Capsid assembly is a critical step in the hepatitis B virus (HBV) life cycle, mediated by the core protein. Core is a potential target for new antiviral therapies, the capsid assembly modulators (CAMs). JNJ-56136379 (JNJ-6379) is a novel and potent CAM currently in phase II trials. We evaluated the mechanisms of action (MOAs) and antiviral properties of JNJ-6379 in vitro. Size exclusion chromatography and electron microscopy studies demonstrated that JNJ-6379 induced the formation of morphologically intact viral capsids devoid of genomic material (primary MOA).
KEYWORDS: capsid, capsid assembly modulator, cccDNA, hepatitis, hepatitis B virus, primary human hepatocytes
ABSTRACT
Capsid assembly is a critical step in the hepatitis B virus (HBV) life cycle, mediated by the core protein. Core is a potential target for new antiviral therapies, the capsid assembly modulators (CAMs). JNJ-56136379 (JNJ-6379) is a novel and potent CAM currently in phase II trials. We evaluated the mechanisms of action (MOAs) and antiviral properties of JNJ-6379 in vitro. Size exclusion chromatography and electron microscopy studies demonstrated that JNJ-6379 induced the formation of morphologically intact viral capsids devoid of genomic material (primary MOA). JNJ-6379 accelerated the rate and extent of HBV capsid assembly in vitro. JNJ-6379 specifically and potently inhibited HBV replication; its median 50% effective concentration (EC50) was 54 nM (HepG2.117 cells). In HBV-infected primary human hepatocytes (PHHs), JNJ-6379, when added with the viral inoculum, dose-dependently reduced extracellular HBV DNA levels (median EC50 of 93 nM) and prevented covalently closed circular DNA (cccDNA) formation, leading to a dose-dependent reduction of intracellular HBV RNA levels (median EC50 of 876 nM) and reduced antigen levels (secondary MOA). Adding JNJ-6379 to PHHs 4 or 5 days postinfection reduced extracellular HBV DNA and did not prevent cccDNA formation. Time-of-addition PHH studies revealed that JNJ-6379 most likely interfered with postentry processes. Collectively, these data demonstrate that JNJ-6379 has dual MOAs in the early and late steps of the HBV life cycle, which is different from the MOA of nucleos(t)ide analogues. JNJ-6379 is in development for chronic hepatitis B treatment and may translate into higher HBV functional cure rates.
INTRODUCTION
Hepatitis B is a major global health problem, and at least 2 billion people have had an acute infection with the hepatitis B virus (HBV). Globally, approximately 292 million people have chronic HBV infection, i.e., 3.9% of the world’s population (1). Approximately 20 to 30% of individuals with chronic hepatitis B (CHB) eventually develop cirrhosis, liver failure, or hepatocellular carcinoma (2).
Currently, there are two main treatment options for CHB (3, 4). The standard of care is treatment with a nucleos(t)ide analogue (NA; e.g., tenofovir disoproxil [TDF] or entecavir [ETV]), which is generally given lifelong and is well tolerated (5). In some parts of the world or for specific populations, finite treatment with pegylated interferon is also used, but this has significant side effects (5). Current CHB treatments rarely result in a functional cure, which is defined as sustained loss of hepatitis B surface antigen (HBsAg) and undetectable HBV DNA in serum off treatment for ≥6 months, with or without HBsAg seroconversion (6). Thus, novel therapeutic approaches with different mechanisms of action (MOAs) are required for CHB treatment; such agents, likely used in combination, may provide finite treatment options with increased functional cure rates.
A promising target for novel CHB therapies is HBV capsid assembly, a critical step in the HBV life cycle. One of the seven proteins encoded by the HBV DNA is core, a protein which self-assembles to form the viral nucleocapsid. Core plays a role in most steps of the HBV life cycle (e.g., subcellular traficking/transport, release of the HBV genome, capsid assembly, and reverse transcription) (7). This protein was reported to be involved in modulation of covalently closed circular DNA (cccDNA); e.g., alpha interferon (IFN-α) was shown to inhibit HBV replication and to result in HBV cccDNA deamination and degradation through a process that involved the association of HBV core protein with apolipoprotein B mRNA-editing catalytic polypeptide 3G and HBV cccDNA (8). Core has also been reported to interfere with innate signaling (9). The HBV life cycle has been well described (7). In brief, the formation of a capsid containing pregenomic RNA (pgRNA) and viral polymerase is the first cytoplasmic step, initiated by the association of polymerase-bound pgRNA (pol-pgRNA) with three core protein dimers (nucleation). Single dimers are rapidly added to this nucleus to form the nucleic acid-containing capsid, or nucleocapsid (10). Within the nucleocapsid, reverse transcription of the pgRNA by the polymerase produces relaxed circular DNA (rcDNA); other products of reverse transcription include single-stranded DNA and double-stranded linear DNA. The rcDNA-containing capsid either reshuttles to the nucleus to replenish cccDNA or is enveloped, thereby forming infectious viral particles that are released from the cell (11). Furthermore, HBV-like particles containing RNA are also produced, the release of which has been linked to the efficacy and prognosis of CHB treatment (12).
Capsid assembly modulators (CAMs) accelerate the kinetics of capsid assembly, whereby they prevent pol-pgRNA complex encapsidation and block HBV replication (13, 14). CAMs also interfere with cccDNA transcription/de novo formation during the early steps of infection (15–17). Such characteristics differentiate CAMs from NAs, as the latter interfere solely with the reverse transcription and polymerase process, inhibiting HBV replication. There are several types of CAMs that impact pol-pgRNA encapsidation at the nucleation step and can be grouped into at least two classes. CAMs whose MOA results in the formation of empty, morphologically intact capsids are referred to as CAM-N (“N” indicates normal structure; this kind of mechanism was formerly referred to as class I MOA by Janssen). Examples of CAM-Ns include phenylpropenamide derivatives, such as AT130 (18, 19), and sulfamoylbenzamide derivatives (20), such as JNJ-632 (16, 21) and JNJ-827 (17). CAMs whose MOA results in the formation of pleiomorphic noncapsid structures (i.e., aberrant particles) are referred to as CAM-A (“A” indicates aberrant structure; this kind of mechanism was formerly referred to as class II MOA by Janssen). Examples of CAM-As include BAY41-4109 (22, 23) and JNJ-890 (17). Several CAMs have entered clinical trials.
CAM JNJ-6379 (Fig. S1 in the supplemental material) demonstrated proof of mechanism in a 4-week phase 1b clinical trial by reducing serum HBV DNA and HBV RNA levels in treatment-naive hepatitis B e antigen (HBeAg)-positive and -negative patients with CHB (24). A phase 2 clinical trial with JNJ-6379 is currently ongoing in HBeAg-positive and -negative patients with CHB (ClinicalTrials registration no. NCT03361956).
In the current paper, we have profiled JNJ-6379 in biochemical and cellular assays, using evaluations similar to those previously conducted with CAM JNJ-632 (16, 21), to demonstrate its dual MOAs of accelerating capsid assembly kinetics and preventing encapsidation of pol-pgRNA (primary MOA) late in the viral life cycle and preventing cccDNA formation during the early steps of viral infection (secondary MOA).
RESULTS
Biochemical MOA studies.
To categorize the primary MOA of JNJ-6379, size exclusion chromatography (SEC) and electron microscopy (EM) studies were conducted versus the positive-control compounds AT130 (CAM-N) (18, 19) and BAY41-4109 (CAM-A) (22, 23).
Positive-control compound AT130 induced the formation of morphologically intact capsids, and positive-control compound BAY41-4109 induced the formation of aberrant structures (Fig. S2 in the supplemental material). JNJ-6379 induced a dose-dependent increase in morphologically intact capsids (Fig. S2) as determined by SEC. These results were confirmed by EM studies (Fig. S3), i.e., morphologically intact capsids were observed with JNJ-6379 (Fig. S3C) and AT130 (Fig. S3D) and aberrant structures with BAY41-4109 (Fig. S3B). Thus, JNJ-6379 is classified as a CAM-N.
To evaluate whether JNJ-6379 had an effect on HBV capsid assembly kinetics, a fluorescence quenching assay was conducted (25). The results of a representative experiment are shown in Fig. S4. The positive controls showed reduced fluorescence with the full-assembly control (Cp*150 protein in high salt concentration and no JNJ-6379) and maximal fluorescence with the no-assembly control (Cp*150 protein without salt and no JNJ-6379). The addition of JNJ-6379 (0.625, 1.25, 2.5, 5.0, or 10 μM) to Cp*150 protein in the presence of 200 mM sodium chloride (NaCl) showed a sigmoidal pattern of reducing fluorescence. With increasing JNJ-6379 concentrations, the lag phase became shorter, the slope of the fast phase became steeper, and the asymptotic phase was achieved earlier than with low JNJ-6379 concentrations (Fig. S4). Thus, JNJ-6379 dose dependently increased the rate and extent of HBV capsid assembly in vitro.
