A strain of extensively drug-resistant (XDR) Salmonella enterica serovar Typhi has caused a large ongoing outbreak in Pakistan since 2016. In Ontario, Canada, 10 cases of mainly bloodstream infections (n = 9) were identified in patients who traveled to Pakistan. Whole-genome sequencing showed that Canadian cases were genetically related to the Pakistan outbreak strain. The appearance of XDR typhoid cases in Ontario prompted a provincial wide alert to physicians to recommend treatment with carbapenems or azithromycin in suspected typhoid cases with travel history to Pakistan.
KEYWORDS: whole genome, extensively drug resistant, XDR, antibiotic resistance, ceftriaxone, typhoid fever
ABSTRACT
A strain of extensively drug-resistant (XDR) Salmonella enterica serovar Typhi has caused a large ongoing outbreak in Pakistan since 2016. In Ontario, Canada, 10 cases of mainly bloodstream infections (n = 9) were identified in patients who traveled to Pakistan. Whole-genome sequencing showed that Canadian cases were genetically related to the Pakistan outbreak strain. The appearance of XDR typhoid cases in Ontario prompted a provincial wide alert to physicians to recommend treatment with carbapenems or azithromycin in suspected typhoid cases with travel history to Pakistan.
TEXT
Typhoid fever, caused by Salmonella enterica subsp. enterica serovar Typhi (S. Typhi) remains a significant public health concern, particularly in developing countries. S. Typhi infection is restricted to humans and is acquired through the fecal-oral route and often via contaminated water. A recent study estimated that 12 to 21 million cases of typhoid fever occur each year, resulting in 129,000 to 223,000 deaths, primarily in developing countries (1). Most of the cases are in children and adolescents living in southern and southeastern Asia. Between 2013 and 2018, 60 to 107 (0.4 to 0.7/100,000) cases of typhoid fever were diagnosed each year in Ontario, Canada. The vast majority of these patients reported recent travel to countries of endemicity. The Public Health Agency of Canada recommends that travelers visiting areas of endemicity be vaccinated against typhoid fever and follow safe food and water practices (2).
Over the years, widespread emergence of multidrug-resistant (MDR) strains has been documented in regions of endemicity. S. Typhi is commonly resistant to first-line drugs (ampicillin, trimethoprim-sulfamethoxazole, and chloramphenicol). As a result, fluoroquinolones became the drug of choice in the 1990s. Since then, fluoroquinolone resistance has increased, necessitating the use of the third-generation cephalosporin ceftriaxone for treatment of typhoid fever (3). Over the last decade, sporadic cases of ceftriaxone- or azithromycin-resistant strains have been reported (4). In Ontario, ceftriaxone is the first-line treatment for hospitalized patients with suspected typhoid fever.
Since November 2016, an outbreak of S. Typhi has been ongoing in two cities in Pakistan (Karachi and Hyderabad). From 2016 to 2018, >5,372 cases have been reported in these regions (5). The outbreak strain was found to be resistant to the first-line treatment (ampicillin, trimethoprim-sulfamethoxazole, and chloramphenicol) and to fluoroquinolones and third-generation cephalosporins, including ceftriaxone, and thus considered an extensively drug-resistant (XDR) strain (6). The XDR strain remained susceptible to azithromycin and carbapenems. In the United States, 5 cases of XDR S. Typhi related to the outbreak in Pakistan were reported to the Centers for Disease Control and Prevention between 2016 and 2018 (7). Here, we report 10 cases of mostly bloodstream infections (n = 9; stool, n = 1) caused by XDR S. Typhi identified in Ontario from July 2018 to May 2019 in patients who had recently traveled to Pakistan. Using whole-genome sequencing (WGS), we demonstrated that all Canadian cases were highly related to the XDR outbreak strain in Pakistan.
The study.
