In this study, the plasmid content of clinical and commensal strains was analyzed and compared. The replicon profile was similar in both populations, except for L, M, A/C, and N (detected only in clinical strains) and HI1 (only in commensal strains). Although I1 and F were the most frequent replicons, only IncI1, sequence type 12 (ST12) was associated with blaCMY-2 in both populations. In contrast, the widespread resistant IncF plasmids were not linked to a single epidemic plasmid.
KEYWORDS: pMLST, replicon, antimicrobial resistance, Enterobacteriaceae, plasmid epidemiology
ABSTRACT
In this study, the plasmid content of clinical and commensal strains was analyzed and compared. The replicon profile was similar in both populations, except for L, M, A/C, and N (detected only in clinical strains) and HI1 (only in commensal strains). Although I1 and F were the most frequent replicons, only IncI1, sequence type 12 (ST12) was associated with blaCMY-2 in both populations. In contrast, the widespread resistant IncF plasmids were not linked to a single epidemic plasmid.
TEXT
The most prevalent mechanism in antimicrobial resistance gene (ARG) acquisition by bacterial pathogens is horizontal gene transfer by plasmids (1, 2). PCR-based replicon typing (PBRT) based on plasmid incompatibility (Inc) is currently the standard method for plasmid identification (3, 4). Plasmid multilocus sequence typing (pMLST) schemes allow researchers to differentiate between plasmids within incompatibility groups and to define epidemiological and evolutionary relatedness (5–10) (http://pubmlst.org/plasmid/).
Several plasmids carrying ARGs have been characterized, most of them recovered from clinically relevant bacteria (11–14). In contrast, there is limited information on plasmids in the commensal microbiota of healthy humans without a selection bias for antimicrobial-resistant bacteria. In this scenario, the aim of this study was to provide a better understanding of resistant plasmid diffusion in a clinical context by comparing plasmids within Escherichia coli and Klebsiella pneumoniae strains isolated from healthy human feces and patients with bloodstream infection.
One hundred and fifty fecal samples were collected during 2014 and 2015 from healthy humans who did not consume antibiotics and were not hospitalized. A total of 145 E. coli strains and 12 K. pneumoniae strains were isolated. In addition, 202 strains from blood cultures, 99 E. coli strains and 103 K. pneumoniae strains, from three hospitals in Barcelona were analyzed (one per patient). (The study was approved by the Hospital de la Santa Creu i Sant Pau ethics committee [13/051/1439].) All strains underwent antimicrobial susceptibility testing using disk diffusion (see Table S1 in the supplemental material), and the results were interpreted according to CLSI guidelines (15). The characterization of extended-spectrum β-lactamases (ESBLs), AmpCs, and carbapenemases (16–20) detected in both populations is shown in Table 1. The prevalence of ESBL-producing E. coli in healthy carriers (4.7%) was higher than that in a previous study in Barcelona in 2005 (3.3%) (21) but still within the 3 to 6% average of Europe (22).
TABLE 1.
ESBL, AmpC, and carbapenemase genes detected in E. coli and K. pneumoniae from fecal and blood samples
| Gene type (no.) | Gene (no.) detected in: |
|||
|---|---|---|---|---|
|
E. coli (n = 244) |
K. pneumoniae (n = 115) |
|||
| Fecal samples (n = 13/145)a | Blood cultures (n = 19/99)b | Fecal samples (n = 0/12) | Blood cultures (n = 17/103)c | |
| ESBLs (43) | n = 8; 5.5% | n = 17; 17.2% | n = 17; 16.5% | |
| blaCTX-M-15 (4) | blaCTX-M-15 (10) | blaCTX-M-15 (9) | ||
| blaCTX-M-14 (3) | blaCTX-M-14 (2) | blaCTX-M-14 (2) | ||
| blaCTX-M-27 (1) | blaCTX-M-27 (2) | blaSHV-28 (5) | ||
| blaSHV-12 (1) | blaCTX-M-32 (1) | blaSHV-2 (1) | ||
| blaSHV-12 (2) | ||||
| AmpCs (11) | n = 5; 3.5% | n = 5; 5.0% | n = 1; 1.0% | |
| blaCMY-2 (5) | blaCMY-2 (4) | blaDHA-1 (1) | ||
| blaDHA-1 (1) | ||||
| Carbapenemases (2) | n = 2; 1.9% | |||
| blaKPC-3 (2) | ||||
| Total genes (56) | 14 | 22 | 0 | 20 |
One E. coli strain had blaCTX-M-15 and blaCTX-M-14.
