LETTER
In a recent issue of Antimicrobial Agents and Chemotherapy, Reingewertz et al. (1) report on sensitization of slow-growing, nontuberculous mycobacteria (NTM) to the anti-tubercular drug isoniazid (INH) upon expression of Mycobacterium bovis KatG.
KatG functions as a catalase-peroxidase (2, 3) and activates INH, which then inhibits InhA, an enzyme involved in mycolic acid synthesis (4). However, a surge in INH-resistant Mycobacterium tuberculosis clinical isolates is jeopardizing the role of INH as a first-line drug (5). In general, resistance to INH can be acquired by mutations in katG or, less frequently, in the promoter region of inhA (6, 7). In the first case, KatG no longer activates INH, while in the second case, a higher tolerance to the drug is conferred by increased InhA expression (8).
The aim of the aforementioned study was to elucidate the differences between KatG-dependent INH activation in mycobacteria and its effect on INH susceptibility, focusing on the opportunistic pathogens Mycobacterium avium subsp. paratuberculosis and Mycobacterium marinum (both NTM and naturally refractory to INH).
NTM are mycobacteria not belonging to the M. tuberculosis complex and encompass both slowly and rapidly growing mycobacterial species (SGM and RGM) (9). As shown by our colleagues and other groups (1, 10–12), NTM usually show an innate decreased susceptibility toward INH. Within NTM, RGM have significantly higher INH MICs than SGM (11–13). The reason for this increased resistance is likely the result of several factors, including a failure to activate the prodrug, target-level mutations, differences in the C-terminal domain of KatG (3), the reduction of intracellular concentration (by means of efflux pumps or decreased permeability) (14), and/or the possible nonessentiality of the mycolic acid synthesis pathway in NTM (12). However, essentiality has been proven by identifying pyridomycin as a specific inhibitor of InhA preventing growth of both M. tuberculosis (MIC = 0.39 mg/liter) and NTM (M. marinum MIC = 3.13 mg/liter) (15).
In the context of NTM, no other mycobacteria have proven to be as resilient as the emerging opportunistic pathogens from the Mycobacterium abscessus complex (16). As members of the RGM (9), their intrinsic antibiotic resistance through drug- and target-modifying enzymes (17) has rendered M. abscessus complex infections extremely challenging to treat. Lengthy regimens on multiple drugs with severe side effects are the norm (11, 13). KatG phylogeny pinpointed M. abscessus complex as being closer to M. tuberculosis than rpoB-based phylogeny did (1). Alignment of KatGMabs with KatGMtb shows that the most common INH resistance-conferring clinical M. tuberculosis mutations (18) are absent in M. abscessus ATCC 19977 and sequence identity is high (approximately 72%).
Our research group has taken advantage of the resistance of M. abscessus complex to INH in a manner similar to that used by Reingewertz and coworkers. In a proof-of-concept study, we heterologously expressed KatGMtb in M. abscessus complex from a pMV361 (19) attB-integrative vector containing an apramycin (APR) resistance cassette for selection (aac). This allowed us to develop new tools for the genetic manipulation of M. abscessus complex and to use INH as an effective counterselection marker for allelic replacements (20–24), even if the MIC was well above achievable therapeutical concentrations (MIC = 32 mg/liter) (Fig. 1). Our observations are in strong agreement with the work from Reingewertz et al., showing a drop in MIC of approximately 30-fold, and are proof that INH susceptibility can be successfully exploited. However, the comparatively high MIC of recombinant M. abscessus complex pMV361-aac-katG indicates that besides poor KatG-dependent INH activation, other factors contribute to the high level INH resistance of M. abscessus.
FIG 1.
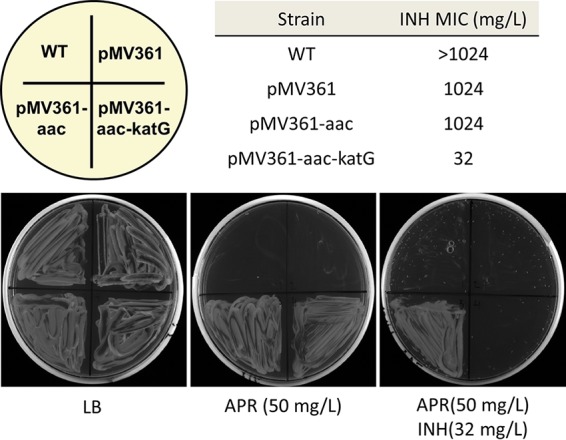
INH susceptibility of M. abscessus complex strains upon KatGMtb expression. MICs (day 7) for the strains used to generate M. abscessus complex pMV316-aac-katG (unpublished data) are shown. MICs were measured by broth microdilution assay (25). Luria-Bertani (LB) agar with APR and with APR-INH shows selective growth of M. abscessus complex.
ACKNOWLEDGMENTS
This work was supported by the University of Zurich and the Institute of Medical Microbiology. We acknowledge financial support from Lungen Liga Schweiz/Georg and Bertha Schwyzer-Winiker Stiftung (SLA-2018-02) and Foundation for Research in Science and the Humanities at the University of Zurich (STWF-18-011).
Ed. Note: The authors of the published article did not feel that a response was necessary.
