Antibiotics revolutionized the treatment of infectious diseases; however, it is now clear that broad-spectrum antibiotics alter the composition and function of the host’s microbiome. The microbiome plays a key role in human health, and its perturbation is increasingly recognized as contributing to many human diseases. Widespread broad-spectrum antibiotic use has also resulted in the emergence of multidrug-resistant pathogens, spurring the development of pathogen-specific strategies such as monoclonal antibodies (MAbs) to combat bacterial infection.
KEYWORDS: microbiome, antibiotics, monoclonal antibodies
ABSTRACT
Antibiotics revolutionized the treatment of infectious diseases; however, it is now clear that broad-spectrum antibiotics alter the composition and function of the host’s microbiome. The microbiome plays a key role in human health, and its perturbation is increasingly recognized as contributing to many human diseases. Widespread broad-spectrum antibiotic use has also resulted in the emergence of multidrug-resistant pathogens, spurring the development of pathogen-specific strategies such as monoclonal antibodies (MAbs) to combat bacterial infection. Not only are pathogen-specific approaches not expected to induce resistance in nontargeted bacteria, but they are hypothesized to have minimal impact on the gut microbiome. Here, we compare the effects of antibiotics, pathogen-specific MAbs, and their controls (saline or control IgG [c-IgG]) on the gut microbiome of 7-week-old, female, C57BL/6 mice. The magnitude of change in taxonomic abundance, bacterial diversity, and bacterial metabolites, including short-chain fatty acids (SCFA) and bile acids in the fecal pellets from mice treated with pathogen-specific MAbs, was no different from that with animals treated with saline or an IgG control. Conversely, dramatic changes were observed in the relative abundance, as well as alpha and beta diversity, of the fecal microbiome and bacterial metabolites in the feces of all antibiotic-treated mice. Taken together, these results indicate that pathogen-specific MAbs do not alter the fecal microbiome like broad-spectrum antibiotics and may represent a safer, more-targeted approach to antibacterial therapy.
INTRODUCTION
Until recently, microbiology research has focused primarily on the bacterial pathogens responsible for infection or chronic disease, ignoring the 100 trillion microbes making up the microbiome that occupies our skin and mucosal surfaces (1). In the last decade, the microbiome has become increasingly recognized as an integral part of human health, immunity, and the response to certain therapeutics. In particular, the gastrointestinal tract microbiome has been reported to play a key role in maintaining health and homeostasis. In fact, disruption of the microbiome, or dysbiosis, has been linked to diseases such as diabetes, obesity, and asthma (2, 3). Although many factors such as diet, travel, and where we live can affect the bacterial composition of our microbiota, it is unlikely that anything directly impacts its composition as much as broad-spectrum antibiotic therapy does (4).
Antibiotics have saved countless lives and have enabled modern medical treatments, including organ transplants, joint replacement surgeries, and immunosuppressive cancer therapies (5). However, broad-spectrum antibiotic use has expanded beyond treating serious bacterial infections into agriculture to promote livestock growth and are often prescribed to treat colds and upper respiratory tract infections that are likely caused by a virus (6, 7). Such widespread antibiotic use has fueled the current resistance epidemic, which some fear will lead us back into a preantibiotic era (8). Additionally, recent studies have demonstrated both an acute and sustained impact of broad-spectrum antibiotics on the composition and function of the human microbiome which can adversely affect human health (9). For example, patients exposed to broad-spectrum antibiotics are at increased risk of Salmonella enterica serovar Typhimurium-induced colitis or recurrent Clostridioides difficile infections (10, 11). Also, children administered antibiotics during the first year of life were found to exhibit an increased risk for developing asthma and childhood obesity (12, 13). Antibiotic use by patients suffering from intestinal diseases such as inflammatory bowel disease (IBD) has been correlated with decreased bacterial diversity in the gut and increased intestinal inflammation (14, 15), highlighting a need for alternatives to broad-spectrum antibiotics.
Antibiotic-mediated dysbiosis not only alters the bacterial population resulting in reduced bacterial diversity in the gut, but it can also alter levels of key metabolites, such as short-chain fatty acids (SCFAs), or the conversion of primary bile acids into secondary bile acids (16, 17). SCFAs are produced by fermenting gut bacteria and play an integral role regulating the intestinal epithelial barrier through tight junction proteins (TJP), influence immunity by driving regulatory T cell (Treg) differentiation, and affect satiety and insulin production (18–21). Constituents of a healthy microbiome also convert primary bile acids produced by the liver into secondary bile acids, which impact host inflammation, immunity, and lipid and glucose metabolism (22, 23). Dysbiosis of the gut microbiome following antibiotic exposure has been shown to impact bile acid metabolism and, consequently, affect immune tone (24, 25). It is therefore clear that alterations in gut bacterial populations can lead to changes in key metabolic pathways that impact human health and well being.
This greater understanding of the impact antibiotic-mediated dysbiosis has on human health along with the ongoing antibiotic resistance epidemic has led to the exploration of new methods to prevent or treat bacterial infections. Monoclonal antibodies (MAbs) are an attractive option due to their target specificity, long half-life, and ability to synergize with the host’s immune response (26–28). Here, we studied the gut microbiome of specific-pathogen-free (SPF) mice following treatment with antibacterial MAbs or broad-spectrum antibiotics to determine if pathogen-specific MAbs alter the host microbiome or its metabolic products.
RESULTS
Antibacterial MAbs do not change the bacterial density in feces.
