Chromosomal resistance islands containing the methicillin resistance gene mecD (McRImecD) have been reported in Macrococcus caseolyticus. Here, we identified novel macrolide resistance genes in Macrococcus canis on similar elements, called McRImsr. These elements were also integrated into the 3′ end of the 30S ribosomal protein S9 gene (rpsI), delimited by characteristic attachment (att) sites, and carried a related site-specific integrase gene (int) at the 5′ end.
KEYWORDS: McRImsr, antibiotic resistance, chromosomal resistance island, erythromycin esterase, macrolides, mef(D)-msr(F), msr(H)
ABSTRACT
Chromosomal resistance islands containing the methicillin resistance gene mecD (McRImecD) have been reported in Macrococcus caseolyticus. Here, we identified novel macrolide resistance genes in Macrococcus canis on similar elements, called McRImsr. These elements were also integrated into the 3′ end of the 30S ribosomal protein S9 gene (rpsI), delimited by characteristic attachment (att) sites, and carried a related site-specific integrase gene (int) at the 5′ end. They carried novel macrolide resistance genes belonging to the msr family of ABC subfamily F (ABC-F)-type ribosomal protection protein [msr(F) and msr(H)] and the macrolide efflux mef family [mef(D)]. Highly related mef(D)-msr(F) fragments were found on diverse McRImsr elements in M. canis, M. caseolyticus, and Staphylococcus aureus. Another McRImsr-like element identified in an M. canis strain lacked the classical att site at the 3′ end and carried the msr(H) gene but no neighboring mef gene. The expression of the novel resistance genes in S. aureus resulted in a low-to-moderate increase in the MIC of erythromycin but not streptogramin B. In the mef(D)-msr(F) operon, the msr(F) gene was shown to be the crucial determinant for macrolide resistance. The detection of circular forms of McRImsr and the mef(D)-msr(F) fragment suggested mobility of both the island and the resistance gene subunit. The discovery of McRImsr in different Macrococcus species and S. aureus indicates that these islands have a potential for dissemination of antibiotic resistance within the Staphylococcaceae family.
INTRODUCTION
The genus Macrococcus was first described in 1998 as a group of bacteria closely related to Staphylococcus, which currently includes 8 species already recognized by the nomenclature and 3 novel species not yet recognized (1–4). Macrococcus species are mainly animal commensals with low pathogenic potential, even though isolates from human clinical samples have been reported (4). Of concern is rather the fact that Macrococcus species can carry antibiotic resistance genes, including the alternative methicillin resistance genes mecB and mecD, on mobile genetic elements (5–7). The recent detection of highly similar mecB-carrying plasmids in both S. aureus and M. canis suggests that horizontal gene transfer may occur between Macrococcus and Staphylococcus species (8, 9). The chromosomal M. caseolyticus resistance island containing mecD (McRImecD) also has the potential to be site-specifically integrated into Staphylococcus chromosomes (10). Other clinically relevant resistance genes often present in Staphylococcus species were also detected in Macrococcus species, including the macrolide resistance genes erm(B) and erm(C) (11–13).
Three main mechanisms leading to macrolide resistance in Gram-positive bacteria are currently known, as follows: (i) inhibition of drug binding to the ribosome by target modification or protection, (ii) active efflux, and (iii) enzymatic inactivation (14). The erythromycin ribosome methylation (erm) genes are responsible for adenine methylation at position 2058 (A2058) in the 23S rRNA, which prevents drug binding and confers cross-resistance to macrolides, lincosamides, and streptogramin B (MLSB). Antibiotic resistance ATP-binding cassette subfamily F (ARE ABC-F) proteins protect the ribosomal function by actively displacing antibiotics from their target sites (15, 16). ARE ABC-F genes specifically responsible for macrolide resistance include the plasmid-carried staphylococcal msr(A) gene (17), the intrinsic chromosomal msr(C) gene in Enterococcus faecium (18), as well as msr(D) (formerly called mel) in streptococci (19–21) and msr(E) in Gram-negative bacteria (22). In streptococci, the msr(D) gene is found together with the macrolide efflux (mef) gene on an operon located on accessory chromosomal elements (23–25). In contrast to msr(A) and msr(C), msr(D) confers lower levels of resistance to 14- and 15-membered macrolides but no resistance to streptogramin B (19, 26). The cotranscribed mef(A) gene [or highly related variants also called mef(E), mef(I), or mef(G); (Fig. 1A)] encodes a major facilitator superfamily (MFS) membrane transporter involved in macrolide efflux (20, 21, 27). Other mef genes, mef(B) and mef(C), were not associated with an msr gene and were only found in Gram-negative bacteria. Macrolide antibiotics can also be inactivated enzymatically by macrolide phosphotransferases (mph) and macrolide esterases (ere). Several different mph genes have been reported, with mph(C) commonly found in Staphylococcus species often on plasmids carrying msr(A) (28). Of the four ere genes mainly found in Gram-negative bacteria, ere(A) and ere(B) genes were detected in some Staphylococcus species in rare cases (29, 30).
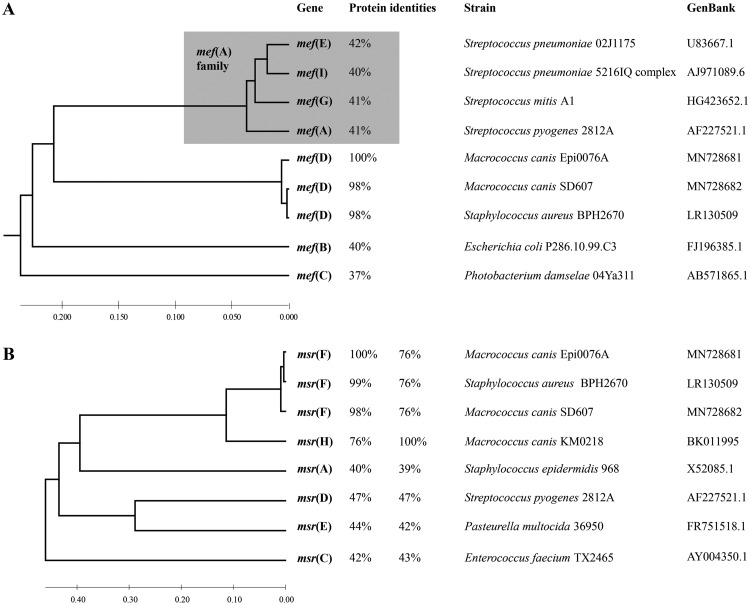
FIG 1.
Phylogenetic trees of macrolide resistance genes belonging to the mef and msr families. Evolutionary analysis was performed for nucleotide sequences using the unweighted pair group method using average linkages (UPGMA) method in MEGA7 (56). The percentages of the deduced amino acid identities between the novel gene and the genes recognized by the MLS nomenclature (https://faculty.washington.edu/marilynr/) were determined by alignment with Clustal OMEGA (https://www.ebi.ac.uk/Tools/msa/clustalo/). (A) Comparison of mef macrolide efflux genes encoding major facilitator superfamily (MFS) membrane transporters. Note that mef(E), mef(I), mef(G), and mef(A) (gray shading) shared more than 80% aa identities with each other (87 to 94% for the selected representatives), and are all assigned to the mef(A) class by the MLS nomenclature. (B) Comparison of msr genes encoding ABC-F-type ribosomal protection proteins. The amino acid identities between the recognized genes by the MLS nomenclature and the two novel genes msr(F) and msr(H) are indicated.
