Delamanid (DLM), a nitro-dihydroimidazooxazole derivative currently approved for pulmonary multidrug-resistant tuberculosis (TB) therapy, is a prodrug activated by mycobacterial 7,8-didemethyl-8-hydroxy 5-deazaflavin electron transfer coenzyme (F420)-dependent nitroreductase (Ddn). Despite inhibiting the biosynthesis of a subclass of mycolic acids, the active DLM metabolite remained unknown. Comparative liquid chromatography-mass spectrometry (LC-MS) analysis of DLM metabolites revealed covalent binding of reduced DLM with a nicotinamide ring of NAD derivatives (oxidized form) in DLM-treated Mycobacterium tuberculosis var.
KEYWORDS: Mycobacterium, delamanid, drug, tuberculosis
ABSTRACT
Delamanid (DLM), a nitro-dihydroimidazooxazole derivative currently approved for pulmonary multidrug-resistant tuberculosis (TB) therapy, is a prodrug activated by mycobacterial 7,8-didemethyl-8-hydroxy 5-deazaflavin electron transfer coenzyme (F420)-dependent nitroreductase (Ddn). Despite inhibiting the biosynthesis of a subclass of mycolic acids, the active DLM metabolite remained unknown. Comparative liquid chromatography-mass spectrometry (LC-MS) analysis of DLM metabolites revealed covalent binding of reduced DLM with a nicotinamide ring of NAD derivatives (oxidized form) in DLM-treated Mycobacterium tuberculosis var. Bacille de Calmette et Guérin. Isoniazid-resistant mutations in the type II NADH dehydrogenase gene (ndh) showed a higher intracellular NADH/NAD ratio and cross-resistance to DLM, which were restored by complementation of the mutants with wild-type ndh. Our data demonstrated for the first time the adduct formation of reduced DLM with NAD in mycobacterial cells and its importance in the action of DLM.
INTRODUCTION
Tuberculosis (TB), caused by Mycobacterium tuberculosis infection, remains the most prevalent cause of death due to a single infectious agent (1). The emergence of TB resistant to current first-line drugs (multidrug-resistant TB [MDR-TB] with resistance to the first-line drugs isoniazid [INH] and rifampin [RIF]) and MDR-TB with additional resistance to any of the fluoroquinolones and to at least one of the three injectable second-line drugs (extensively drug-resistant TB [XDR-TB]) is problematic in various clinical settings.
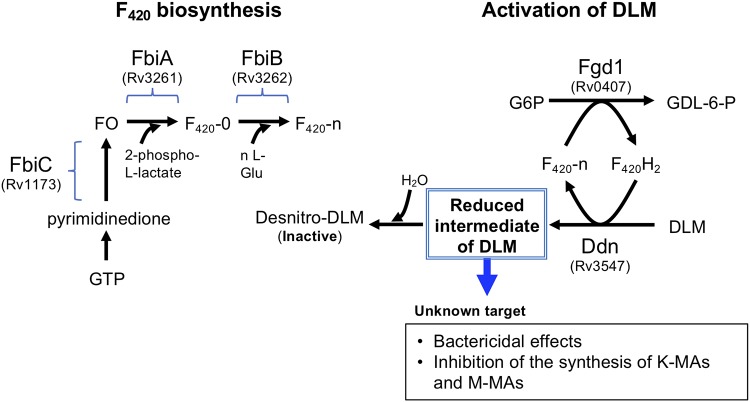
Delamanid (DLM) is a key drug in the treatment of MDR-TB and XDR-TB, which was approved in Europe and Japan in April and July 2014, respectively. DLM is a prodrug that is metabolized in M. tuberculosis (Fig. 1). In M. tuberculosis, coupled with oxidation of glucose-6-phosphate (G6P) to 6-phospho-gluconolactone (GDL-6-P), 7,8-didemethyl-8-hydroxy 5-deazaflavin electron transfer coenzyme (F420) is reduced to F420H2 by F420-dependent G6P dehydrogenase (Fgd1, Rv0407). DLM is then reduced by F420H2-dependent nitroreductase (Ddn, Rv3547) coupled with F420H2 oxidation (2–4). Actually, mutations in the genes encoding F420 synthesizing enzymes, including FbiA (Rv3261), FbiB (Rv3262), and FbiC (Rv1173) (5–7), as well as Fgd1 and Ddn, lead to resistance to DLM (2, 4, 8, 9). However, the critical metabolite of DLM that expresses antimycobacterial activity remains unclear.
FIG 1.
Biosynthesis of coenzyme F420 and F420H2-dependent reduction of DLM in M. tuberculosis. The 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) is synthesized from pyrimidinedione by a series of reactions catalyzed by enzymes, including FbiC (Rv1173) (5). FbiA (Rv3261) transfers a 2-phospho-l-lactate moiety to FO, resulting in the synthesis of F420 with no poly-γ-glutamate tail (F420-0) (6). FbiB (Rv3262, γ-glutamyl ligase) catalyzes the addition of l-Glu to F420-0, resulting in the synthesis of F420-n (6, 7). Coupled with oxidation of G6P and reduction of F420 by Fgd1, DLM is reduced by F420H2-dependent Ddn (Rv3547) (2–4). The reduced intermediate of DLM is deduced to attack an unknown target. A major part of the reduced intermediate is further converted in mycobacteria to desnitro-DLM (inactive form) by H2O quenching.