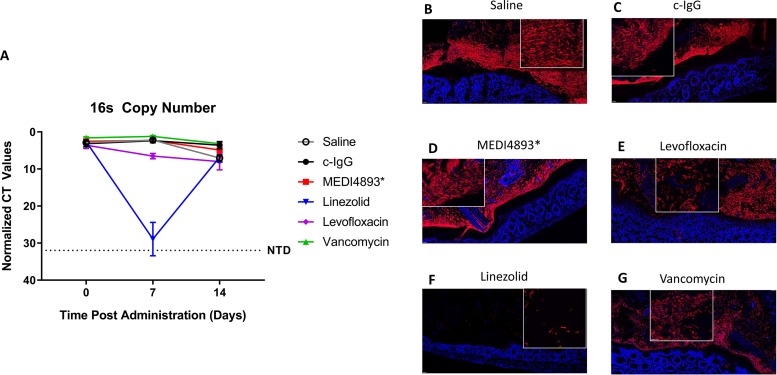
We hypothesized that unlike antibiotics (Abx), pathogen-specific MAbs would not alter the number or composition of bacteria in the intestinal microbiome. Specific-pathogen-free (SPF) mice were treated with a single dose of antibacterial MAb on day 0 or with a human-equivalent dose of broad-spectrum antibiotics for 5 days. A single dose of MAb was given because the half-life of human IgG in mice is 7 to 10 days, and MAbs such as MEDI4893 and MEDI3902 are administered as a single dose in human clinical trials (29, 30). Fecal samples were collected on days 0, 7, and 14 from mice treated with a clinical candidate MAb targeting Staphylococcus aureus (MEDI4893*) or human-equivalent doses of antibiotics used to treat S. aureus infections (vancomycin [VAN], levofloxacin [LVX], and linezolid [LZD]). Relative concentrations of bacteria, as measured by 16S rRNA gene quantitative PCR (qPCR), in fecal samples from mice treated with control IgG (c-IgG), MEDI4893*, or vancomycin were unchanged relative to the concentrations in saline controls at all time points. Conversely, the number of fecal bacteria was significantly reduced on day 7 in mice treated with levofloxacin or linezolid (P = 0.0083 and 0.0126, respectively) compared with saline-treated controls but was not significantly different than saline-treated controls on day 14 (Fig. 1A). Sections of colon containing a fecal pellet were removed, and fluorescent in situ hybridization (FISH) with a 16S rRNA gene probe was used to image bacteria within the colon. Bacterial density along the colonic epithelium was similar in mice treated with c-IgG, MED4893*, and saline. Antibiotic treatments noticeably affected the bacteria in the colon at day 7 posttreatment, with levofloxacin and linezolid reducing the bacterial burden and vancomycin eliminating all rod-shaped bacteria (Fig. 1B to G). Treatment of mice with anti-Pseudomonas aeruginosa MAb MEDI3902 or anti-Klebsiella pneumoniae MAb KPE33 also did not alter the overall bacterial content, while the antibiotic meropenem significantly (P = 0.0003) reduced bacterial load (see Fig. S1A and B in the supplemental material). These data demonstrate that unlike most of the antibiotics tested, the pathogen-specific MAbs have a negligible effect on the overall size of the bacterial population present in murine feces.
FIG 1.
Antibacterial MAbs do not change the microbial concentration in feces. (A) The relative concentrations of bacteria, as measured by 16S qPCR, in fecal samples of mice at day 0 (prior to treatment) and at 7 and 14 days posttreatment with saline, c-IgG, MEDI4893*, levofloxacin, linezolid, or vancomycin. (B to G) Segments of mouse colon with fecal pellet at 7 days posttreatment fixed in Carnoy’s solution and stained with red 16S probe for bacteria in the feces and lumen; blue indicates DAPI staining the nuclei of the epithelium. Inset shows bacteria at increased magnification. All data are representative of at least two independent experiments, n = 5/group.
Antibacterial MAbs do not change the bacterial composition of feces.
The 16S rRNA gene V4 region was sequenced to determine the effect the different antibacterial therapies had on the overall microbial community structure and the abundances of individual taxa.
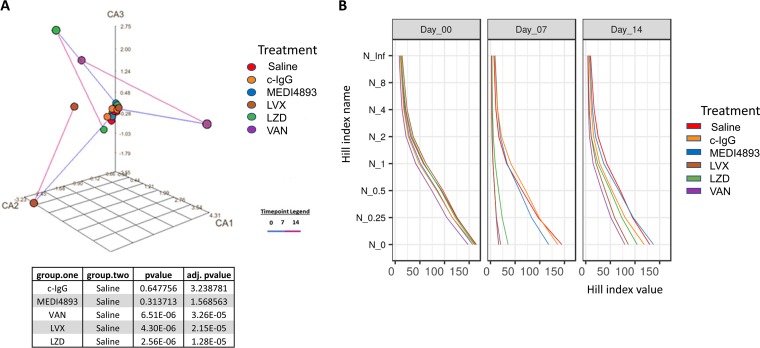
Correspondence analysis (CA) plots of the Bray-Curtis dissimilarity between the geometric medians of the taxonomic profiles of different treatment groups (Fig. 2A and S2A and B) revealed dramatic changes in the overall taxonomic composition on day 7 in antibiotic-treated groups. That was followed by a shift closer to the original state on day 14 in the levofloxacin and linezolid groups, while the vancomycin group remained in a more perturbed state. In contrast, treatment with pathogen-specific MAbs MEDI4893*, MEDI3902, or KPE33 resulted in much smaller compositional shifts during the course of treatment, comparable to those of the saline or c-IgG controls.
FIG 2.
Antibacterial MAbs do not change the overall microbial composition of feces. (A) Correspondence analysis (CA) plot of microbiota in fecal samples treated with saline, c-IgG, MEDI4893*, levofloxacin (LVX), linezolid (LZD), or vancomycin (VAN). Each point represents the geometric median of the genus taxonomic profiles within each treatment group at days 0 (prior to treatment), 7, and 14. The time point legend indicates the period of collection, with the blue line extending from time points 0 to 7 and the purple line extending from time points 7 to 14. adj., adjusted. (B) Plots of the abundance-based diversity indices (Hill numbers), also known as the effective numbers of species. The vertical axis corresponds to different values of the order parameter q that defines the Hill index Nq (66). The horizontal axis shows the respective index values. Taxonomic profiles of individual observations were aggregated into geometric medians for each combination of treatment and time of sample collection. Each such aggregated community is represented by one Hill index series shown as a line. One community is more diverse than the other if the values in the entire Hill series are higher for the first community (i.e., the lines for the two communities do not cross). All data are representative of at least two independent experiments, n = 5/group.
In the richness and alpha-diversity analyses, we observed that the administration of the broad-spectrum antibiotics levofloxacin, linezolid, vancomycin, ciprofloxacin, or meropenem dramatically reduced the Hill numbers in the treated samples on day 7, whereas the Hill numbers from the antibacterial MAb-treated samples remained similar to those of the controls (Fig. 2B and S2C and D).