In a previous study, two M. canis strains, Epi0076A and SD607, isolated from dogs were found to be resistant to macrolides but contained none of the known resistance genes (12), prompting us to search for the underlying resistance mechanism. We used whole-genome sequencing for the identification of a new mef-msr operon as well as recombinant gene expression in S. aureus for proof of functionality. In addition, comparative sequence analysis by BLAST search allowed the identification of the same mef-msr operon in M. caseolyticus and S. aureus strains deposited in the NCBI GenBank database, as well as another novel msr gene integrated similarly into the chromosome of the erm(B)-carrying M. canis strain KM0218 obtained from a dog (9, 12).
RESULTS
New mef(D)-msr(F) genes in Macrococcus canis.
Both M. canis strains, Epi0076A and SD607 (12), showed increased MICs of erythromycin (4 μg/ml and 16 μg/ml, respectively) compared to the M. canis type strain KM45013 (0.25 μg/ml) (Table 1). The MICs of clindamycin were identical in all three strains (0.25 μg/ml).
TABLE 1.
Antimicrobial susceptibilities of Staphylococcus aureus and Macrococcus canis to erythromycin and pristinamycin IA, as determined by broth microdilution
| Strain/vector | Reference(s) or source | Characteristicsa | MIC (μg/ml)b
|
||
|---|---|---|---|---|---|
| ERY | PIA | iPIAc | |||
| S. aureus | |||||
| RN4220 | 44 | NCTC8325-4 derivative, antibiotic susceptible | 0.25 | 8 | NA |
| RN4220/pTSSCm | 36 | Plasmid with tet(L) | 0.25 | 4 | NA |
| RN4220/pTSSCm-Pcap | 36 | Plasmid with tet(L) | 0.25 | ND | ND |
| RN4220/pTSSCm::mef(D)-msr(F) | This study | Plasmid with mef(D)-msr(F) genes, including upstream sequence (493 bp), tet(L) | 16 | 8 | 8 |
| RN4220/pTSSCm::mef(D) | This study | Plasmid with mef(D), including upstream sequence (493 bp), tet(L) | 1 | 8 | NA |
| RN4220/pTSSCm::msr(F) | This study | Plasmid with msr(F), including 217 bp upstream sequence [3′ end of mef(D) and intergenic region], tet(L) | 0.5 | 4 | NA |
| RN4220/pTSSCm::Δmef(D)-msr(F) | This study | Plasmid with msr(F) fused to upstream sequence of mef(D) (493 bp), tet(L) | 32 | 8 | 8 |
| RN4220/pTSSCm::msr(H) | This study | Plasmid with msr(H), including upstream sequence (698 bp), tet(L) | 2 | 8 | 4 |
| RN4220/pTSSCm::ere-like | This study | Plasmid with ere-like, including upstream sequence containing a transcriptional regulator (tr) (710 bp), tet(L) | 0.25 | ND | ND |
| RN4220/pTSSCm::Δtr-ere-like | This study | Plasmid with ere-like, including tr-deleted upstream sequence (345 bp), tet(L) | 0.25 | ND | ND |
| RN4220/pTSSCm-Pcap::ere-like | This study | Plasmid with ere-like gene controlled by promoter Pcap, tet(L) | 0.25 | ND | ND |
| M. canis | |||||
| KM45013T | 3, 43 | mecB, blaZm | 0.25 | 2 | NA |
| Epi0076A | 9, 12 | mef(D), msr(F), mecB | 4 | 2 | 2 |
| SD0607 | 12 | mef(D), msr(F), ere-like | 16 | 2 | 2 |
| KM0218 | 9, 12 | msr(H), mecB, blaZm, aac(6′)-aph(2ʺ), sat4, aph(3′)-IIIa, erm(B), ant(6)-Ia, tet(S) | >64 | >64 | ND |
Antibiotic resistance genes and their functions are as follows: mef(D), macrolide efflux protein; msr(F) and msr(H), macrolide ABC-F ribosomal protection protein; ere-like, putative erythromycin esterase; tet(L), tetracycline efflux protein; tet(S), ribosome protection tetracycline resistance gene; mecB, penicillin-binding protein 2a (PBP2a) with low affinity for β-lactams; blaZm, β-lactamase; aac(6′)-aph(2ʺ), gentamicin and kanamycin acetyltransferase and phosphotransferase; sat4, streptothricin acetyltransferase; erm(B), macrolide, lincosamide, and streptogramin B 23S rRNA methylase gene; ant(6)-Ia, streptomycin nucleotidyltransferase gene; aph(3′)-III, kanamycin phosphotransferase gene.
ERY, erythromycin; PIA, pristinamycin IA; iPIA, inducible PIA; ND, not determined; NA, not applicable.
iPIA, inducible resistance to pristinamycin IA was measured in the presence of 1 μg/ml erythromycin, except for RN4220/pTSSCm::msr(H), for which the erythromycin concentration was 0.25 μg/ml.
The genome sequences of Epi0076A and SD607 analyzed using TBLASTN with protein query of known macrolide resistance determinants listed in the MLS nomenclature (https://faculty.washington.edu/marilynr/) revealed the presence of putative new mef and msr genes. In both strains, the putative mef gene was situated directly upstream of the msr gene on 3.6-kb DNA fragments, which shared 98% nucleotide (nt) identity. The new putative mef gene was only distantly related to other mef family members, with the deduced amino acid (aa) sequence sharing less than 43% identity to all other Mef proteins (Fig. 1A). It represented a new mef gene, named mef(D), according to the MLS nomenclature that defines a new gene if the aa sequence shares less than 80% identity to any previously characterized MLS genes (31). The Mef(D) protein possessed the major facilitator superfamily (MFS) profile (PROSITE entry PS50850) that covers the 12 putative transmembrane helices typically found in MFS efflux pumps. A MefA-like domain (Conserved Domain Database [CDD] entry cd06173) suggested that this new Mef protein could be involved in macrolide efflux. The mef(D) genes of Epi0076A and SD607 differed from each other by a 6-bp deletion in the mef gene of SD607, generating 399-aa and 397-aa proteins with 98% identities. Downstream of mef(D), Epi0076A and SD607 encoded a similar new 486-aa Msr protein that contained the features of ARE ABC-F family proteins with two nucleotide-binding domains separated by a 100-aa linker (PROSITE entry PS50893). This new putative msr gene shared 47% aa identity to the closest related msr(D) of streptococci and was named msr(F), following the MLS nomenclature (Fig. 1B).
The mef(D)-msr(F) genes were organized in tandem similar to mef(A)-msr(D) genes in streptococci with a short intergenic region of 111 bp in Epi0076A and 112 bp in SD607. Both mef(D) genes contained identical upstream sequences possibly involved in controlling mef(D)-msr(F) expression. A promoter sequence (−35 box [5′-TTGACT] and −10 box [5′-GTTTATAAT]) was identified 35 bp and 15 bp upstream of the putative transcription start site represented by a guanine located 414 bp upstream of mef(D). The 414-bp region upstream of mef(D)-msr(F) contained imperfect inverted repeats capable of folding into stem-loops, two ribosomal binding sites (RBS), a coding sequence for a small leader peptide [Mef(D)L, MTHAMKLRF], and a rho-independent terminator sequence (see Fig. S1 in the supplemental material). Similar features were observed in the regulatory 5′ region upstream of mef(A)-msr(D) of streptococci and erm(K) of Bacillus licheniformis and were shown to be involved in transcriptional attenuation of the antibiotic resistance genes (32–34). Even though the overall sequence identity of the putative cotranscribed 5′ region upstream of mef(D)-msr(F) (414 bp) to those of mef(A)-msr(D) (327 bp; 49% nt identity) and erm(K) (357 bp; 56% nt identity) was low, the structural similarity suggests possible transcriptional attenuation control of mef(D)-msr(F).
