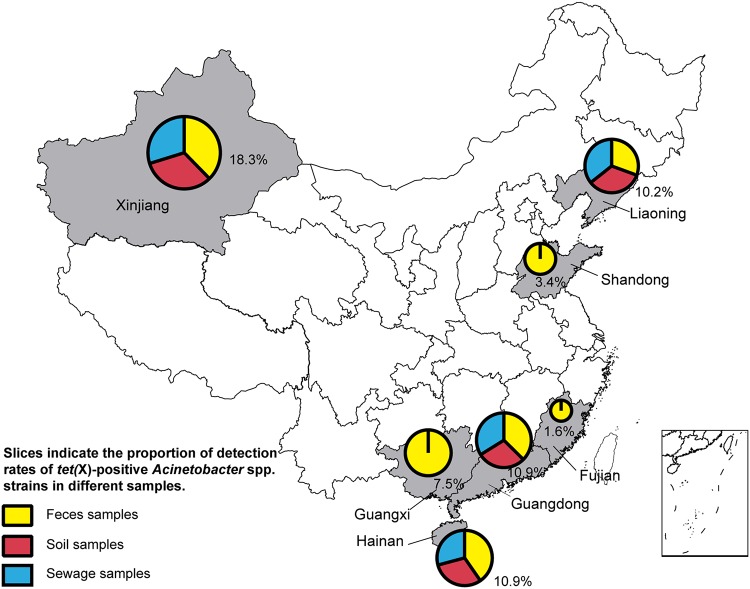
The mobile tigecycline resistance gene tet(X) was detected in 180 (8.9%) Acinetobacter isolates from 2,018 samples collected from avian farms and the adjacent environment, including isolates from all seven sampled provinces, namely, Guangdong, Hainan, Guangxi, Fujian, Shandong, Xinjiang, and Liaoning (Fig. 1 and Table 1). The prevalence of tet(X)-positive Acinetobacter species isolates ranged from 1.6% to 18.3% among the seven provinces, with Xinjiang province having the highest (18.3%) (Table 1). The tet(X)-positive Acinetobacter species isolates were mainly detected in pigeons (37.5%) and waterfowls (geese, 15.0%; ducks, 8.8%), followed by chickens (8.2%) and environmental samples (sewage, 7.5%; soil, 6.7%) (Table 1).
FIG 1.
Map of tet(X)-positive Acinetobacter species strain sampling areas in China. For the seven provinces, the area of each pie graph represents the total detection rate of tet(X)-positive Acinetobacter species strains in all samples in the respective region. Slices indicate the proportion of detection rates of tet(X)-positive Acinetobacter species strains in different samples.
TABLE 1.
Prevalence of tet(X)-positive Acinetobacter species strains in samples from avian farms and adjacent environments in this study
| Province | Origin | Sampling period | No. (%) of tet(X)-positive strains by sample type (no. positive samples/no. of tested samples)a
|
||||
|---|---|---|---|---|---|---|---|
| Feces | Soil | Sewage | Dust | Total overall | |||
| Hainan | Goose farm | 2017 | 8/42 (7.1) (3) | 1/5 (20.0) | 0/5 | 0/5 | 9/57 (15.8) |
| Chicken farm | 2017–2018 | 16/149 (10.7) | 1/16 (6.3) | 3/28 (10.7) | 0/17 | 20/210 (9.5) | |
| Guangdong | Pigeon farm | 2016 | 15/40 (37.5) | 3/10 (30) | 0/4 | 18/54 (29.6) | |
| Chicken farm | 2017 | 3/182 (1.6) | 1/14 (7.1) | 0/6 | 0/6 | 4/208 (1.9) | |
| Duck farm | 2017–2018 | 62/471 (13.2) (8) | 10/107 (9.3) (3) | 10/104 (9.6) | 0/13 | 82/695 (11.9) | |
| Goose farm | 2018 | 5/40 (12.5) (3) | 0/3 | 1/13 (7.7) | 6/56 (10.7) | ||
| Guangxi | Chicken farm | 2017 | 8/60 (13.3) | 0/5 | 0/4 | 8/69 (11.6) | |
| Goose farm | 2017 | 3/25 (12.0) | 3/25 (12.0) | ||||
| Duck farm | 2017 | 2/80 (2.5) | 2/80 (2.5) | ||||
| Xinjiang | Chicken farm | 2017–2018 | 8/38 (20.1) | 2/11 (18.2) | 1/6 (16.7) (1) | 0/5 | 11/60 (18.3) |
| Shandong | Duck farm | 2018 | 9/193 (4.7) | 0/36 | 0/34 | 9/263 (3.4) | |
| Fujian | Duck farm | 2018 | 3/120 (2.5) | 0/36 | 0/36 | 3/192 (1.6) | |
| Liaoning | Chicken farm | 2018 | 2/21 (9.5) | 2/19 (10.5) | 1/9 (11.1) | 5/49 (10.2) | |
| Total | 144/1,461 (9.9) (14) | 17/252 (6.7) (3) | 19/255 (7.5) (1) | 0/50 | 180/2,018 (8.9) (18) | ||
Parenthetical numbers in boldface represent the numbers of tet(X)- and blaNDM-1-coharboring Acinetobacter species strains.
Antimicrobial susceptibility testing revealed that all 180 tet(X)-positive Acinetobacter species isolates were resistant to tigecycline and tetracycline and exhibited high MICs to the newly FDA-approved eravacycline (MIC range, 1 to 4 mg/liter) and omadacycline (MIC, 4 to 16 mg/liter). In addition, 98.9% (n = 178), 97.8% (n = 176), 36.1% (n = 65), 22.8% (n = 41), and 21.7% (n = 39) of isolates also showed resistance to trimethoprim-sulfamethoxazole, ciprofloxacin, ceftazidime, cefotaxime, and gentamicin, respectively. Alarmingly, 10.0% (n = 18) of Acinetobacter species isolates were also resistant to meropenem.
Characteristics of Acinetobacter species isolates coharboring tet(X) and blaNDM-1.