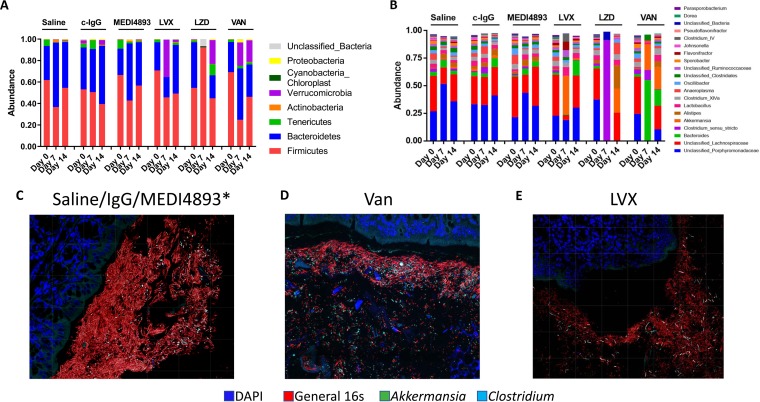
Prior to treatment, the fecal microbiota of naive mice was mostly composed of bacteria from the Bacteroidetes and Firmicutes phyla (Fig. 3A). Porphyromonadaceae (∼25%) and Lachnospiraceae (∼25%) had the highest abundances at the family level (Fig. 3B). Similar taxonomic composition was observed in day 7 and 14 fecal samples from mice treated with MEDI4893*, c-IgG, or saline. Levofloxacin most strongly affected Lachnospiraceae on day 7, but that treatment group returned to the taxonomic distribution observed in the saline-treated controls by day 14. Linezolid resulted in a taxonomic profile dominated by Clostridium sensu stricto on day 7 and remained perturbed on day 14. Vancomycin reduced the relative abundance of the Gram-positive Firmicutes phylum on day 7, corresponding to a reduction in Porphyromonadaceae and Lachnospiraceae families and an increase in the relative abundance of members of the Akkermansia genus. The taxonomic profile of that treatment group remained perturbed on day 14. FISH was used to probe for specific taxa that were observed to be high in abundance within the fecal microbiota. Relatively high abundances of Akkermansia and Bacteroides spp. were observed in the colon sections collected from the mice treated with levofloxacin and vancomycin, respectively. These relative abundance proportions were not observed in the mice treated with saline, c-IgG, or MEDI4893* (Fig. 3C to E). Similar outcomes were observed with MAbs MEDI3902 and KPE33, which behaved like the saline controls, whereas additional antibiotics, ciprofloxacin and meropenem, were disrupted in the microbiota, particularly on day 7 (Fig. S3A to D).
FIG 3.
Antibacterial MAbs do not change bacterial taxonomic abundances in feces. (A) Relative abundances of major bacterial phyla of mice treated with saline, c-IgG, MEDI4893*, levofloxacin (LVX), linezolid (LZD), or vancomycin (VAN) at day 0 (prior to treatment) and days 7 and 14. (B) The relative abundance of major bacterial genera of treated mice at days 0 (prior to treatment), 7, and 14. (C to E) Segments of mouse colon with fecal pellet 7 days posttreatment stained with red 16S probe for bacteria, green for Akkermansia spp., and cyan for Clostridium sensu stricto; blue indicates DAPI staining. All data are representative of at least two independent experiments, n = 5/group.
Antibacterial MAbs do not affect the levels of SCFAs or bile acids.
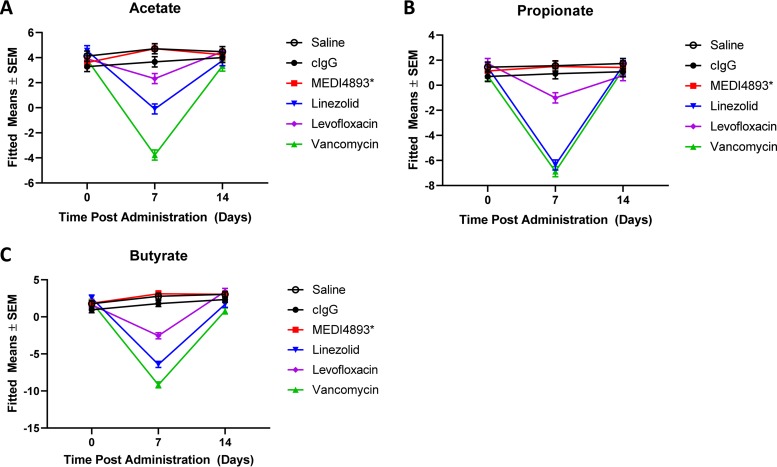
SCFAs, notably, acetate, propionate, or butyrate, can be produced or influence several bacteria within the Firmicutes phylum (Lachnospiraceae and Lactobacillus) and the Bacteroidetes phylum (Porphyromonadaceae and Alistipes spp.) (31–34). Decreases in the relative abundances of acetate, propionate, and butyrate were observed in fecal samples from the mice treated with vancomycin, linezolid, or levofloxacin on day 7 in comparison to the controls. Consistent with pathogen-specific MAbs not affecting the fecal microbiome, the SCFA levels in the MEDI4893*-treated samples remained similar to those of the saline control (Fig. 4A to C).
FIG 4.
Antibacterial MAbs do not affect the levels of SCFAs. (A to C) Relative abundances of acetate (A), propionate (B), and butyrate (C) in fecal samples of mice treated with saline, MEDI4893*, levofloxacin, linezolid, or vancomycin at day 0 and days 7 and 14 posttreatment. All data are representative of at least two independent experiments, n = 5/group. SEM, standard error of the mean.