The association of mef(D) and msr(F) from M. canis Epi0076A with antimicrobial resistance was analyzed by cloning the genes with their putative regulatory DNA region into the promoterless plasmid pTSSCm and subsequent expression in S. aureus RN4220. Thus, S. aureus RN4220 was transformed with pTSSCm carrying both mef(D) and msr(F) [plasmid pTSCCm::mef(D)-msr(F)], as well as with pTSSCm containing either mef(D) [pTSSCm::mef(D)] or msr(F) [pTSSCm::msr(F)] alone (see Fig. 2 for plasmid constructs). Susceptibility testing showed that RN4220 containing pTSCCm::mef(D)-msr(F) exhibited a 64-fold increased erythromycin MIC (16 μg/ml) compared to those of the parental strain and RN4220 containing only the empty vector (0.25 μg/ml) (Table 1). RN4220 containing pTSSCm::mef(D) exhibited only a modestly increased (4-fold) erythromycin MIC value of 1 μg/ml. S. aureus RN4220 containing pTSSCm::msr(F) did not exhibit a clear increase in phenotype, with an MIC for erythromycin of 0.5 μg/ml. Of note, the cloned region preceding msr(F) consisted of the 111-bp intergenic region between mef(D) and msr(F) and the 106-bp 3′ end of mef(D). A promoter search in the proximate upstream region of msr(F) predicted a possible −10 box (5′-TTTTATTAT) and a −35 box (5′-TAGGAG) with a low probability score, suggesting that msr(F) is also under the control of the native regulator of the mef(D), with the two genes forming a mef(D)-msr(F) operon. The mef(D) gene was therefore deleted from plasmid pTSCCm::mef(D)-msr(F) (Fig. 2). The resulting plasmid, pTSCCm::Δmef(D)-msr(F), conferred a high MIC value of 32 μg/ml when inserted into RN4220 (Table 1), indicating that DNA upstream of mef(D) is necessary for msr(F) expression and that msr(F) is the predominant gene of the mef(D)-msr(F) operon causing macrolide resistance. Resistance to the streptogramin B pristinamycin IA was not observed in S. aureus strains containing any of the mef(D)-msr(F)-expressing plasmids nor in M. canis strains carrying mef(D)-msr(F) (Table 1). The MIC for pristinamycin IA also remained unchanged in the presence of 1 μg/ml erythromycin used for the induction of gene expression.
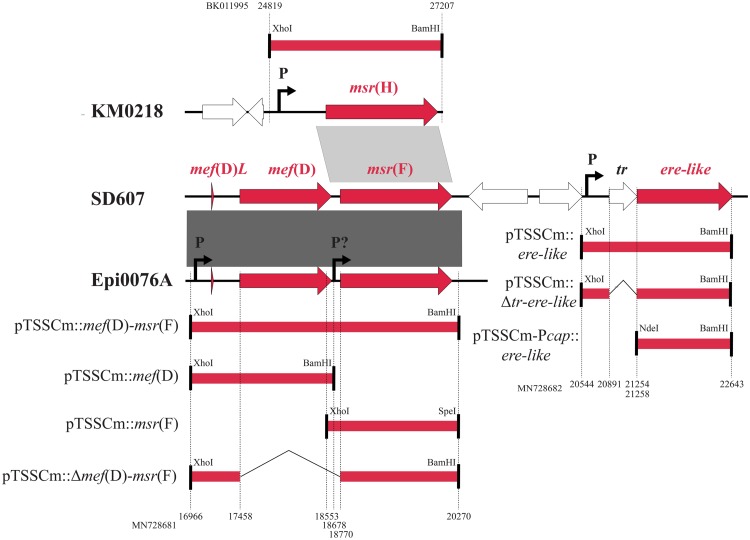
FIG 2.
Cloning of msr(H), mef(D)-msr(F), and ere-like DNA fragments of Macrococcus canis strains KM0218, Epi0076A, and SD607 into the E. coli-S. aureus shuttle vector pTSSCm or pTSSCm-Pcap. The fragments are represented by thick lines below/above the gene structure of the strain used for PCR amplification (see Table S1 for PCR conditions). Flanking restriction sites are indicated, and nucleotide positions are given for the corresponding GenBank accession numbers, BK011995 for KM0218, MN728682 for SD607, and MN728681 for Epi0076A. Gray areas between the gene structures indicate regions with nucleotide sequence identities of 81% (light gray) and 98% (dark gray). The positions of promoter (P) or possible promoter (P?) are indicated.
Diverse structures of McRImsr macrolide resistance islands.
In M. canis strains Epi0076A and SD607, the mef(D)-msr(F) operon was located in the chromosome downstream of the 30S ribosomal protein S9 gene (rpsI) (Fig. 3). This locus has already been reported to contain the integration site for resistance islands carrying mecD (McRImecD) in methicillin-resistant M. caseolyticus strains (7, 13). Similar to McRImecD, the mef(D)-msr(F)-containing element in Epi0076A and SD607 was delimited at both sides by characteristic attachment (att) sites of 61 bp and carried a related site-specific integrase gene (int) of the tyrosine recombinase family preceded by its regulator’s intR and xis (Fig. 3). The integrase gene of McRImecD-1 has been shown to be responsible for the mobilization of att-delimited elements (10). Based on these similar features to McRImecD, the mef(D)-msr(F)-containing elements were called Macrococcus resistance island msr (McRImsr) and represent inserts of 18,834 bp in Epi0076A and 20,836 bp in SD607. The int of McRImsr of Epi0076A showed 96% nt identity to the int0473 of McRImecD-2/3 but only 75% to the int of McRImsr of SD607, which, on the other hand, was more similar to the int0819 of McRImecD-1 (90% nt identity). The McRImsr elements of Epi0076A and SD607 were different from the integrase gene, the mef(D)-msr(F) fragment, and unrelated types of genes coding for DNA methylase (mtase) and a putative AAA-type ATPase (aase) (Fig. 3).
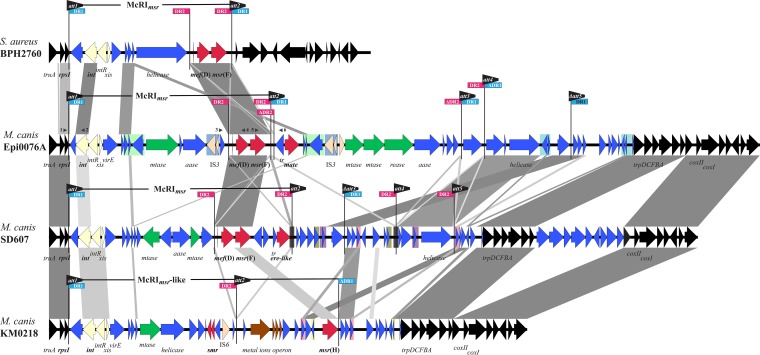
FIG 3.
Structures of Macrococcus resistance islands (McRI) chromosomally integrated into the 3′ end of the 30S ribosomal protein S9 gene (rpsI) of Staphylococcus aureus and Macrococcus canis. Three different McRImsr containing the macrolide resistance genes mef(D)-msr(F) are shown, all containing a related integrase (int) at the left end and are flanked by characteristic attachment (att) sites. The macrolide resistance gene msr(H) is found downstream of an att-flanked island in a McRImsr-like structure. Imperfect direct repeats (DR1 and DR2) often associated with att sites are indicated. More extended direct repeats are shaded equally. Genes are represented by arrows and colored by putative function, as follows: resistance genes are shown in red, genes involved in the excision and integration of McRI are in light yellow, recombinase genes are in darker yellow, genes of DNA restriction-modification systems are in green, genes involved in heavy metal metabolism are in brown, other possible accessory genes are shown in blue, and those assumed to be usually present in the chromosome are in black. Arrowheads labeled with numbers indicate primers used to detect circular excision of mef(D)-msr(F) subunits. Primer names are as follows: 1, rpsI-MC-F; 2, int-0473-F; 3, Epi0076A_18-2-F; 4, mefD-R; 5, msrF_F; and 6, Epi0076A_22-2-F. Gray areas indicate regions with between 75% and 100% nucleotide sequence identity. The figure was generated using the Easyfig software (57) and the sequences of the S. aureus strains BPH2760 (GenBank accession no. NZ_LR130509.1, positions 2178720 to 2208720), M. canis Epi0076A (accession no. MN728681), M. canis SD607 (accession no. MN728682), and M. canis KM0218 (accession no. BK011995).