The major metabolite of DLM in DLM-treated Mycobacterium was a desnitro-imidazooxazole form (desnitro-DLM; Fig. 1) that had no antimycobacterial activity, suggesting that a nitro radical anion form of DLM that may be an intermediate metabolite between DLM and the desnitro-DLM may possibly target cellular components (2). It is well known that the radicals of INH and ethionamide (ETH) generated in mycobacterial cells formed the adducts with NADH, which interfere with the activity of essential NADH-dependent trans-2-enoyl-acyl carrier protein reductase (InhA) (10–13). Our previous study revealed that around 20% of the radioactivity of 14C-labeled DLM (14C-DLM) was distributed to cellular components of wild-type (WT) BCG (2). In contrast, radioactivity was almost background level in a DLM-resistant (DLMr) mutant in which DLM is not reduced due to a mutation in ddn (2, 4). In this study, we demonstrated an adduct formation of activated DLM with NAD derivatives (oxidized form) by comprehensively analyzed DLM-derived components specifically generated in DLM-susceptible mycobacteria using liquid chromatography-mass spectrometry (LC-MS) and suggested an important role of NAD-DLM adduct formation in the bactericidal action of DLM.
RESULTS
Detection of DLM metabolites specifically generated in a DLM-susceptible strain.
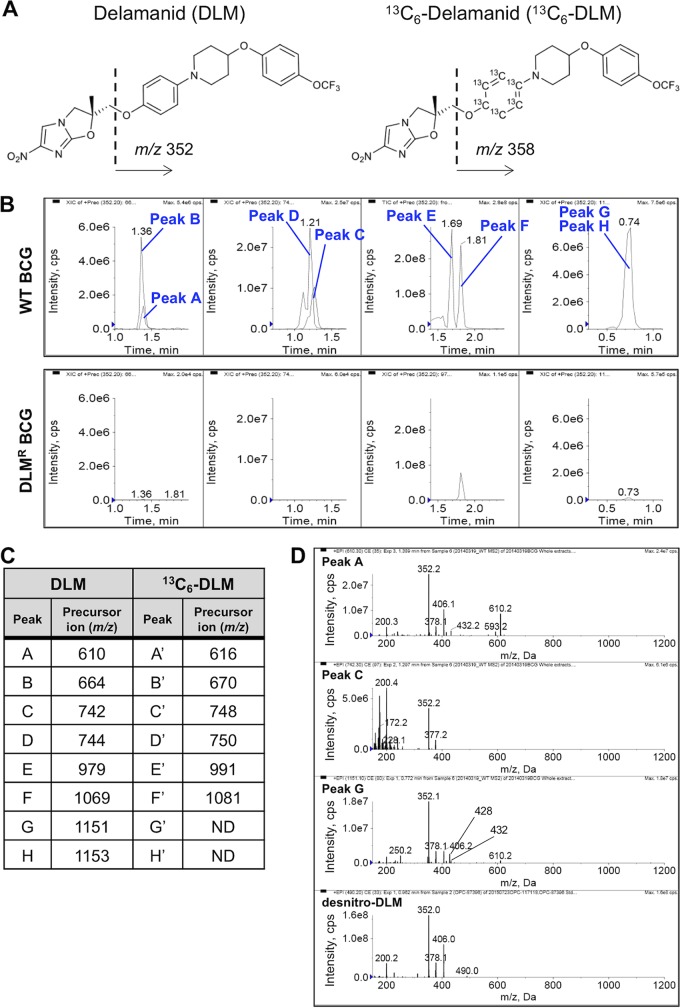
We analyzed cellular extracts isolated from DLM-treated mycobacterial cells by LC-MS to identify DLM metabolites specifically generated in a DLM-susceptible strain. Figure 2A shows the chemical structure of DLM. Based on our previous observations (2), we predicted that activated DLM binds to cellular targets through a reduced nitroimidazole ring. Resultant DLM adducts should always contain the 4-(4-[4-trifluoromethoxyphenoxy]piperidin-1-yl)phenoxy group (m/z 352). We thus analyzed the extracts with a precursor ion scan for m/z 352 and compared the spectrum data between WT Mycobacterium tuberculosis var. Bacille de Calmette et Guérin (BCG; formerly called Mycobacterium bovis BCG) (14) and DLM-resistant (DLMr) BCG. To confirm whether these peaks were actually derived from DLM metabolites, a precursor ion scan for m/z 358 was also performed after treatment of WT BCG with stable isotope-labeled DLM (13C6-DLM; Fig. 2A), whose fragment ion would be 6 Da larger than that of DLM.
FIG 2.
Identification of DLM-binding molecules, comparing the WT and DLMr BCG strains using a precursor ion scan. (A) Chemical structures of DLM and 13C6-DLM. When DLM or 13C6-DLM was analyzed with a product ion scan, a fragment ion of m/z 352 or 358 was identified as the characteristic major fragment ion. (B) LC peak profiles obtained using LC-MS analysis of DLM-binding molecules using a precursor ion scan. Cellular components extracted from WT and DLMr BCG strains were analyzed with LC-MS analysis using a precursor ion scan for m/z 352. Each panel indicates the extracted ion chromatograms of components identified by the precursor ion scan for m/z 352 in the ranges of m/z 500 to 700 (left), m/z 700 to 800 (second from left), m/z 900 to 1,100 (second from right), and m/z 1,100 to 1,200 (right). Upper panels, components extracted from WT BCG; lower panels, components extracted from DLMr BCG. (C) Summary of precursor ions specifically detected in the extracts of DLM- or 13C6-DLM-treated WT BCG using a precursor ion scan for m/z 352 and 358, respectively. Original precursor ion spectra of the extracts are shown in Fig. S1. ND, not detected because the intensity was weak. (D) Product ion spectra of peaks A, C, G, and desnitro-DLM. Common fragment ions with m/z 378 (or 377), 352, and 200 are detected in each spectrum.
Specific peaks detected in the extract from DLM-treated WT BCG are shown in Fig. 2B. Eight components (peaks A to H; refer to Fig. 2C and Fig. S1A, C, E, and G in the supplemental material) were detected in precursor ion chromatograms for m/z 352 of the WT BCG but not in those of DLMr BCG (Fig. 2B; DLMr). As for peaks A to D, similar peaks (A′ to D′) whose precursor ions were 6 more than those of peaks A to D were detected in precursor ion chromatograms for m/z 358 of 13C6-DLM-treated WT BCG (Fig. 2C; Fig. S1B and D). Regarding peaks A, C, and G, the common fragment ions of m/z 378 (or 377), 352, and 200 were detected in their product ion spectra and in that of desnitro-DLM, (Fig. 2D), suggesting that a desnitro-DLM moiety is a common partial structure of peaks A, C, and G.