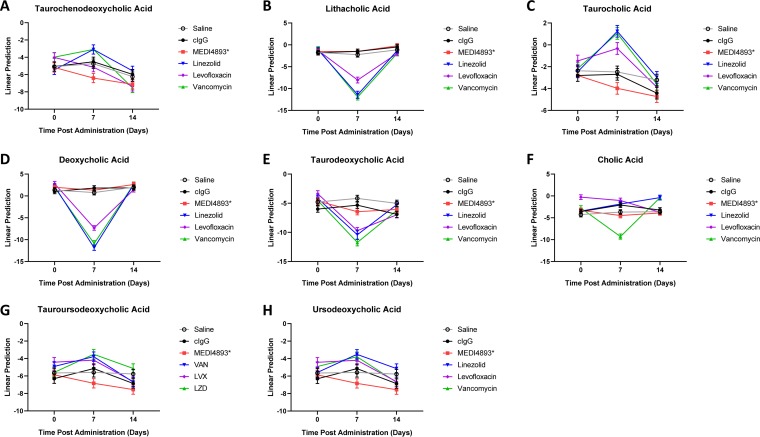
Recent studies acknowledged the role of bile acids in health and disease (35). Changes were observed in the conversion of primary bile acids (BAs) into secondary BAs in mice treated with vancomycin, linezolid, or levofloxacin. Antibiotic-treated samples exhibited reduced the conversion of taurochenodeoxycholic acid (TCDCA) into the secondary BAs lithocholic acid (LA) or taurine-conjugated lithocholic acid (TLCA) and taurocholic acid (TCA) into deoxycholic acid (DCA) or taurodeoxycholic acid (TDCA) in comparison to the controls. In contrast, conversion of the primary BAs into secondary BAs in the MAb-treated samples remained similar to that of the controls (Fig. 5A to F).
FIG 5.
Antibacterial MAbs do not affect bile acid metabolism. (A to H) Relative abundances of taurochenodeoxycholic acid (A), lithocholic acid (B), taurine-conjugated lithocholic acid (C), deoxycholic acid (D), taurodeoxycholic acid (E), cholic acid (F), tauroursodeoxycholic acid (G), and ursodeoxycholic acid (H) in fecal samples of mice treated with saline, MEDI4893*, levofloxacin, linezolid, or vancomycin at day 0 and days 7 and 14 posttreatment. All data are representative of at least two independent experiments, n = 5/group.
DISCUSSION
Antibiotics save countless lives; however, recent evidence demonstrates that antibiotics can have major off-target effects on the gut microbiome. The human gut microbiome is an “external organ” that is integral to human health and should be considered as part of the risk assessment process for new drugs. Antibiotic perturbation of a healthy microbiome coupled with the emergence and expansion of antibiotic-resistant bacterial pathogens have necessitated the development of novel antibacterial strategies that limit some of these adverse effects. Pathogen-specific MAbs have emerged as one such strategy due to their precision targeting and low risk for off-target effects. Antibodies have unique properties which enable them to promote pathogen clearance through multiple mechanisms, including virulence factor neutralization, inhibition of bacterial clumping and biofilm formation, and engagement of the immune system through Fc-dependent interactions (36). MAbs targeting the major human pathogens S. aureus, P. aeruginosa, and K. pneumoniae have a minimal effect on the gut microbiome compared with standard-of-care antibiotics.
Alterations to the microbiome have been linked to numerous diseases, as dysbiosis can influence key functions of the microbiome (3). Bacteria in the gut act not just as a physical barrier against colonization by pathogenic organisms but also by metabolizing small molecules which then circulate throughout the host, influencing numerous host processes. Our data demonstrate that pathogen-specific MAbs do not reduce the absolute abundance, diversity, or taxonomic composition of the gut microbiome compared to those with the effects of antibiotic treatment. Such effects observed in the antibiotic treatment groups are indicative of a reduced bacterial burden in the gut and diminished capacity to prevent infection by opportunistic pathogens (37–40).
Additionally, metabolic functions of the microbiome were not altered in the presence of MAbs as they were with antibiotics. The host relies on bacterial metabolism not only to process food but also to break down metabolites such as SCFAs and bile acids (41, 42). Altered processing of either SCFAs or bile acids due to restructuring of the microbiome has been linked to metabolic diseases such as diabetes and nonalcoholic fatty liver disease (NAFLD) (42–46). Functional consequences of microbiome disruption are not limited to metabolic disease. Autoimmune disease, graft-versus-host disease, allergies, and inflammatory bowel syndrome have all been linked to microbiome dysbiosis (47–50). Furthermore, both the development of different types of cancer and the response rates to current immune-targeted cancer therapies have been linked to the structure and function of the microbiome (51–54). Many of these microbiome-related diseases are associated with bacterial infection, and we postulate that antibiotic treatment of concurrent infections could exacerbate the underlying disease by modifying the microbiome. Due to their minimal impact on the microbiome, pathogen-specific MAbs would avoid such a complication.
The symbiotic relationship between our immune system and the bacteria which colonize us is increasingly recognized as having influence over a wide array of biological processes. This occurs through microbe-dependent production of SCFAs, bile acids, and other metabolites. We demonstrate that unlike standard-of-care antibiotics, pathogen-specific approaches for the treatment of infectious diseases do not alter the composition or function of the gut microbiome. MAb-based approaches have the potential to work synergistically with the host, reducing the duration and/or amount of overall antibiotic use, resulting in reduced impacts on human health.
MATERIALS AND METHODS
Reagents.
MAbs were diluted and prepared fresh daily from refrigerated or frozen stocks into sterile phosphate buffer saline (PBS; pH 7.2). The anti-S. aureus alpha toxin MAb MEDI4893*, anti-P. aeruginosa MAb MEDI3902, and anti-K. pneumoniae MAbs KPE33 and KPN42 were previously described (26, 55, 56). Isotype control human IgG1 (c-IgG) was included as a control. Analytical-grade vancomycin, linezolid, levofloxacin, ciprofloxacin, and meropenem were corrected for potency and prepared fresh daily.
Mice.
All animal studies were approved by the AstraZeneca Institutional Animal Care and Use Committee and were conducted in an Association for Accreditation and Assessment Laboratory Animal Care (AAALAC)-accredited facility in compliance with U.S. regulations governing the housing and use of animals. Specific-pathogen-free 7- to 8-week-old female C57BL/6J mice (The Jackson Laboratory) were ear tagged, randomized, and housed in sterile cages with autoclaved mouse chow (LabDiet 5K52) and water. At least 5 mice per group were used in each experiment.
Study design.