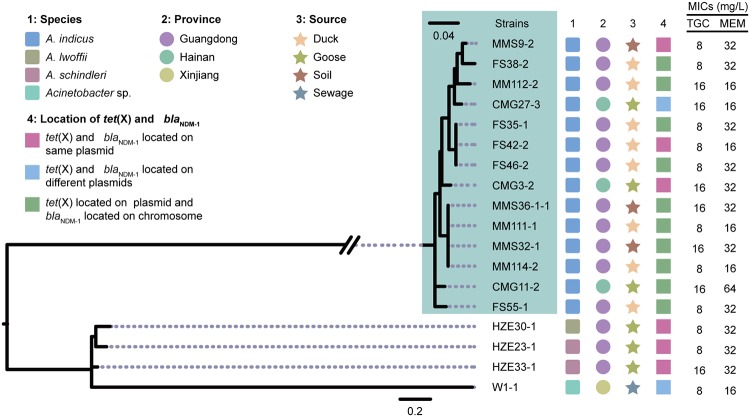
The tigecycline degradation and carbapenem inactivation method (CIM) assays showed that all 18 meropenem-resistant Acinetobacter species isolates could hydrolyze both tigecycline and meropenem, and subsequent PCR and Sanger sequencing indicated that they all carried blaNDM-1 (Fig. 2 and 3). The 18 tet(X)- and blaNDM-1-coharboring strains showed multidrug resistance phenotypes, including resistance to tigecycline, meropenem, ceftazidime, cefotaxime, ampicillin, ciprofloxacin, trimethoprim-sulfamethoxazole, and florfenicol (Table 2). The isolates were discovered from three provinces (Guangdong, Hainan, and Xinjiang) and were identified as Acinetobacter indicus (n = 14), Acinetobacter schindleri (n = 2) and Acinetobacter lwoffii (n = 1) and one unknown Acinetobacter species (n = 1) (Fig. 2). They were mainly isolated from waterfowls (1.4%, 14/971) and their neighboring environmental soil samples (1.6%, 3/187), except that one isolate was from a sewage sample next to a chicken farm (Fig. 2).
FIG 2.
Genetic characteristics of isolates coharboring tet(X) and blaNDM-1. (Left) Core genome single nucleotide polymorphism (CG-SNP) phylogenetic tree of 18 Acinetobacter spp. coharboring tet(X) and blaNDM-1. (Right) The species, isolate source, sampling province, and the locations of tet(X) and blaNDM-1 in the genomes are indicated according to the color legend. TGC, tigecycline; MEM, meropenem.
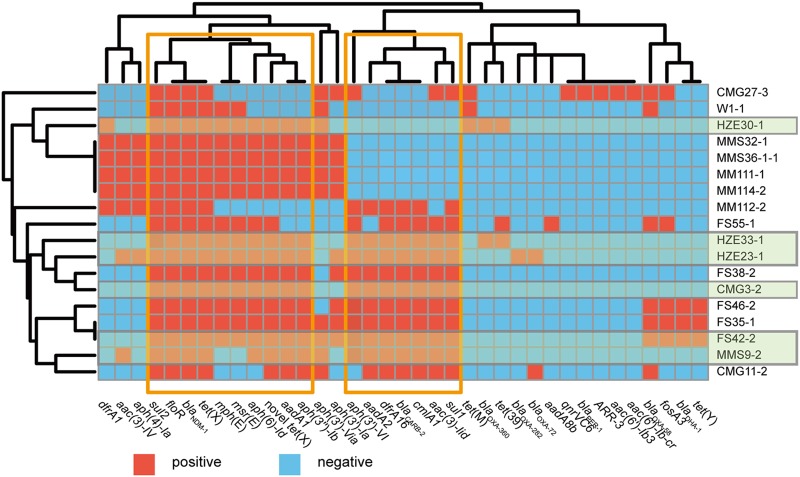
FIG 3.
A heat map of the antimicrobial resistance genes for 18 Acinetobacter species isolates cocarrying tet(X) and blaNDM-1. Genes positive and negative for resistance are indicated. Green shading indicates the strains in which tet(X) and blaNDM-1 are located on the same plasmids. Areas boxed in orange indicate the two major resistance gene clusters. The heat map was generated using R, version 3.3.2 (R Foundation for Statistical Computing) using the pheatmap package (https://cran.r-project.org/web/packages/pheatmap/).
TABLE 2.
MICs of antimicrobials for 18 Acinetobacter species isolates coharboring tet(X) and blaNDM-1
| Strain | Province | MIC (mg/liter)a
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TGC | ERA | OMA | TET | MEM | CAZ | CTX | AMP | FFC | CIP | S/T | GEN | AMK | FOS | CS | ||
| FS35-1 | Guangdong | 8 | 4 | 8 | >256 | 32 | >256 | >256 | 256 | 64 | 32 | >320 | 32 | 32 | >256 | 0.25 |
| FS38-2 | Guangdong | 8 | 4 | 8 | >256 | 32 | >256 | >256 | 256 | 64 | 32 | >320 | 128 | 32 | 256 | 0.25 |
| FS42-2 | Guangdong | 8 | 4 | 8 | 256 | 16 | >256 | 128 | 256 | 256 | 16 | >320 | 1 | 8 | >256 | 0.25 |
| FS46-2 | Guangdong | 8 | 4 | 16 | >256 | 32 | >256 | >256 | >256 | 256 | 64 | >320 | 128 | 8 | >256 | 0.25 |
| FS55-1 | Guangdong | 8 | 4 | 8 | 256 | 32 | >256 | >256 | 128 | >256 | 128 | 320 | 1 | 32 | >256 | 0.25 |
| CMG3-2 | Hainan | 16 | 4 | 16 | 128 | 32 | >256 | 256 | 128 | >256 | 256 | >320 | 16 | 8 | 256 | 0.5 |
| CMG11-2 | Hainan | 16 | 4 | 16 | 128 | 64 | >256 | 256 | 256 | 256 | 32 | >320 | 128 | 16 | 256 | 0.5 |
| CMG27-3 | Hainan | 16 | 4 | 16 | 128 | 16 | >256 | >256 | 32 | 256 | 128 | >320 | 64 | 8 | 256 | 0.25 |
| HZE23-1 | Guangdong | 8 | 4 | 16 | 64 | 32 | >256 | >256 | 256 | 256 | 64 | 320 | 16 | 32 | 256 | 0.25 |
| HZE30-1 | Guangdong | 8 | 4 | 16 | >256 | 32 | >256 | >256 | >256 | >256 | 64 | >320 | 0.25 | 64 | 256 | 0.25 |
| HZE33-1 | Guangdong | 16 | 4 | 16 | 256 | 32 | >256 | >256 | >256 | 256 | 32 | >320 | 32 | 32 | 256 | 0.5 |
| MM111-1 | Guangdong | 8 | 4 | 16 | 128 | 16 | >256 | 256 | 64 | >256 | 32 | 160 | 8 | 4 | 128 | 0.25 |
| MM112-2 | Guangdong | 16 | 4 | 16 | 256 | 16 | >256 | 128 | 64 | >256 | 32 | 160 | 64 | 4 | 64 | 0.25 |
| MM114-2 | Guangdong | 8 | 4 | 16 | 128 | 16 | >256 | 256 | 64 | >256 | 32 | 160 | 4 | 4 | 128 | 0.25 |
| MMS9-2 | Guangdong | 8 | 4 | 16 | 128 | 32 | >256 | >256 | 64 | >256 | 16 | 160 | 8 | 4 | 128 | 0.25 |
| MMS36- | Guangdong | 16 | 4 | 16 | 256 | 32 | >256 | 256 | 64 | >256 | 32 | 160 | 8 | 4 | 128 | 0.25 |
| MMS32-1 | Guangdong | 16 | 4 | 16 | 128 | 64 | >256 | >256 | 128 | 128 | 16 | 160 | 2 | 16 | 256 | 0.25 |
| W1-1 | Xinjiang | 8 | 4 | 16 | 128 | 16 | >256 | >256 | 128 | 256 | 64 | 160 | 0.5 | 16 | 256 | 0.25 |
TGC, tigecycline; ERA, eravacycline; OMA, omadacycline; TET, tetracycline; MEM, meropenem; CAZ, ceftazidime; CTX, cefotaxime; AMP, ampicillin; FFC, florfenicol; CIP, ciprofloxacin; S/T, trimethoprim-sulfamethoxazole; GEN, gentamicin; AMK, amikacin; FOS, fosfomycin; CS, colistin.