In vitro antiviral properties and cytotoxicity of JNJ-6379.
The antiviral properties of JNJ-6379 were evaluated in stably HBV-replicating HepG2.117 and HepG2.2.15 cells. With HepG2.117 cells, the median 50% effective concentration (EC50) and 90% effective concentration (EC90) of JNJ-6379 were 54 and 226 nM, respectively (Table 1). The median EC50 values for the reference CAMs were 69 nM (BAY41-4109; CAM-A), 415 nM (JNJ-632; CAM-N), and 1,540 nM (AT-130; CAM-N) in HepG2.117 cells. In the presence of 40% human serum, the median EC50 and EC90 values shifted to 205 (3.8-fold) and 842 (3.7-fold) nM, respectively, indicative of JNJ-6379 binding to proteins.
TABLE 1.
Antiviral properties and cytotoxicity of JNJ-6379
| Cell line | Antiviral activitya
|
Cytotoxicityb
|
SIc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of expts | Median EC50 (range) (nM) | No. of expts | Median EC90 (range) (nM) | No. of expts | CC50 (nM) | No. of expts | CC90 (nM) | ||
| HepG2 | 21 | >25,000 | 21 | >25,000 | >463 | ||||
| HepG2.117 | 38 | 54 (14 to 115) | 36 | 226 (152 to >500) | 5 | >25,000 | 5 | >25,000 | >463 |
| HepG2.2.15 | 15 | 69 (28 to 121) | 12 | 241 (212 to 606) | |||||
The antiviral properties of JNJ-6379 were tested in a dose-response assay in stably HBV-replicating HepG2.117 and HepG2.2.15 cells. Intracellular (HepG2.117) or extracellular (HepG2.2.15) HBV DNA was extracted from cell lysates and assessed using quantitative PCR (qPCR). In each experiment, EC50 values were determined based on the mean inhibition from two wells per compound concentration. EC50, 50% effective concentration; EC90, 90% effective concentration.
Cytotoxicity was assessed in HepG2 cells using a resazurin readout. CC50, 50% cytotoxic concentration; CC90, 90% cytotoxic concentration.
SI, selectivity index (CC50/EC50) calculated with EC50 values from HepG2.117 cells.
A dose-dependent reduction of capsid-associated DNA was observed in the presence of JNJ-6379 using HBV-replicating HepG2.117 (Fig. S5). There was also a dose-dependent shift from T = 4 capsids (120 core protein dimers) to T = 3 capsids (90 core protein dimers), indicating that JNJ-6379 modulates capsid assembly in HepG2.117 cells (Fig. S5). For the CAM-A compound, BAY41-4109, there was a dose-dependent decrease in both T = 4 and T = 3 capsids (Fig. S6) due to the formation of aggregated core/capsid structures. For both reference CAM-N compounds, JNJ-632 and AT-130, there was a dose-dependent shift from T = 4 capsids to T = 3 capsids (Fig. S6), consistent with their MOA. Compared with the results for JNJ-6379 (median EC50 of 54 nM), higher JNJ-632 and AT130 concentrations were needed to observe similar effects on capsids, which is in line with the less potent antiviral activity of these CAM-Ns (median EC50s of 415 and 1,540 nM, respectively).
The antiviral property of JNJ-6379 was specific to HBV in vitro, as no antiviral properties were seen against a panel of representative DNA and RNA viruses (Table S1). JNJ-6379 also demonstrated broad antiviral properties against HBV clinical isolates and site-directed core mutants (26, 27).
JNJ-6379 did not show cytotoxicity up to a concentration of 25 μM in HepG2 cells (selectivity index of >463) (Table 1), which was the highest concentration tested. The median 50% cytotoxic concentration (CC50) values for JNJ-6379 in other human cell lines (A549, HEL299, HeLa, Huh7, and MT4) ranged from >25 to >100 μM (data not shown). With animal cell lines MDCK and Vero, the median CC50 values for JNJ-6379 ranged from 49 to >100 μM (data not shown).
Antiviral properties of JNJ-6379 in combination with an NA.
To evaluate the antiviral properties of JNJ-6379 in combination with an NA, synergy experiments were performed. The antiviral properties were assessed using the Bliss-Independence model based on the algorithm of Prichard and Shipman (28), using MacSynergy II software. In stably HBV-replicating HepG2.2.15 cells, combining JNJ-6379 with either TDF or ETV resulted in additive-to-synergistic antiviral properties (Fig. S7). No antagonist effects were seen. These results confirm the potential utility of JNJ-6379 for use in combination regimens with NAs.
Antiviral properties in HBV-infected PHHs.
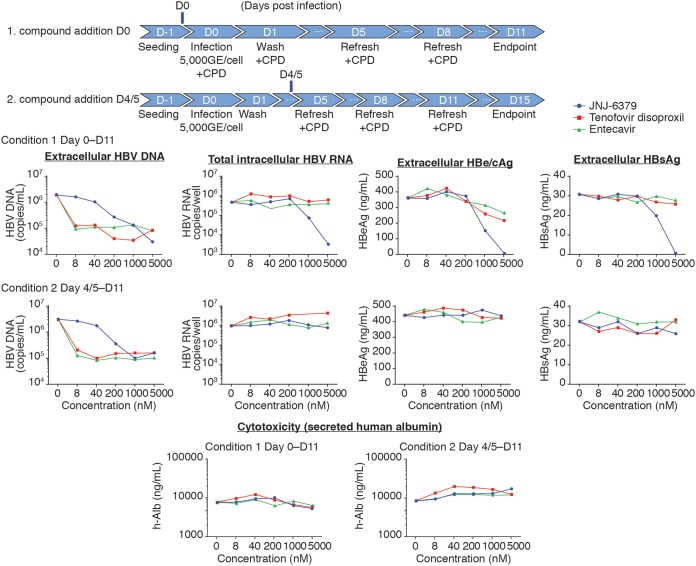
JNJ-6379, TDF, and ETV were tested in HBV-infected primary human hepatocytes (PHHs) to evaluate the MOA under the following two conditions: condition 1, compounds were added together with the viral inoculum; condition 2, compounds were added on day 4 or 5 postinfection.
Under both conditions, all three compounds reduced HBV DNA levels in the cell culture supernatant (Fig. 1). The median EC50s for conditions 1 and 2, respectively, were 93 and 102 nM (JNJ-6379), <8 and <8 nM (TDF), and 0.023 and 0.028 nM (ETV) (Table 2). The median EC90s are listed in Table S2.
FIG 1.
Antiviral properties in HBV-infected PHHs. Cryopreserved primary human hepatocytes (PHHs) were infected with purified HBV (from stably HBV-replicating HepG2.2.15 cells). Compounds were either added together with the viral inoculum (day zero) or on day 4 or 5 postinfection. Infected cells were incubated with compounds for a total of 11 days, and the compounds replenished when the culture medium was changed. The antiviral properties of compounds were tested in dose-response assays. HBV DNA (extracellular) and HBV RNA (intracellular) levels were assessed using quantitative PCR (qPCR). Extracellular HBe/cAg and HBsAg were evaluated using AlphaLISA. Cytotoxicity was evaluated by assessing extracellular human albumin levels. The graphs show the results from a representative experiment. CPD, compound; GE, genome equivalents; D, day; HBe/cAg, hepatitis B e antigen and hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen.
TABLE 2.