Three E. coli strains had blaCTX-M-15 and blaCTX-M-32, blaCTX-M-27 and blaDHA-1, and blaSHV-12 and blaCMY-2.
One K. pneumoniae strain had blaCTX-M-15 and blaCTX-M-14, and two strains had blaCTX-M-15 and blaSHV-28.
Plasmid identification was performed using the PBRT-kit (Diatheva) and simplex PCR for ColE, X3, X4, L, and M replicons (23–25). Twenty-nine replicons were analyzed, and only FIIS, W, T, U, and HI2 were not detected in any strain. A total of 978 replicons were identified in the 359 studied strains; 84.1% (302/359) harbored from one to four replicons, and 10.9% (39/359) harbored from five to seven. In 5% (18/359) of the strains, no replicon was detected. Overall, the results suggest that the replicon content of E. coli strains followed a similar trend in patients and healthy individuals, and the most prevalent in both sample groups were ColE, FII, and FIB (Fig. 1A). Nevertheless, replicons M, A/C, and N were only detected in clinical strains in accordance with the literature (26, 27), while FIIK and HI1 were observed only in fecal strains (Fig. 1A). Hence, it might be hypothesized that the hospital environment, where there is high antimicrobial use and intense interhuman transmission, selects for plasmids more adapted to these settings. The plasmid content in K. pneumoniae isolates seems to follow similar trends (Fig. 1B) to that of E. coli, but this could not be confirmed due to the low number of strains obtained from fecal samples. Notably, both the diversity and frequency of replicons were higher in E. coli than in K. pneumoniae, except for those of R and FIIK (Fig. 1C).
FIG 1.
Replicon prevalence. (a) Comparison between replicons detected in E. coli from clinical and fecal samples. (b) Comparison between replicons detected in K. pneumoniae from clinical and fecal samples. (c) Comparison between replicons detected in E. coli and K. pneumoniae from the total strains. Tables show the number of each replicon detected. ND, not detected. *, Statistical differences (P < 0.05) between each population, fecal and blood samples or E. coli and K. pneumoniae.
IncF and IncI1 plasmids have been reported in Enterobacteriaceae as promoters of beta-lactamase gene dissemination in multiple environments, specifically blaCTXM-15 and blaCMY-2 (13, 28–32). In this study, 56 beta-lactamase genes were detected (Table 1). After S1 pulsed-field gel electrophoresis (PFGE) and Southern hybridization (19, 33), 75% of ESBL, AmpC, and carbapenemase genes identified in E. coli (27/36) and K. pneumoniae (15/20) were located on a plasmid, the most prevalent being IncF and IncI1 (37% for both) in E. coli and IncF (47%) and IncR (20%) in K. pneumoniae. The predominant genes were blaCTX-M-15/14/27 and blaCMY-2 in IncF and InI1 plasmids of E. coli (see Fig. S1A in the supplemental material) and blaCTX-M-15 in IncF plasmids of K. pneumoniae (see Fig. S1B). Figure 2 summarizes the 49 ESBL-, AmpC-, and/or carbapenemase-producing strains detected in the study, the plasmids they harbored, and the location of the beta-lactamase genes.
FIG 2.