Peaks E and F (m/z 979 and 1,069) were assigned to [2M+H]+ (M = 489, desnitro-DLM and M = 534, DLM, respectively), a finding supported by the 12-Da greater m/z values of peaks E′ and F′ in the sample treated with 13C6-DLM compared to peaks E and F (Fig. S1E and F). Additional evidence was that a peak showing a similar retention time as peak F was also detected in the high-pressure liquid chromatography (HPLC) chromatogram of a cellular extract isolated from DLM-treated DLMr BCG (Fig. 2B).
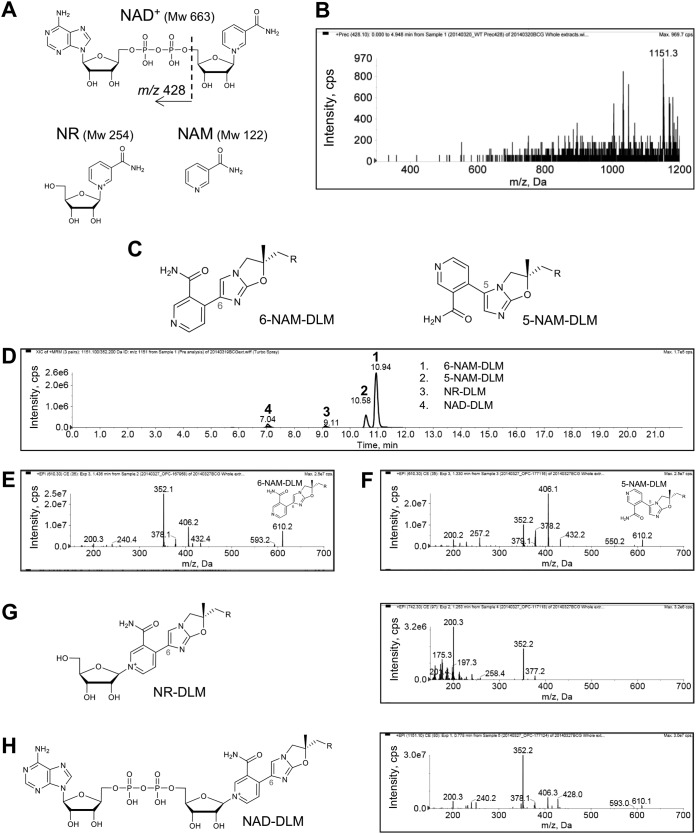
Identification of NAD+ during activation of DLM.
Based on the equation described in Materials and Methods, molecular weights (MW) of partner molecules in peaks A, C, and G were calculated as MW 122, 254, and 663, respectively. In the Reaxys database, the MW 663 and 122 molecules were predicted, respectively, as NAD+ and nicotinamide (NAM), which is a part of NAD+ (Fig. 3A). Although there was no candidate for the MW 254 molecule in the Reaxys database, we hypothesized that ADP was dissociated from NAD+ to yield a nicotinamide-riboside (NR) (MW 254; Fig. 3A). In the product ion scan of a commercially available NAD+, a fragment ion of m/z 428 (Fig. S2A), namely, ADP, was detected (15). Both detection of a precursor ion of m/z 1,151, which corresponded to peak G, in the precursor ion spectrum for m/z 428 (Fig. 3B) and detection of m/z 428 in the product ion spectrum of peak G (Fig. 2D) strongly supported this prediction.
FIG 3.
Comparative analysis of predicted DLM-binding molecule candidates using chemically synthesized 5-NAM-DLM, 6-NAM-DLM, 6-NR-DLM, and 6-NAD-DLM. (A) Chemical structures of DLM-binding molecular candidates: NAD+ (MW, 663) and its component molecules, nicotinamide-riboside (NR; MW, 254) and nicotinamide (NAM; MW, 122) predicted with the Reaxys database and based on mass sizes of the peaks. The structure of ADP (m/z 428) is indicated. (B) Precursor ion scan for m/z 428 of DLM-treated WT BCG extract. The precursor ion of m/z 1,151, which corresponds to peak G, is indicated. (C) Chemical structures of 6-NAM-DLM (chemical formula C31H30F3N5O5; exact mass, 609.2199 Da) and 5-NAM-DLM (chemical formula C31H30F3N5O5; and exact mass, 609.2199 Da). (D) Ion chromatogram of synthesized 6-NAM-DLM (1), 5-NAM-DLM (2), 6-NR-DLM (3), and 6-NAD-DLM (4). (E and F) Product ion spectra of synthesized 6-NAM-DLM (E) and 5-NAM-DLM (F). (G) Chemical structure and product ion spectrum of 6-NR-DLM (chemical formula C36H39F3N5O9+; exact mass, 742.2700 Da). (H) Chemical structure and product ion spectrum of 6-NAD-DLM (chemical formula C46H52F3N10O18P2+; exact mass, 1,151.2888 Da). R = 4-(4-[4-trifluoromethoxyphenoxy]piperidin-1-yl)phenoxy group.