Analysis of the localization of tet(X) and blaNDM-1.
S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blot hybridization results of the 18 tet(X)- and blaNDM-1-coharboring isolates showed that in 10 isolates the tet(X) gene was located on a plasmid and that the blaNDM-1 gene was on the chromosome, while in 2 isolates the two genes were located on two different plasmids. Interestingly, we found six isolates, including three A. indicus, one A. lwoffii, and two A. schindleri, in which the tet(X) and blaNDM-1 genes were located on the same plasmids (∼110 kb to ∼140 kb) (Fig. 2 and Fig. S1 in the supplemental material).
Transfer of tet(X)-carrying plasmids.
Conjugation experiments were then conducted in 30 selected tet(X)-positive Acinetobacter species strains, including the 18 aforementioned tet(X)- and blaNDM-1-coharboring isolates. The tigecycline resistance in the 18 tet(X)- and blaNDM-1-coharboring isolates failed to transfer to Acinetobacter baylyi ADP1 via conjugation. In contrast, the tet(X) gene was successfully transferred into A. baylyi ADP1 through conjugation in 2 of the remaining 12 tet(X)-positive isolates.
Subsequently, we performed natural transformation experiments on 18 tet(X)- and blaNDM-1-coharboring isolates in which tigecycline resistance failed to transfer by conjugation. The tigecycline resistance of 2 of 18 strains was successfully transferred to A. baylyi ADP1 by natural transformation, including a strain (MMS9-2) in which tet(X) and blaNDM-1 were located on the same plasmid.
Whole-genome sequencing (WGS) analysis of Acinetobacter species isolates coharboring tet(X) and blaNDM-1.
Core-genome phylogenetic analysis of the 18 tet(X)- and blaNDM-1-coharboring isolates showed that the 14 A. indicus isolates clustered together, while the other 4 isolates were distinct from A. indicus isolates (Fig. 2). Among the A. indicus strains, we identified two small clusters of three (FS35-1, FS42-2, and FS46-2) and four (MMS36-1-1, MM111-1, MMS32-1, and MM114-2) strains (Fig. 2) with limited core single nucleotide polymorphism (SNP) differences (n ≤ 10). Notably, the four strains within one small A. indicus cluster were isolated from both waterfowls (MM111-1 and MM114-2) and their adjacent environmental soils (MMS36-1-1 and MMS32-1), and they shared identical resistance genotypes (Fig. 2 and 3). Further analysis of resistance genes showed that the 18 isolates carried some similar resistance genes, such as sul, floR, blaNDM-1, two tet(X) genes, mph(E), msr(E), aph, aadA, aac(3), dfrA, blaCARB, and cmlA; most of them were divided into mainly two clusters, indicating that these genes may be cotransferred with tet(X) and blaNDM-1 (Fig. 3).
Identification of a novel tet(X) orthologue.
The novel tet(X) orthologue encodes a putative monooxygenase enzyme of 378 amino acids, displaying 84.39%, 84.66%, 79.63%, and 87.30% amino acid identities, respectively, to the previously reported tigecycline resistance proteins, all of which are now named Tet(X): GenBank accession no. M37699.1; AJ311171, formerly named Tet(X2); MK134375, formerly named Tet(X3); and CP037909, formerly named Tet(X4). To verify the function of this novel tet(X) orthologue in tigecycline resistance, this tet(X) gene from pCMG3-2-1 was cloned into plasmid vector pBAD24 as described previously (9). The construct demonstrated a 32-fold increase in the tigecycline MIC (8 mg/liter) in comparison to that of the empty vector control (Table S2), indicating that this newly identified tet(X) gene was active and conferred high-level tigecycline resistance. Among the 18 tet(X)- and blaNDM-1-coharboring isolates, 15 were found to carry this novel tet(X) gene (Fig. 3), including all six strains in which tet(X) and blaNDM-1 were located on the same plasmids. PCR mapping was performed on the remaining 9 strains cocarrying tet(X) and the novel tet(X) orthologue using primers hp2-F and tnpF-R (Table S1). It was confirmed that tet(X) genes were located downstream of the novel orthologue of tet(X) in four strains, as with pCMG3-2-1, while the other five strains could not be amplified by PCR, suggesting that they may have different genetic contexts or two tet(X) genes not on the same plasmid.
Characteristics of the genetic contexts of tet(X) and blaNDM-1 genes.
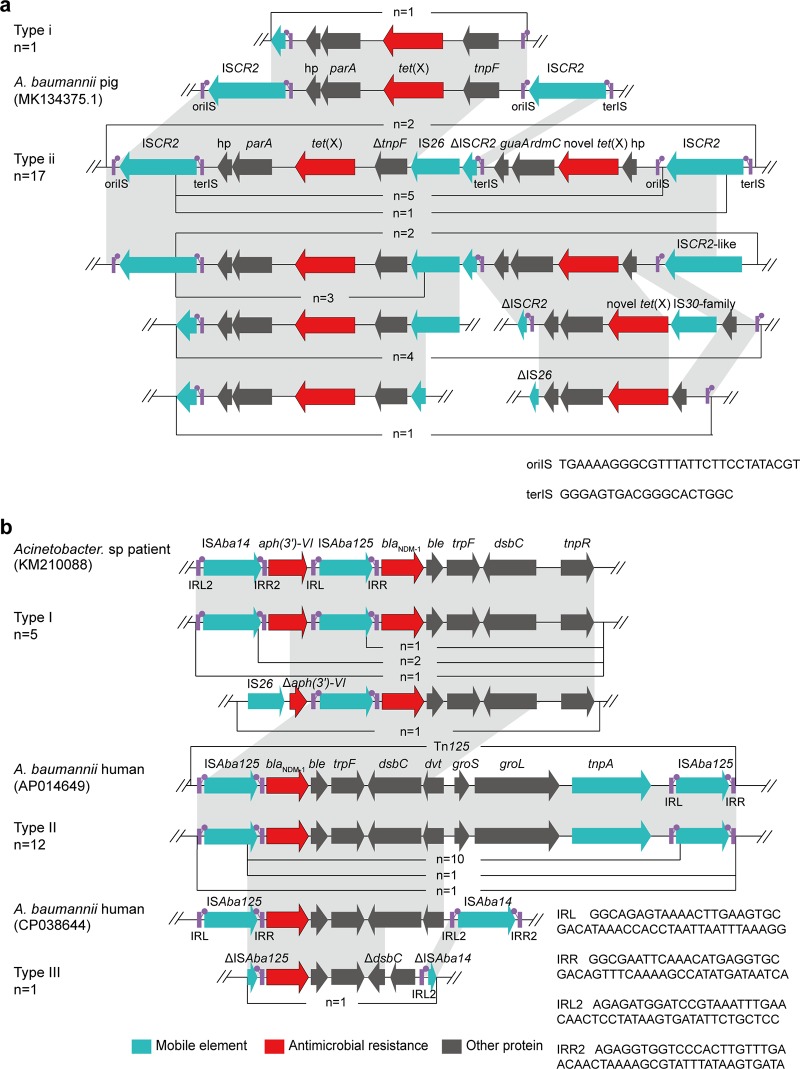
We analyzed the 18 tet(X)-carrying contigs showing two types of genetic profiles (type i, n = 1; type ii, n = 17) (Fig. 4a). Only one isolate, W1-1 belonged to type i, and inverse PCR showed that it formed a 5.1-kb circular intermediate of the structure of tet(X)-parA-hp-ISCR2-tnpF, which was the same as that of A. baumannii plasmid 34AB isolated from a pig at slaughter in China (GenBank accession no. MK134375). Type ii contained the sequence ΔISCR2-IS26-ΔtnpF-tet(X)-parA-hp-ISCR2 which was found in isolates from duck feces (n = 8), soil of duck farms (n = 3), and goose feces (n = 6). Notably, ISCR2 was located downstream of the tet(X) gene among all 18 strains, which has been reported to be associated with the mobilization of tet(X) in previous studies (8).
