The RESTORE-IMI 1 phase 3 trial demonstrated the efficacy and safety of imipenem-cilastatin (IMI) combined with relebactam (REL) for treating imipenem-nonsusceptible infections. The objective of this analysis was to compare the outcomes among patients meeting eligibility requirements based on central laboratory susceptibility versus local laboratory susceptibility. Patients with serious infections caused by imipenem-nonsusceptible, colistin-susceptible, and imipenem-REL-susceptible pathogens were randomized 2:1 to IMI-REL plus placebo or colistin plus IMI for 5 to 21 days.
KEYWORDS: carbapenem resistant, supplemental analysis population, local microbiology data, β-lactamase inhibitor
ABSTRACT
The RESTORE-IMI 1 phase 3 trial demonstrated the efficacy and safety of imipenem-cilastatin (IMI) combined with relebactam (REL) for treating imipenem-nonsusceptible infections. The objective of this analysis was to compare the outcomes among patients meeting eligibility requirements based on central laboratory susceptibility versus local laboratory susceptibility. Patients with serious infections caused by imipenem-nonsusceptible, colistin-susceptible, and imipenem-REL-susceptible pathogens were randomized 2:1 to IMI-REL plus placebo or colistin plus IMI for 5 to 21 days. The primary endpoint was a favorable overall response. Key endpoints included the clinical response and all-cause mortality. We compared outcomes between the primary microbiological modified intent-to-treat (mMITT) population, where eligibility was based on central laboratory susceptibility testing, and the supplemental mMITT (SmMITT) population, where eligibility was based on local, site-level testing. The SmMITT (n = 41) and MITT (n = 31) populations had similar baseline characteristics, including sex, age, illness severity, and renal function. In both analysis populations, favorable overall response rates in the IMI-REL treatment group were >70%. Favorable clinical response rates at day 28 were 71.4% for IMI-REL and 40.0% for colistin plus IMI in the mMITT population, whereas they were 75.0% for IMI-REL and 53.8% for colistin plus IMI in the SmMITT population. Day 28 all-cause mortality rates were 9.5% for IMI-REL and 30.0% for colistin plus IMI in the mMITT population, whereas they were 10.7% for IMI-REL and 23.1% for colistin plus IMI in the SmMITT population. The outcomes in the SmMITT population were generally consistent with those in the mMITT population, suggesting that outcomes may be applicable to the real-world use of IMI-REL for treating infections caused by imipenem-nonsusceptible Gram-negative pathogens. (This study has been registered at ClinicalTrials.gov under identifier NCT02452047.)
INTRODUCTION
Carbapenems are a mainstay of treatment in patients with serious Gram-negative bacterial infections (1). Infections caused by multidrug-resistant Pseudomonas spp. and carbapenem-resistant Enterobacteriaceae (new taxonomy, Enterobacterales), including carbapenemase-producing Klebsiella pneumoniae strains, are associated with high morbidity and mortality (2–4), and these bacterial pathogens are considered serious and urgent threats to public health, respectively (1). Conventional treatment options for carbapenem-nonsusceptible infections include polymyxins (polymyxin B and colistin), fosfomycin, tigecycline, and aminoglycosides; however, the causative pathogens are frequently resistant to one or more of these antibacterial agents (5–7). In addition, safety issues associated with these therapies can present a challenge, particularly in patients with complex conditions at high risk of adverse clinical outcomes. Tigecycline therapy carries an elevated risk of all-cause mortality, and the nephrotoxicity of polymyxins is associated with a high risk of acute kidney injury (7–10). Similarly, aminoglycosides may cause acute kidney injury or ototoxicity (11, 12). Despite advantages over the conventional therapy used to treat carbapenem-resistant infections, β-lactam–β-lactamase inhibitor combinations, including ceftazidime-avibactam, may not be effective against pathogens that develop certain resistance mechanisms (13). New antibacterial agents or combinations effective against carbapenem-resistant bacteria with favorable safety profiles are urgently needed.
Unlike other carbapenem antibacterial agents, imipenem induces the overproduction of Pseudomonas-derived cephalosporinase (PDC), a chromosomal AmpC β-lactamase, in Pseudomonas aeruginosa. Imipenem resistance development in this pathogen is due to the overproduction of PDC with the concomitant loss of the imipenem entry porin OprD (14, 15). Relebactam (REL) restores imipenem susceptibility in imipenem-nonsusceptible P. aeruginosa isolates and potentiates the activity of imipenem in imipenem-susceptible P. aeruginosa strains via inhibition of PDC, and unlike other β-lactamase–β-lactamase inhibitor combinations, neither imipenem nor REL is a substrate for the major efflux mechanisms of P. aeruginosa (15, 16). REL can also restore the activity of imipenem against many imipenem-nonsusceptible isolates of Enterobacteriaceae through its inhibition of class A carbapenemases (such as K. pneumoniae carbapenemases) (17, 18). Therefore, the combination of REL with imipenem-cilastatin (IMI), a well-established carbapenem for the treatment of serious infections, is a potential treatment option for infections caused by carbapenem-resistant pathogens (19, 20). In global surveillance studies, REL restored imipenem susceptibility to 53.5% to 84.9%, 71.4%, and 23.9% to 100.0% of imipenem-nonsusceptible isolates of P. aeruginosa, K. pneumoniae, and other Enterobacteriaceae, respectively (15, 21–23).