While the McRImsr of SD607 seems to be unique, two elements similar to McRImsr of Epi0076A were found by a nucleotide similarity search in the GenBank database. The first strain, M. caseolyticus DaniaSudan, isolated from a wound from a donkey in Sudan (GenBank accession no. NZ_RBVL01000008), carried an McRImsr that lacks only the IS3 element of Epi0076A. Second, M. caseolyticus strain CCM7927, isolated from human clinical material in the Czech Republic (accession no. NZ_MJBJ02000004), differed additionally by an alternative first open reading frame (ORF) of an island previously named McRICCM7927 (4) (Fig. S2). The McRImsr elements of the two M. caseolyticus strains were also delimited by the characteristic att sites. A second direct repeat (DR), DR2, flanking the 3,886-bp mef(D)-msr(F) segment was identified in all Epi0076A-like McRImsr sequences. These imperfect 93-bp DR2 elements contained at the 3′-end 7 bases (5′-GAACGTA) overlap the 5′ end of the att site (Fig. S2). Interestingly, such a DR2-flanked mef(D)-msr(F) segment (99% nt identity) was also detected in S. aureus strain BPH2760, isolated from human blood in Australia (accession no. NZ_LR130509). In this S. aureus strain, the mef(D)-msr(F) operon was also part of an McRImsr. The 15,178-bp element of BPH2760 was also integrated into the chromosome at the 3′ end of the rpsI gene, carried int-intR-xis genes at the 5′ end, and was flanked by att sites (Fig. 3). It formed a third type of McRImsr that contained an int gene that shared 97% nt identity with that of Epi0076A and unique genes (e.g., ORF1 and a helicase) not present in the McRImsr elements of M. canis strains Epi0076A and SD607. A DR2-flanked segment containing mef(D)-msr(F) was also observed in M. canis SD607, but this segment was larger (7,173 bp) and contained a macrolide esterase-like gene (ere-like) (see below) (Fig. 3).
Mobility of McRImsr.
The presence of an integrase gene (int) related to that of McRImecD and intact flanking att sites suggests that McRImsr could also be mobilized by an Int-catalyzed site-specific recombination reaction as observed for mecD elements (7, 10, 13). To determine whether McRImsr was able to excise itself, the genomic DNA of Epi0076A was analyzed for the presence of circular forms. Circular McRImsr was detected by PCR using divergent primers specific for int and msr(F) (primers 2 and 5 in Fig. 3; see Fig. S3 for PCR products). A PCR product corresponding to the chromosomal segment after McRImsr deletion was also obtained using short extension time and convergent primers specific for rpsI and a transcriptional regulator (tr) downstream of msr(F) (Fig. 3, primers 1 and 6, and Fig. S3). Sequencing confirmed that circular excision of McRImsr results from recombining the att1 and att2 sequences in Epi0076A.
The highly similar DR2-flanked mef(D)-msr(F) segments seen in a different genetic context in McRImsr of M. canis Epi0076A and McRImsr of S. aureus BPH2670 indicate potential mobility of this subunit. Excision and circularization of the mef(D)-msr(F) subunit were tested using pairs of divergent and convergent primers to detect the circular form and chromosomal deletion of the mef(D)-msr(F) subunit, respectively (Fig. 3 and S3). The sequence of the amplicon obtained with the mef(D) reverse and msr(F) forward primers (primers 4 and 5 in Fig. 3) showed the circular mef(D)-msr(F) subunit joined by a DR2 identical to DR2-2 (Fig. S3). The sequence chromatograms showed no double peaks at the DR2-1/DR2-2 mismatch position, indicating that a site-specific strand exchange occurred at the end of the DR2 sequence. However, the chromosomal segment that remained after excision of the mef(D)-msr(F) subunit amplified with primers Epi0076A_18-2-F and Epi0076A_22-2-F (primers 3 and 6 in Fig. 3) showed ambiguous bases at the mismatch position within the joining DR2 (Fig. S3). This result suggests that deletion of the mef(D)-msr(F) subunit can also occur by random strand exchange within the DR2 sequences. The McRImsr and downstream region in Epi0076A contain 5 DR2 that could also become involved in homologous recombination. The att sequences were often found embedded in more extended repeats with the DR2 sequence upstream and another partially overlapping repeat (49-bp overlap with att) called DR1 downstream (see Fig. S2 for sequences). Additional repetitive sequences formed by an insertion sequence (IS) of the IS3 family or other long imperfect duplicated sequences were also detected (indicated by shading in Fig. 3). Such extended repeats may also play a role in recombination leading to loss or gain of fragments within McRImsr.
Putative resistance genes associated with McRImsr.
The region downstream of McRImsr in Epi0076A and SD607 carried unique sequences organized in segments flanked by the att sites (Fig. 3). In Epi0076A, the rpsI downstream region was divided into the 18,834-bp McRImsr, followed by a 17,789-bp segment (att2-att3), a 2,137-bp segment (att3-att4), and an 8,044-bp segment (att4-Δatt5) flanked at the right side by a 5′-truncated att site. On these segments, putative genes for enzymes of restriction-modification systems (mtase and restriction endonuclease [rease]), a helicase, and a multidrug and toxic compound extrusion (MATE)-like proteins were identified (Fig. 3). The putative 446-aa MATE protein of Epi0076A contains the MepA (cd13143) and the MATE-like superfamily domain (cl09326) found in integral membrane proteins involved in low-level resistance against a broad spectrum of compounds, including several monovalent and divalent biocides (35).
M. canis SD607 contained, downstream of McRImsr, further att-flanked segments of 4,896 bp (att2-Δatt3), 4,782 bp (Δatt3-att4), and 5,418 bp (att4-att5) (Fig. 3). An ere-like gene was found downstream of mef(D)-msr(F) within the DR2-flanked subunit and was analyzed further for macrolide resistance by recombinant gene expression. The ere-like gene of SD607, including 710 bp upstream of its translational start site, was cloned into pTSSCm, and the resulting pTSSCm::ere-like plasmid was introduced into S. aureus RN4220 (see Fig. 2 for plasmid constructs). The measured MIC of erythromycin remained unchanged for RN4220/pTSSCm::ere-like compared to that of RN4220 without plasmid and with pTSSCm (Table 1). The cloned fragment contained a putative transcriptional regulator (tr) that overlapped by 4 bp with the 5′ end of the ere-like gene. To exclude the possibility that tr represses ere-like expression, the tr sequence was deleted in pTSSCm::ere-like. However, the RN4220 strain containing pTSSCm::Δtr-ere-like also did not exhibit erythromycin resistance (Table 1). It included an upstream region containing possible native promoter sequences with a −35 box (5′-TGGATA) and a −10 box (5′-TACTATCAT) located 327 bp and 303 bp upstream of the start codon, respectively. Additionally, the ere-like gene alone was placed under the control of the strong constitutive S. aureus type 1 capsule gene 1A promoter (Pcap) to ensure expression in S. aureus (36). The lack of a phenotype observed in RN4220/pTSSCm-Pcap::ere-like indicated that this ere-like gene is probably not involved in erythromycin inactivation (Table 1). The 415-aa Ere-like protein of SD607 contains the Ere-like domain (cd14728), including strictly conserved residues in the active-site cleft (E63, H66, E94, H276, and H279), suggesting that the enzyme functions as a hydrolase (37). The similarity of the Ere-like protein of SD607 to the erythromycin esterases recognized by the MLS nomenclature [Ere(A) and Ere(B) enzymes of Escherichia coli and Ere(C) and Ere(D) proteins of Riemerella anatipestifer] was low and less than 21% overall aa identity.