Antiviral properties measured as EC50 values for different markers in HBV-infected PHHs
| Day, compound | Median EC50 (range) (nM) for indicated marker; no of exptsa
|
|||
|---|---|---|---|---|
| Extracellular HBV DNA | Total intracellular HBV RNA | HBe/cAg | HBsAg | |
| Zero | ||||
| JNJ-6379 | 93 (46 to 188); 8 | 876 (535 to 1,339); 6 | 960 (794 to 1,326); 8 | 1,608 (916 to 3,827); 6 |
| TDF | <8.0 (ND); 1 | >5,000 (ND); 1 | >5,000 (ND); 1 | >5,000 (ND); 1 |
| ETV | 0.023 (0.014 to 0.023); 3 | >5,000 (>1.0 to >5,000); 17 | >5,000 (>1.0 to >5,000); 19 | >5,000 (>1.0 to >5,000); 22 |
| 4 or 5 | ||||
| JNJ-6379 | 102 (52 to 137); 8 | >25,000 (>5,000 to >25,000); 8 | >25,000 (>5,000 to >25,000); 8 | >25,000 (>5,000 to >25,000); 8 |
| TDF | <8.0 (ND); 1 | >5,000 (ND); 1 | >5,000 (ND); 1 | >5,000 (ND); 1 |
| ETV | 0.028 (0.012 to 0.028); 3 | >5,000 (>1.0 to >5,000); 23 | >5,000 (>1.0 to >5,000); 23 | >5,000 (>1.0 to >5,000); 23 |
Stably HBV-replicating HepG2.2.15 cells were the source of HBV for the infection of cryopreserved PHHs. The antiviral properties of each compound were tested in a dose-response assay in HBV-infected PHHs. HBV DNA (extracellular) and HBV RNA (intracellular) levels were assessed using quantitative PCR (qPCR). HBe/cAg and HBsAg in the supernatants were evaluated by AlphaLISA. HBV, hepatitis B virus; PHHs, primary human hepatocytes; EC50, 50% effective concentration; HBe/cAg, hepatitis B e antigen and hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen; ETV, entecavir; TDF, tenofovir disoproxil; ND, not determined.
Dose-dependent reductions in total intracellular HBV RNA, hepatitis B e antigen and hepatitis B core-related antigen (HBe/cAg), and HBsAg levels in the cell culture supernatant were observed with JNJ-6379 when added to PHHs at the same time as the viral inoculum (condition 1) (Fig. 1). The median EC50s of JNJ-6379 for these endpoints were, respectively, 876 nM, 960 nM, and 1,608 nM (Table 2). In contrast, JNJ-6379 had no effect on total intracellular HBV RNA, HBe/cAg, and HBsAg levels in the cell culture supernatant when added at concentrations up to 25 μM on day 4 or 5 postinfection (condition 2) (Fig. 1, Table 2). The median EC90s are listed in Table S2. TDF and ETV had no effect on total intracellular HBV RNA, HBe/cAg, and HBsAg levels in the cell culture supernatant under either condition (Fig. 1, Table 2, Table S2). Collectively, these results demonstrated that JNJ-6379, a CAM, but not TDF and ETV, which are both NAs, interferes with early events in the life cycle of HBV when added together with the inoculum.
JNJ-6379, TDF, and ETV in HBV-infected PHHs did not affect human albumin levels in the cell culture supernatant at any of the concentrations evaluated under either experimental condition (Fig. 1).
Thus, based on these results in PHHs, we concluded that CAM JNJ-6379 has a dual MOA, acting on early events, as well as on capsid assembly during the late stages of HBV infection.
Capsid stability assay with JNJ-6379.
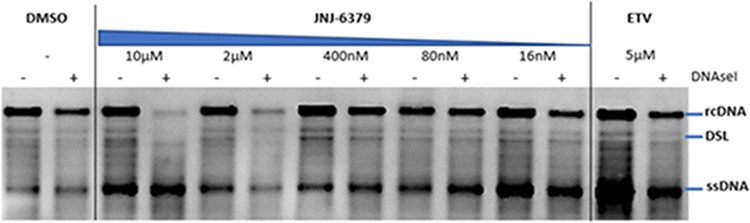
To further explore the effects of JNJ-6379 on preformed capsids during the early stages of HBV infection, a capsid stability assay was conducted. Intracellular preformed capsids were established in HBV-replicating HepG2.117 cells and exposed for 30 min to dimethyl sulfoxide (DMSO), ETV, and various doses of JNJ-6379. Cell lysates containing capsids were prepared and incubated with or without DNase I, and the impact of DNase I on encapsidated HBV DNA was assessed by Southern blotting. DNase I treatment did not have any impact on encapsidated HBV DNA when intracellular preformed capsids were treated with DMSO or ETV, whereas treatment with JNJ-6379 (2 and 10 μM) resulted in reduced relaxed circular and double-stranded linear HBV DNA signals (Fig. 2). These findings suggest that JNJ-6379 damages mature capsids, making them DNase I sensitive, and has a minimal effect on immature capsids (which contain single-stranded DNA) (29).
FIG 2.
Effect of JNJ-6379 on preformed mature capsids. HBV-replicating HepG2.117 cells were cultured for 3 days without doxycycline, and on day 4, cells were treated with JNJ-6379 (1:5 serial dilution at 5 different concentrations) or ETV for 30 min in the presence of doxycycline. Cell lysates were incubated with or without DNase I. HBV DNA was assessed by Southern blotting. DMSO, dimethyl sulfoxide; ETV, entecavir; rcDNA, relaxed circular DNA; ssDNA, single-stranded DNA; DSL, double-stranded linear DNA.
Impacts on capsid assembly were also observed with other CAMs (BAY-41-4109, JNJ-632, and AT130) (Fig. S8), which served as controls. Compared with the results for JNJ-6379, higher JNJ-632 and AT130 concentrations were required to induce similar effects on the capsids, which is in line with the less potent antiviral activity of these compounds.
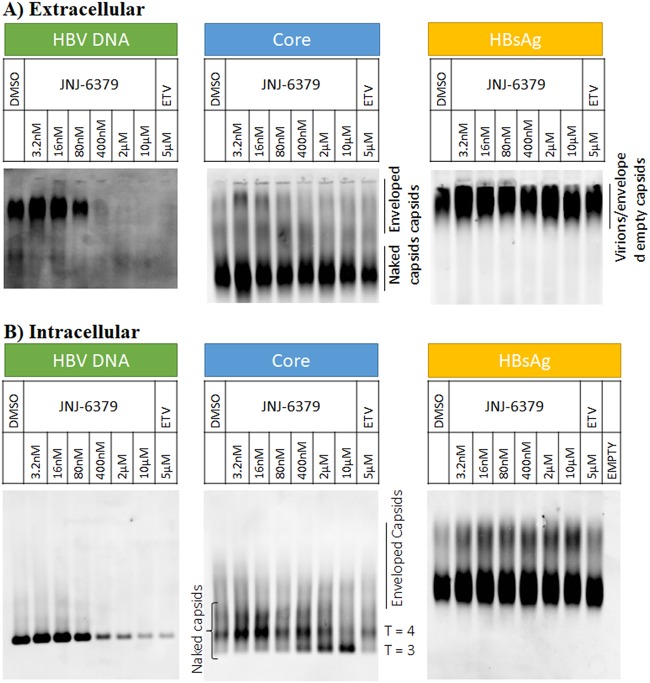
Effects of JNJ-6379 and ETV on viral particles.
HBV-infected PHHs were treated with JNJ-6379 or ETV from 5 days postinfection for 7 days. HBV DNA was evaluated by Southern blotting, and core and HBsAg were assessed by Western blotting. In the supernatant of DMSO-treated PHHs, both enveloped and naked capsids were present, and the viral DNA was mainly present in the enveloped capsids (Fig. 3A). In contrast, JNJ-6379 reduced the presence of enveloped capsids in the supernatant and had no effect on the secretion of naked capsids (Fig. 3A). JNJ-6379 resulted in a potent and dose-dependent reduction of HBV DNA in the enveloped and naked capsids in the supernatant (Fig. 3A). There was no effect of JNJ-6379 on extracellular HBsAg levels (Fig. 3A). ETV also reduced the presence of HBV DNA in the enveloped and naked capsids in the medium, with no effect on extracellular HBsAg levels (Fig. 3A).
FIG 3.
Effect of JNJ-6379 or ETV on viral particles. HBV-infected PHHs were incubated with compound from day 5, and samples were harvested on day 12. Extracellular and intracellular HBV DNA (Southern blot), HBV core (Western blot), and HBsAg (Western blot) were assessed. DMSO, dimethyl sulfoxide; ETV, entecavir; HBsAg, hepatitis B surface antigen.
Regarding intracellular levels of core, HBV DNA, and HBsAg, treatment with JNJ-6379 resulted in a dose-dependent shift from T = 4 to T = 3 capsids, reduced intracellular capsid-associated HBV DNA, and had no effect on HBsAg (Fig. 3B). Thus, the predominant intracellular capsid form following JNJ-6379 exposure was naked nucleocapsids, with a dose-dependent reduction of HBV DNA.
Time-of-addition studies in HBV-infected PHHs.