Possible locations for the coupling reaction on the nitroimidazooxazole ring of DLM were predicted to be at the 6- or 5-positions. To determine the exact structure of NAD-DLM adducts, we chemically synthesized the two derivatives 6-NAM-DLM and 5-NAM-DLM (Fig. 3C). These compounds were analyzed, and their product ion spectra were compared with that of peak A (m/z 610, corresponding to the m/z of NAM-DLM). The retention times of 6-NAM-DLM and 5-NAM-DLM in HPLC were 10.94 and 10.58 min, respectively (Fig. 3D, peaks 1 and 2), and the retention time of peak A (10.95 min) was almost identical to that of 6-NAM-DLM (Fig. S2B to D, peak 1). The fragment ions of m/z 593, 432, 406, 378, 352, 240, and 200 detected in the product ion spectrum of 6-NAM-DLM (Fig. 3E) were also detected in that of peak A (Fig. 2D). In contrast, the fragment ion pattern of 5-NAM-DLM (Fig. 3F) was not similar to that of peak A. Taken together, these data suggest that NAM binds covalently at the 6-position in the nitroimidazooxazole ring of DLM.
We next synthesized 6-NR-DLM and 6-NAD-DLM (desnitro-DLM bound to NR or NAD+ at the 6-position, shown in Fig. 3G and H, respectively). The retention times of 6-NR-DLM and 6-NAD-DLM were 9.11 and 7.04 min, respectively (Fig. 3D, peaks 3 and 4) and were almost identical to those of peaks C (9.13 min) and G (7.05 min) (Fig. S2B to D, peaks 2 and 3, respectively). The fragment ions of m/z 377, 352, and 200 were detected in the product ion spectrum of 6-NR-DLM (Fig. 3G) and in the product ion spectrum of peak C (Fig. 2D). The fragment ions of m/z 610, 593, 428, 406, 378, 352, and 200 were detected in the product ion spectrum of NAD-DLM (Fig. 3H) and in the product ion spectrum of peak G (Fig. 2D). Judging from the corresponding retention times and MS patterns, peaks A, C, and G were identical to 6-NAM-DLM, 6-NR-DLM, and 6-NAD-DLM, respectively. From this result, we anticipate that peak D and peak H can be assigned to reduced 6-NR-DLM and 6-NADH-DLM (Fig. S1C and G).
In vitro antimycobacterial activity of chemically synthesized DLM adducts.
We examined the antimycobacterial effects of DLM adducts (6-NAM-DLM, 6-NR-DLM, and 6-NAD-DLM) to M. tuberculosis and BCG strains and found that none of these synthesized adducts showed antimycobacterial activity (MICs, >25 μg/ml; Table S1). This is presumably due to a lack of membrane permeability described as follows. Localized charge or highly polar groups can significantly decrease the passive permeation (16). Both NAD-DLM and NR-DLM have pyridinium moiety which has a positive charge, and NAD-DLM has negatively charged phosphate moiety as well. It has been reported that membrane permeability is typically limited when total polar surface area (tPSA) exceeds 140 Å (17). Lipinski’s “Rule of 5” has also been used to predict drug bioavailabilities and has been generally successful at predicting membrane permeability (18). A compound with fewer than 5 hydrogen bond donors and 10 hydrogen bond acceptors, a molecular weight of less than 500, and an octanol-water partition coefficient (ClogP) of less than 5 would be membrane permeable. Having 10 or fewer rotatable bonds is also known as a good criterion for better permeability across artificial membranes (19). As shown in Table S2, these criteria suggest that at least NAD-DLM and NR-DLM might not permeate the membranes of mycobacteria. On the other hand, NAM-DLM had the same range of these values as DLM, suggesting that NAM-DLM may permeate the membrane but not affect the drug target of DLM directly.
Isolation of thermosensitive (TS) mutants showing cross-resistance to INH and DLM.
We further evaluated the role of DLM adducts in the antimycobacterial action of DLM. Mycolicibacterium smegmatis (formerly called Mycobacterium smegmatis) (14), a saprophytic, fast-growing mycobacterial species widely used as a model for the study of mycobacteria (20, 21), is naturally resistant to DLM (MIC, ≥16 μg/ml; Fig. S3A). The expression of BCG-derived fbiA (JTY_3286), fbiB (JTY_3287), fbiC (JTY_1209), fgd1 (JTY_0416), and ddn (JTY_3612) was enough for M. smegmatis (called strain shi) to be susceptible to DLM (MIC, 0.063 to 0.125 μg/ml; Fig. S3A), suggesting that the target pathway of DLM is conserved among M. tuberculosis, BCG, and M. smegmatis. NAD-DLM was actually detected in chloroform/methanol extract of DLM-treated shi strain (Fig. S3B). Thus, we decided to use this DLM-susceptible M. smegmatis strain shi to evaluate the role of adduct formation in DLM action.
Currently, it is known that adducts of INH and ETH with NADH inhibit a target enzyme InhA competitively to NADH (10–13). Vilchèze et al. and Miesel et al. reported the tolerance to INH and ETH due to elevated cellular NADH levels caused by the TS mutations of the type II NADH dehydrogenase (Ndh) gene (ndh, MSMEG_3621) (12, 22), suggesting that similar mutations in ndh may also alter bacterial susceptibility to DLM. From spontaneous TS INH-resistant (INHr) mutant colonies, we established four DLM-resistant (DLMr) mutants (shi-4-1, shi-8-1, shi-10-1, and shi-11-1; Fig. S3C), which grew in the presence of 1 μg/ml DLM. After exposure of these strains to 25 μg/ml DLM, we further established four mutant derivatives (shi-4-25, shi-8-25, shi-10-25, and shi-11–25; Fig. S3C).
As we expected, the analysis of single nucleotide polymorphisms (SNPs) using next-generation sequencing revealed that all TS INHr DLMr mutants contained nonsynonymous nucleotide substitutions in ndh (shown in Table 1 and Fig. S3D). Of the four original mutants (shi-4-1, shi-8-1, shi-10-1, and shi-11-1), two (shi-10-1 and shi-11-1) contained the same SNP (T662G [Met221Arg]; Table 1 and Fig. S3D). Strain shi-8-1 contained a SNP in the NAD-binding domain of Ndh (G523A [Ala175Thr]; Table 1 and Fig. S3D) (23). Strain shi-4-1 contained another SNP (G251T [Gly84Val]; Table 1 and Fig. S3D). Strains shi-8-25, shi-10-25, and shi-11–25 contained the same ndh SNPs found in their respective parental strains. In contrast, strain shi-4-25 contained an additional SNP (G1297A [Ala433Thr]; Table 1 and Fig. S3D) and the G251T of its parent, shi-4-1. No SNP was found in either the flavin adenine dinucleotide (FAD)-binding domain or the transmembrane domain of ndh (23, 24).