Heat-map summary of the sources, phylogenetic groups, β-lactamase resistance genes, and the corresponding Inc plasmid types and their sizes for 49 β-lactam-resistant E. coli (n = 32) and K. pneumoniae (n = 17) strains from fecal and blood samples. Black and white squares denote the presence and absence of a particular feature, respectively. ND, not determined; CP, carbapenemases. a, Plasmids where the replicon hybridization occurred in the same plasmid size (hybrid plasmids). b, bla Genes detected in the same plasmid.
As IncF and IncI1 were two of the most frequently detected plasmids in both fecal and clinical samples (27, 34, 35), they were further characterized using the pMLST method (5, 10). In E. coli strains, 29 different IncI1 sequence types (STs) were detected, 59% of which were assigned as new STs. This result reflects the great diversity within this plasmid family, with only ST12 and ST36 being present in both clinical and fecal populations (see Fig. S2 in the supplemental material). Moreover, some of the most frequently reported STs worldwide (ST2, ST12, ST26, and ST36) (36) were only found in E. coli from healthy humans. The detection of many newly assigned STs in the clinical isolates and a scarce number of the most reported STs suggest that the latter may have been overreported due to their involvement in ARG dissemination, resulting in an epidemiological bias.
In addition, IncI1 plasmids have been associated with the carriage of blaCMY-2, particularly IncI1 ST2, ST12, and ST23 (5, 36–38). In the current study, all identified ST12 plasmids harbored blaCMY-2 and were detected in E. coli from both populations (Fig. 2). These results support the suggestion that some IncI1 plasmids have been able to evolve and persist in clinical settings thanks to particular features that provide resistance, persistence, and adaptive success, which would explain why they are more frequently reported and described as epidemic plasmids (36–38).
After defining the final number of IncF plasmids (n = 279; 211 in E. coli strains and 68 in K. pneumoniae strains) by Southern hybridization of F replicons within each strain, subtypification using replicon sequence typing (RST) was performed. In E. coli strains, 86 different FAB (FII, FIA, FIB) formulas (indicating the allele type and number identified for each replicon according to RST scheme [10]) from 205 typeable plasmids (45 from fecal samples, 24 from blood, and 17 from both) were defined, where F29:A−:B10, F2:A−:B1, F2:A−:B−, and F24:A−:B1 were the most frequent (see Fig. S3 in the supplemental material). Some of these formulas have been previously identified in different environments, such as avian-pathogenic E. coli strains and uropathogenic and extraintestinal pathogenic E. coli strains (27, 39), indicating a broad distribution. In K. pneumoniae, 16 different FAB formulas from 68 typeable plasmids (12 from blood and 4 from both) were detected, with K1:A−:B− being the most frequent (see Fig. S3).
IncF plasmids, including F2:A−:B−, F2:A1:B−, F31:A4:B1, and F1:A2:B20, have been associated with the worldwide emergence of CTX-M-15 (10, 12, 40–42). Although these four plasmids were detected in our study, only F2:A−:B− harbored the blaCTX-M-15 gene in a clinical strain and blaCTX-M-27 in a fecal strain. All of the other CTX-M-encoding genes were located in different IncF plasmids (Fig. 2). Thus, according to our results and in agreement with those of previous reports (32, 34), there is no evidence for the persistence of a unique IncF. These highly versatile plasmids are able to adapt to intracellular environments by the rapid evolution of replicon regulatory sequences (10), and they were widely distributed in the Enterobacteriaceae before antimicrobial use, facilitating the persistence and spread of beta-lactamases (10, 32). Their coexistence with other resistance determinants also contributes to the dissemination of IncF-CTX-M plasmids (43).