A comparison of the McRImsr elements of Epi0076A and SD607 with other Macrococcus species sequences from the GenBank database revealed another McRImsr-like structure with a putative novel msr gene in M. canis KM0218 (Fig. 3). The novel msr(H) gene of KM0218 encoded a 486-aa ARE ABC-F protein (PROSITE entry PS50893) that shared 76% aa identity to the closest related msr(F) gene, hence representing a new msr gene named msr(H), according to MLS nomenclature (Fig. 1B). KM0218 showed high-level resistance to erythromycin and pristinamycin IA due to an erm(B) gene carried on a multidrug resistance plasmid (9) (Table 1). Whether the msr(H) gene detected in the rpsI region also contributed to the macrolide and streptogramin B phenotype was therefore not clear. A complete mef gene was not present in KM0218 but the upstream region of msr(H) contained an 81-bp remnant resembling the 3′ end of mef(D) as well as a 109-bp region similar to the intergenic region of mef(D)-msr(F). Features found in the 5′ region of inducibly expressed mef, msr, and erm genes were not identified in the msr(H) upstream region. The expression of msr(H) from native promoter [msr(H), including 698 bp upstream of its translational start site of KM0218 cloned into pTSSCm::msr(H) (Fig. 2)] in S. aureus mediated low-level resistance to erythromycin, with an MIC value of 2 μg/ml, but not resistance to pristinamycin IA (Table 1).
M. canis KM0218 contained only one att-delimited segment of 15,707 bp (att1-att2) downstream of the rpsI gene that also carried the int-intR-xis genes at the 5′ end (Fig. 3). The msr(H) gene was found outside this segment and therefore reported to be part of an McRImsr-like region (att1-ΔDR1). A unique helicase and a DNA methylase gene were also identified in this accessory DNA of strain KM0218. This region also contains genes involved in heavy metal metabolism and two genes belonging to the small multidrug resistance (SMR) family (Fig. 3).
DISCUSSION
Three new macrolide resistance genes were characterized in this study, msr(F) and msr(H), encoding ARE ABC-F family proteins, and mef(D), encoding an MFS protein. While msr(H) was found alone, mef(D) and msr(F) were present in tandem within the same operon. The expression of these genes in S. aureus resulted in a low-to-moderate increase in erythromycin resistance but did not affect streptogramin B resistance, even under inducing conditions. Such an M-type resistance phenotype has been described for streptococci (38) associated with the mef(A)-msr(D) operon. The mef(D)-msr(F) operon identified in M. canis also showed structural similarity to the mef(A)-msr(D) operon of streptococci, even though the overall nucleotide sequence identity was low (53%), and elements carrying the genes in Macrococcus and Streptococcus species were unrelated. The 414-bp sequence upstream of mef(D)-msr(F), suggested to be cotranscribed with the two genes, also encodes a leader peptide [Mef(D)L, MTHAMKLRF] that contains the possible pause sequence M(R/K)L(R/K) (32, 39). The mef(A)-msr(D) genes might therefore be regulated by a macrolide-dependent ribosome stalling mechanism which could lead to reorganization of the mRNA structure between the leader ORF and the mef(D) gene. The presence of a rho-independent terminator sequence in this region suggests a transcriptional attenuation control as found for mef(A)-msr(D) and erm(K) (32, 33). Cloning experiments in S. aureus showed that the promoter sequence upstream of mef(D) is also needed for the expression of msr(F). Moreover, the msr(F) gene is the principal determinant of the mef(D)-msr(F) operon responsible for macrolide resistance and also functions without the presence of mef(D). This finding is in accordance with the current model of ARE ABC-F proteins that protect ribosomes independently of a partner protein (15, 16). The mef(D) gene alone only mediated a slight decrease in susceptibility to erythromycin, and its presence upstream of msr(F) seems even to diminish the effect of msr(F), as a 2-fold-lower MIC value was repeatedly obtained with a plasmid containing the complete mef(D)-msr(F) operon. In streptococci, msr(D) also has a predominant role over mef(A) in macrolide resistance, but an additive effect was observed for the two genes (20, 21).
The msr(H) gene was most closely related to the msr(F) gene and was not preceded by a mef gene. However, the presence of an upstream region exhibiting similarity to the end of mef(D) and the intergenic region of mef(D)-msr(F) suggests that the 5′ region of a former mef-msr operon could have been truncated. This would also explain the lack of structural similarity of the upstream region of msr(H) to the 5′ regulatory region of other msr, mef, and erm genes. The expression of msr(H) under the control of its own promoter in S. aureus mediated only a moderate increase in the erythromycin MIC, which could also reflect suboptimal gene expression rather than a per se less-effective ARE ABC-F protein.
The putative ere gene of strain SD607 cloned under the control of Pcap did not affect the erythromycin susceptibility of S. aureus. The capability of ere(A) and ere(B) of E. coli to hydrolyze the macrolactone rings has been proven experimentally (37, 40, 41), but their functionality in a staphylococcal host has not yet been definitively clarified. Li and colleagues found ere(A) in both macrolide-resistant and -susceptible strains (29). Another protein related to Ere(A) and Ere(B), Bcr136 of Bacillus cereus, has been shown to possess esterase activity but was unable to inactivate macrolides (37). Current annotation tools are assigning many proteins, including the Bcr136 and Ere-like proteins of SD607, to the erythromycin esterase family. These proteins might possess esterase activity, but it seems that their specificity for erythromycin is not always guaranteed, as seen for Bcr136 and the putative macrolide esterase of SD607.
The McRImsr genetic elements carrying mef(D)-msr(F) detected in M. canis and S. aureus showed, despite an overall high diversity, some common important features, as follows: they share the same DR2-flanked mef(D)-msr(F) fragment, possess highly related integrases and flanking att sites, and are found to be site-specifically integrated into the chromosome at the rpsI locus. Similarly, the msr(H) gene of M. canis KM0218 was also associated with an McRImsr-like structure integrated at the rpsI locus. The 3′ end of the rpsI gene, a highly conserved locus in Staphylococcaceae, has already been shown to hold integrated elements carrying an int gene or small accessory islands without an obvious mobilization function (4, 7, 10, 13). The att-flanked segments found in M. canis Epi0076A and SD607 suggests that the site-specific int gene is active in accumulating accessory DNA and diversifying the rpsI locus. Circular excision of McRImsr was demonstrated for Epi0076A. In addition, a circular intermediate of the DR2-flanked mef(D)-msr(F) subunit was detected, emphasizing the mobility potential of this subunit also present in S. aureus. Analysis of the joining DR2 sequence in the circular mef(D)-msr(F) subunit indicated that strand cleavage occurred at the 3′ end of the homologous DR2 sequences. Since the 3′ end of the second DR2 overlaps the 5′ end of the att2 site, the int of McRImsr could mediate this excisive recombination reaction. Analysis of recombination products of McRImecD-1-McCIIMD0819 subunits indicated that strand cleavage occurred within the first 8 bases of the core att site (7). The int of rpsI-associated islands seems to be flexible in att site selection and be influenced by regulators like xis (10). Furthermore, M. canis has previously been shown to have potential to use imperfect direct repeats for the formation of unconventional circularizable structures (9, 42, 43).