TABLE 1.
SNPs found in TS INHr DLMr mutant strains and drug susceptibility
| Strain | SNPs in ndh |
Susceptibility (MIC, μg/ml) to: |
||||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide change | Amino acid change | DLM | INH | ETH | RIF | SM | EB | |
| shi | 0.063 | 16 | 64 | 16 | 0.625 | 0.031 | ||
| shi-4-1 | G251T | Gly84Val | ≥8 | 256 | ≥1024 | 4 | 0.625 | 0.016 |
| shi-4-25 | G251T, G1297A | Gly84Val, Ala433Thr | ≥8 | 128 | ≥1024 | 16 | 0.625 | 0.031 |
| shi-8-1 | G523A | Ala175Thr | ≥8 | 128 | ≥1024 | 16 | 0.625 | 0.031 |
| shi-8-25 | G523A | Ala175Thr | ≥8 | 128 | ≥1024 | 16 | 0.625 | 0.031 |
| shi-10-1a | T662G | Met221Arg | 0.25 | 128 | ≥1024 | 8 | 0.625 | 0.031 |
| shi-10-25a | T662G | Met221Arg | ≥8 | 128 | ≥1024 | 16 | 0.625 | 0.031 |
Strains shi-11-1 and shi-11-25 indicated same genotypes as those of strains shi-10-1 and shi-10-25, respectively.
All mutants showed high INH MIC levels (128 to 256 μg/ml; Table 1) compared to that of the parental strain shi (16 μg/ml). Similarly, all mutants except for shi-10-1 showed high DLM MIC levels (≥8 μg/ml; Table 1) compared to that of strain shi (0.063 μg/ml). The strain shi-10-1 showed moderate resistance to DLM (MIC, 0.25 μg/ml; Table 1). As reported by Vilchèze et al. (12), cross-resistance of all mutants to ETH was also observed (≥1,024 μg/ml, compared to 64 μg/ml of strain shi; Table 1). In contrast, mutations in ndh did not affect the susceptibility to other drugs, such as RIF, streptomycin (SM), and ethambutol (EB) (Table 1). Taken together, all TS INHr DLMr mutants contained SNPs in ndh and showed cross-resistance to DLM, INH, and ETH, all which form an adduct with NADH or NAD in mycobacteria.
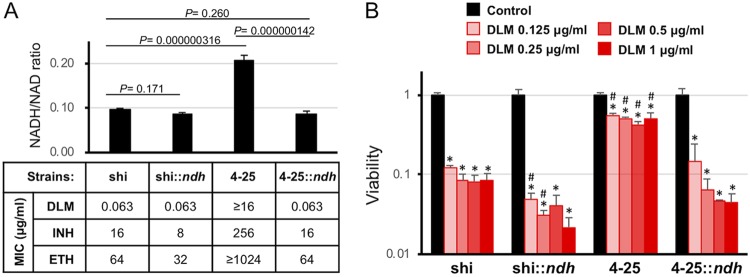
The mutations in ndh affected cellular redox balance and DLM susceptibility.
We then examined the intracellular NADH/NAD redox balance of the mutant. As expected, the shi-4-25 strain showed a significantly higher NADH/NAD ratio (0.207 ± 0.012, P = 0.000000316) than that of the WT shi strain (0.0984 ± 0.0012; Fig. 4A), suggesting the shift of cellular redox balance to a reduced environment by the reduction of Ndh activity. In fact, we found reduced NAD-DLM formation in such reduced intracellular environments of ndh mutants (Fig. S3B). We complemented shi-4-25 with WT ndh (shi-4-25::ndh; Fig. S3E). The mutation in ndh and complementation with WT ndh did not significantly alter the expression levels of other proteins, such as InhA (Fig. S3E). Expression of WT Ndh restored the NADH/NAD ratio (0.0868 ± 0.0064) to levels comparable to that of the WT strain (Fig. 4A; P = 0.260). Concomitantly with the NADH/NAD ratio, bacterial susceptibility to DLM, INH, and ETH was also restored by complementation of WT Ndh (Fig. 4A). These findings suggest the role of NAD-DLM adduct in the antimycobacterial action of DLM.
FIG 4.
Complementation of the TS INHr DLMr mutant with WT ndh restored to its DLM susceptibility. (A) Intracellular NADH/NAD balance and drug susceptibility of the above-named strains under aerobic conditions. Regarding NADH/NAD balance, data are presented as the mean ± standard deviation (SD) (n = 3). The result shown is representative of at least three independent experiments. P values are indicated in a panel. (B) Drug susceptibility of the above-named strains under anaerobic conditions. For each strain, CFU after drug exposure were normalized by CFU of the control culture (no drug) and shown as viability. Data are presented as the mean ± SD (n = 3). Data were analyzed using analysis of variance (ANOVA). *, P < 0.05 compared to control; #, P < 0.05 compared to shi at the same DLM concentration.
DLM can kill not only replicating mycobacteria, but also hypoxic nonreplicating mycobacteria (25). Therefore, we further tested DLM susceptibility of the established ndh mutants under hypoxic nonreplicating conditions. Even under hypoxic conditions, the viability of the WT strain shi was significantly reduced by exposure to DLM at ≥0.125 μg/ml (Fig. 4B; P < 0.05). In contrast, reduction of viability of the ndh mutant (shi-4-25) was only slight compared to shi (Fig. 4B; P < 0.05). Complementation of shi-4-25 with WT ndh (shi-4-25::ndh) restored susceptibility to DLM (P < 0.05). In addition, shi::ndh (which expresses additional WT ndh inserted in a plasmid) tended to be more susceptible to DLM than the original shi. These findings suggest the same antimycobacterial action of DLM even under hypoxic nonreplicating conditions.