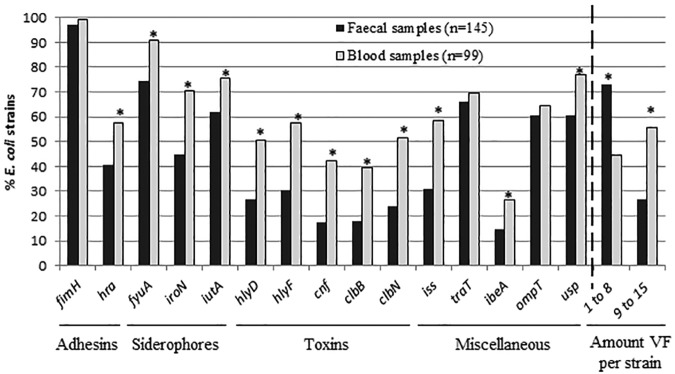
Additionally, E. coli strains were assigned to phylogenetic groups following the procedure of Clermont et al. (44) (Table 2), and the presence of 15 virulence factors (VFs) was determined (45, 46) (Fig. 3). In commensal E. coli, the prevalence of phylogenetic groups varies among studies (35, 47). It has been reported that the highly diverse hosts and environmental factors, the determinants of virulence, and the antimicrobial pressure can modify prevalence for a better adaptation to commensal habitats (47). In our study, even though commensal E. coli presented a higher diversity of phylogroups compared to that of the clinical samples (Table 2), a predominance of the phylogroup B2 carrying high rates of VFs was found in both populations. Although no evident association has been reported between plasmids and phylogroups (34), our results indicate a possible association of HI1 plasmids with phylogroup A (P ≤ 0.007, Bonferroni’s correction was applied).
TABLE 2.
Phylogenetic groups detected in E. coli strains isolated from fecal and blood samples
| E. coli isolate typea | No. (%) detected of each phylogenetic group |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total no. | A | B1 | B2 | C | D | E | F | Clade I | Unknown | |
| Total E. coli isolates | 244 | 26 (10.6) | 22 (9) | 124 (50.8) | 3 (1.2) | 32 (13.1) | 15 (6.1) | 17 (6.9) | 4 (1.6) | 1 (0.41) |
| Fecal samples | 145 | 23 (15.9) | 11 (7.6) | 59 (40.7) | 3 (2.1) | 23 (15.9) | 8 (5.5) | 13 (8.9) | 4 (2.7) | 1 (0.7) |
| Susceptible | 71 | 16 (22.5) | 1 (1.4) | 24 (33.8) | 3 (4.2) | 10 (14.1) | 7 (9.9) | 9 (12.6) | 1 (1.4) | 0 |
| Resistant | 74 | 7 (9.5) | 10 (13.5) | 35 (47.3) | 0 | 13 (17.6) | 1 (1.3) | 4 (5.4) | 3 (4.1) | 1 (1.3) |
| Blood samples | 99 | 3 (3)b | 11 (11.1) | 65 (65.7)c | 0 | 9 (9.1) | 7 (7.1) | 4 (4) | 0 | 0 |
| Susceptible | 9 | 0 | 1 (11.1) | 6 (66.7) | 0 | 2 (22.2) | 0 | 0 | 0 | 0 |
| Resistant | 90 | 3 (3.4) | 10 (11.1) | 59 (65.5)d | 0 | 7 (7.8) | 7 (7.8) | 4 (4.4) | 0 | 0 |
Susceptible indicates susceptibility to all antimicrobials tested. Resistant indicates resistance to at least one of the antimicrobials tested.
Statistical differences (P = 0.002) between A phylogroup strains from fecal and blood samples.
Statistical differences (P < 0.001) between B2 phylogroup strains from fecal and blood samples.
Statistical differences (P < 0.001) between resistant B2 phylogroup strains from fecal and blood samples.
FIG 3.
Frequency of virulence factors (VFs) analyzed in E. coli from fecal and blood sample strains and percentages of E. coli strains according to the number of VFs they carry. Adhesins include fimH (mannose-specific adhesin of type 1 fimbriae) and hra (heat-resistant agglutinin); siderophores include fyuA (yersiniabactin), iutA (aerobactin), and iroN (salmochelin receptor); toxins include hlyD (α-hemolysin), hlyF (hemolysin F), cnf1 (cytotoxic necrotizing factor 1), and clbB and clbN (colibactin); and the miscellaneous VF genes include iss (surface exclusion serum survival protein), traT (serum resistance), ompT (outer membrane protease), ibeA (invasion of brain endothelium), and usp (uropathogenic-specific protein). *, Statistical differences (P < 0.05) between each population, fecal and blood samples.