Novel erythromycin resistance genes, mef(D), msr(F), and msr(H), were found on the McRImsr and McRImsr-like chromosomal resistance islands, site-specifically integrated into the rpsI locus. The same type of resistance islands, McRImecD carrying the methicillin resistance gene mecD, were described before. While McRImecD was so far only found in M. caseolyticus, diverse McRImsr islands containing the same mef(D)-msr(F) fragment were detected in both Macrococcus species and S. aureus in this study. This finding showed that McRImsr elements play an important role in the distribution of the M-type phenotype in both commensal and opportunistic pathogens of the Staphylococcaceae family.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Three erythromycin-resistant M. canis strains from dogs, KM0218 [sequence type 12 (ST12); resistance genes mecB, blaZm, aac(6′)-aph(2ʺ), sat4, aph(3′)-IIIa, erm(B), ant(6)-Ia, and tet(S)], Epi0076A (ST2; mecB and grlA [Ser80Leu]), and SD607 (ST21), were obtained from a study by Cotting and colleagues (12). The E. coli strains TOP10 (Thermo Fisher Scientific [Invitrogen], Waltham, MA) and DH5α were used for cloning and plasmid amplifications. S. aureus strain RN4220 (44) was used for plasmid transformation and phenotypic gene expression. The strains were routinely cultivated under aerobic conditions at 37°C on either Trypticase soy agar plates containing 5% sheep blood (TSA-SB; Becton, Dickinson Company, Franklin Lakes, NJ, USA) in Luria-Bertani (LB) broth with shaking or on LB agar plates. DH5α and RN4220 strains containing pTSSCm-derived plasmids [tet(L)] were selected and grown in LB medium containing 10 μg/ml tetracycline (36). TOP10 cells carrying pCRII-TOPO-derived plasmids [with aph(3′)-II and blaTEM] were cultivated in LB medium containing 50 μg/ml kanamycin.
DNA preparation and PCR.
High-quality genomic DNA was extracted from pure cultures using the either peqGOLD bacterial DNA kit (Peqlab Biotechnologie GmbH, Jena, Germany) or the DNeasy blood and tissue kit (Qiagen GmbH, Hilden, Germany). Lysis of M. canis cells was improved through a 30-min initial incubation at 37°C in the presence of 50 μg/ml lysostaphin and 2 mg/ml lysozyme. Chelex-based genomic DNA extractions were prepared without enzymatic predigestion of cell walls. To do so, M. canis or S. aureus cells taken from the plate with a 1-μl loop were incubated in 200 μl of 10% (wt/vol) Chelex 100 molecular biology-grade resin (Bio-Rad, Hercules, CA, USA) solution (100 mM Tris-HCl [pH 8], 0.5 mM EDTA) at 56°C for 30 min, followed by a 15-min boiling step. The DNA-containing supernatant was separated from Chelex resins by centrifugation (5 min at 10,000 × g). Plasmid DNA from E. coli was isolated using the peqGOLD plasmid miniprep kit I (Peqlab Biotechnologie GmbH).
PCRs were performed according to the manufacturer’s instructions using FIREPol DNA polymerase (Solis BioDyne, Tartu, Estonia) for analytical PCRs, Phusion Hot Start II high-fidelity DNA polymerase (Thermo Fisher Scientific) for cloning, and GoTaq long PCR master mix (Promega, Madison, WI, USA) for the detection of circular excision fragments. All relevant primers and PCR conditions are listed in Table S1. PCR fragments were purified using the High Pure PCR product purification kit (Roche Diagnostics, Rotkreuz, Switzerland) before incubation with restriction endonucleases, T4 DNA ligase, and Sanger sequencing.
Construction of recombinant plasmids.
Target sequences for cloning were amplified from high-quality genomic DNA using Phusion Hot Start II high-fidelity DNA polymerase. The resulting blunt-end PCR products were introduced into the pCR-Blunt II-TOPO vector using the topoisomerase-based Zero Blunt TOPO PCR cloning kits (Thermo Fisher Scientific [Invitrogen]) and transformed into One Shot TOP10 chemically competent E. coli cells (Thermo Fisher Scientific [Invitrogen]), according to the manufacturer’s protocol. TOP10 colonies obtained on selective agar were screened by colony-PCR for insert orientation using combinations of vector and insert primers. Inserts were subcloned into the pTSSCm or pTSSCm-Pcap vector (36) (see Fig. 2 for plasmid constructs) using endonuclease restriction and T4 DNA ligation (Promega). Restriction sites in the flanking pCRII-TOPO sequence or incorporated in the 5′ overhangs of cloning primers were used for that purpose (Table S1). The generated pTSSCm/-Pcap constructs were transformed through heat shock into chemically competent DH5α cells (45). To construct the plasmids pTSSCm::mef(D)-msr(F), pTSSCm::mef(D), and pTSSCm::msr(F), inserts were amplified from Epi0076A DNA using the PCR conditions listed in Table S1. Inserts for pTSSCm::ere-like and pTSSCm-Pcap::ere-like were amplified from SD607 DNA, and that for pTSSCm::msr(H) was obtained using KM0218 DNA (Table S1). pCRII-TOPO constructs containing the inserted genes in antisense orientation respective to backbone genes were used for restriction endonuclease-based subcloning into the shuttle vectors pTSSCm and pTSSCm-Pcap. The enzymes XhoI and BamHI were used to generate pTSSCm::mef(D)-msr(F), pTSSCm::mef(D), pTSSCm::ere-like, and pTSSCm::msr(H) (Fig. 2). The enzymes XhoI and SpeI were used to obtain pTSSCm::msr(F). The enzymes NdeI and BamHI were used to construct pTSSCm-Pcap::ere-like. Plasmids pTSSCm::Δmef(D)-msr(F) and pTSSCm::Δtr-ere-like were generated through PCR-based mutagenesis of pTSSCm::mef(D)-msr(F) and pTSSCm::ere-like, respectively (Table S1 and Fig. 2). Deletion of the mef(D) gene in the template plasmid pTSSCm::mef(D)-msr(F) and of the tr gene in pTSSCm::ere-like was obtained using primers that excluded amplification of mec(D) or tr and that contained homologous 5′ ends. The PCR products were treated with the restriction enzyme DpnI and directly transformed into E. coli DH5α cells, which allowed recombining their homologous ends in vivo. All plasmid structures were verified by restriction digestion, and the sequences of the DNA inserts were confirmed by Sanger sequencing. All of the generated plasmids were transformed into RN4220 strain bacteria through electroporation using the protocol of Schenk and Laddaga (46).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility was determined for M. canis strains and S. aureus RN4220 strains containing pTSSCm/-Pcap-derived plasmids. The MICs were measured by the microdilution method in Mueller-Hinton broth, according to CLSI guidelines (47), using both Sensititre EUST plates (Thermo Fisher Scientific) and 96-well plates containing serial 2-fold dilutions ranging from 0.125 μg/ml to 64 μg/ml erythromycin or pristinamycin IA. Inducible resistance to pristinamycin IA was measured in the presence of 1 or 0.25 μg/ml erythromycin. MICs were determined in duplicate.
Assembly, annotation, and analysis of genome sequences.