The efficacy and safety of IMI-REL have been investigated in several randomized, controlled clinical trials. Results from 2 phase 2, dose-ranging studies demonstrated that IMI-REL is well tolerated and noninferior to IMI alone for the treatment of adults with complicated urinary tract infection (cUTI) and complicated intra-abdominal infection (cIAI) (24, 25). A recent phase 3 trial, RESTORE-IMI 1, investigated IMI-REL for the treatment of serious, imipenem-nonsusceptible bacterial infections and demonstrated that IMI-REL was effective and generally well tolerated (20). RESTORE-IMI 1 was a small, descriptive study conducted in patients with infections confirmed to be caused by imipenem-nonsusceptible pathogens, without formal hypothesis testing for efficacy endpoints.
Identification of appropriate study populations using microbiological criteria presents a challenge to conducting clinical trials for antibacterial agents. To initiate effective treatment as rapidly as possible, which has demonstrated clinical benefit, including improved survival rates, patients potentially eligible for such trials need to be identified at the investigational site level, using results from local susceptibility testing (4). However, to standardize trial data appropriately for regulatory review, confirmation of local test results by a central laboratory is a requirement (26, 27). Repeat susceptibility testing of isolates at different facilities can contribute to complexity, as the results between tests for any isolate may vary by ≥4 dilutions, even under standardized conditions within the same facility, which may impact the designation of the susceptibility or the nonsusceptibility of isolates to the tested agents (28). Determination of patient eligibility for inclusion in the primary analysis population of our trial was based on centrally assessed susceptibility results. In this study, we evaluated the outcomes for a study population in which IMI-REL was used in a manner that more closely represents the real-world use of IMI-REL, where treatment decisions are made based on local laboratory results. We addressed this objective by comparing the results from the RESTORE-IMI 1 microbiological modified intent-to-treat (mMITT) population with those from an expanded supplemental microbiological modified intent-to-treat (SmMITT) population that included those patients who were eligible based on local laboratory results but not central laboratory results. Expanding the eligibility criteria for the study population also allowed the evaluation of RESTORE-IMI 1 results in a more traditional intent-to-treat population.
RESULTS
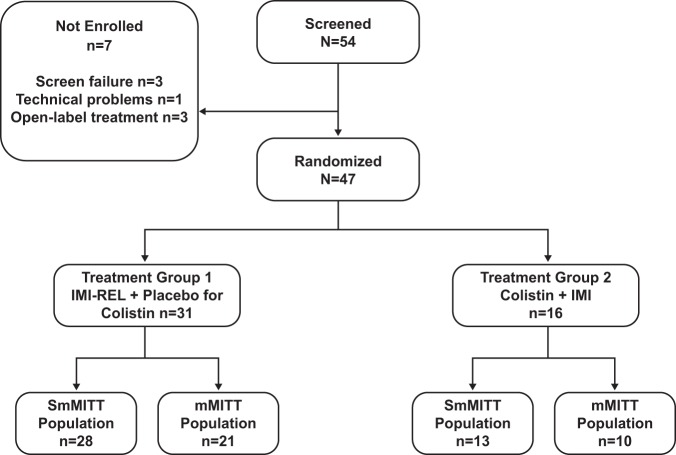
A total of 41 and 31 patients were included in the SmMITT and mMITT populations, respectively (Fig. 1). Baseline characteristics and demographics grouped by population and treatment arm are summarized in Table 1. Overall, the SmMITT and mMITT populations were comparable in terms of demographic and clinical characteristics, including age, infection type, Acute Physiologic Assessment and Chronic Health Evaluation II scores, and renal function. Within the SmMITT population, 28 patients were treated with IMI-REL (8 for hospital-acquired bacterial pneumonia [HABP]/ventilator-associated bacterial pneumonia [VABP], 5 for cIAI, 15 for cUTI) and 13 were treated with colistin plus IMI (4 for HABP/VABP, 3 for cIAI, 6 for cUTI). Within the mMITT population, 21 patients were treated with IMI-REL (8 for HABP/VABP, 2 for cIAI, 11 for cUTI) and 10 were treated with colistin plus IMI (3 for HABP/VABP, 2 for cIAI, 5 for cUTI). P. aeruginosa was the most common qualifying baseline pathogen (SmMITT population, 27/42 [64.3%]; mMITT population, 24/31 [77.4%]), followed by K. pneumoniae (SmMITT population, 8/42 [19.0%]; mMITT population, 4/31 [12.9%]) and other Enterobacteriaceae (SmMITT population, 7/42 [16.7%]; mMITT population, 3/31 [9.7%]). Polymicrobial Gram-negative bacterial infections were uncommon (SmMITT population, 3/41 [7.3%]; mMITT population, 1/31 [3.2%]). One patient (2.4%) in the SmMITT population had 2 qualifying baseline pathogens.