All VFs studied were detected in both populations. As expected, the clinical strains had a higher diversity (9 to 15 VFs) and frequency of VFs compared to those of the fecal samples (1 to 8 VFs) (Fig. 3). Finally, as supported by other authors (48), an association between some VFs (fyuA, iutaA, hlyF, iss, and traT) and strains carrying F plasmids was determined (P ≤ 0.003, Bonferroni’s correction was applied).
In conclusion, new information is provided about the plasmid background in strains isolated in a nonhospital setting. Although a similar trend was observed in the Inc groups from both populations, IncL/M, IncA/C, and IncN plasmids were only detected in clinical strains, whereas HI1 was only present in fecal strains. Also, two different evolutionary pathways followed by plasmids were observed as follows: specific IncI1 plasmids, such as IncI1 ST12, seem to have evolved by acquiring persistence, adaptive, and antibiotic resistance features relevant in clinical settings, whereas the more widespread multireplicon IncF plasmids have randomly acquired resistance genes. Additionally, the findings from this study confirm that strains from healthy individuals have less antimicrobial resistance and fewer VFs and display a higher diversity of phylogenetic lineages (in E. coli) than strains causing infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Plan Nacional de I+D+i and Instituto de Salud Carlos III (grant PI13/00329) and the Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015/0017) and cofinanced by European Development Regional Fund “A way to achieve Europe.” P. Espinal is funded by the Instituto de Salud Carlos III (“Sara Borrell” contract number CD15/00017).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wright GD. 2011. Molecular mechanisms of antibiotic resistance. Chem Commun (Camb) 47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- 2.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Carattoli A. 2011. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol 301:654–658. doi: 10.1016/j.ijmm.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 5.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother 61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 6.Hancock SJ, Phan MD, Peters KM, Forde BM, Chong TM, Yin WF, Chan KG, Paterson DL, Walsh TR, Beatson SA, Schembri MA. 2017. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother 61:e01740-16. doi: 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Fernández A, Carattoli A. 2010. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum β-lactamase and quinolone resistance genes. J Antimicrob Chemother 65:1155–1161. doi: 10.1093/jac/dkq101. [DOI] [PubMed] [Google Scholar]
- 8.García-Fernández A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A. 2011. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother 66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- 9.Phan MD, Kidgell C, Nair S, Holt KE, Turner AK, Hinds J, Butcher P, Cooke FJ, Thomson NR, Titball R, Bhutta ZA, Hasan R, Dougan G, Wain J. 2009. Variation in Salmonella enterica serovar typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob Agents Chemother 53:716–727. doi: 10.1128/AAC.00645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa L, García-Fernández A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 11.Argente M, Miró E, Martí C, Vilamala A, Alonso-Tarrés C, Ballester F, Calderón A, Gallés C, Gasós A, Mirelis B, Morta M, Olsina M, Sauca G, Sierra M, Rivera A, Navarro F. 2019. Molecular characterization of OXA-48 carbapenemase-producing Klebsiella pneumoniae strains after a carbapenem resistance increase in Catalonia. Enferm Infecc Microbiol Clin 37:82–88. doi: 10.1016/j.eimc.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Burke L, Humphreys H, Fitzgerald-Hughes D. 2016. The molecular epidemiology of resistance in cefotaximase-producing Escherichia coli clinical isolates from Dublin, Ireland. Microb Drug Resist 22:552–558. doi: 10.1089/mdr.2015.0154. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro TG, Novais Â, Rodrigues C, Nascimento R, Freitas F, Machado E, Peixe L. 2019. Dynamics on clonal and plasmid backgrounds of Enterobacteriaceae producing acquired AmpC in Portuguese clinical settings throughout time. Int J Antimicrob Agents 53:650–656. doi: 10.1016/j.ijantimicag.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Wang P, Cheng J, Qin S, Xie W. 2019. Characterization of a novel blaNDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect Drug Resist 12:511–519. doi: 10.2147/IDR.S192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing: twenty-fourth informational supplement M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother 47:3724–3732. doi: 10.1128/aac.47.12.3724-3732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Kim JY, Lee SK, Jin W, Kang SG, Lee KJ. 2000. Discriminatory detection of extended-spectrum beta-lactamases by restriction fragment length dimorphism-polymerase chain reaction. Lett Appl Microbiol 31:307–312. doi: 10.1046/j.1472-765x.2000.00806.x. [DOI] [PubMed] [Google Scholar]
- 18.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother 59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 19.Espinal P, Miró E, Segura C, Gómez L, Plasencia V, Coll P, Navarro F. 2018. First description of blaNDM-7 carried on an IncX4 plasmid in Escherichia coli ST679 isolated in Spain. Microb Drug Resist 24:113–119. doi: 10.1089/mdr.2017.0039. [DOI] [PubMed] [Google Scholar]
- 20.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Miró E, Mirelis B, Navarro F, Rivera A, Mesa RJ, Roig MC, Gómez L, Coll P. 2005. Surveillance of extended-spectrum β-lactamases from clinical samples and faecal carriers in Barcelona, Spain. J Antimicrob Chemother 56:1152–1155. doi: 10.1093/jac/dki395. [DOI] [PubMed] [Google Scholar]
- 22.Karanika S, Karantanos T, Arvanitis M, Grigoras C, Mylonakis E. 2016. Fecal colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin Infect Dis 63:310–318. doi: 10.1093/cid/ciw283. [DOI] [PubMed] [Google Scholar]
- 23.Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. 2015. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Fernández A, Fortini D, Veldman K, Mevius D, Carattoli A. 2009. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother 63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 25.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. 2012. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Weingarten RA, Johnson RC, Conlan S, Ramsburg AM, Dekker JP, Lau AF, Khil P, Odom RT, Deming C, Park M, Thomas PJ, Henderson DK, Palmore TN, Segre JA, Frank KM. 2018. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio 9:e02011-17. doi: 10.1128/mBio.02011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK. 2007. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983. doi: 10.1128/AEM.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irrgang A, Falgenhauer L, Fischer J, Ghosh H, Guiral E, Guerra B, Schmoger S, Imirzalioglu C, Chakraborty T, Hammerl JA, Käsbohrer A. 2017. CTX-M-15-producing E. coli isolates from food products in Germany are mainly associated with an IncF-type plasmid and belong to two predominant clonal E. coli lineages. Front Microbiol 8:2318. doi: 10.3389/fmicb.2017.02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao WH, Hu ZQ. 2013. Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit Rev Microbiol 39:79–101. doi: 10.3109/1040841X.2012.691460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dupouy V, Abdelli M, Moyano G, Arpaillange N, Bibbal D, Cadiergues MC, Lopez-Pulin D, Sayah-Jeanne S, De Gunzburg J, Saint-Lu N, Gonzalez-Zorn B, Andremont A, Bousquet-Mélou A. 2019. Prevalence of beta-lactam and quinolone/fluoroquinolone resistance in Enterobacteriaceae from dogs in France and Spain—characterization of ESBL/pAmpC isolates, genes, and conjugative plasmids. Front Vet Sci 6:279. doi: 10.3389/fvets.