The genome of M. canis strain KM0218 was obtained from GenBank (accession no. CP035309.1). Strain Epi0076A was sequenced using Illumina MiSeq technology within a previous study (9). During this study, strain SD607 was sequenced using Illumina HiSeq technology (2 × 150-bp paired-end; Eurofins, Constance, Germany), and both strains, SD607 and Epi0076A, were sequenced with MinION Oxford Nanopore Technology (ONT). The ONT library was prepared from mechanically fragmented DNA (g-TUBE; Covaris) using ONT 1D ligation sequencing kits (SQK-LSK108 for Epi0076A and SQK-LSK109 for SD607) with the native barcoding expansion kit (EXP-NBD103; Oxford Nanopore). MinION sequencing was done on an R9.4 SpotON flow cell with a MinION MK1b device. The generated fast5 ONT reads were base called and demultiplexed using the ONT Albacore software (v2.0.1) for Epi0076A and the Guppy software (v3.2.4) for SD607. The software Cutadapt (v2.5) was used for end trimming and size sorting (48). The complete genomes were de novo assembled using the Unicycler assembly pipeline (v0.4.4) run with default parameters, paired-end Illumina reads, and ONT reads with a minimum length of 12 kb for Epi0076A and 10 kb for SD607 (49). The Illumina HiSeq reads of SD607 were used without processing, while the MiSeq reads of Epi0076A were prior quality filtered using Trimmomatic v0.36 to ensure removal of Illumina adaptor sequences (parameter, ILLUMINACLIP: NexteraPE-PE.fa:2:30:10), an average quality per base of at least 15 (parameters, LEADING:3, TRAILING:3, and SLIDINGWINDOW:4:15), and a minimum length of 36 bases (parameter, MINLEN:36) (50). Illumina reads of SD607 were additionally assembled using SPAdes (v3.12.0), with the mismatch careful option (51). Calculations were performed on UBELIX (https://www.unibe.ch/universitaet/campus__und__infrastruktur/rund_um_computer/soft_und_hardware/hardware/hochleistungsrechner_hpc_grid/index_ger.html), the High-Performance Computing (HPC) cluster at the University of Bern. The complete genomes of Epi0076A and SD607 were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) service. Additionally, features present in the McRImsr islands of strains Epi0076A, SD607, and KM0218 were annotated with Prokka (v1.13) (52) and edited manually. Repeat sequences were identified by BLASTN, promoters and transcription start sites by PBROM (53), and transcription terminators by ARNOLD (54). The secondary structure of the nucleotide sequence upstream of mef(D)-msr(F) was predicted using the RNAfold Web server (55). Putative protein function was analyzed by searching against PROSITE entries and Conserved Domains Databases (Conserved Domain Search Service [CD] search with NCBI-curated position-specific scoring matrices [PSSMs]) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Detection of recombination products.
Circular excision of mef(D)-msr(F) fragments in strain Epi0076A was analyzed by PCR (Table S1). A high-quality genomic DNA template (around 200 ng per reaction) was used for the detection of chromosomal deletion products and Chelex extracts (2 μl per reaction) for the detection of circular forms. The obtained PCR products were analyzed on agarose gels and confirmed by Sanger sequencing (Fig. S3).
Data availability.
The nucleotide sequences of rpsI-downstream regions with manually curated features were deposited in GenBank under the accession numbers MN728681, MN728682, and BK011995 for M. canis strains Epi0076A, SD607, and KM0218, respectively. The complete genome sequences of Epi0076A and SD607 are available under GenBank accession numbers CP047363 to CP047365 and CP047361 and CP047362, respectively (NCBI BioProject no. PRJNA596733).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by internal funds from the Institute of Veterinary Bacteriology, University of Bern, and by the Swiss Federal Food Safety and Veterinary Office FSVO (grant no. 1.18.10).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kloos WE, Ballard DN, George CG, Webster JA, Hubner RJ, Ludwig W, Schleifer KH, Fiedler F, Schubert K. 1998. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., and Macrococcus bovicus sp. no. and Macrococcus carouselicus sp. nov. Int J Syst Bacteriol 48:859–877. doi: 10.1099/00207713-48-3-859. [DOI] [PubMed] [Google Scholar]
- 2.Mannerová S, Pantůček R, Doskar J, Svec P, Snauwaert C, Vancanneyt M, Swings J, Sedlácek I. 2003. Macrococcus brunensis sp. nov., Macrococcus hajekii sp. nov. and Macrococcus lamae sp. nov., from the skin of llamas. Int J Syst Evol Microbiol 53:1647–1654. doi: 10.1099/ijs.0.02683-0. [DOI] [PubMed] [Google Scholar]
- 3.Gobeli Brawand S, Cotting K, Gómez-Sanz E, Collaud A, Thomann A, Brodard I, Rodriguez-Campos S, Strauss C, Perreten V. 2017. Macrococcus canis sp. nov., a skin bacterium associated with infections in dogs. Int J Syst Evol Microbiol 67:621–626. doi: 10.1099/ijsem.0.001673. [DOI] [PubMed] [Google Scholar]
- 4.Mašlaňová I, Wertheimer Z, Sedláček I, Švec P, Indráková A, Kovařovic V, Schumann P, Spröer C, Králová S, Šedo O, Krištofová L, Vrbovská V, Füzik T, Petráš P, Zdráhal Z, Ružičková V, Doškař J, Pantuček R. 2018. Description and comparative genomics of Macrococcus caseolyticus subsp. hominis subsp. nov., Macrococcus goetzii sp. nov., Macrococcus epidermidis sp. nov., and Macrococcus bohemicus sp. nov., novel macrococci from human clinical material with virulence potential and suspected uptake of foreign DNA by natural transformation. Front Microbiol 9:1178. doi: 10.3389/fmicb.2018.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. 2009. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci. J Bacteriol 191:1180–1190. doi: 10.1128/JB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. 2010. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother 54:1469–1475. doi: 10.1128/AAC.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwendener S, Cotting K, Perreten V. 2017. Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Sci Rep 7:43797. doi: 10.1038/srep43797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker K, van Alen S, Idelevich EA, Schleimer N, Seggewiß J, Mellmann A, Kaspar U, Peters G. 2018. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 24:242–248. doi: 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chanchaithong P, Perreten V, Schwendener S. 2019. Macrococcus canis contains recombinogenic methicillin resistance elements and the mecB plasmid found in Staphylococcus aureus. J Antimicrob Chemother 74:2531–2536. doi: 10.1093/jac/dkz260. [DOI] [PubMed] [Google Scholar]
- 10.Schwendener S, Perreten V. 2018. The integrase of the Macrococcus caseolyticus resistance island mecD (McRImecD) inserts DNA site-specifically into Staphylococcus and Bacillus chromosomes. Mol Microbiol 110:455–468. doi: 10.1111/mmi.14112. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, Wu C, Shen J. 2012. Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis. J Antimicrob Chemother 67:1824–1827. doi: 10.1093/jac/dks163. [DOI] [PubMed] [Google Scholar]
- 12.Cotting K, Strauss C, Rodriguez-Campos S, Rostaher A, Fischer NM, Roosje PJ, Favrot C, Perreten V. 2017. Macrococcus canis and M. caseolyticus in dogs: occurrence, genetic diversity and antibiotic resistance. Vet Dermatol 28:559–e133. doi: 10.1111/vde.12474. [DOI] [PubMed] [Google Scholar]