FIG 4.
Genetic features of tet(X) and blaNDM-1. (a) Linear sequence comparison of genetic context of tet(X) in the 18 isolates in this study carrying plasmid 34AB (GenBank accession no. MK134375.1) from A. baumannii isolated from a pig in China. (b) Linear sequence comparison of genetic context of blaNDM-1 in this study with that of an Acinetobacter species isolate from human in China (KM210088) and from chromosome of a clinical A. baumannii IOMTU 433 isolate from Vietnam (AP014649). The arrows indicate the positions and directions of transcription for the genes. Regions of >99.0% nucleotide sequence identity are shaded in gray. The delta (Δ) symbol indicates a truncated gene. oriIS, the origin of replication; terIS, the termination sequence; IRL, terminal inverted repeat, left; IRR, terminal inverted repeat, right.
From analysis of the 18 contigs carrying blaNDM-1, three genomic backbone profiles were obtained (type I, n = 5; type II, n = 12; type III, n = 1), and they all belong to the Tn125 or truncated Tn125 structures. Notably, 3 isolates including A. indicus CMG3-2 and A. lwoffii HZE30-1 within type I [ISAba14-aph(3′)-VI-ISAba125-blaNDM-1-ble-trpF-dsbC-tnpR], 12 isolates within type II (ΔISAba125-blaNDM-1-ble-trpF-dsbC-dvt-groS-groL-tnpA-ISAba125), and 1 isolate within type III (ΔISAba125-blaNDM-1-ble-trpF-ΔdsbC-hp-ΔISAba14) were highly similar to the corresponding regions on the plasmid from a human Acinetobacter species isolate from China (GenBank accession no. KM210088), on the chromosome of a clinical A. baumannii IOMTU 433 isolate from Vietnam (AP014649), and on the chromosome of a human A. baumannii ACN21 isolate from India (CP038644) (Fig. 4b), respectively.
Characteristics of tet(X)- and blaNDM-1-coharboring plasmids.
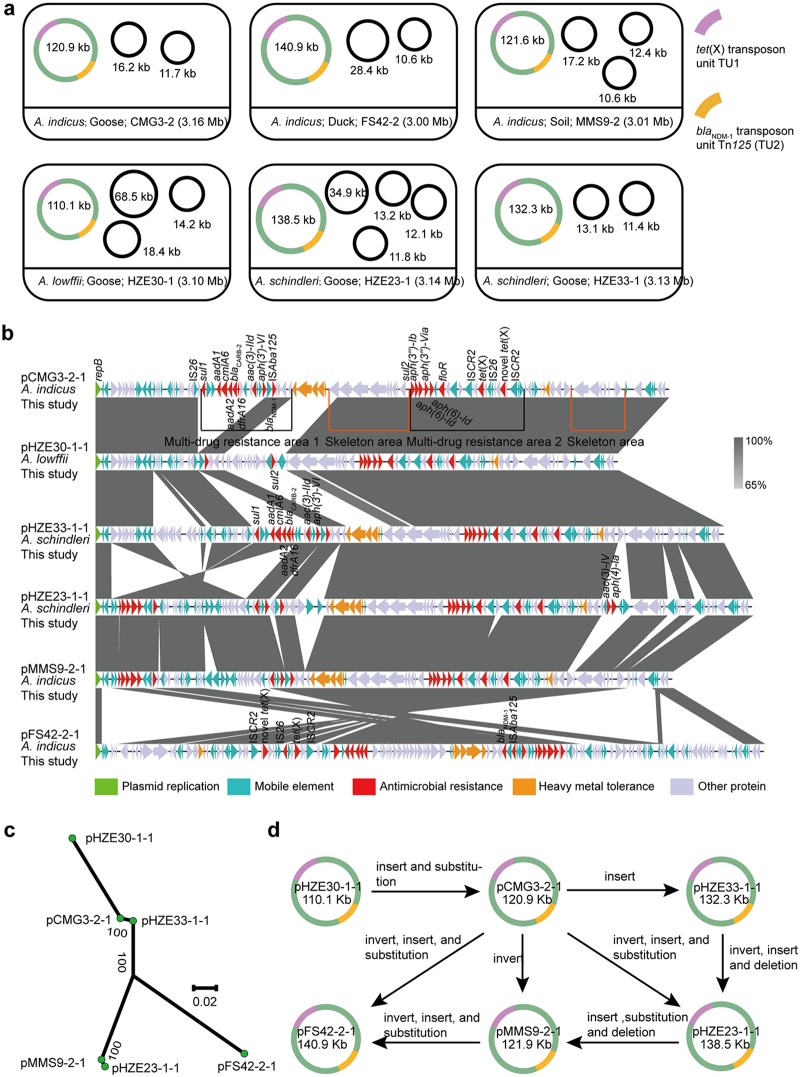
To obtain the complete chromosome and plasmid structures, the six tet(X)- and blaNDM-1-coharboring isolates, including three A. indicus (MMS9-2, FS42-2, and CMG3-2), one A. lwoffii (HZE30-1), and two A. schindleri (HZE23-1 and HZE33-1) with plasmids coharboring tet(X) and blaNDM-1 were subsequently sequenced by a PacBio RSII system (Nextomics) to obtain the complete chromosome and plasmid structures. The genomes of these 6 isolates varied from 3.00 to 3.16 Mb in chromosome size and carried between three and five plasmids ranging from 10.6 to 140.2 kb (Fig. 5a). In A. indicus CMG3-2, tet(X) and blaNDM-1 genes were found to be on a 120,957-bp untypeable plasmid, pCMG3-2-1, along with multiple resistance genes: ble, sul1, sul2, blaCARB-2, cmlA6, aadA1, aadA2, aac(3)-IId, aph(3′)-VI, aph(3′)-Ib, aph(6)-Id, aph(3′)-Via, floR, dfrA16, and the novel tet(X) orthologue (Fig. 3 and 5b).