From July 2018 to May 2019, 10 XDR typhoid cases were identified in Ontario. All but 1 patient resided in south-central Ontario in the Greater Toronto area. All patients had recent travel history to Pakistan. Patients ranged in age from 3 to 52 years. Four patients were male, 4 were female, and 2 did not report gender. Isolates were sent to Public Health Ontario Laboratory as part of a surveillance program and for additional susceptibility testing (i.e., azithromycin) and WGS. Phenotypic susceptibility testing was carried out by the broth microdilution method, and MIC results were interpreted according to Clinical and Laboratory Standards Institute guidelines (8). All isolates were resistant to ampicillin, ceftriaxone, chloramphenicol, ciprofloxacin, nalidixic acid, trimethoprim-sulfamethoxazole, and streptomycin. All isolates were susceptible to amoxicillin-clavulanate, cefoxitin, gentamicin, tetracycline, azithromycin, and meropenem (Table 1).
TABLE 1.
Antimicrobial susceptibility profiles of S. Typhi isolates
| Sample | Collection date (mo-yr) | Specimen type | MIC (mg/liter) and susceptibility ofa
: |
Genotypic resistance | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AZM | AMC | AMP | FOX | CRO | CHL | CIP | GEN | MEM | STR | TET | SXT | ||||
| PHL5950 | Jul-18 | Blood | 4 (S) | 16/8 (I) | >32 (R) | 4 (S) | >64 (R) | >32 (R) | 2 (R) | 0.5 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL4653 | Sep-18 | Stool | 4 (S) | 8/4 (S) | >32 (R) | 8 (S) | >64 (R) | >32 (R) | 4 (R) | ≤0.25 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL3183 | Jan-19 | Blood | 4 (S) | 8/4 (S) | >32 (R) | 4 (S) | >64 (R) | >32 (R) | 2 (R) | 0.5 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL9922 | Dec-18 | Blood | 4 (S) | 16/8 (S) | >32 (R) | 8 (S) | >64 (R) | >32 (R) | 2 (R) | ≤0.25 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7, tet(A) |
| PHL3127 | Jan-19 | Blood | 4 (S) | 8/4 (S) | >32 (R) | 4 (S) | >64 (R) | >32 (R) | 2 (R) | 0.5 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL4981 | Apr-19 | Blood | 4 (S) | 8/4 (S) | >32 (R) | 4 (S) | >64 (R) | >32 (R) | 2 (R) | 0.5 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL4949 | Sep-18 | Blood | 4 (S) | 8/4 (S) | >32 (R) | 4 (S) | >64 (R) | >32 (R) | 2 (R) | 0.5 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL8802 | Apr-19 | Blood | 4 (S) | 16/8 (I) | >32(R) | 8 (S) | >64 (R) | >32 (R) | 2 (R) | 0.5 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL3828 | May-19 | Blood | 4 (S) | 16/8 (I) | >32 (R) | 8 (S) | >64 (R) | >32 (R) | 2 (R) | ≤0.25 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
| PHL4989 | Apr-19 | Blood | 4 (S) | 16/8 (I) | >32 (R) | 8 (S) | >64 (R) | >32 (R) | 2 (R) | ≤0.25 (S) | ≤0.06 (S) | >64 (R) | ≤4 (S) | >4/76 (R) | aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, blaCTX-M-15, qnrS1, S83F, catA1, sul1, sul2, dfrA7 |
CLSI document M100-S29 clinical interpretative breakpoints criteria were used to determine susceptibility profile (8). AZM, azithromycin; AMC, amoxicillin-clavulanate; AMP, ampicillin; FOX, cefoxitin; CRO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; MEM, meropenem; STR, streptomycin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; S, susceptible; I, intermediate; R, resistant.
WGS was performed on all isolates using the Nextera XT DNA library preparation kit followed by short-read sequencing on the Illumina MiSeq platform (Illumina, Inc., San Diego, CA) using 300-bp paired-end reads (v3 chemistry) and aiming to achieve >100-fold coverage. In addition, the Ontario isolate, PHL5950, was subjected to long-read sequencing in the MinION platform (Oxford Nanopore Technologies, Oxford, UK). Briefly, the DNA library was prepared for long-read sequencing using a 1D ligation sequencing kit (SQK-LSK-108; Nanopore) without fragmentation and included the DNA repair step with the NEBNext FFPE DNA repair module (New England Biolabs, Ipswich, MA).