DLM is not metabolized by NAD-related mechanisms in host cells.
Because NAD is a coenzyme that is generally found in all living cells, we quantitatively measured NAD-DLM, NR-DLM, and NAM-DLM in WT BCG and human hepatocytes using LC-MS. WT BCG or human hepatocytes (8.3 × 106 cells) were treated with 10 μg/ml DLM for 1 or 24 h, followed by quantitative measurement of NAD-DLM, NR-DLM, and NAM-DLM in cellular extracts using LC-MS as described for the previous experiment. As shown in Table 2, the concentrations of NAM-DLM, NR-DLM, and NAD-DLM in the DLM-treated BCG extract were 1.51, 16.5, and 52.0 ng/ml, respectively, after 1 h of incubation and increased further after 24 h of incubation (65.1, 524, and 182 ng/ml, respectively). On the other hand, in human hepatocytes, concentrations of these complexes were below the lower limit of quantification after either 1 h or 24 h of incubation.
TABLE 2.
Cellular concentrations of NAM-DLM, NR-DLM, and NAD-DLM in BCG and human hepatocytes after treatment with DLM
| Treatment |
Cellular concn of DLM metabolites (ng/ml)a
|
||||||
|---|---|---|---|---|---|---|---|
| NAM-DLM |
NR-DLM |
NAD-DLM |
|||||
| Time (h) | IDc | BCG Tokyo | Human hepatocytes | BCG Tokyo | Human hepatocytes | BCG Tokyo | Human hepatocytes |
| 1 | 1 | 1.52 | <0.104b | 16.0 | <3.35b | 47.8 | <2.65b |
| 2 | 1.49 | <0.104b | 16.9 | <3.35b | 56.1 | <2.65b | |
| Mean | 1.51 | <0.104b | 16.5 | <3.35b | 52.0 | <2.65b | |
| 24 | 1 | 64.2 | <0.104b | 520 | <3.35b | 170 | <2.65b |
| 2 | 66.0 | <0.104b | 528 | <3.35b | 194 | <2.65b | |
| Mean | 65.1 | <0.104b | 524 | <3.35b | 182 | <2.65b | |
The concentrations were measured in duplicate.
Below lower limit of quantification.
ID, measurement number. Mean, mean of 1 and 2.
DISCUSSION
Our previous observation (2) suggests that, after reduction of DLM by Ddn in mycobacterial cells, reduced DLM chemically reacts with a molecule which is involved in a target cellular reaction, eventually resulting in diminished synthesis of keto- and methoxy-mycolic acids (K-MAs and M-MAs) and bacterial death. Based on LC-MS analysis of the extracts of DLM-treated BCG and molecular prediction in this study, we predicted NAD+ to be a partner molecule that covalently binds to reduced DLM in mycobacteria. Based on data obtained from comprehensive LC-MS analysis using synthesized DLM adduct with NAD derivatives, the proposed mechanism for DLM metabolization and subsequent adduct formation with NAD+ is shown in Fig. 5. In M. tuberculosis and BCG, the electron-deficient 6-position carbon on the nitroimidazooxazole ring of DLM is attacked by a hydride from F420H2 of Ddn (Rv3547) to form a carbanion next to the nitro group (26). If the carbanion is quenched by water, the intermediate is converted to desnitro-DLM (Fig. 1). The 4-position of the pyridinium of NAD receives two electrons from the anion, followed by elimination of nitrous acid from the nitroimidazooxazole ring of DLM. Therefore, the possible initial form is NADH-DLM. Reduced NR-DLM would be obtained by decomposition of NADH-DLM or direct reaction of the reduced carbanion intermediate with reduced NR. Eventually, NADH-DLM would be further oxidized to NAD-DLM. Oxidized NR-DLM and NAM-DLM would be obtained by decomposition of NAD-DLM or direct reaction of the reduced carbanion intermediate with oxidized NR or NAM.
FIG 5.
Proposed DLM metabolism in mycobacteria based on the findings of this study. The nitroimidazooxazole ring of DLM is reduced by Ddn (Rv3547). R, 4-(4-[4-trifluoromethoxyphenoxy]piperidin-1-yl)phenoxy group. Asterisks (*) indicate the structures identified with synthesized compounds. Corresponding peaks detected in the precursor ion scan for m/z 352 are also indicated. [O] indicates oxidation.
The analyses of TS INHr DLMr ndh mutants and the WT ndh-complemented strain (Table 1 and Fig. 4) suggested an important role of DLM adducts in the antimycobacterial activity. Antimycobacterial action of synthesized NAD-DLM, NR-DLM, and NAM-DLM has not been seen so far. As described above (Table S2), our prediction using tPSA and Lipinski’s Rule of 5 strongly supported low membrane permeability of NAD-DLM and NR-DLM. However, NAM-DLM was predicted to be as membrane permeable as DLM, suggesting that antimycobacterial forms of DLM might be NAD-DLM (and/or NR-DLM), rather than NAM-DLM. Unlike a partner molecule of INH or ETH (NADH), DLM forms an adduct with NAD+. Therefore, a possible explanation of DLM tolerance is that depletion of NAD due to TS mutations in ndh prevents adduct formation of reduced DLM with NAD. Our LC-MS data also suggest the formation of NADH-DLM (Fig. S1, peak H), suggesting a tolerance mechanism similar to those of INH and ETH (12, 22). However, NADH-DLM is an unstable intermediate of NAD-DLM, since reduced NAM-DLM (whose protonated molecule should be m/z 612) was not detected in this study.