FIG 5.
Characterization of the six tet(X)- and blaNDM-1-coharboring plasmids by the PacBio RSII platform. (a) Species, source, and chromosome size of each strain are indicated. The transposition unit TU1 of tet(X) and the transposition unit Tn125 (TU2) of blaNDM-1 are indicated. (b) Linear sequence comparison of the six tet(X)- and blaNDM-1-coharboring plasmids. The results of sequence alignment were generated with Easyfig, version 2.1. Regions of homology are marked by shading. The delta (Δ) symbol indicates a truncated gene. Genes associated with plasmid replication, antimicrobial resistance, heavy metal resistance, and mobility are marked in green, red, orange, and blue, respectively, while other gene are shown in gray. (c) A radial representation of a phylogenetic tree of six tet(X)- and blaNDM-1-coharboring plasmids and homologs. (d) Hypothesis of the molecular evolution of the six tet(X)- and blaNDM-1-coharboring plasmids.
The complete sequences of the six tet(X)- and blaNDM-1-coharboring plasmids were further compared. The results showed that the six plasmids contained two similar multidrug resistance regions (pHZE30-1-1 contained only one) and plasmid backbone structures, including genes encoding plasmid replication, maintenance, and stability (Fig. 5b). Plasmid pHZE33-1-1 (from an A. schindleri isolate from goose), pMMS9-2-1 (A. indicus isolate from soil), pHZE23-1-1 (A. schindleri isolate from goose), pHZE30-1-1 (A. lwoffii isolate from goose), and pFS42-2-1 (A. indicus isolate from duck) showed 84% to 99% query coverage and 98% to 100% nucleotide identity to the sequence of pCMG3-2-1. Notably, the six plasmids were isolated from different Acinetobacter species (A. lwoffii, A. indicus, and A. schindleri) and from various sources (goose, duck, and soil samples) from different regions of China (Guangdong and Hainan). On the basis of pCMG3-2-1, the other five plasmids may be formed through sequence insertion, substitution, deletion, and inversion (Fig. 5d).
DISCUSSION
The genus Acinetobacter, including non-baumannii Acinetobacter species, includes opportunistic pathogens and commonly causes nosocomial infections (10). Worrisomely, the tet(X) genes have already been reported in A. baumannii strains isolated from inpatients. By BLAST analysis, we also detected the tet(X)-carrying contigs in Acinetobacter species from clinical samples, including A. pittii (GenBank accession no. JRQZ01000088.1), A. nosocomialis (NNSH01000121.1), and A. baumannii (UFMQ01000023.1). All of these indicated the importance of tet(X)-positive Acinetobacter species strains for human diseases, challenging the clinical effectiveness of the last-resort tigecycline.
In the present study, during our recent antimicrobial surveillance on avian farms and adjacent environments, the tet(X) gene was found in 180 (8.9%) Acinetobacter species isolates from all seven sampled provinces (Fig. 1), which was significantly higher than the rate (0.1%) in avian samples from a previous report (8). This indicated widespread presence of tet(X)-positive Acinetobacter spp. in avian farms and their surrounding environments. Surprisingly, 10% (n = 18) of these tet(X)-bearing strains also carried the carbapenem resistance gene blaNDM-1, mainly from waterfowls and the soils of their neighboring environments (1.6%), suggesting that the environmental soils adjacent to farms may be contaminated by avian feces and could serve as a reservoir for the spread of tigecycline- and carbapenem-resistant Acinetobacter spp. Indeed, previous studies showed that the soils from greenhouse and open-field agricultural soil can often contain tet(X) genes (12), likely from organic fertilizer with animal feces (13). Additionally, one sample of sewage (0.4%) adjacent to a chicken farm carried one tet(X)- and blaNDM-1-coharboring Acinetobacter species isolate (Fig. 2); however, none of the chicken samples from the farm were positive for such Acinetobacter spp. (Table 1). Therefore, we suspected that this tet(X)- and blaNDM-1-coharboring isolate may have been transmitted from somewhere through contaminated water (14). In fact, the high detection rate among tet(X)- and blaNDM-1-coharboring isolates in waterfowl samples suggested the transmission of these bacteria through contaminated water systems. Tigecycline is a novel glycylcycline, only approved for the treatment of complex skin and intra-abdominal infections and community-acquired pneumonia, and its use has never been allowed in animal husbandry. However, recent epidemiological studies show that the detection rate of tet(X)-positive strains in farmed animals was significantly higher than that in humans (8, 9). Some studies suggest that this is related to the large-scale use of tetracyclines (e.g., tetracycline, oxytetracycline, chlortetracycline, and doxycycline) in the livestock industry (8, 9, 15). The selection pressure from tetracyclines may promote the enrichment and transmission of tet(X) genes. Further studies are required to reveal how these clinically significant ARGs [tet(X) and blaNDM] were transferred into waterfowl farms.
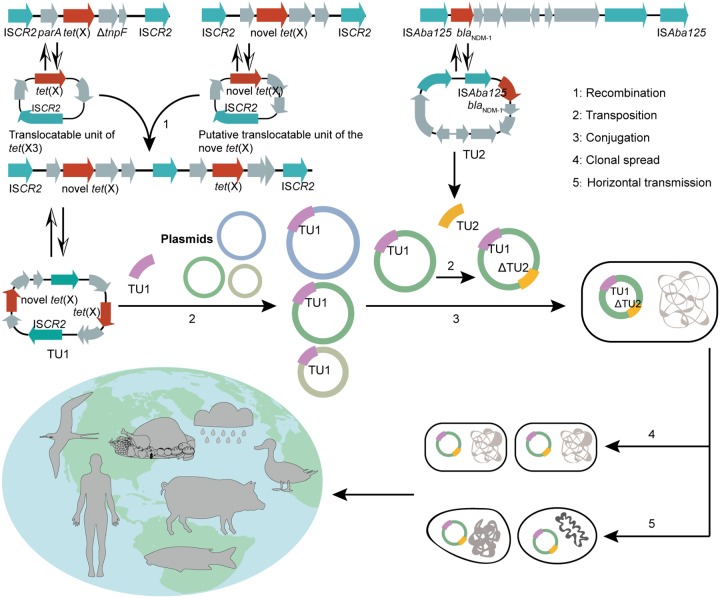
Notably, in the present study, we found common tet(X)- and blaNDM-1-coharboring vectors, pCMG3-2-1-like plasmids, which have been disseminated among three Acinetobacter species isolates from diverse sources, including waterfowls, soil, and sewage, in three geographic areas in China. Plasmid sequence phylogenetic analysis showed that the six plasmids have a very close evolutionary relationship with each other (Fig. 5c). Additionally, it is also noted that a pCMG3-2-1-like plasmid carried two copies of tet(X) genes, and these two tet(X) genes were located in a composite transposon-like unit (TU1), flanked by two intact copies of ISCR2 in the same orientation, and the gene arrangement of TU1 was ISCR2-hp-tet(X)-rmdc-guaA-ΔISCR2-IS26-ΔtnpF-tet(X)-parA-hp-ISCR2. However, a tet(X)-only circular intermediate failed to be amplified by inverse PCR. In contrast, two tet(X) genes coharboring a circular intermediate were obtained. Analysis of the blaNDM-1-containing elements among the six plasmids revealed that blaNDM-1 genes were harbored by similar Tn125 elements (1 intact and 5 partial), located approximately 35 to 45 kb upstream of tet(X), further confirming that the Tn125 transposon is mainly responsible for the horizontal transmission of blaNDM-1 in Acinetobacter spp. (16). These results prompted us to propose a hypothesis about the formation, evolution, and spread of the six tet(X)- and blaNDM-1-coharboring plasmids (Fig. 6). We speculate that the aforementioned TU1 might have originated by the integration of two tet(X) translocatable units together (Fig. 6). The blaNDM-1-harboring Tn125 may be transposed independently into the same pCMG3-2-1-like plasmid background, and under the pressure of antibiotic selection, this type of multidrug-resistant plasmid can be transferred between various pathogens and ecosystems through clonal spread or horizontal transmission of strains.