Animals were injected intraperitoneally (i.p.) with either a single 0.5-ml MAb dose (15 mg/kg of body weight) or administered human-equivalent doses of vancomycin (100 mg/kg twice a day [BID]), linezolid (60 mg/kg BID), ciprofloxacin (10 mg/kg BID), meropenem (66.7 mg/kg BID), or levofloxacin (235 mg/kg once a day [QID]) in 0.2 ml via subcutaneous (s.c.) injection for 5 consecutive days. Animals treated with PBS vehicle (BID) were included in each study as a control. Two fecal pellets (∼40 mg total) were collected from each animal on days 0 (prior to treatment), 7 (after a 36-h washout period), and 14 and immediately processed for DNA extraction (see below). A single fecal pellet was collected from the same animals and immediately stored at –80°C for SCFA and bile acid analyses. In select experiments, an additional cohort of mice from each group was euthanized by CO2 asphyxiation on day 7 for intestinal fluorescent in situ hybridization (FISH) staining.
DNA extraction and quantification.
DNA was extracted from mouse feces using the PowerSoil DNA isolation kit (Mo Bio, West Carlsbad CA, USA), according to the manufacturer’s protocol, and DNA concentrations were determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Purified DNA was stored at −80°C until use. The universal qPCR 16S rRNA primers U16SRT-F (5′-ACTCCTACGGGAGGCAGCGT-3′) and U16SRT-R (5′-TATTACCGCGGCTGCTGCTGGC-3′) were used to quantify total bacterial 16S rRNA genes from the purified fecal DNA (19). qPCRs were performed using 2 μl of template DNA from 2 fecal pellets per mouse, 2 pmol each primer, 3 μl of deionized water, and 5 μl of Sybr green master mix (Promega). The cycling conditions were as follows: 50°C for 2 min, 95°C for 2 min, 95°C for 15 s, and 60°C for 1 min, followed by a dissociation stage for 40 cycles. Differences in threshold cycle (CT) values were compared following normalization of DNA input in each qPCR.
Library preparation and sequencing.
16S rRNA gene PCR was carried out as described previously (57). Briefly, V4 16S rRNA gene libraries were constructed using AccuPrime Taq high-fidelity DNA polymerase (Invitrogen). Library cleanup and normalization were performed using the SequalPrep normalization plate (96-well) kit (Invitrogen). Libraries were then pooled, and the final concentration of the library was determined using a Qubit fluorometer (Thermo Fisher Scientific). Libraries were mixed with PhiX control v3 (Illumina) and denatured using fresh 0.04 N NaOH. Pooled libraries were sequenced on an Illumina MiSeq instrument using a 2 × 250-bp MiSeq reagent kit v2.
Sequence analysis.
Sequences were trimmed from adaptors using BBTools (58) and processed into amplicon sequence variants (ASVs) using the R package Dada2 (59), with taxonomy assigned from the RDP (60) training set v.14, as maintained by the Dada2 repository.
Fluorescence in situ hybridization of colonic microbiome.
To visualize the colonic microbiome, colon sections containing a fecal pellet were processed as previously described (61). Briefly, the tissue was fixed in Carnoy’s solution for 2 weeks and paraffin embedded. Slides were made from 5-μm slices, which were deparaffinized in xylene (20 min at room temperature), followed by a 5-min incubation in 99.5% ethanol. The tissue was then incubated overnight at 50°C in hybridization solution (20 mM Tris-HCl, 0.9 M NaCl, 0.1% SDS, 25% formamide) containing the following FISH probes: 16S rRNA, GCTGCCTCCCGTAGGAGT; Akkermansia spp., GGTTCCCCTCCATTAC; Clostridium sensu stricto, GCCGTGGCTTCCTCCTY; Porphyromonas spp., CTCGTTATGGCACTTAAGCCGA; and Bacteroides spp., CGCAATCGGAGTTCTTCGTGATATC (62). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and the slides were washed 3× with PBS and coverslipped with ProLong Gold mountant (Invitrogen). Images were collected using a Zeiss LSM 880 Airyscan or Leica SP8 microscope. Following acquisition, images were deconvolved using Airyscan processing or Leica Lightning deconvolution software.
Metabolite extraction from fecal samples.
Fecal samples were stored at –80°C prior to metabolite extraction. Metabolite extraction was performed with 50:50 acetonitrile-water. The amount of extraction solvent added to each sample was normalized to sample mass at a ratio of 10 μl extraction solvent to 1 mg sample. After the addition of extraction solvent, samples were manually homogenized and mixed at 2,000 rpm for 15 min at room temperature. Samples were centrifuged for 10 min at 18,000 × g and 4°C. One hundred microliters of supernatant was transferred to new tubes and centrifuged for 10 min at 18,000 × g and 4°C. Twenty microliters of supernatant was transferred to new tubes for derivatization of SCFAs as described below. Ten microliters of unlabeled metabolite extract was combined with 90 μl bile acid internal standard composed of a mixture of 15 bile acids at 1 μM in 50:50 MeOH-H2O. The bile acid internal standard mixture contains the following deuterated bile acids: lithocholic acid-d4, chenodeoxycholic acid-d4, deoxycholic acid-d4, ursodeoxycholic acid-d4, cholic acid-d4, glycolithocholic acid-d4, glycochenodeoxycholic acid-d4, glycodeoxycholic acid-d4, glycoursodeoxycholic acid-d4, glycocholic acid-d4, taurolithocholic acid-d4, taurochenodeoxycholic acid-d4, taurodeoxycholic acid-d4, tauroursodeoxycholic acid-d4, and taurocholic acid-d4.
Short-chain fatty acid derivatization.
SCFAs were derivatized with 3-nitrophenylhydrazine (3-NPH) based on a method previously described by Borchers and coworkers (63). Fecal extracts (20 μl) were sequentially mixed with 10 μl of 200 mM 3-NPH–HCl in 50:50 acetonitrile-water (ACN-H2O) solution and 10 μl of 120 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide–HCl (EDC) in 50:44:6 ACN-H2O-pyridine solution. Mixtures were reacted at 40°C with 800 rpm mixing for 30 min. 3-NPH-derivatized samples were diluted with 960 μl of 50:50 ACN-H2O. One hundred microliters of 3-NPH-derivatized samples was mixed with 100 μl 13C6-3-NPH-derivatized SCFA internal standard containing derivatized acetic acid, propionic acid, isobutyric acid, butyric acid, 2-methyl butyric acid, isovaleric acid, and valeric acid.
Liquid chromatography-mass spectrometry.