- 13.Schwendener S, Nigg A, Collaud A, Overesch G, Kittl S, Phumthanakorn N, Perreten V. 2019. Typing of mecD islands in genetically diverse methicillin-resistant Macrococcus caseolyticus strains from cattle. Appl Environ Microbiol 85:e01496-19. doi: 10.1128/AEM.01496-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fyfe C, Grossman TH, Kerstein K, Sutcliffe J. 2016. Resistance to macrolide antibiotics in public health pathogens. Cold Spring Harb Perspect Med 6:a025395. doi: 10.1101/cshperspect.a025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharkey LKR, Edwards TA, O'Neill AJ. 2016. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 7:e01975-15. doi: 10.1128/mBio.01975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su W, Kumar V, Ding Y, Ero R, Serra A, Lee BST, Wong ASW, Shi J, Sze SK, Yang L, Gao YG. 2018. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc Natl Acad Sci U S A 115:5157–5162. doi: 10.1073/pnas.1803313115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross JI, Eady EA, Cove JH, Cunliffe WJ, Baumberg S, Wootton JC. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol 4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 18.Singh KV, Malathum K, Murray BE. 2001. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob Agents Chemother 45:263–266. doi: 10.1128/AAC.45.1.263-266.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daly MM, Doktor S, Flamm R, Shortridge D. 2004. Characterization and prevalence of MefA, MefE, and the associated msr(D) gene in Streptococcus pneumoniae clinical isolates. J Clin Microbiol 42:3570–3574. doi: 10.1128/JCM.42.8.3570-3574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrose KD, Nisbet R, Stephens DS. 2005. Macrolide efflux in Streptococcus pneumoniae is mediated by a dual efflux pump (mel and mef) and is erythromycin inducible. Antimicrob Agents Chemother 49:4203–4209. doi: 10.1128/AAC.49.10.4203-4209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Tatsuno I, Okada R, Hata N, Matsumoto M, Isaka M, Isobe KI, Hasegawa T. 2016. Predominant role of msr(D) over mef(A) in macrolide resistance in Streptococcus pyogenes. Microbiology 162:46–52. doi: 10.1099/mic.0.000206. [DOI] [PubMed] [Google Scholar]
- 22.Kadlec K, Brenner Michael G, Sweeney MT, Brzuszkiewicz E, Liesegang H, Daniel R, Watts JL, Schwarz S. 2011. Molecular basis of macrolide, triamilide, and lincosamide resistance in Pasteurella multocida from bovine respiratory disease. Antimicrob Agents Chemother 55:2475–2477. doi: 10.1128/AAC.00092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santagati M, Iannelli F, Oggioni MR, Stefani S, Pozzi G. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob Agents Chemother 44:2585–2587. doi: 10.1128/aac.44.9.2585-2587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gay K, Stephens DS. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J Infect Dis 184:56–65. doi: 10.1086/321001. [DOI] [PubMed] [Google Scholar]
- 25.Mingoia M, Vecchi M, Cochetti I, Tili E, Vitali LA, Manzin A, Varaldo PE, Montanari MP. 2007. Composite structure of Streptococcus pneumoniae containing the erythromycin efflux resistance gene mefI and the chloramphenicol resistance gene catQ. Antimicrob Agents Chemother 51:3983–3987. doi: 10.1128/AAC.00790-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds ED, Cove JH. 2005. Resistance to telithromycin is conferred by msr(A), msrC and msr(D) in Staphylococcus aureus. J Antimicrob Chemother 56:1179–1180. doi: 10.1093/jac/dki378. [DOI] [PubMed] [Google Scholar]
- 27.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath AV, Bergeron J, Retsema JA. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol 22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 28.Feßler AT, Wang Y, Wu C, Schwarz S. 2018. Mobile macrolide resistance genes in staphylococci. Plasmid 99:2–10. doi: 10.1016/j.plasmid.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Feng W, Zhang Z, Xue H, Zhao X. 2015. Macrolide-lincosamide-streptogramin resistance phenotypes and genotypes of coagulase-positive Staphylococcus aureus and coagulase-negative staphylococcal isolates from bovine mastitis. BMC Vet Res 11:168. doi: 10.1186/s12917-015-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz FJ, Petridou J, Milatovic D, Verhoef J, Fluit AC, Schwarz S. 2002. In vitro activity of new ketolides against macrolide-susceptible and -resistant Staphylococcus aureus isolates with defined resistance gene status. J Antimicrob Chemother 49:580–582. doi: 10.1093/jac/49.3.580. [DOI] [PubMed] [Google Scholar]
- 31.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. doi: 10.1128/AAC.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chancey ST, Bai X, Kumar N, Drabek EF, Daugherty SC, Colon T, Ott S, Sengamalay N, Sadzewicz L, Tallon LJ, Fraser CM, Tettelin H, Stephens DS. 2015. Transcriptional attenuation controls macrolide inducible efflux and resistance in Streptococcus pneumoniae and in other Gram-positive bacteria containing mef/mel(msr(D)) elements. PLoS One 10:e0116254. doi: 10.1371/journal.pone.0116254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak JH, Choi EC, Weisblum B. 1991. Transcriptional attenuation control of ermK, a macrolide-lincosamide-streptogramin B resistance determinant from Bacillus licheniformis. J Bacteriol 173:4725–4735. doi: 10.1128/jb.173.15.4725-4735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon AR, Min YH, Yoon EJ, Kim JA, Shim MJ, Choi EC. 2006. ErmK leader peptide: amino acid sequence critical for induction by erythromycin. Arch Pharm Res 29:1154–1157. doi: 10.1007/bf02969307. [DOI] [PubMed] [Google Scholar]
- 35.Kaatz GW, McAleese F, Seo SM. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob Agents Chemother 49:1857–1864. doi: 10.1128/AAC.49.5.1857-1864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwendener S, Perreten V. 2015. New shuttle vector-based expression system to generate polyhistidine-tagged fusion proteins in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol 81:3243–3254. doi: 10.1128/AEM.03803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morar M, Pengelly K, Koteva K, Wright GD. 2012. Mechanism and diversity of the erythromycin esterase family of enzymes. Biochemistry 51:1740–1751. doi: 10.1021/bi201790u. [DOI] [PubMed] [Google Scholar]
- 38.Sutcliffe J, Tait-Kamradt A, Wondrack L. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother 40:1817–1824. doi: 10.1128/AAC.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramu H, Mankin A, Vazquez-Laslop N. 2009. Programmed drug-dependent ribosome stalling. Mol Microbiol 71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 40.Arthur M, Autissier D, Courvalin P. 1986. Analysis of the nucleotide sequence of the ereB gene encoding the erythromycin esterase type II. Nucleic Acids Res 14:4987–4999. doi: 10.1093/nar/14.12.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ounissi H, Courvalin P. 1985. Nucleotide sequence of the gene ereA encoding the erythromycin esterase in Escherichia coli. Gene 35:271–278. doi: 10.1016/0378-1119(85)90005-8. [DOI] [PubMed] [Google Scholar]
- 42.Gómez-Sanz E, Schwendener S, Thomann A, Gobeli Brawand S, Perreten V. 2018. Correction for Gómez-Sanz et al., First staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a canine infection. Antimicrob Agents Chemother 62:e01916-18. doi: 10.1128/AAC.01916-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gómez-Sanz E, Schwendener S, Thomann A, Gobeli Brawand S, Perreten V. 2015. First staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus canis isolate from a canine infection. Antimicrob Agents Chemother 59:4577–4583. doi: 10.1128/AAC.05064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreiswirth BN, Löfdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. 1998. Molecular cloning. A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 46.Schenk S, Laddaga RA. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett 94:133–138. doi: 10.1111/j.1574-6968.1992.tb05302.x. [DOI] [PubMed] [Google Scholar]
- 47.Clinical and Laboratory Standards Institute. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 11th ed. CLSI document M07 Clinical and Laboratory Standards Institute, Wayne, PA, USA. [Google Scholar]
- 48.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 49.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 53.Solovyev V, Salamov SA. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW. (ed), Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 54.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. 2001. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res 29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lorenz WA, Clote P. 2011. Computing the partition function for kinetically trapped RNA secondary structures. PLoS One 6:e16178. doi: 10.1371/journal.pone.0016178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequences of rpsI-downstream regions with manually curated features were deposited in GenBank under the accession numbers MN728681, MN728682, and BK011995 for M. canis strains Epi0076A, SD607, and KM0218, respectively. The complete genome sequences of Epi0076A and SD607 are available under GenBank accession numbers CP047363 to CP047365 and CP047361 and CP047362, respectively (NCBI BioProject no. PRJNA596733).