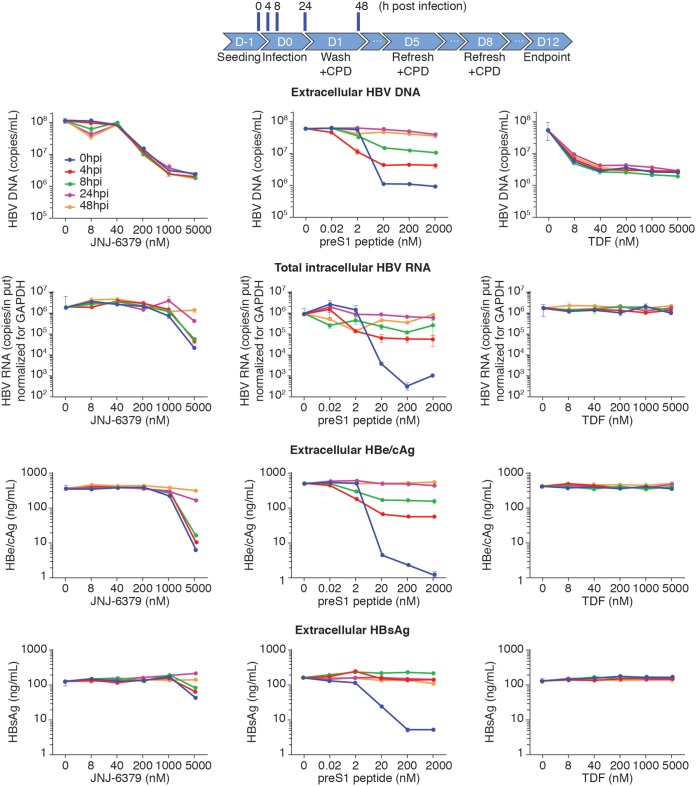
Detailed time-of-addition experiments were conducted to characterize the inhibitory effect of JNJ-6379 on early steps of the viral life cycle and the impact on de novo cccDNA formation. Two controls were used, the HBV preS1 peptide, which blocks HBV from binding to the Na+-taurocholate-cotransporting polypeptide (NTCP) receptor, and TDF. A range of concentrations of each compound was added to PHHs either with the viral inoculum (0 h) or at 4, 8, 24, or 48 h postinfection.
The addition of JNJ-6379 to PHHs at 0 h resulted in dose-related inhibition of extracellular HBV DNA, intracellular HBV RNA, extracellular HBe/cAg, and extracellular HBsAg (Fig. 4). Regardless of the time of addition of JNJ-6379 to PHHs postinfection, strong inhibition of extracellular HBV DNA was seen. For intracellular HBV RNA levels, JNJ-6379 resulted in strong inhibition when added at 4 and 8 h postinfection. Inhibition of intracellular HBV RNA levels was much reduced by the addition of JNJ-6379 at 24 h postinfection and was not observed when the compound was added at 48 h postinfection (Fig. 4). The impacts of JNJ-6379 addition postinfection on extracellular HBe/cAg levels were strong inhibition at 4 and 8 h and limited/no effect at 24 and 48 h (Fig. 4). For extracellular HBsAg levels, JNJ-6379’s inhibitory effect was limited when added at 4 or 8 h postinfection and was not observed when the compound was added at 24 or 48 h postinfection (Fig. 4).
FIG 4.
Time-of-addition studies in HBV-infected PHHs. Cryopreserved primary human hepatocytes (PHHs) were infected with HBV (derived from HepG2.2.15 cell culture supernatant). Compounds were added at 0, 4, 8, 24, or 48 h postinfection at the concentration ranges shown and replenished as indicated. Cells were treated until day 11 postinfection. Extracellular HBV DNA levels and intracellular HBV RNA levels were quantified by qPCR. The HBe/cAg and HBsAg levels in the cell culture supernatant were detected by AlphaLISA. The data represent mean values from at least 2 replicates. Error bars show standard deviations. CPD, compound; D, day; HBe/cAg, hepatitis B e antigen and hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen; hpi, hours postinfection; TDF, tenofovir disoproxil.
In keeping with its MOA, addition of the preS1 peptide at 0 h resulted in marked, dose-related inhibition of extracellular HBV DNA, intracellular HBV RNA, extracellular HBe/cAg, and extracellular HBsAg (Fig. 4). When preS1 peptide was added 4 and 8 h postinfection, the inhibitory effects were strong for extracellular HBV DNA and intracellular HBV RNA levels, moderate for extracellular HBe/cAg levels, and very limited for extracellular HBsAg levels (Fig. 4). Addition of preS1 peptide at 24 and 48 h postinfection had limited effects (extracellular HBV DNA and intracellular HBV RNA) or no effects (extracellular HBe/cAg and extracellular HBsAg) (Fig. 4).
TDF exhibited potent and dose-dependent reductions in extracellular HBV DNA levels regardless of when the compound was added postinfection (Fig. 4). However, TDF had a very limited effect on intracellular HBV RNA levels and no effect on extracellular HBe/cAg or extracellular HBsAg (Fig. 4).
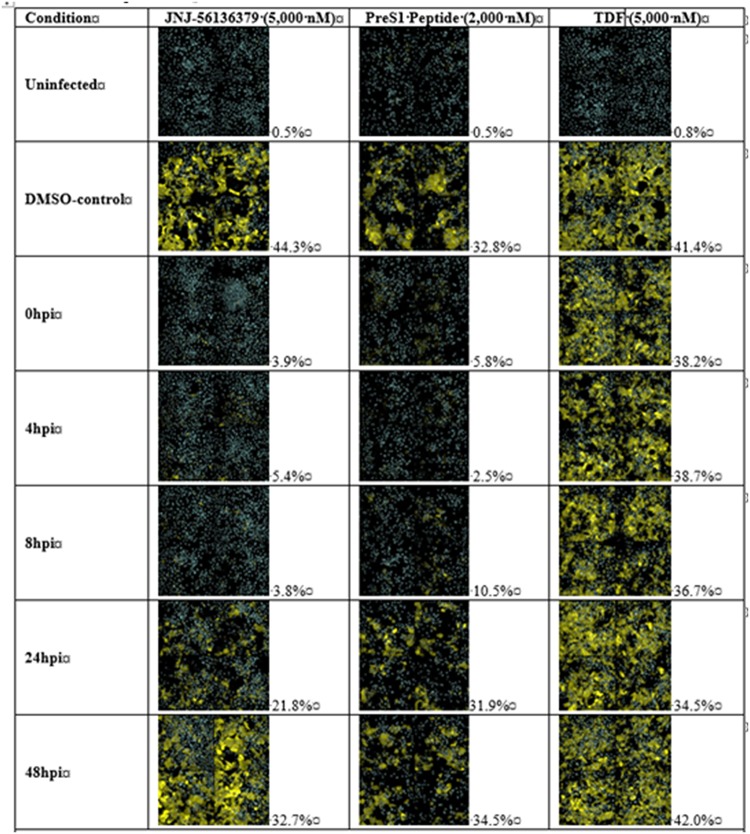
The effects of JNJ-6379 (5,000 nM), preS1 peptide (2,000 nM), and TDF (5,000 nM) were also evaluated at the single-cell level using an immunofluorescence stain against HBsAg. As expected, a minor background of HBsAg was detected in the noninfected cells due to the image analysis settings used and automated counting. With DMSO, the percentages of infected cells ranged from 32.8% to 44.3% (Fig. 5). For preS1 peptide-treated cells, the percentage of infected cells started to increase when the preS1 peptide was added 8 h postinfection and reached the level of the DMSO-treated control when preS1 peptide was added 24 or 48 h postinfection (Fig. 5). With JNJ-6379 addition, the percentages of infected cells only increased at 24 h and 48 h postinfection (Fig. 5), indicative of JNJ-6379 acting at a step after HBV’s binding to the NTCP receptor. TDF did not reduce the intracellular levels of HBsAg at any time point evaluated (Fig. 5).
FIG 5.
Time-of-addition studies in HBV-infected primary human hepatocytes (PHHs). Cryopreserved PHHs were infected with HBV. Compounds were added at 0, 4, 8, 24, or 48 h postinfection and replenished when culture medium was changed. Immunofluorescence analysis of intracellular HBsAg at the single-cell level was conducted on day 12. Automated image analysis was used to determine the number of infected cells. The mean percentage of intracellular HBsAg-positive cells is listed for each compound and time point (mean of 2 wells/condition and compound). CPD, compound; D, day; hpi, hours postinfection; TDF, tenofovir disoproxil.
Effects of JNJ-6379 and TDF on cccDNA formation.