FIG 6.
Schematic diagram of the formation, evolution, and spread of the six tet(X)- and blaNDM-1-coharboring plasmids.
The discovery of a highly similar pCMG3-2-1-like plasmid in different species, sources, and geographical regions suggested the horizontal gene transfer (HGT) of this plasmid, even though several attempts have failed to demonstrate conjugation of a pCMG3-2-1-like plasmid into the recipient A. baylyi ADP1. Accordingly, the plasmid sequence analysis failed to identify any plasmid transfer operons. Other mechanisms, including natural transformation, transduction, and/or outer membrane vesicle (OMV)-mediated transfer, may contribute to the HGT of this plasmid (17–19), which deserves further study. Nevertheless, our study clearly demonstrated that these antibiotic-resistant plasmids or strains are spreading in waterfowls and their neighboring environments in China. A formidable concern is that these resistance genes, plasmids, and strains may eventually spread into human populations via direct contact or consumption of contaminated food products.
It is noteworthy that only the term of tet(X) is currently accepted by the nomenclature center, and according to the standard of nomenclature, a series of recently discovered tet(X) genes [formerly named tet(X3), tet(X4), tet(X5), and other variants] can only be called tet(X). However, the initial tet(X) gene is mainly located in obligate anaerobic bacteria (20–23), which could not confer tigecycline resistance due to its requirement for O2 and NADPH (24). Although this gene was subsequently detected in aerobic bacteria, there is no evidence that it can convey tigecycline resistance at the resistance breakpoint (EUCAST, >2 mg/liter; FDA, ≥8mg/liter). In contrast, one study cloned the initial tet(X) into an Escherichia coli expression system and found that the MIC of tigecycline is only 0.25 mg/liter (25), which is much lower than that of the recently reported tet(X) genes (8 or 16 mg/liter) (8, 9). Similarly, the research by Xu and coworkers (26) showed that different tet(X) orthologues have different levels of resistance to tigecycline, so we think that it is very confusing to use only the name tet(X) to refer to all tet(X) genes. Here, we ask that the nomenclature standard for tet(X) be improved as soon as possible to facilitate further research on this important resistance gene.
Conclusion.
Our study reported the relative prevalence of tigecycline-resistant Acinetobacter species isolates coharboring tet(X) genes and blaNDM-1 in avian samples, especially in waterfowls and samples from their adjacent environments, in China. In addition, we identified a novel plasmid-borne tet(X) gene in Acinetobacter spp. which can mediate high levels of resistance to tigecycline. More importantly, we described for the first time a common tet(X)- and blaNDM-1-coharboring plasmid in Acinetobacter species isolates in avian and adjacent environmental samples from different regions in China, highlighting the mobile nature of these highly resistant genes. Considering the close relationship among animals, environments, and humans, the further spread of these strains and plasmids into human health care settings should be closely monitored. Coordinated efforts should be taken to control the spread of these antibiotic-resistant bacteria in different sectors.
MATERIALS AND METHODS
Sample collection and identification of tet(X)-positive strains.
From July 2016 to November 2018, a total of 2,018 nonduplicate samples were collected from 34 avian farms in seven provinces of China (Table 1). Briefly, the fecal samples were randomly collected from ducks (n = 864), geese (n = 107), chickens (n = 450), and pigeons (n = 40). Samples from the environments surrounding the farms were also collected (Table 1), including soil (n = 252), sewage (n = 255), and dust (n = 50) samples (Table 1). Following selection on CHROMagar Acinetobacter plates containing tigecycline (2 mg/liter), the tigecycline-nonsusceptible isolates were screened for tet(X) genes using previously reported primers (8). The species of tet(X)-positive Acinetobacter strains were further determined by 16S rRNA and rpoB sequencing analyses (27). This study protocol was reviewed and approved by the South China Agriculture University Animal Ethics Committee.
Antimicrobial susceptibility testing.
We determined the MICs of amikacin, gentamicin, tetracycline, meropenem, ceftazidime, cefotaxime, ciprofloxacin, trimethoprim-sulfamethoxazole, and florfenicol for all tet(X)-positive strains using the agar dilution method, and the results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). Fresh Mueller-Hinton (MH) broth was prepared, and the MICs of tigecycline, eravacycline, omadacycline, and colistin were examined by the broth microdilution method. The breakpoints for tigecycline and colistin were interpreted according to the FDA criteria for Enterobacteriaceae bacteria (resistant, ≥8 mg/liter) and the European Committee on Antimicrobial Susceptibility Testing (resistant, >2 mg/liter), respectively. No MIC breakpoints exist of eravacycline and omadacycline for Acinetobacter species strains. E. coli ATCC 25922 was used as a quality control strain.
Microbiological degradation assays and PCR testing.
The tigecycline degradation assays were determined as previously described (9), with little change. First, the MH broth suspension of the test strains was prepared with an optical density of 2.0 at 600 nm. Then, tigecycline was added to the suspension (2.5 mg/liter) and incubated at 37°C for 8 h. Meanwhile, an overnight culture of Bacillus stearothermophilus 7953 was spread on an MH agar plate, and three 6-mm-diameter holes were punched. Finally, the culture solution of the previous step was centrifuged and filtered through a 0.22-μm-pore-size filter; 20 μl was dispensed into plate wells and incubated at 60°C for 16 h to observe the results. The A. baumannii ATCC 19606 strain was used as a negative control.