Relative quantitation of SCFAs and bile acids was performed by liquid chromatography-mass spectrometry (LC-MS) on an Agilent 1290 Infinity II LC coupled to a 6560 quadrupole time of flight (Q-TOF) instrument equipped with an Agilent Jet Stream (AJS) source operated in negative-ion mode. Chromatographic separation was performed using a Waters Acquity ultraperformance liquid chromatography (UPLC) BEH C18 column (1.7 μm, 2.1 by 100 mm). Mobile phases A and B were 0.01% formic acid in H2O and 0.01% formic acid in acetonitrile, respectively. SCFA analysis was performed with a flow rate of 0.3 ml·min−1, and the column temperature was maintained at 50°C. The initial solvent composition of 15% B was maintained for 2 min before increasing to 55% B at 11 min. The column was washed with 100% B for 3 min and equilibrated at initial solvent composition between injections for 5 min. MS spectra were recorded from m/z 50 to 1,000 at a rate of 1.5 spectra·s−1. Bile acid analysis was performed with a flow rate of 0.3 ml·min−1, and the column temperature was maintained at 50°C. The initial solvent composition of 5% B was maintained for 2.4 min before increasing to 30% B over 1 min and 70% B over 12 min. The column was washed with 100% B for 2.5 min and equilibrated at the initial solvent composition between injections for 4 min. MS spectra were recorded from m/z 50 to 1,200 at a rate of 1.5 spectra·s−1. The mass spectra for both methods were recalibrated to reference masses of m/z 112.9856 and 966.0007.
LC-MS data analysis and metabolite quantitation.
The Agilent Profinder software was used for retention time alignment and feature extraction by batch recursive feature extraction method. Compound groups were annotated by their m/z and LC retention times compared to isotopically labeled internal standards, saved as Profinder Archive files, and exported with the integrated intensities from each sample. Relative quantitation was based on the integrated ion intensities of the features and internal standards. To test for the significance of the changes in the metabolite abundances, the ion intensities from each compound were log transformed and subtracted from the corresponding internal controls. The results (logR) were fitted to a mixed-effect model considering the interactions among the compounds, treatments, and time points. The between-subject effects and the dilution factors for each sample were modeled as “random” factors, while the mean-subtracted background level (logMedian.BK2) for each sample estimated from the median (logMedian.BK) of the unassigned features was incorporated as a covariate. The data are available at MetaboLights (study number MTBLS1257; https://www.ebi.ac.uk/metabolights/).
Statistical analysis.
Microbiome analysis was performed in R. The majority of the analyses were done using the open-source package MGSAT, which wraps several R packages in order to perform -omics analyses (https://github.com/andreyto/mgsat). The scripts which generated the results presented in this paper are located in a directory, examples/16S/projects/mab_abx, versioned with Git tag mab_abx. Figures were generated with the R package ggplot2 (64). Richness and alpha- and beta-diversity metrics were calculated with the R package vegan (65) at the ASV level; all ASVs were included, regardless of abundance. To control for differences in sequencing depth per sample, samples were randomly rarefied to the minimum sample read count, and then each richness, alpha-diversity, or beta-diversity index was calculated. For each index, this process was repeated 400 times, and the results were averaged. Beta diversity was assessed using the Bray-Curtis dissimilarity index.
To estimate the statistical significance of the differences in compositional shifts caused by different treatments, we computed for each mouse the Bray-Curtis dissimilarity between day 7 and day 0 and used this within-mouse compositional shift in the t test for the difference in means between the pairs of treatment groups.
Richness and other abundance-based measures of alpha diversity in each sample were assessed using a series of Hill numbers (66), Nq, where q was the order parameter ranging from zero to infinity. Hill numbers are a unified family of diversity indices (differing among themselves only by an exponent, q) that incorporate other measures of diversity and richness, expressing them on a uniform scale of the effective number of species (67). In particular, N0 is equivalent to richness, and N1 and N2 are equivalent to the exponentials of the Shannon index and the inverted Simpson index, respectively. Following the diversity analysis, sequence counts were normalized into simple proportions of taxa per sample to obtain the relative abundance profiles. For the genus-level analysis, genera from the lower quartiles of the mean relative abundance and mean incidence over all samples were discarded. The dynamic of the overall abundance profiles was plotted in the coordinates obtained from a correspondence analysis of a Bray-Curtis dissimilarity index computed between the geometric medians of samples in each treatment group at each time point.
Data availability.
Data are available through the European Nucleotide Archive under accession number PRJEB34462.
Supplementary Material
ACKNOWLEDGMENTS
Funding was provided by AstraZeneca.
All authors were employed by AstraZeneca while the work was performed.