We evaluated whether the reductions in intracellular HBV RNA, extracellular HBe/cAg, and extracellular HBsAg induced by JNJ-6379 when added at the same time as the viral inoculum were due to the inhibition of cccDNA formation. HBV-infected PHHs were treated with JNJ-6379, preS1 peptide, or TDF either at the same time as the viral inoculum (Fig. 6, top) or 5 days postinfection (Fig. 6, bottom). At the end of treatment, a Hirt DNA extraction was performed and the extract evaluated by Southern blotting (30).
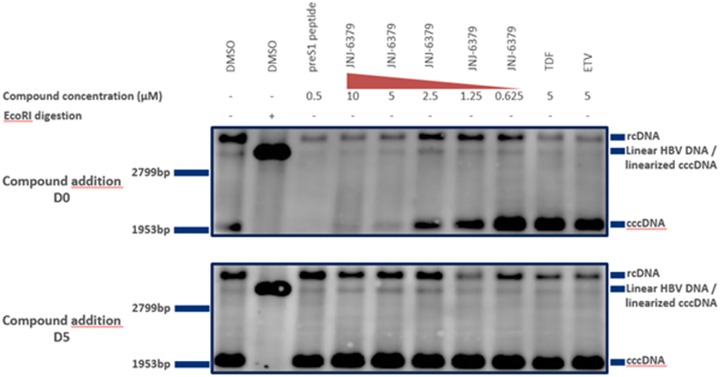
FIG 6.
HBV cccDNA Southern blotting. HBV-infected primary human hepatocytes (PHHs) were treated with compounds either together with the viral inoculum (day zero) (top) or on day 4 or 5 postinfection (bottom). Ten-microgram amounts of DNA extracted using the Hirt method (30) were subjected to agarose gel electrophoresis and Southern blotting to detect HBV cccDNA with an HBV-specific probe. HBV cccDNA contains a single EcoRI restriction site that can be used for linearization. cccDNA, covalently closed circular DNA; D, day; DMSO, dimethyl sulfoxide; ETV, entecavir; rcDNA, relaxed circular DNA; TDF, tenofovir disoproxil.
A cccDNA band and linear and rcDNA HBV DNA bands were seen for both experimental conditions in the DMSO-treated control (Fig. 6, 1st lane, top and bottom), indicative of infected cells with the establishment of cccDNA. Digestion of the Hirt extract with EcoRI in the DMSO-treated control resulted in linearization of cccDNA into a unit-length species migrating at a different height (Fig. 6, 2nd lane, top and bottom), confirming that the previous band was cccDNA.
The addition of preS1 peptide with the viral inoculum prevented the formation of cccDNA, as expected, since this peptide prevents viral entry (Fig. 6, 3rd lane, top), whereas no effect of this peptide on cccDNA was seen when it was added 5 days postinfection (Fig. 6, 3rd lane, bottom).
JNJ-6379 resulted in a dose-dependent reduction of cccDNA when added at the same time as the viral inoculum (Fig. 6, 4th to 8th lanes, top) and had no effect when added 5 days postinfection (Fig. 6, 4th to 8th lanes, bottom). These observations suggest that JNJ-6379 prevented cccDNA formation during the early steps of viral infection and was without effect on preformed cccDNA.
TDF and ETV had no effect on cccDNA under either experimental condition (Fig. 6, 9th and 10th lanes, top and bottom).
DISCUSSION
New CHB treatments are required to achieve treatments of finite duration that bring about high HBV functional cure rates. Targets for direct-acting antivirals may be limited, as the condensed HBV genome only encodes seven proteins (31). However, each of these proteins, including core, serves many roles in the HBV life cycle (7). For example, core is a building block of the capsid shell, and capsid formation is critical for the creation of infectious virions and cccDNA amplification (11). Core is also recruited onto viral chromatin (32, 33) and modulates host promoter regions (34). These multiple roles make core an interesting target for the development of new CHB therapies.
In the current MOA and antiviral studies with JNJ-6379, we used standard techniques and the appropriate control compounds that have been used to assess the properties of other CAMs, such as JNJ-632 (16, 21), JNJ-0440 (35), JNJ-827 (17), JNJ-890 (17), and NVR 3-778 (36). Our studies clearly demonstrated that JNJ-6379 exhibited antiviral properties in vitro that were specific to HBV, as this CAM had no antiviral effect on a wide range of other RNA and DNA viruses. Moreover, cytotoxicity studies in an HBV-replicating cell line, HBV-infected PHHs, and other human and animal cell lines confirmed that the antiviral properties of JNJ-6379 occurred at concentrations (EC50 of 54 nM in HepG2.117 cells) much lower than those resulting in cytotoxic effects (CC50s of >25 μM). Detailed MOA studies also showed that, as expected, JNJ-6379 has a different MOA than NAs, which is supported by the additive/synergistic antiviral effects when JNJ-6379 was combined with either TDF or ETV.
Our SEC and EM studies demonstrated that the primary MOA of JNJ-6379 classifies this compound as a CAM-N (formerly referred to as class I mechanism by Janssen), i.e., it induces the formation of morphologically normal (N) HBV capsids, similar to the action of the positive control AT130 (18, 19) and in contrast to the action of BAY41-4109, which is a CAM-A (formerly referred to as class II mechanism by Janssen) that induces the formation of aberrant (A) capsid structures (22, 23). Moreover, the fluorescence-quenching assay showed that JNJ-6379 shortened the lag phase (related to the kinetics of the rate-limiting nucleation step, i.e., the formation of the initial trimer of core protein dimers) and increased the slope of the fast phase (represents the elongation reaction, i.e., the addition of subsequent core protein dimers to the growing polymer) and the asymptotic (plateau) phase was reached earlier. These observations demonstrated that JNJ-6379 also increased the rate and extent of HBV capsid assembly in vitro.
Initially, the evaluation of HBV capsid formation in HepG2.117 cells indicated that JNJ-6379 shifted capsid formation from T = 4 to T = 3 capsids and reduced capsid-associated HBV DNA, confirming that this CAM modulates HBV nucleocapsid assembly in vitro. These observations were extended by evaluating the effect of JNJ-6379 treatment on viral particles in PHHs (particle gel assays). The supernatant of PHHs contained both enveloped and naked capsids and HBV DNA was present in both capsid types, although mainly in enveloped capsids. The addition of JNJ-6379 to HBV-infected PHHs did reduce the presence of enveloped capsids, but not that of naked capsids. Furthermore, the PHH experiments confirmed the dose-dependent shift from T = 4 to T = 3 capsids in response to JNJ-6379, with a dose-dependent decrease in intracellular capsid-associated HBV DNA and no intracellular accumulation of core and HBsAg. Collectively, these results confirm that JNJ-6379 modulates HBV capsid assembly and that naked capsids (i.e., no surface HBsAg) with no HBV DNA occur within the cell, as well as being secreted.
Our observations also indicate that JNJ-6379 inhibits the early steps of the HBV cycle and interferes with postentry processes prior to the formation of cccDNA. MOA studies in HBV-infected PHHs showed that JNJ-6379 inhibited HBV replication and reduced intracellular HBV RNA and secreted antigen levels, with inhibition of cccDNA formation, when added to the PHHs at the same time as the viral inoculum. In contrast, adding JNJ-6379 to PHHs when the HBV infection was established only reduced extracellular HBV DNA levels and had a minimal effect on the other markers evaluated. The two NAs, TDF and ETV, only reduced extracellular HBV DNA levels in these cultures, and neither NA had any inhibitory effects against HBV RNA and secreted antigens when added either at the time of infection or postinfection.
In the time-of-addition studies, the control preS1 peptide fully inhibited infection of the PHHs when added at time zero (addition of the viral inoculum), as demonstrated by the inhibition of HBV DNA, intracellular HBV RNA, antigen secretion, and cccDNA formation. These inhibitory effects of preS1 peptide were reduced when added postinfection, in keeping with its MOA, i.e., blocking HBV binding to the NTCP receptor (37). Inhibition of cccDNA formation and transcription by JNJ-6379 occurs at a postentry step, as the addition of this CAM was able to suppress intracellular HBV RNA and antigen production when added to PHHs at 0, 4, and 8 h postinfection, whereas these inhibitory effects were diminished or lost between 8 and 24 h postinfection. In contrast, the NA TDF had no effects on intracellular HBV RNA and antigen secretion at any time point evaluated. The consistent inhibition of extracellular HBV DNA by JNJ-6379 (as expected from the primary MOA of this CAM) and TDF regardless of time of addition confirms the effect of these compounds on the late step of the HBV life cycle. Thus, we demonstrated that JNJ-6379 has a dual MOA. In addition to an effect on late steps of the viral life cycle (inhibition of HBV replication), JNJ-6379 also inhibits early steps of the viral life cycle (cccDNA formation). Therefore, CAMs are differentiated from both entry inhibitors and NAs.