Paired-end Illumina reads were assembled using the de novo assembler in CLC Genomics Workbench. Hybrid de novo assembly was performed in Unicycler (v0.4.8-beta) using both Illumina and Nanopore sequences to circularize the chromosome and plasmids. De novo-assembled contigs were analyzed in silico with the Center for Genomic Epidemiology Web-based tools (http://www.genomicepidemiology.org/), including ResFinder, MLST, core genome MLST (cgMLST), and PlasmidFinder (9–12).
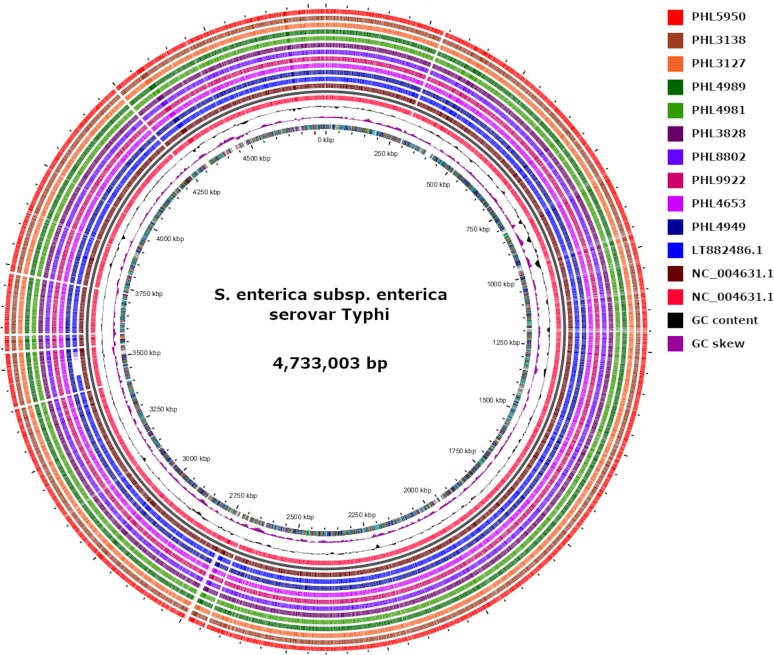
WGS analysis showed that all 10 isolates were serotype 9(O):D(H1) with MLST 1 and cgMLST 83019. To assess genetic relatedness, short-read sequences were compared against the genome of strain 22420_110_pak6006_2016 (GenBank accession no. LT882486), which was characterized from the Pakistan outbreak (4). Single-nucleotide variant (SNV) analysis showed high similarity among Ontario isolates (1 or 2 SNVs) and between Ontario isolates and the outbreak reference strain 22420_110_pak6006_2016 (3 or 4 SNVs), suggesting that all 10 isolates recovered in Ontario were closely related to the XDR strain circulating in Pakistan (Fig. 1).
FIG 1.
Multiple genome comparison of XDR S. Typhi from this study and the 22420_1_10_Pak60006_2016 strain (GenBank accession no. LT882486.1) against reference S. enterica subsp. enterica serovar Typhi strain Ty2 (GenBank accession no. NC_004631.1). The innermost ring indicates the genomic position followed by GC skewed (purple) and plot of GC content (black). The outer 10 circles represent strains from this study. The gap in strain 22420_1_10_Pak60006_2016 represents a missing region of ∼46 kb of chromosomal genes (46,317 bp) compared with the reference NC_004631.1 and Ontario strains.
All isolates harbored several antimicrobial resistance genes and mutations consistent with the observed phenotypic resistance to β-lactams (blaCTX-M-15, blaTEM-1B), sulfonamides (sul1/sul2), streptomycin [aph(6)-Id, aac(6′)-laa, aph(3″)-lb], chloramphenicol (catA1), trimethoprim (dfrA7), and fluoroquinolones (qnrS1), as well as S83F mutation in gyrA (Table 1). Extended-spectrum β-lactamase (ESBL) molecular class A genes, including blaCTX-M-15, are known to confer resistance to ampicillin and third-generation cephalosporins (e.g., ceftriaxone) but to remain susceptible to the cephamycin class of drugs (e.g., cefoxitin) and β-lactam–β-lactamase inhibitor combination antibody.