PA-824 with a bicyclic nitroimidazooxazine core structure is also activated by Ddn in mycobacteria, followed by release of nitro group (3). Previously, a significant role of reactive nitrogen oxide (NO) generated from the released nitro group in the killing of hypoxic, nonreplicating mycobacteria was reported (26). However, we found that the ndh mutant M. smegmatis was resistant to DLM and that complementation with WT ndh also restored susceptibility to DLM under anaerobic conditions (Fig. 4B). Furthermore, an NO scavenger has no significant effect on antimycobacterial activity of DLM against hypoxic nonreplicating BCG or M. smegmatis shi (Fig. S4), suggesting the role of NAD-DLM adduct formation, rather than NO generation, in DLM activity even under anaerobic conditions.
In conclusion, we propose that the active DLM that is generated on reduction by Ddn forms an adduct with NAD+. Although we could not demonstrate inhibitory activity of DLM adducts against mycobacterial growth, our data obtained using ndh mutants suggest the importance of NAD-DLM formation in the expression of antimycobacterial activity under both normoxic and hypoxic conditions. This DLM metabolism is seen only in mycobacterial cells, not in host cells. In humans, two main nitroreductases were identified, NQO1 (DT-diaphorase) and NQO2 (NAD[P]H quinone oxidoreductase 2) (27); however, DLM is not metabolized by these two enzymes (28). Indeed, the concentrations of NAD-DLM, NR-DLM, and NAM-DLM were below the detection limits of quantification in DLM-treated human hepatocytes (Table 2). This fact supports the selective toxicity of DLM to mycobacteria. The identification of substantial activity of DLM adducts in mycobacteria (a principal target of the adducts) is awaited to reveal the entire mode of action of DLM.
MATERIALS AND METHODS
Bacterial strains and culture media.
DLM-susceptible WT BCG Tokyo strain was purchased from the Institute of Medical Science, the University of Tokyo. A DLMr mutant BCG Tokyo strain that contains a mutation in ddn (JTY_3612) was developed previously (2). M. smegmatis mc2_155 was kindly provided by Thomas Dick (Novartis Institute for Tropical Diseases).
All BCG strains were cultured in Middlebrook 7H9 broth (Becton, Dickinson, San Jose, CA) supplemented with 10% ADC enrichment (Becton, Dickinson) and 0.05% (vol/vol) Tween 80 (7H9-ADC broth) or on Middlebrook 7H11 agar (Becton, Dickinson) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment (Becton, Dickinson) (7H11-OADC agar). If necessary, ADC-free 7H9 broth was also used. All M. smegmatis strains were cultured in Mueller-Hinton II broth (Becton, Dickinson) containing 0.05% (vol/vol) Tween 80 (MHB II-Tween 80) or on Mueller-Hinton agar (MHA) (Becton, Dickinson) supplemented with appropriate antibiotics necessary to maintain individual genetic characteristics. For preparation of hypoxic nonreplicating cells, all BCG and M. smegmatis strains were cultured in modified Dubos-bovine serum albumin (BSA) broth (0.5 g/liter pancreatic digest of casein, 2 g/liter asparagine, 1 g/liter monopotassium phosphate, 2.5 g/liter disodium phosphate, 50 mg/liter ferric ammonium citrate, 10 mg/liter magnesium sulfate, 0.5 mg/liter calcium chloride, 0.1 mg/liter zinc sulfate, 0.1 g/liter copper sulfate, and 0.7 g/liter Tween 80, 0.5% bovine serum albumin, 0.75% d-glucose, and 0.085% sodium chloride) supplemented with methylene blue (final amount, 1.5 μg/ml) and appropriate antibiotics.
Compounds synthesized in this study.
The compounds synthesized in this study are listed in Table S3. Test compounds and DLM derivatives were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 and 1 mg/ml, respectively. DLM derivatives were appropriately diluted with methanol/sample buffer (1:1). The ClogP and tPSA of DLM, NAD-DLM, NR-DLM, and NAM-DLM were calculated using ChemDraw version 16.0 (PerkinElmer, Waltham, MA).
Drug preparation.
INH, ETH, RIF, and hygromycin B (HYG) were purchased from Sigma-Aldrich (Tokyo, Japan). SM and kanamycin (KM) were purchased from Wako Pure Chemical Industries (Osaka, Japan). EB was purchased from LKT Laboratories (St. Paul, MN, USA). DLM, ETH, and RIF were dissolved in DMSO. INH, HYG, KM, and SM were dissolved in distilled water.
Other methods.
LC-MS analysis of mycobacterial extracts and synthesized compounds, construction of DLM-susceptible M. smegmatis, isolation of TS INHr DLMr mutants, drug susceptibility tests, Western blot analysis, measurement of cellular NADH/NAD concentrations, analysis of DLM metabolism in mycobacteria and human hepatocytes, and statistical analysis are described in detail in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Teruo Kirikae (National Center for Global Health and Medicine, Tokyo, Japan) for kindly providing antibody specific to InhA. We thank Yuichiro Kawasaki and Fumina Ono (Niigata University) for technical support with drug susceptibility testing. We thank Noriyuki Koyama and Takashi Hayashi (Otsuka Pharmaceutical Co. Ltd., Tokushima, Japan), Takahashi Hideyuki, and Ueno Yuichi (Niigata University) for discussing the procedure and results of LC-MS-MS analysis. We also thank Chemical Biology Unit, Niigata University. We are also grateful to Yuko Kobayashi, Haruka Kobayashi, Sara Matsumoto, and Satoko Matsumoto for their assistance and heartfelt encouragement.
This work was in part supported by grants from Japan Agency for Medical Research and Development (AMED; JP19fk0108089 to S. Matsumoto and JP19fk0108090 to A. Nishiyama).