Our evaluations demonstrated that higher JNJ-6379 concentrations were required to inhibit cccDNA formation (e.g., the median EC50/EC90 values of 876 nM/4,019 nM in PHHs for intracellular HBV RNA inhibition, indicative of cccDNA formation) compared with the reduction of HBV DNA in HBV-infected PHHs (e.g., median EC50/EC90 values of 93 nM/378 nM), which may reflect subtle structural differences in the binding pocket when the core protein is in a dimer/dimer versus capsid state. The effect of JNJ-6739 on the early step in the HBV life cycle is likely mediated by binding to the preformed capsids. Indeed, the capsid stability assay in HBV-replicating HepG2.117 cells demonstrated that intracellular preformed mature capsids were damaged by JNJ-6379 (only a minimal effect was seen on immature capsids) and that this effect happened fast (within a 30-min incubation). Consequently, rcDNA might be released into the cytoplasm, where it cannot be converted to cccDNA.
Interest in CAMs as a potential CHB therapy has emerged recently with reports of various compounds in different chemical classes, such as phenylpropenamide derivatives (AT130) (18, 19), sulfamoylbenzamide derivatives (20), e.g., JNJ-632 (16, 21), JNJ-0440 (35), JNJ-827 (17), and AB-423 (38), heteroarylpyrimidines (e.g., BAY41-4109) (22, 23, 39), and NVR3-778 (36). Indeed, a high-throughput screen has been developed to rapidly identify potential CAMs (40). Importantly, several CAMs, such as NVR3-778, RO7049389, JNJ-0440, JNJ-6379, ABI-H0731, and AB-423, were well tolerated with favorable pharmacokinetics in healthy volunteers, and several of these compounds have showed antiviral properties in patients with CHB in early clinical trials (24, 41–46).
The present results on the MOA of JNJ-6379 reported herein are in keeping with and extend previously published in vitro data on other CAM-Ns, e.g., NVR3-778 (36), JNJ-632 (16, 21), JNJ-0440 (35), and AB-423 (38).
A phase 1 trial (ClinicalTrials registration no. NCT02662712) with JNJ-6739 has been conducted. The phase 1a part reported that single and multiple doses of JNJ-6379 were well tolerated in healthy adults, with dose-proportional pharmacokinetics (44). The 4-week phase 1b part in treatment-naive HBeAg-positive and -negative patients with CHB showed that all test doses of JNJ-6379 reduced serum HBV DNA, with a mean 2.70 log10 reduction from baseline with the 250-mg once-daily dose (highest tested dose) and a mean 1.43 log10 reduction from baseline in HBV RNA (24), which is expected from the MOA of JNJ-6739 in preventing encapsidation of HBV RNA. A phase 2 clinical trial (ClinicalTrials registration no. NCT03361956) is ongoing in HBeAg-positive and -negative CHB patients to evaluate the efficacy, safety, and pharmacokinetics of JNJ-6379 alone and in combination with an NA.
In summary, our studies demonstrated the MOA and antiviral properties of the novel and potent CAM, JNJ-6379, in several in vitro systems. Collectively, the data showed that JNJ-6379 has the primary MOA of a CAM-N of accelerating the rate and extent of HBV capsid assembly in vitro and forming empty, morphologically intact viral capsids and a secondary MOA of inhibiting de novo cccDNA formation. Therefore, JNJ-6379 has a dual MOA impacting early and late steps of the HBV life cycle, which is different from the action of NAs. JNJ-6739 is being further developed as a potential CHB treatment.
MATERIALS AND METHODS
Compounds.
JNJ-6379 was synthesized in-house with a purity of >99% (47). AT130, BAY41-4109 (WuXi AppTech, Shanghai, China), TDF (WuXi AppTech, Shanghai, China), and ETV (Sequoia Research Products, Pangbourne, UK) each had a purity of >95%.
SEC and EM studies.
The details of performing size exclusion chromatography (SEC) and electron microscopy (EM) studies have been described previously (16), and a brief summary is provided in the supplemental material.
Biochemical fluorescence quenching assay.
The modified recombinant HBV core protein Cp*150 (48) was made according to the method of Zlotnick et al. (49). Following desalting of Cp*150 proteins, the C-terminal residue of the Cp*150 protein was labeled with maleimidyl BODIPY-FL, and the unreacted dye was removed. Serial dilutions of test compounds were added to black view, flat-bottom plates, and labeled Cp*150 protein added to all wells. Full-assembly and no-assembly controls were incubated with NaCl and HEPES, respectively. The fluorescence signal was measured (extinction, 480 nm; emission, 540 nm) (FDSS6000 system; Hamamatsu) every minute for 5 min. Assembly was initiated in all compound-containing wells with NaCl, and the fluorescence signal was measured every 10 s for 30 min. Fluorescence data were plotted as a function of time.
Antiviral assay with HepG2.117 and HepG2.2.15 cells and cytotoxicity assay (HepG2 and other cell types).
Details of the antiviral and cytotoxicity (HepG2) assays have been described previously (16); see the supplemental material for a brief summary. Cytotoxicity was also assessed in several human cell lines (A549, HEL299, HeLa, Huh7, and MT4) and in animal cell lines MDCK and Vero. Cell viability was measured by an indicator dye (resazurin), ATP-based bioluminescence readout, and/or green fluorescent protein (GFP) reporter gene readout.
Antiviral assay with a panel of DNA and RNA viruses.
The antiviral properties of JNJ-6379 were assessed against five positive-sense single-stranded RNA viruses, four negative-sense single-stranded RNA viruses, and one DNA virus in cell lines supporting viral replication (Table S1 in the supplemental material). Positive controls for each virus were used to ensure the validity of the specific antiviral assays (Table S1). The antiviral properties of JNJ-6379 against human rhinovirus, Coxsackie virus, and influenza A and B viruses were assessed by a cytopathic-effect inhibition assay using ATPLite (Perkin Elmer, Waltham, USA), and the readout was based on the bioluminescence measurement of ATP in metabolically active cells. With respiratory syncytial virus, human parainfluenza virus, human immunodeficiency virus (HIV), Dengue virus, and cytomegalovirus, infectious-replication inhibition assays using recombinant virus harboring enhanced GFP (eGFP) reporter genes were used; eGFP was quantified by assessing fluorescence with a laser microscope. For hepatitis C virus (HCV), the HCV replicon assay was used with a luciferase reporter assay (SteadyLite plus) and measuring luminescence with a ViewLux reader. EC50 values were calculated, and the specificity of antiviral properties was checked by assessing cellular toxicity (CC50).
Detection of HBV capsids by Western blotting.
HepG2.117 cells were lysed, and cell lysates were diluted in 4× native polyacrylamide gel electrophoresis (PAGE) sample buffer and subjected to native PAGE, followed by protein transfer to a polyvinylidene difluoride membrane, according to the manufacturer’s protocol. Proteins were fixed on the membrane, rinsed, and blocked. Capsid particles were detected by incubation with a diluted polyclonal rabbit HBV core antibody overnight, followed by washing and incubation with a horseradish peroxidase-linked anti-rabbit antibody. Washed membranes were treated with SuperSignal West femto maximum sensitivity substrate according to the manufacturer’s recommendations (Thermo Fisher Scientific). The blots were imaged with an ImageQuant LAS 4000.
Synergy experiments (antiviral properties of JNJ-6379 and an NA).
The effect of JNJ-6379 combined with either TDF or ETV in HBV-expressing HepG2.2.15 cells was determined across 3 individual experiments. Combinations of JNJ-6379 and NA were added to the cells at the following concentration ranges: JNJ-6739, 0.24 to 1,000 nM; TDF, 0.24 to 250 nM; and ETV, 0.024 to 25 nM. The percentage of inhibition for each combination was calculated by averaging the values from 3 to 5 wells/experiment. The antiviral properties were assessed using the Bliss independence model, based on the algorithm of Prichard and Shipman (28), using MacSynergy II software. If the theoretical additive surface subtracted from the experimental surface was 0%, the combination was considered to be additive. Any peak above this plain indicated synergy, and depression below this plain was considered antagonism between the two compounds.
Antiviral studies in HBV-infected PHHs have been described by Berke et al. (16) (see the supplemental material for a brief summary), including quantification of extracellular HBV DNA, intracellular HBV RNA, extracellular antigens, and human albumin and immunofluorescence analysis of HBsAg.
Capsid stability assay with JNJ-6379.