Testing using the carbapenem inactivation method (CIM) was performed as previously reported (28) with slight modification. Briefly, a loopful of bacteria (∼10 μl) was suspended in 400 μl of sterile water with one 10-μg imipenem disc and incubated for 2 h. The disc was then placed on a Mueller-Hinton agar plate coated with a susceptible E. coli indicator strain (ATCC 29522) and reincubated for 18 h. If the bacterial isolate produced carbapenemase, then the antibiotic in the disc would be inactivated, leading to the appearance of an obviously diminished zone of inhibition. E. coli ATCC 25922 was used as a negative-control strain. Moreover, carbapenemase-producing strains were screened for the carbapenemase-encoding genes blaNDM, blaIMP, blaVIM, blaSPM, blaAIM, blaDIM, blaGIM, blaSIM, blaKPC, blaBIC, blaOXA-48, blaOXA-23, blaOXA-51, and blaOXA-58 using previously described primers (29, 30). Strains positive for related genes were used as positive controls.
Conjugation experiments and natural transformation experiments.
The transferability of tigecycline resistance in selected tet(X)-positive Acinetobacter isolates was examined by the filter mating method using rifampin-resistant A. baylyi ADP1 as the recipient strain. Briefly, the donor and recipient strains were mixed at a ratio of 1:3 and applied to a sterilized 0.22-μm-pore-size filter, followed by culturing at 30°C for 24 h. The transconjugants were selected on selective LB agar containing tigecycline (4 mg/liter) together with rifampin (150 mg/liter). We further confirmed the transconjugants by PCR for tet(X) and by M13 PCR (PCR-based fingerprinting for Acinetobacter spp.) (31).
For 18 tet(X)- and blaNDM-1-coharboring isolates for which tigecycline resistance was not transferred by conjugation, natural transformation experiments were performed as described previously (32). Briefly, the naturally competent strain A. baylyi ADP1 was cultured in LB broth at 30°C to 1 × 109 cells ml−1, concentrated to 1 × 1010 cells ml−1 in LB broth with 10% glycerol, and stored at –80°C until use. For transformation assays, a fresh bacterial suspension was diluted to 2.5 × 108 cells ml−1 in LB broth containing 100 ng ml−1 donor DNA. After incubation at 30°C for 2 h, the bacterial suspension was appropriately diluted and plated on LB agar plates containing 4 mg/liter tigecycline. The colonies were selected for PCR screening after 24 h at 30°C.
Localization of tet(X) and blaNDM-1.
To analyze the chromosomal and plasmid localizations of tet(X) and blaNDM-1 genes, we performed S1 nuclease-PFGE as previously described (33). Subsequently, Southern blot hybridization was conducted using DNA probes specific for tet(X) and blaNDM-1 nonradioactively labeled with a DIG (digoxigenin) High Prime DNA labeling and detection kit (Roche Diagnostics, Mannheim, Germany).
Whole-genome sequencing.
Total DNA of the 18 Acinetobacter species isolates coharboring tet(X) and blaNDM-1 was extracted using a TIANamp Bacteria DNA kit (Tiangen, China) and then sequenced using an Illumina HiSeq 2500 platform (Bionova Biotech Co.) (Illumina). The sequences were assembled using SPAdes, version 3.12.0 (34), followed by antibiotic resistance gene prediction using ResFinder, version 3.1 (https://cge.cbs.dtu.dk/services/ResFinder/), and transposon and insertion (IS) element mining using ISfinder (https://www-is.biotoul.fr). Functional annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline server and RAST server (35). Core genome SNPs were analyzed using a previously described method (36). In brief, the trimmed, paired-end sequences were mapped to A. indicus genome SGAir0564 (GenBank accession number CP024620), followed by the removal of SNPs in prophages and repeated and recombination regions. Concatenated core genome SNP sites were extracted from the recombination-free alignment, and a maximum likelihood phylogenetic tree was generated by IQ-TREE (37). The blaNDM-1 and tet(X) gene flanking regions were compared by blastn.
To characterize the complete sequences of plasmids cocarrying tet(X) and blaNDM-1, isolates with plasmids coharboring the blaNDM-1 and tet(X) genes were subjected to single-molecule real-time sequencing using a PacBio RSII system (Nextomics), followed by assembly using Unicycler, version 0.4.1 (38). ClustalW was utilized to generate a nucleotide-guided multiple sequence alignment, which was utilized for the subsequent phylogenetic analysis. A maximum likelihood tree with 500 bootstrap replicates was generated by MEGA, version 7 (39). Comparison of these plasmids was performed by Easyfig, version 2.1 (40). To determine the potential transferability of the two tet(X)-cocarrying segments, inverse PCR was performed for isolate CMG3-1 using P1 primers (see Table S1 in the supplemental material).
Data availability.
Genome assemblies of 18 strains coharboring tet(X) and blaNDM-1 were deposited in GenBank under BioProject accession number PRJNA558439.
Supplementary Material
ACKNOWLEDGMENTS
This work was jointly supported by the National Key Research and Development Program of China (2016YFD0501300), the Program for Innovative Research Team in the University of Ministry of Education of China (IRT_17R39), the 111 Project (D20008), the Foundation for Innovation and Strengthening School Project of Guangdong, China (2016KCXTD010), and the U.S. National Institutes of Health (R01AI090155).
We declare that we have no competing interests.