REFERENCES
- 1.Reingewertz TH, Meyer T, McIntosh F, Sullivan J, Meir M, Chang Y-F, Behr MA, Barkan D. 2019. Differential sensitivity of mycobacteria to isoniazid is related to differences in KatG-mediated enzymatic activation of the drug. Antimicrob Agents Chemother 64:e01899-19. doi: 10.1128/AAC.01899-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Heym B, Allen B, Young D, Cole S. 1992. The catalase–peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 3.Heym B, Zhang Y, Poulet S, Young D, Cole ST. 1993. Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol 175:4255–4259. doi: 10.1128/jb.175.13.4255-4259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K, Wilson T, Collins D, de Lisle G, Jacobs W. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 5.Gumbo T, Louie A, Liu W, Ambrose PG, Bhavnani SM, Brown D, Drusano GL. 2007. Isoniazid’s bactericidal activity ceases because of the emergence of resistance, not depletion of Mycobacterium tuberculosis in the log phase of growth. J Infect Dis 195:194–201. doi: 10.1086/510247. [DOI] [PubMed] [Google Scholar]
- 6.Larsen MH, Vilchèze C, Kremer L, Besra GS, Parsons L, Salfinger M, Heifets L, Hazbon MH, Alland D, Sacchettini JC, Jacobs WR Jr. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol Microbiol 46:453–466. doi: 10.1046/j.1365-2958.2002.03162.x. [DOI] [PubMed] [Google Scholar]
- 7.Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, Hill-Cawthorne G, Mallard K, Nair M, Miranda A, Alves A, Perdigão J, Viveiros M, Portugal I, Hasan Z, Hasan R, Glynn JR, Martin N, Pain A, Clark TG. 2015. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med 7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Yew WW, Barer MR. 2012. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother 56:2223–2230. doi: 10.1128/AAC.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tortoli E, Fedrizzi T, Meehan CJ, Trovato A, Grottola A, Giacobazzi E, Serpini GF, Tagliazucchi S, Fabio A, Bettua C, Bertorelli R, Frascaro F, De Sanctis V, Pecorari M, Jousson O, Segata N, Cirillo DM. 2017. The new phylogeny of the genus Mycobacterium: the old and the news. Infect Genet Evol 56:19–25. doi: 10.1016/j.meegid.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Brown-Elliott BA, Nash KA, Wallace RJ. 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev 25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffith DE, Infectious Disease Society of America, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 12.van Ingen J, Boeree MJ, van Soolingen D, Mouton JW. 2012. Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria. Drug Resist Updat 15:149–161. doi: 10.1016/j.drup.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Haworth CS, Banks J, Capstick T, Fisher AJ, Gorsuch T, Laurenson IF, Leitch A, Loebinger MR, Milburn HJ, Nightingale M, Ormerod P, Shingadia D, Smith D, Whitehead N, Wilson R, Floto RA. 2017. British Thoracic Society guideline for the management of non-tuberculous mycobacterial pulmonary disease (NTM-PD). BMJ Open Respir Res 4:e000242. doi: 10.1136/bmjresp-2017-000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams KN, Szumowski JD, Ramakrishnan L. 2014. Verapamil, and its metabolite norverapamil, inhibit macrophage-induced, bacterial efflux pump-mediated tolerance to multiple anti-tubercular drugs. J Infect Dis 210:456–466. doi: 10.1093/infdis/jiu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartkoorn RC, Sala C, Neres J, Pojer F, Magnet S, Mukherjee R, Uplekar S, Boy-Röttger S, Altmann K-H, Cole ST. 2012. Towards a new tuberculosis drug: pyridomycin—nature’s isoniazid. EMBO Mol Med 4:1032–1042. doi: 10.1002/emmm.201201689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 17.Luthra S, Rominski A, Sander P. 2018. The role of antibiotic-target-modifying and antibiotic-modifying enzymes in Mycobacterium abscessus drug resistance. Front Microbiol 9:2179. doi: 10.3389/fmicb.2018.02179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Hersleth H-P, Zhu J, Andersson KK, Magliozzo RS. 2013. Access channel residues Ser315 and Asp137 in Mycobacterium tuberculosis catalase-peroxidase (KatG) control peroxidatic activation of the pro-drug isoniazid. Chem Commun (Camb) 49:11650–11652. doi: 10.1039/c3cc47022a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 20.Rominski A, Roditscheff A, Selchow P, Böttger EC, Sander P. 2017. Intrinsic rifamycin resistance of Mycobacterium abscessus is mediated by ADP-ribosyltransferase MAB_0591. J Antimicrob Chemother 72:376–384. doi: 10.1093/jac/dkw466. [DOI] [PubMed] [Google Scholar]
- 21.Becker K, Haldimann K, Selchow P, Reinau LM, Dal Molin M, Sander P. 2017. Lipoprotein glycosylation by protein-O-mannosyltransferase (MAB_1122c) contributes to low cell envelope permeability and antibiotic resistance of Mycobacterium abscessus. Front Microbiol 8:2123. doi: 10.3389/fmicb.2017.02123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rominski A, Schulthess B, Muller DM, Keller PM, Sander P. 2017. Effect of beta-lactamase production and beta-lactam instability on MIC testing results for Mycobacterium abscessus. J Antimicrob Chemother 72:3070–3078. doi: 10.1093/jac/dkx284. [DOI] [PubMed] [Google Scholar]
- 23.Rominski A, Selchow P, Becker K, Brulle JK, Dal Molin M, Sander P. 2017. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J Antimicrob Chemother 72:2191–2200. doi: 10.1093/jac/dkx125. [DOI] [PubMed] [Google Scholar]
- 24.Dal Molin M, Gut M, Rominski A, Haldimann K, Becker K, Sander P. 2018. Molecular mechanisms of intrinsic streptomycin resistance in Mycobacterium abscessus. Antimicrob Agents Chemother 62:e01427-17. doi: 10.1128/AAC.01427-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reller LB, Weinstein MP, Woods GL. 2000. Susceptibility testing for mycobacteria. Clin Infect Dis 31:1209–1215. doi: 10.1086/317441. [DOI] [PubMed] [Google Scholar]