HepG2.117 cells were cultured in supplemented Dulbecco’s modified Eagle medium (DMEM) without doxycycline. After 3 days, the culture medium was replaced with doxycycline-supplemented culture medium. On day 4, cells were treated with JNJ-6379 (1:5 serial dilution at 5 different concentrations) or ETV for 30 min in the presence of doxycycline. The supernatant was then removed, and cells were lysed. Cell lysates were split into two aliquots and incubated for 30 min at 37°C with or without DNase I and then inactivated by heat. Viral DNA was extracted using the QIAamp DNA microkit (Qiagen) according to the manufacturer’s protocol. HBV DNA was subjected to Southern blotting as previously described (16), with a minor change: the resolution on agarose gel was performed for 3 h (not 16 h). For each sample, the agarose gels were loaded with 2 μg DNA (quantified using a NanoDrop instrument).
Particle gel assay.
Cryopreserved PHHs (Life Technologies) were plated on day –1 (2 × 106 cells/well of 6-well collagen-coated plates [Biocoat]) in Universal Cryopreservation Recovery Medium (UCRM) and subsequently infected with HBV on day zero in supplemented DMEM culture medium (500 copies of genome equivalent/cell). On day 1, cells were supplied with fresh culture medium (after washing 3 times). On day 5, culture medium was exchanged with medium containing either DMSO or compound diluted in DMSO, and medium was replenished every 3 to 4 days. On day 12, the supernatant was collected, cell debris was removed, and cells were washed once and lysed. After 10 min at room temperature, cells were collected and centrifuged, and the cell lysate was stored. HBV particles were precipitated from the cell culture supernatants, and the resulting pellet was incubated overnight to dissolve the viral particles.
Cell lysates and dissolved virus particle samples were subjected to Southern blotting to detect HBV DNA according to the same method described above for the capsid stability assay. For Western blotting, intra- and extracellular HBV particles were resolved on a 1% agarose gel and transferred to a nitrocellulose membrane. After fixation and blocking, membranes were incubated with a polyclonal rabbit HBV core antibody (Dako) or an anti-HBs antibody (in-house; for HBsAg, the antibody used binds to an epitope localized in the small HBsAg and, thus, detects all forms of HBsAg, i.e., small, middle, and large, as well as all glycosylated forms), followed by an anti-rabbit horseradish peroxidase-linked antibody (Amersham/GE Healthcare) in blocking buffer for HBV core detection or a horseradish peroxidase-linked, species-specific anti-human IgG whole antibody (from sheep) (Amersham/GE Healthcare) for HBsAg. Washed membranes were treated with SuperSignal west femto maximum-sensitivity substrate according to the manufacturer’s instructions (Thermo Fisher Scientific) and imaged with an ImageQuant LAS 4000.
DNA extraction from HBV-infected PHHs by the Hirt method and Southern blotting details have been described previously (16), and a brief summary is provided in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jackie Phillipson from Zoetic Science (Macclesfield, UK), an Ashfield company, part of UDG Healthcare PLC, who provided medical writing support (all drafts, assembling tables and figures, collating author comments, grammatical editing, and referencing); this support was funded by Janssen Pharmaceuticals.
J. M. Berke, P. Dehertogh, K. Vergauwen, W. Mostmans, and F. Pauwels are employees of Janssen Pharmaceuticals and may be Johnson & Johnson stockholders. K. Vandyck and P. Raboisson were employed by Janssen Research and Development at the time of the studies and may be Johnson & Johnson stockholders.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Polaris Observatory Collaborators. 2018. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 18 July 2019. Hepatitis B. www.who.int/mediacentre/factsheets/fs204/en/index.html. Accessed 27 August 2019.
- 3.Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, Brown RS Jr, Bzowej NH, Wong JB. 2018. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 67:1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Association for the Study of the Liver. 2017. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 67:370–398. doi: 10.1016/j.jhep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Brahmania M, Feld J, Arif A, Janssen HL. 2016. New therapeutic agents for chronic hepatitis B. Lancet Infect Dis 16:e10–e21. doi: 10.1016/S1473-3099(15)00436-3. [DOI] [PubMed] [Google Scholar]
- 6.Lok AS, Zoulim F, Dusheiko G, Ghany MG. 2017. Hepatitis B cure: from discovery to regulatory approval. Hepatology 66:1296–1313. doi: 10.1002/hep.29323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diab A, Foca A, Zoulim F, Durantel D, Andrisani O. 2018. The diverse functions of the hepatitis B core/capsid protein (HBC) in the viral life cycle; implications for the development of HBC-targeting antivirals. Antiviral Res 149:211–220. doi: 10.1016/j.antiviral.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruffaz M, Testoni B, Luangsay S, Fusil F, Ait-Goughoulte M, Isorce N, Petit MA, Klumpp K, Ma H, Javanbakht H, Loic Cosset F, Zoulim F, Durantel D. 2013. Hepatitis B core protein is a key and very early negative regulator of IFN response. J Hepatol 58(Suppl 1):S155–S156. doi: 10.1016/S0168-8278(13)60380-3. [DOI] [Google Scholar]
- 10.Zlotnick A. 2003. Are weak protein-protein interactions the general rule in capsid assembly? Virology 315:269–274. doi: 10.1016/s0042-6822(03)00586-5. [DOI] [PubMed] [Google Scholar]
- 11.Kock J, Rosler C, Zhang JJ, Blum HE, Nassal M, Thoma C. 2010. Generation of covalently closed circular DNA of hepatitis B viruses via intracellular recycling is regulated in a virus specific manner. PLoS Pathog 6:e1001082. doi: 10.1371/journal.ppat.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Shen T, Huang X, Kumar GR, Chen X, Zeng Z, Zhang R, Chen R, Li T, Zhang T, Yuan Q, Li PC, Huang Q, Colonno R, Jia J, Hou J, McCrae MA, Gao Z, Ren H, Xia N, Zhuang H, Lu F. 2016. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 65:700–710. doi: 10.1016/j.jhep.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 13.Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. 2008. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J Virol 82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XY, Wei ZM, Wu GY, Wang JH, Zhang YJ, Li J, Zhang HH, Xie XW, Wang X, Wang ZH, Wei L, Wang Y, Chen HS. 2012. In vitro inhibition of HBV replication by a novel compound, GLS4, and its efficacy against adefovir-dipivoxil-resistant HBV mutations. Antivir Ther 17:793–803. doi: 10.3851/IMP2152. [DOI] [PubMed] [Google Scholar]
- 15.Belloni L, Li L, Palumbo SR, Chirapu SR, Calvo L, Finn MG, Lopatin U, Zlotnick A, Levrero M. 2014. HAPs hepatitis B virus (HBV) capsid inhibitors prevent HBc interaction with the viral minichromosome and selected host cemm genes to inhibits transcription and affect cccDNA stability. Dig Liver Dis 46:e9. doi: 10.1016/j.dld.2014.01.024.24629821 [DOI] [Google Scholar]
- 16.Berke JM, Dehertogh P, Vergauwen K, Van Damme E, Mostmans W, Vandyck K, Pauwels F. 2017. Capsid assembly modulators have a dual mechanism of action in primary human hepatocytes infected with hepatitis B virus. Antimicrob Agents Chemother 61:e00560-17. doi: 10.1128/AAC.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahlali T, Berke JM, Vergauwen K, Foca A, Vandyck K, Pauwels F, Zoulim F, Durantel D. 2018. Novel potent capsid assembly modulators regulate multiple steps of the hepatitis B virus life cycle. Antimicrob Agents Chemother 62:e00835-18. doi: 10.1128/AAC.00835-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perni RB, Conway SC, Ladner SK, Zaifert K, Otto MJ, King RW. 2000. Phenylpropenamide derivatives as inhibitors of hepatitis B virus replication. Bioorg Med Chem Lett 10:2687–2690. doi: 10.1016/s0960-894x(00)00544-8. [DOI] [PubMed] [Google Scholar]
- 19.Katen SP, Chirapu SR, Finn MG, Zlotnick A. 2010. Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, Chang J, Xu X, Block TM, Guo JT. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87:6931–6942. doi: 10.1128/JVI.00582-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandyck K, Rombouts G, Stoops B, Tahri A, Vos A, Verschueren W, Wu Y, Yang J, Hou F, Huang B, Vergauwen K, Dehertogh P, Berke JM, Raboisson P. 2018. Synthesis and evaluation of N-phenyl-3-sulfamoyl-benzamide derivatives as capsid assembly modulators inhibiting hepatitis B birus (HBV). J Med Chem 61:6247–6260. doi: 10.1021/acs.jmedchem.8b00654. [DOI] [PubMed] [Google Scholar]
- 22.Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, Graef E, Koletzki D, Masantschek RN, Reimann A, Jaeger R, Gross R, Beckermann B, Schlemmer KH, Haebich D, Rubsamen-Waigmann H. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- 23.Stray SJ, Zlotnick A. 2006. BAY 41-4109 has multiple effects on hepatitis B virus capsid assembly. J Mol Recognit 19:542–548. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- 24.Zoulim F, Yogaratnam JZ, Vandenbossche JJ, Moscalu I, Strienu-Cercel A, Lenz O, Bourgeois S, Buti M, Pascasio JM, Sarrazin C, Vanwolleghem T, Vistuer C, Blatt L, Fry J. 2018. Safety, pharmacokinetics and antiviral activity of a novel capsid assembly modulator JNJ-56136379 in patient with chronic hepatitis B patients. Hepatology 68(Suppl 1):47A. [Google Scholar]
- 25.Stray SJ, Johnson JM, Kopek BG, Zlotnick A. 2006. An in vitro fluorescence screen to identify antivirals that disrupt hepatitis B virus capsid assembly. Nat Biotechnol 24:358–362. doi: 10.1038/nbt1187. [DOI] [PubMed] [Google Scholar]
- 26.Berke JM, Verbinnen T, Tan Y, Dehertogh P, Vergauwen K, Vandyck K, Lenz O. 2017. The HBV capsid assembly modulator JNJ-379 is a potent inhibitor of viral replication across full length genotype A-H clinical isolates in vitro. Hepatology 66(Suppl 1):503A. [Google Scholar]
- 27.Verbinnen T, Berke JM, Tan Y, Wang G, Vergauwen K, Dehertogh P, Vandyck K, Lenz O. 2018. Anti-HBV activity of capsid assembly modulators JNJ-56136397 (JNJ-6379) and BAY41-4109 on an extensive panel of clinical isolates and HBV core mutants, poster 18. Int HBV Meet, Taormina, Italy, October 2018.