O.J.-N., A.T., P.W., T.S.C., M.S.G., J.J.H., S.H., and B.R.S. conceived and designed the study; O.J.-N., M.S.G., A.T., F.F., U.R., H.L., D.E.T., J.B., W.Y., G.D., S.H., P.W., C.K.S., T.S.C., and B.R.S. analyzed and interpreted the data; and O.J.-N., T.S.C., and B.R.S. drafted the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Doré J, Ehrlich SD, MetaHIT Consortium, Bork P, Wang J. 2014. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 2.Kamada N, Seo S-U, Chen GY, Núñez G. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. 2016. The human intestinal microbiome in health and disease. N Engl J Med 375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 4.Langdon A, Crook N, Dantas G. 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adedeji WA. 2016. The treasure called antibiotics. Ann Ib Postgrad Med 14:56–57. [PMC free article] [PubMed] [Google Scholar]
- 6.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, Laxminarayan R. 2014. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis 14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 7.Imanpour S, Nwaiwu O, McMaughan DK, DeSalvo B, Bashir A. 2017. Factors associated with antibiotic prescriptions for the viral origin diseases in office-based practices, 2006–2012. JRSM Open 8:2054270417717668. doi: 10.1177/2054270417717668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshikawa TT. 2002. Antimicrobial resistance and aging: beginning of the end of the antibiotic era? J Am Geriatr Soc 50:S226–S229. doi: 10.1046/j.1532-5415.50.7s.2.x. [DOI] [PubMed] [Google Scholar]
- 9.Doan T, Hinterwirth A, Worden L, Arzika AM, Maliki R, Abdou A, Kane S, Zhong L, Cummings SL, Sakar S, Chen C, Cook C, Lebas E, Chow ED, Nachamkin I, Porco TC, Keenan JD, Lietman TM. 2019. Gut microbiome alteration in MORDOR I: a community-randomized trial of mass azithromycin distribution. Nat Med 25:1370–1376. doi: 10.1038/s41591-019-0533-0. [DOI] [PubMed] [Google Scholar]
- 10.Angulo FJ, Molbak K. 2005. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infect Dis 41:1613–1620. doi: 10.1086/497599. [DOI] [PubMed] [Google Scholar]
- 11.Brown KA, Khanafer N, Daneman N, Fisman DN. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 57:2326–2332. doi: 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozyrskyj AL, Ernst P, Becker AB. 2007. Increased risk of childhood asthma from antibiotic use in early life. Chest 131:1753–1759. doi: 10.1378/chest.06-3008. [DOI] [PubMed] [Google Scholar]
- 13.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe EL, Crother TR, Rabizadeh S, Hu B, Wang H, Chen S, Shimada K, Wong MH, Michelsen KS, Arditi M. 2010. Toll-like receptor 2 signaling protects mice from tumor development in a mouse model of colitis-induced cancer. PLoS One 5:e13027. doi: 10.1371/journal.pone.0013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nitzan O, Elias M, Peretz A, Saliba W. 2016. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol 22:1078–1087. doi: 10.3748/wjg.v22.i3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Høverstad T, Carlstedt-Duke B, Lingaas E, Midtvedt T, Norin KE, Saxerholt H, Steinbakk M. 1986. Influence of ampicillin, clindamycin, and metronidazole on faecal excretion of short-chain fatty acids in healthy subjects. Scand J Gastroenterol 21:621–626. doi: 10.3109/00365528609003109. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Limaye PB, Renaud HJ, Klaassen CD. 2014. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol Appl Pharmacol 277:138–145. doi: 10.1016/j.taap.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, Wang RX, Onyiah JC, Kominsky DJ, Colgan SP. 2017. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of claudin-2. J Immunol 199:2976–2984. doi: 10.4049/jimmunol.1700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo T, Kishi M, Fushimi T, Ugajin S, Kaga T. 2009. Vinegar intake reduces body weight, body fat mass, and serum triglyceride levels in obese Japanese subjects. Biosci Biotechnol Biochem 73:1837–1843. doi: 10.1271/bbb.90231. [DOI] [PubMed] [Google Scholar]
- 21.Morrison DJ, Preston T. 2016. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan C. 2014. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A 111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan PM, Stanton C, Caplice NM. 2017. Bile acids at the cross-roads of gut microbiome–host cardiometabolic interactions. Diabetol Metab Syndr 9:102. doi: 10.1186/s13098-017-0299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antunes LCM, Han J, Ferreira RBR, Lolić P, Borchers CH, Finlay BB. 2011. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother 55:1494–1503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan W, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF. 2018. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360:eaan5931. doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiGiandomenico A, Keller AE, Gao C, Rainey GJ, Warrener P, Camara MM, Bonnell J, Fleming R, Bezabeh B, Dimasi N, Sellman BR, Hilliard J, Guenther CM, Datta V, Zhao W, Gao C, Yu X-Q, Suzich JA, Stover CK. 2014. A multifunctional bispecific antibody protects against Pseudomonas aeruginosa. Sci Transl Med 6:262ra155. doi: 10.1126/scitranslmed.3009655. [DOI] [PubMed] [Google Scholar]
- 27.Cohen TS, Boland ML, Boland BB, Takahashi V, Tovchigrechko A, Lee Y, Wilde AD, Mazaitis MJ, Jones-Nelson O, Tkaczyk C, Raja R, Stover CK, Sellman BR. 2018. S. aureus evades macrophage killing through NLRP3-dependent effects on mitochondrial trafficking. Cell Rep 22:2431–2441. doi: 10.1016/j.celrep.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen TS, Pelletier M, Cheng L, Pennini ME, Bonnell J, Cvitkovic R, Chang CS, Xiao X, Cameroni E, Corti D, Semenova E, Warrener P, Sellman BR, Suzich J, Wang Q, Stover CK. 2017. Anti-LPS antibodies protect against Klebsiella pneumoniae by empowering neutrophil-mediated clearance without neutralizing TLR4. JCI Insight 2:e92774. doi: 10.1172/jci.insight.92774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali SO, Yu XQ, Robbie GJ, Wu Y, Shoemaker K, Yu L, DiGiandomenico A, Keller AE, Anude C, Hernandez-Illas M, Bellamy T, Falloon J, Dubovsky F, Jafri HS. 2018. Phase 1 study of MEDI3902, an investigational anti-Pseudomonas aeruginosa PcrV and Psl bispecific human monoclonal antibody, in healthy adults. Clin Microbiol Infect 25:629.e1–629.e6. doi: 10.1016/j.cmi.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Ruzin A, Wu Y, Yu L, Yu XQ, Tabor DE, Mok H, Tkaczyk C, Jensen K, Bellamy T, Roskos L, Esser MT, Jafri HS. 2018. Characterisation of anti-alpha toxin antibody levels and colonisation status after administration of an investigational human monoclonal antibody, MEDI4893, against Staphylococcus aureus alpha toxin. Clin Transl Immunology 7:e1009. doi: 10.1002/cti2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biddle A, Stewart L, Blanchard J, Leschine S. 2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5:627–640. doi: 10.3390/d5030627. [DOI] [Google Scholar]