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietsch M, RESET Study Group, Irrgang A, Roschanski N, Brenner Michael G, Hamprecht A, Rieber H, Käsbohrer A, Schwarz S, Rösler U, Kreienbrock L, Pfeifer Y, Fuchs S, Werner G, Bühling A, Domurath B, Wendt C, Valenza G, Wahl HG, Hellkamp J, Arvand M, Kresken M, Podschun R, Schneider S, Tobisch S, Witt M, Eckmanns T, Hachmann U, Bührlen U, von Salviati-Claudius C, Lu HM, Laube H, Hering J. 2018. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics 19:601. doi: 10.1186/s12864-018-4976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robin F, Beyrouthy R, Bonacorsi S, Aissa N, Bret L, Brieu N, Cattoir V, Chapuis A, Chardon H, Degand N, Dubois V, Fortineau N, Grillon A, Lanotte P, Leyssene D, Patry I, Podglajen I, Recule C, Ros A, Ponties V, Ploy MC, Bonnet R. 2017. Inventory of extended-spectrum-β-lactamase-producing Enterobacteriaceae in France as assessed by a multicenter study. Antimicrob Agents Chemother 61:e01911-16. doi: 10.1128/AAC.01911-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 34.Branger C, Ledda A, Billard-Pomares T, Doublet B, Fouteau S, Barbe V, Roche D, Cruveiller S, Médigue C, Castellanos M, Decré D, Drieux-Rouze L, Clermont O, Glodt J, Tenaillon O, Cloeckaert A, Arlet G, Denamur E. 2018. Extended-spectrum beta-lactamase-encoding genes are spreading on a wide range of Escherichia coli plasmids existing prior to the use of third-generation cephalosporins. Microb Genom 4:e000203. doi: 10.1099/mgen.0.000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moran RA, Anantham S, Pinyon JL, Hall RM. 2015. Plasmids in antibiotic susceptible and antibiotic resistant commensal Escherichia coli from healthy Australian adults. Plasmid 80:24–31. doi: 10.1016/j.plasmid.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Carattoli A, Villa L, Fortini D, García-Fernández A. 5 December 2018. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid doi: 10.1016/j.plasmid.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 38.Smith H, Bossers A, Harders F, Wu G, Woodford N, Schwarz S, Guerra B, Rodríguez I, van Essen-Zandbergen A, Brouwer M, Mevius D. 2015. Characterization of epidemic IncI1-Iγ plasmids harboring Ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob Agents Chemother 59:5357–5365. doi: 10.1128/AAC.05006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang QE, Sun J, Li L, Deng H, Liu BT, Fang LX, Liao XP, Liu YH. 2015. IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front Microbiol 6:964. doi: 10.3389/fmicb.2015.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumura Y, Kyoto-Shiga Clinical Microbiology Study Group, Yamamoto M, Nagao M, Ito Y, Takakura S, Ichiyama S. 2013. Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob Agents Chemother 57:4736–4742. doi: 10.1128/AAC.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho WS, Yap KP, Yeo CC, Rajasekaram G, Thong KL. 2016. The complete sequence and comparative analysis of a multidrug-resistance and virulence multireplicon IncFII plasmid pEC302/04 from an extraintestinal pathogenic Escherichia coli EC302/04 indicate extensive diversity of IncFII plasmids. Front Microbiol 6:1547. doi: 10.3389/fmicb.2015.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh H, Doijad S, Falgenhauer L, Fritzenwanker M, Imirzalioglu C, Chakraborty T. 2017. blaCTX-M-27-encoding Escherichia coli sequence type 131 lineage C1-M27 clone in clinical isolates. Emerg Infect Dis 23:1754–1756. doi: 10.3201/eid2310.170938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 45.Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 46.Oteo J, Spanish Network for Research in Infectious Diseases (REIPI), González-López JJ, Ortega A, Quintero-Zárate JN, Bou G, Cercenado E, Conejo MC, Martínez-Martínez L, Navarro F, Oliver A, Bartolomé RM, Campos J. 2014. Inhibitor-resistant TEM- And OXA-1-producing Escherichia coli isolates resistant to amoxicillin-clavulanate are more clonal and possess lower virulence gene content than susceptible clinical isolates. Antimicrob Agents Chemother 58:3874–3881. doi: 10.1128/AAC.02738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 48.Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev 73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.