- 28.Prichard MN, Shipman C Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antiviral Res 14:181–205. doi: 10.1016/0166-3542(90)90001-n. [DOI] [PubMed] [Google Scholar]
- 29.Hu J, Liu K. 2017. Complete and incomplete hepatitis B virus particles: formation, function, and application. Viruses 9:56. doi: 10.3390/v9030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 31.Li S, Wang Z, Li Y, Ding G. 2017. Adaptive evolution of proteins in hepatitis B virus during divergence of genotypes. Sci Rep 7:1990. doi: 10.1038/s41598-017-02012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. 2001. Structural organization of the hepatitis B virus minichromosome. J Mol Biol 307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 33.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y, Kang W, Lei X, Li Y, Xiang A, Liu Y, Zhao J, Zhang J, Yan Z. 2012. Hepatitis B viral core protein disrupts human host gene expression by binding to promoter regions. BMC Genomics 13:563. doi: 10.1186/1471-2164-13-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berke JM, Dehertogh P, Vergauwen K, Verbinnen T, Vendeville S, Raboisson P, Pauwels F. 2018. In vitro antiviral activity and mode of action of JNJ-64530440, a novel potent hepatitis B virus capsid assembly modulator in clinical development. Hepatology 68(Suppl 1):239A. [Google Scholar]
- 36.Lam AM, Espiritu C, Vogel R, Ren S, Lau V, Kelly M, Kuduk SD, Hartman GD, Flores OA, Klumpp K. 2018. Preclinical characterization of NVR 3-778, a first-in-class capsid assembly modulator against hepatitis B virus. Antimicrob Agents Chemother 63:e01734-18. doi: 10.1128/AAC.01734-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye X, Zhou M, He Y, Wan Y, Bai W, Tao S, Ren Y, Zhang X, Xu J, Liu J, Zhang J, Hu K, Xie Y. 2016. Efficient inhibition of hepatitis B virus infection by a preS1-binding peptide. Sci Rep 6:29391. doi: 10.1038/srep29391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mani N, Cole AG, Phelps JR, Ardzinski A, Cobarrubias KD, Cuconati A, Dorsey BD, Evangelista E, Fan K, Guo F, Guo H, Guo JT, Harasym TO, Kadhim S, Kultgen SG, Lee ACH, Li AHL, Long Q, Majeski SA, Mao R, McClintock KD, Reid SP, Rijnbrand R, Snead NM, Micolochick Steuer HM, Stever K, Tang S, Wang X, Zhao Q, Sofia MJ. 2018. Preclinical profile of AB-423, an inhibitor of hepatitis B virus pregenomic RNA encapsidation. Antimicrob Agents Chemother 62:e00082-18. doi: 10.1128/AAC.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. 2005. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A 102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei Y, Wang C, Ben H, Wang L, Ma Y, Ma Q, Xiang Y, Zhang L, Liu G. 2019. Discovery of new hepatitis B virus capsid assembly modulators by an optimal high-throughput cell-based assay. ACS Infect Dis 5:778–787. doi: 10.1021/acsinfecdis.9b00030. [DOI] [PubMed] [Google Scholar]
- 41.Yuen MF, Gane EJ, Kim DJ, Weilert F, Yuen Chan HL, Lalezari J, Hwang SG, Nguyen T, Flores O, Hartman G, Liaw S, Lenz O, Kakuda TN, Talloen W, Schwabe C, Klumpp K, Brown N. 2019. Antiviral activity, safety, and pharmacokinetics of capsid assembly modulator NVR 3-778 in patients with chronic HBV infection. Gastroenterology 156:1392–1403. doi: 10.1053/j.gastro.2018.12.023. [DOI] [PubMed] [Google Scholar]
- 42.Gane E, Liu A, Yuen M-F, Schwabe C, Bo Q, Das S, Gao L, Zhou X, Wang Y, Coakley E, Jin Y. 2018. RO7049389, a core protein allosteric modulator, demonstrates robust anti-HBV activity in chronic hepatitis B patients and is safe and well tolerated. J Hepatol 68(Suppl 1):S101. doi: 10.1016/S0168-8278(18)30422-7. [DOI] [Google Scholar]
- 43.Kakuda TN, Yogaratnam JZ, Westland C, Gane EJ, Schwabe C, Patel M, Talloen W, Lenz O, van Remoortere P. 2019. JNJ-64530440, a novel capsid assembly modulator: single- and multiple-ascending dose safety, tolerability and pharmacokinetics in healthy volunteers. J Hepatol 70(Suppl 1):e469. doi: 10.1016/S0618-8278(19)30925-9. [DOI] [PubMed] [Google Scholar]
- 44.Vandenbossche J, Jessner W, van den Boer M, Biewenga J, Berke JM, Talloen W, De Zwart L, Snoeys J, Yogaratnam J. 2019. Pharmacokinetics, safety and tolerability of JNJ-56136379, a novel hepatitis B virus capsid assembly modulator, in healthy subjects. Adv Ther 36:2450–2462. doi: 10.1007/s12325-019-01017-1. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, Lalezari J, Nguyen T, Bae H, Schiff ER, Fung S, Yuen M-F, Hassanein R, Hann H-W, Elkhashab M, Dieterich D, Sulkowski M, Kwo P, Nahass R, Agarwal K, Ramji A, Park J, Ravendhran N, Chan S, Weilert F, Han S-H, Ayoub W, Gane E, Jacobson I, Bennett M, Huang Q, Yan R, Huey V, Ruby E, Liaw S, Colonno R, Lopatin U. 2019. Interim safety and efficacy results of the ABI-H0731 phase 2a program exploring the combination of ABI-H0731 with Nuc therapy in treatment-naïve and treatment-supressed chronic hepatitis B patients. J Hepatol 70(Suppl):e130. doi: 10.1016/S0618-8278(19)30230-0. [DOI] [Google Scholar]
- 46.Eley T, Caamano S, Denning J, Sims K, Larouche R, Symonds W, Mendez P. 2017. Single dose safety, tolerability, and pharmacokinetics of AB-423 in healthy volunteers from the ongoing single and multiple ascending dose study AB-423-001. Hepatology 66(Suppl 1):490A. [Google Scholar]
- 47.Vandyck K, Haché G, Geerwin YP, Last SJ, Gowan DC, Rombouts G, Verschueren WG, Raboisson PJ-MB. 20 November 2014. Sulphamoylpyrrolamide derivative and the use thereof as medicaments for the treatment of the treatment of hepatitis. International patent WO/2014/184350.
- 48.Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. 1997. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc Natl Acad Sci U S A 94:9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zlotnick A, Cheng N, Conway JF, Booy FP, Steven AC, Stahl SJ, Wingfield PT. 1996. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry 35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.