C.-Y.C., C.C., and B.-T.L. contributed equally in this study. Y.-H.L., J.S., L.C., and X.-P.L. designed the study. C.-Y.C., C.C., Q.H., X.-T.W., Y.Z., Z.-H.C., W.-Y.G., Q.-.LJ., and C.L. collected the data. C.-Y.C., J.S., L.C., C.C., B.-T.L., and R.-Y.S. analyzed and interpreted the data. Y.-H.L., L.C., J.S., C.C., B.-T.L., X.-P.L., and C.-Y.C. wrote the draft of the manuscript. All authors reviewed, revised, and approved the final report.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Laxminarayan R, Amábile-Cuevas CF, Cars O, Evans T, Heymann DL, Hoffman S, Holmes A, Mendelson M, Sridhar D, Woolhouse M, Røttingen J-A. 2016. UN high-level meeting on antimicrobials–what do we need? Lancet 388:218–220. doi: 10.1016/S0140-6736(16)31079-0. [DOI] [PubMed] [Google Scholar]
- 2.Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulens SN, Yi SH, Walters MS, Jacob JT, Bower C, Reno J, Wilson L, Vaeth E, Bamberg W, Janelle SJ, Lynfield R, Vagnone PS, Shaw K, Kainer M, Muleta D, Mounsey J, Dumyati G, Concannon C, Beldavs Z, Cassidy PM, Phipps EC, Kenslow N, Hancock EB, Kallen AJ. 2018. Carbapenem-nonsusceptible Acinetobacter baumannii, 8 US metropolitan areas, 2012–2015. Emerg Infect Dis 24:727–734. doi: 10.3201/eid2404.171461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kock R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, Schwarz S, Jurke A. 2018. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect 24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 5.van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerson S, Nowak J, Zander E, Ertel J, Wen Y, Krut O, Seifert H, Higgins PG. 2018. Diversity of mutations in regulatory genes of resistance-nodulation-cell division efflux pumps in association with tigecycline resistance in Acinetobacter baumannii. J Antimicrob Chemother 73:1501–1508. doi: 10.1093/jac/dky083. [DOI] [PubMed] [Google Scholar]
- 7.He F, Shi Q, Fu Y, Xu J, Yu Y, Du X. 2018. Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect Genet Evol 66:188–191. doi: 10.1016/j.meegid.2018.09.025. [DOI] [PubMed] [Google Scholar]
- 8.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Chen C, Cui CY, Zhang Y, Liu X, Cui ZH, Ma XY, Feng YJ, Fang LX, Lian XL, Zhang RM, Tang YZ, Zhang KX, Liu HM, Zhuang ZH, Zhou SD, Lv JN, Du H, Huang B, Yu FY, Mathema B, Kreiswirth BN, Liao XP, Chen L, Liu YH. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol 4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almasaudi SB. 2018. Acinetobacter spp. as nosocomial pathogens: epidemiology and resistance features. Saudi J Biol Sci 25:586–596. doi: 10.1016/j.sjbs.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Q, Sun J, Zhu L. 2019. Occurrence and distribution of antibiotics and resistance genes in greenhouse and open-field agricultural soils in China. Chemosphere 224:900–909. doi: 10.1016/j.chemosphere.2019.02.167. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Qiu T, Gao M, Shi M, Zhang H, Wang X. 2019. Inorganic and organic fertilizers application enhanced antibiotic resistome in greenhouse soils growing vegetables. Ecotoxicol Environ Saf 179:24–30. doi: 10.1016/j.ecoenv.2019.04.039. [DOI] [PubMed] [Google Scholar]
- 14.Mikhail AFW, Jenkins C, Dallman TJ, Inns T, Douglas A, Martin AIC, Fox A, Cleary P, Elson R, Hawker J. 2018. An outbreak of Shiga toxin-producing Escherichia coli O157:H7 associated with contaminated salad leaves: epidemiological, genomic and food trace back investigations. Epidemiol Infect 146:187–196. doi: 10.1017/S0950268817002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Liu D, Lv Y, Cui L, Li Y, Li T, Song H, Hao Y, Shen J, Wang Y, Walsh TR. 2019. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline and omadacycline in clinical Acinetobacter baumannii. Antimicrob Agents Chemother 64:e01326-19. doi: 10.1128/AAC.01326-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bontron S, Nordmann P, Poirel L. 2016. Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii. Antimicrob Agents Chemother 60:7245–7251. doi: 10.1128/AAC.01755-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilharm G, Skiebe E. 2019. Methods for natural transformation in Acinetobacter baumannii. Methods Mol Biol 1946:75–85. doi: 10.1007/978-1-4939-9118-1_8. [DOI] [PubMed] [Google Scholar]
- 18.Fulsundar S, Domingues S, Nielsen KM. 2019. Vesicle-mediated gene transfer in Acinetobacter baumannii. Methods Mol Biol 1946:87–94. doi: 10.1007/978-1-4939-9118-1_9. [DOI] [PubMed] [Google Scholar]
- 19.Krahn T, Wibberg D, Maus I, Winkler A, Bontron S, Sczyrba A, Nordmann P, Puhler A, Poirel L, Schluter A. 2016. Intraspecies transfer of the chromosomal Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother 60:3032–3040. doi: 10.1128/AAC.00124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartha NA, Soki J, Urban E, Nagy E. 2011. Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. Int J Antimicrob Agents 38:522–525. doi: 10.1016/j.ijantimicag.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 21.Speer BS, Bedzyk L, Salyers AA. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J Bacteriol 173:176–183. doi: 10.1128/jb.173.1.176-183.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eitel Z, Soki J, Urban E, Nagy E, ESCMID Study Group on Anaerobic Infection. 2013. The prevalence of antibiotic resistance genes in Bacteroides fragilis group strains isolated in different European countries. Anaerobe 21:43–49. doi: 10.1016/j.anaerobe.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Sadarangani SP, Cunningham SA, Jeraldo PR, Wilson JW, Khare R, Patel R. 2015. Metronidazole- and carbapenem-resistant bacteroides thetaiotaomicron isolated in Rochester, Minnesota, in 2014. Antimicrob Agents Chemother 59:4157–4161. doi: 10.1128/AAC.00677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W, Moore IF, Koteva KP, Bareich DC, Hughes DW, Wright GD. 2004. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem 279:52346–52352. doi: 10.1074/jbc.M409573200. [DOI] [PubMed] [Google Scholar]
- 25.Linkevicius M, Sandegren L, Andersson DI. 2016. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother 60:789–796. doi: 10.1128/AAC.02465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu YC, Liu LZ, Sun J, Feng YJ. 2019. Limited distribution and mechanism of the TetX4 tetracycline resistance enzyme. Science Bull 64:1478–1481. doi: 10.1016/j.scib.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 27.Lee MJ, Jang SJ, Li XM, Park G, Kook JK, Kim MJ, Chang YH, Shin JH, Kim SH, Kim DM, Kang SH, Moon DS. 2014. Comparison of rpoB gene sequencing, 16S rRNA gene sequencing, gyrB multiplex PCR, and the VITEK2 system for identification of Acinetobacter clinical isolates. Diagn Microbiol Infect Dis 78:29–34. doi: 10.1016/j.diagmicrobio.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 28.van der Zwaluw K, de Haan A, Pluister GN, Bootsma HJ, de Neeling AJ, Schouls LM. 2015. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS One 10:e0123690. doi: 10.1371/journal.pone.0123690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Fu Y, Jiang J, Zhou H, Jiang Y, Fu Y, Yu Y, Zhou J. 2014. Characterization of a novel plasmid type and various genetic contexts of bla OXA-58 in Acinetobacter spp. from multiple cities in China. PLoS One 9:e84680. doi: 10.1371/journal.pone.0084680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grundmann HJ, Towner KJ, Dijkshoorn L, Gerner-Smidt P, Maher M, Seifert H, Vaneechoutte M. 1997. Multicenter study using standardized protocols and reagents for evaluation of reproducibility of PCR-based fingerprinting of Acinetobacter spp. J Clin Microbiol 35:3071–3077. doi: 10.1128/JCM.35.12.3071-3077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harms K, Schon V, Kickstein E, Wackernagel W. 2007. The RecJ DNase strongly suppresses genomic integration of short but not long foreign DNA fragments by homology-facilitated illegitimate recombination during transformation of Acinetobacter baylyi. Mol Microbiol 64:691–702. doi: 10.1111/j.1365-2958.2007.05692.x. [DOI] [PubMed] [Google Scholar]
- 33.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 34.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, Strydom KA, Ehlers MM, Mbelle NM, Shashkina E, Haslam DB, Dhawan P, Donnelly RJ, Chen L, Kreiswirth BN, Pitout J. 2019. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg Infect Dis 25:739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies of 18 strains coharboring tet(X) and blaNDM-1 were deposited in GenBank under BioProject accession number PRJNA558439.