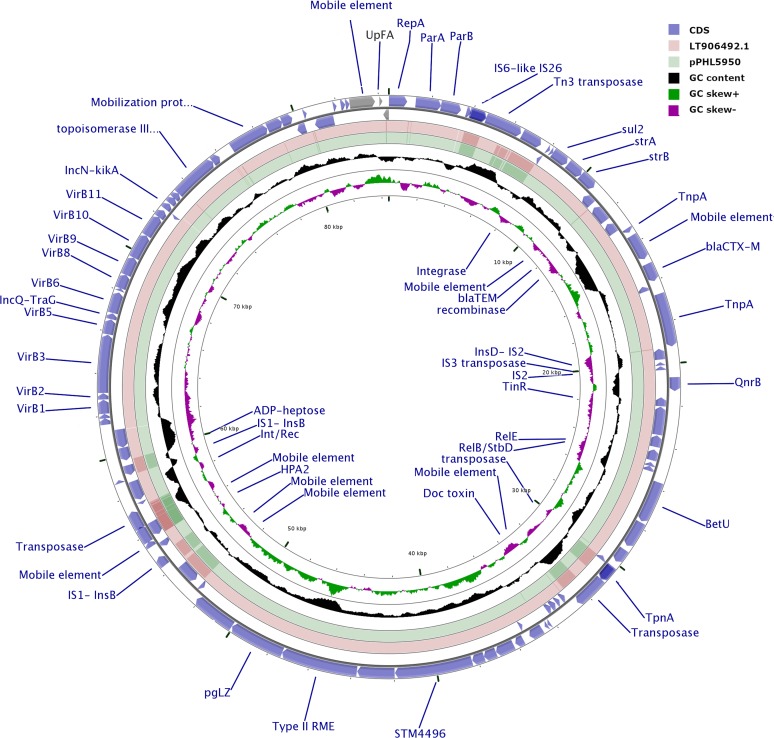
All isolates contained reads for the IncY plasmid that was reported in the outbreak in Pakistan (10). The circularized IncY plasmid (84,492 bp) obtained by the hybrid assembly of both long and short reads of isolate PHL5950 confirmed the presence of a transposon carrying the antimicrobial resistance genes aph(6)-Id, aac(6′)-Iaa, aph(3″)-Ib, blaCTX-M-15, blaTEM-1B, qnrS1, and sul2. The IncY plasmid from the Canadian isolate in this study was almost identical (99.99% with 4 SNVs) to the previously reported plasmid (GenBank accession no. LT882486.1) from the Pakistan outbreak (Fig. 2) (4). The LT882486.1 plasmid was genetically similar to an ESBL-encoding plasmid found in Escherichia coli; it has been hypothesized that the S. Typhi strain acquired this plasmid from E. coli, converting an MDR strain to XDR followed by clonal expansion (4).
FIG 2.
Schematic circular map comparing one representative S. Typhi XDR plasmid from this study (pPHL5950) and plasmid p60006 from the 22420_1_10_Pak60006_2016 (GenBank accession no. LT882486.1) XDR strain identified in Pakistan. The outer two rings (blue arrows) represent all putative open reading frames and their orientation on the positive and negative strand. The outbreak reference plasmid LT906492.1 S. enterica plasmid 2 (p60006) (pink ring) and the closed pPHL5950 plasmid from Canada (green ring) are also shown. GC content is represented by the black ring, and GC skew by the green and purple ring.
Conclusions.