- 32.LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. 2017. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact 16:79. doi: 10.1186/s12934-017-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 34.Tabor DE, Yu L, Mok H, Tkaczyk C, Sellman BR, Wu Y, Oganesyan V, Slidel T, Jafri H, McCarthy M, Bradford P, Esser MT. 2016. Staphylococcus aureus alpha-toxin is conserved among diverse hospital respiratory isolates collected from a global surveillance study and is neutralized by monoclonal antibody MEDI4893. Antimicrob Agents Chemother 60:5312–5321. doi: 10.1128/AAC.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. 2014. Bile acids and the gut microbiome. Curr Opin Gastroenterol 30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichert JM. 2014. Antibody Fc: linking adaptive and innate immunity. mAbs 6:619–621. doi: 10.4161/mabs.28617. [DOI] [Google Scholar]
- 37.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA, Soeters MR, Blaak EE, Dallinga-Thie GM, Reijnders D, Ackermans MT, Serlie MJ, Knop FK, Holst JJ, van der Ley C, Kema IP, Zoetendal EG, de Vos WM, Hoekstra JBL, Stroes ES, Groen AK, Nieuwdorp M. 2014. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol 60:824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 38.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, Pamer EG. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest 120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL Jr, Droit L, Berbers GAM, Kemper EM, van Leeuwen EMM, Boele van Hensbroek M, Wiersinga WJ. 2018. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized-control proof-of-concept trial. Cell Host Microbe 24:197–207.e4. doi: 10.1016/j.chom.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isaac S, Scher JU, Djukovic A, Jimenez N, Littman DR, Abramson SB, Pamer EG, Ubeda C. 2017. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J Antimicrob Chemother 72:128–136. doi: 10.1093/jac/dkw383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wahlström A, Sayin SI, Marschall HU, Backhed F. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, Mujagic Z, Masclee AAM, Jonkers D, Oosting M, Joosten LAB, Netea MG, Franke L, Zhernakova A, Fu J, Wijmenga C, McCarthy MI. 2019. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 51:600–605. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moschen AR, Kaser S, Tilg H. 2013. Non-alcoholic steatohepatitis: a microbiota-driven disease. Trends Endocrinol Metab 24:537–545. doi: 10.1016/j.tem.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Neuschwander-Tetri BA, NASH Clinical Research Network, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, Kowdley KV, McCullough A, Terrault N, Clark JM, Tonascia J, Brunt EM, Kleiner DE, Doo E, Network N. 2015. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajani C, Jia W. 2018. Bile acids and their effects on diabetes. Front Med 12:608–623. doi: 10.1007/s11684-018-0644-x. [DOI] [PubMed] [Google Scholar]
- 46.Chávez-Talavera O, Tailleux A, Lefebvre P, Staels B. 2017. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 152:1679–1694.e3. doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 47.Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, Hofmann J, Raifer H, Vachharajani N, Carrascosa LC, Lamp B, Nist A, Stiewe T, Shaul Y, Adhikary T, Zaiss MM, Lauth M, Steinhoff U, Visekruna A. 2019. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun 10:760. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varelias A, Ormerod KL, Bunting MD, Koyama M, Gartlan KH, Kuns RD, Lachner N, Locke KR, Lim CY, Henden AS, Zhang P, Clouston AD, Hasnain SZ, McGuckin MA, Blazar BR, MacDonald KP, Hugenholtz P, Hill GR. 2017. Acute graft-versus-host disease is regulated by an IL-17-sensitive microbiome. Blood 129:2172–2185. doi: 10.1182/blood-2016-08-732628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, Dekens J, Peters V, Voskuil MD, Visschedijk MC, van Dullemen HM, Keszthelyi D, Swertz MA, Franke L, Alberts R, Festen E, Dijkstra G, Masclee AAM, Hofker MH, Xavier RJ, Alm EJ, Fu J, Wijmenga C, Jonkers D, Zhernakova A, Weersma RK. 2018. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med 10:eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 50.Leung D. 2019. The microbiome and allergic diseases: a struggle between good and bad microbes. Ann Allergy Asthma Immunol 122:231–232. doi: 10.1016/j.anai.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, Ameh S, Sandel D, Liang XS, Mazzilli S, Whary MT, Meyerson M, Germain R, Blainey PC, Fox JG, Jacks T. 2019. Commensal microbiota promote lung cancer development via γδ T cells. Cell 176:998–1013.e16. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, Vaysse T, Marthey L, Eggermont A, Asvatourian V, Lanoy E, Mateus C, Robert C, Carbonnel F. 2017. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 53.Sethi V, Kurtom S, Tarique M, Lavania S, Malchiodi Z, Hellmund L, Zhang L, Sharma U, Giri B, Garg B, Ferrantella A, Vickers SM, Banerjee S, Dawra R, Roy S, Ramakrishnan S, Saluja A, Dudeja V. 2018. Gut microbiota promotes tumor growth in mice by modulating immune response. Gastroenterology 155:33–37.e6. doi: 10.1053/j.gastro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, et al. 2018. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua L, Hilliard JJ, Shi Y, Tkaczyk C, Cheng LI, Yu X, Datta V, Ren S, Feng H, Zinsou R, Keller A, O’Day T, Du Q, Cheng L, Damschroder M, Robbie G, Suzich J, Stover CK, Sellman BR. 2014. Assessment of an anti-alpha-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus-induced pneumonia. Antimicrob Agents Chemother 58:1108–1117. doi: 10.1128/AAC.02190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Chang C-S, Pennini M, Pelletier M, Rajan S, Zha J, Chen Y, Cvitkovic R, Sadowska A, Heidbrink Thompson J, Yu Lin H, Barnes A, Rickert K, Wilson S, Stover CK, Dall'Acqua WF, Chowdhury PS, Xiao X. 2016. Target-agnostic identification of functional monoclonal antibodies against Klebsiella pneumoniae multimeric MrkA fimbrial subunit. J Infect Dis 213:1800–1808. doi: 10.1093/infdis/jiw021. [DOI] [PubMed] [Google Scholar]
- 57.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bushnell B. 2016. BBMap short read aligner. University of California, Berkeley, Berkeley, CA: https://sourceforge.net/projects/bbmap/. [Google Scholar]
- 59.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. 2015. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright ES, Yilmaz LS, Corcoran AM, Ökten HE, Noguera DR. 2014. Automated design of probes for rRNA-targeted fluorescence in situ hybridization reveals the advantages of using dual probes for accurate identification. Appl Environ Microbiol 80:5124–5133. doi: 10.1128/AEM.01685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J, Lin K, Sequeira C, Borchers CH. 2015. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry. Anal Chim Acta 854:86–94. doi: 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 64.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 65.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2014. vegan: community ecology package. R package version 2.0-10. [Google Scholar]
- 66.Hill MO. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432. doi: 10.2307/1934352. [DOI] [Google Scholar]
- 67.Chao A, Gotelli NJ, Hsieh TC, Sander EL, Ma KH, Colwell RK, Ellison AM. 2014. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84:45–67. doi: 10.1890/13-0133.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available through the European Nucleotide Archive under accession number PRJEB34462.