M.H., R.K., X.C., M.F., Y.I., Y.K., M.K., and M.M. are employees of Otsuka Pharmaceutical Co., Ltd., which is the originator and owner of delamanid. All other authors declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2018. Global Tuberculosis Report 2018. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. 2006. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med 3:e466. doi: 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurumurthy M, Mukherjee T, Dowd CS, Singh R, Niyomrattanakit P, Tay JA, Nayyar A, Lee YS, Cherian J, Boshoff HI, Dick T, Barry CE, Manjunatha UH. 2012. Substrate specificity of the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis responsible for the bioreductive activation of bicyclic nitroimidazoles. FEBS J 279:113–125. doi: 10.1111/j.1742-4658.2011.08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiwara M, Kawasaki M, Hariguchi N, Liu Y, Matsumoto M. 2018. Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis (Edinb) 108:186–194. doi: 10.1016/j.tube.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Choi K-P, Kendrick N, Daniels L. 2002. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F(420) and FO biosynthesis. J Bacteriol 184:2420–2428. doi: 10.1128/jb.184.9.2420-2428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi KP, Bair TB, Bae YM, Daniels L. 2001. Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG. J Bacteriol 183:7058–7066. doi: 10.1128/JB.183.24.7058-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashiri G, Rehan AM, Sreebhavan S, Baker HM, Baker EN, Squire CJ. 2016. Elongation of the poly-γ-glutamate tail of F420 requires both domains of the F420:γ-glutamyl ligase (FbiB) of Mycobacterium tuberculosis. J Biol Chem 291:6882–6894. doi: 10.1074/jbc.M115.689026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloemberg GV, Keller PM, Stucki D, Trauner A, Borrell S, Latshang T, Coscolla M, Rothe T, Hömke R, Ritter C, Feldmann J, Schulthess B, Gagneux S, Böttger EC. 2015. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 373:1986–1988. doi: 10.1056/NEJMc1505196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann H, Kohl TA, Hofmann-Thiel S, Merker M, Beckert P, Jaton K, Nedialkova L, Sahalchyk E, Rothe T, Keller PM, Niemann S. 2016. Delamanid and bedaquiline resistance in Mycobacterium tuberculosis ancestral Beijing genotype causing extensively drug-resistant tuberculosis in a Tibetan refugee. Am J Respir Crit Care Med 193:337–340. doi: 10.1164/rccm.201502-0372LE. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 11.Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Sacchettini JC. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 12.Vilchèze C, Weisbrod TR, Chen B, Kremer L, Hazbón MH, Wang F, Alland D, Sacchettini JC, Jacobs WR. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother 49:708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Langley R, Gulten G, Dover LG, Besra GS, Jacobs WR, Sacchettini JC. 2007. Mechanism of thioamide drug action against tuberculosis and leprosy. J Exp Med 204:73–78. doi: 10.1084/jem.20062100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta RS, Lo B, Son J. 2018. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol 9:67. doi: 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Sherwood R, Zhang S, Zhu X, Liu Q, Graeff R, Kriksunov IA, Lee HC, Hao Q, Lin H. 2013. Identification of ADP-ribosylation sites of CD38 mutants by precursor ion scanning mass spectrometry. Anal Biochem 433:218–226. doi: 10.1016/j.ab.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang NJ, Hinner MJ. 2015. Getting across the cell membrane: an overview for small molecules, peptides, and proteins. Methods Mol Biol 1266:29–53. doi: 10.1007/978-1-4939-2272-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsson P, Kihlberg J. 2017. How big is too big for cell permeability? J Med Chem 60:1622–1664. [DOI] [PubMed] [Google Scholar]
- 18.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev 23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 19.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. 2002. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 20.Reyrat J-M, Kahn D. 2001. Mycobacterium smegmatis: an absurd model for tuberculosis? Trends Microbiol 9:472–474. doi: 10.1016/S0966-842X(01)02168-0. [DOI] [PubMed] [Google Scholar]
- 21.Rodionova IA, Schuster BM, Guinn KM, Sorci L, Scott DA, Li X, Kheterpal I, Shoen C, Cynamon M, Locher C, Rubin EJ, Osterman AL. 2014. Metabolic and bactericidal effects of targeted suppression of NadD and NadE enzymes in mycobacteria. mBio 5:e00747-13. doi: 10.1128/mBio.00747-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miesel L, Weisbrod TR, Marcinkeviciene JA, Bittman R, Jacobs WR Jr. 1998. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol 180:2459–2467. doi: 10.1128/JB.180.9.2459-2467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKie JH, Douglas KT. 1991. Evidence for gene duplication forming similar binding folds for NAD(P)H and FAD in pyridine nucleotide-dependent flavoenzymes. FEBS Lett 279:5–8. doi: 10.1016/0014-5793(91)80236-v. [DOI] [PubMed] [Google Scholar]
- 24.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
- 25.Chen X, Hashizume H, Tomishige T, Nakamura I, Matsuba M, Fujiwara M, Kitamoto R, Hanaki E, Ohba Y, Matsumoto M. 2017. Delamanid kills dormant mycobacteria in vitro and in a guinea pig model of tuberculosis. Antimicrob Agents Chemother 61:e02402-16. doi: 10.1128/AAC.02402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh R, Manjunatha U, Boshoff HIM, Ha YH, Niyomrattanakit P, Ledwidge R, Dowd CS, Lee IY, Kim P, Zhang L, Kang S, Keller TH, Jiricek J, Barry CE. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science 322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasiliou V, Ross D, Nebert DW. 2006. Update of the NAD(P)H:quinone oxidoreductase (NQO) gene family. Hum Genomics 2:329–335. doi: 10.1186/1479-7364-2-5-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanaki E, Hayashi M, Matsumoto M. 2017. Delamanid is not metabolized by Salmonella or human nitroreductases: a possible mechanism for the lack of mutagenicity. Regul Toxicol Pharmacol 84:1–8. doi: 10.1016/j.yrtph.2016.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.