In this study, we used WGS to characterize 10 cases of imported XDR S. Typhi isolates in Ontario, Canada, that were linked to an ongoing outbreak in Pakistan. Several genes conferring resistance to commonly used antimicrobials for the treatment of typhoid fever were identified. The acquisition of an ESBL-encoding plasmid by S. Typhi, resulting in an XDR strain, and the subsequent large outbreak in Pakistan is concerning in that this suggests the circulation of a fit clone with the ability to disseminate. Ceftriaxone is the drug of choice for treatment of patients with suspected typhoid infection in Ontario, thus the importation of XDR S. Typhi isolates is also a concern because it limits options for empirical treatment. Approximately 5% of patients may shed the bacteria for a long time after resolution of symptoms (13). Therefore, it is essential to identify cases of infection with the XDR strain, particularly individuals who work in the food industry, to ensure that appropriate follow up is conducted to prevent transmission (13). Our investigation highlights the importance of ongoing surveillance and rapid detection and characterization of XDR strains to inform treatment strategies and infection control measures.
Accession numbers.
The whole-genome sequencing data from this study have been deposited in GenBank under accession numbers RHPM00000000, WSUJ00000000, WSUK00000000, WSUL00000000, WSUM00000000, WSUN00000000, WSUO00000000, WSUP00000000, WSUQ00000000, and WSUR00000000 and BioProject number PRJNA595493.
ACKNOWLEDGMENT
This work was supported by the Public Health Ontario Laboratory as part of routine investigation.
REFERENCES
- 1.Mogasale V, Maskery B, Ochiai RL, Lee JS, Mogasale VV, Ramani E, Kim YE, Park JK, Wierzba TF. 2014. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health 2:e570–e580. doi: 10.1016/S2214-109X(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 2.Government of Canada. 2019. Prevention of typhoid fever. https://www.canada.ca/en/public-health/services/diseases/typhoid-fever/prevention.html.
- 3.Parry CM, Ribeiro I, Walia K, Rupali P, Baker S, Basnyat B. 2019. Multidrug resistant enteric fever in South Asia: unmet medical needs and opportunities. BMJ 364:k5322. doi: 10.1136/bmj.k5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, Saeed DK, Wong VK, Dallman TJ, Nair S, Baker S, Shaheen G, Qureshi S, Yousafzai MT, Saleem MK, Hasan Z, Dougan G, Hasan R. 2018. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9:e00105-18. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization, WHO Regional Office for Eastern Mediterranean. 2018. Disease outbreaks in eastern Mediterranean region (EMR). Wkly Epidemiol Monit 11(12) http://applications.emro.who.int/docs/epi/2018/Epi_Monitor_2018_11_52.pdf?ua=1. [Google Scholar]
- 6.German GJ, Gilmour M, Tipples G, Adam HJ, Almohri H, Bullard J, Dingle T, Farrell D, Girouard G, Haldane D, Hoang L, Levett PN, Melano R, Minion J, Needle R, Patel SN, Rennie R, Reyes RC, Longtin J, Mulvey MR. 2018. Canadian recommendations for laboratory interpretation of multiple or extensive drug resistance in clinical isolates of Enterobacteriaceae, Acinetobacter species and Pseudomonas aeruginosa. Can Commun Dis Rep 44:29–34. doi: 10.14745/ccdr.v44i01a07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatham-Stephens K, Medalla F, Hughes M, Appiah GD, Aubert RD, Caidi H, Angelo KM, Walker AT, Hatley N, Masani S, Nash J, Belko J, Ryan ET, Mintz E, Friedman CR. 2019. Emergence of extensively drug-resistant Salmonella Typhi infections among travelers to or from Pakistan—United States, 2016–2018. MMWR Morb Mortal Wkly Rep 68:11–13. doi: 10.15585/mmwr.mm6801a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing. CLSI document M100-S29. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 9.Alikhan NF, Zhou Z, Sergeant MJ, Achtman M. 2018. A genomic overview of the population structure of Salmonella. PLoS Genet 14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock-Allen J, Cronquist AB, Peden J, Adamson D, Corral N, Brown K. 2016. Notes from the field: typhoid fever outbreak associated with an asymptomatic carrier at a restaurant - Weld County, Colorado, 2015. MMWR Morb Mortal Wkly Rep 65:606–607. doi: 10.15585/mmwr.mm6523a4. [DOI] [PubMed] [Google Scholar]