FIG 1.
Trial enrollment and summary of patients in analysis populations. IMI, imipenem-cilastatin; mMITT, microbiological modified intent-to-treat; REL, relebactam; SmMITT, supplemental microbiological modified intent-to-treat.
TABLE 1.
Baseline demographics and clinical characteristicsc
| Characteristic | SmMITT population |
mMITT population |
||||
|---|---|---|---|---|---|---|
| IMI-REL (n = 28) | Colistin plus IMI (n = 13) | Total (n = 41) | IMI-REL (n = 21) | Colistin plus IMI (n = 10) | Total (n = 31) | |
| No. of patients in population | 28 | 13 | 41 | 21 | 10 | 31 |
| No. of patients by sex | ||||||
| Male | 18 (64.3) | 10 (76.9) | 28 (68.3) | 13 (61.9) | 7 (70.0) | 20 (64.5) |
| Female | 10 (35.7) | 3 (23.1) | 13 (31.7) | 8 (38.1) | 3 (30.0) | 11 (35.5) |
| No. of patients by age (yr) | ||||||
| <65 | 16 (57.1) | 6 (46.2) | 22 (53.7) | 15 (71.4) | 5 (50.0) | 20 (64.5) |
| ≥65 | 12 (42.9) | 7 (53.8) | 19 (46.3) | 6 (28.6) | 5 (50.0) | 11 (35.5) |
| Median (range) age (yr) | 61 (19–77) | 65 (49–80) | 63 (19–80) | 59 (19–75) | 61 (49–80) | 59 (19–80) |
| Median (range) wt (kg) | 79.3 (53.0–140.0) | 75.8 (52.8–117.0) | 78.0 (52.8–140.0) | 75 (53.0–132.3) | 75.6 (52.8–117.0) | 75 (52.8–132.3) |
| No. of patients with the following APACHE II score: | ||||||
| ≤15 | 20 (71.4) | 11 (84.6) | 31 (75.6) | 14 (66.7) | 8 (80.0) | 22 (71.0) |
| >15 | 8 (28.6) | 2 (15.4) | 10 (24.4) | 7 (33.3) | 2 (20.0) | 9 (29.0) |
| No. of patients with the following primary diagnosis: | ||||||
| HABP | 1 (3.6) | 1 (7.7) | 2 (4.9) | 1 (4.8) | 1 (10.0) | 2 (6.5) |
| VABP | 7 (25.0) | 3 (23.1) | 10 (24.4) | 7 (33.3) | 2 (20.0) | 9 (29.0) |
| cIAI | 5 (17.9) | 3 (23.1) | 8 (19.5) | 2 (9.5) | 2 (20.0) | 4 (12.9) |
| cUTI (urinary tract abnormalities) | 8 (28.6) | 3 (23.1) | 11 (26.8) | 5 (23.8) | 3 (30.0) | 8 (25.8) |
| cUTI (acute pyelonephritis) | 7 (25.0) | 3 (23.1) | 10 (24.4) | 6 (28.6) | 2 (20.0) | 8 (25.8) |
| No. of patients with the following creatinine clearance (ml/min): | ||||||
| ≥90 | 8 (28.6) | 5 (38.5) | 13 (31.7) | 8 (38.1) | 3 (30.0) | 11 (35.5) |
| <90 to ≥60 | 14 (50.0) | 4 (30.8) | 18 (43.9) | 8 (38.1) | 4 (40.0) | 12 (38.7) |
| <60 to ≥30 | 4 (14.3) | 3 (23.1) | 7 (17.1) | 3 (14.3) | 2 (20.0) | 5 (16.1) |
| <30 to ≥15 | 1 (3.6) | 1 (7.7) | 2 (4.9) | 1 (4.8) | 1 (10.0) | 2 (6.5) |
| Not available | 1 (3.6) | 0 | 1 (2.4) | 1 (4.8) | 0 | 1 (3.2) |
| No. of patients with the following qualifying baseline pathogens: | ||||||
| Citrobacter freundii | 1 (3.4) | 0 (0.0) | 1 (2.4) | 1 (4.8) | 0 | 1 (3.2) |
| Enterobacter aerogenes | 0 | 1 (7.7) | 1 (2.4) | 0 | 0 | 0 |
| Enterobacter cloacae | 2 (6.9) | 1 (7.7) | 3 (7.1) | 1 (4.8) | 0 | 1 (3.2) |
| Escherichia coli | 1 (3.4) | 0 | 1 (2.4) | 0 | 0 | 0 |
| Klebsiella oxytoca | 0 | 1 (7.7) | 1 (2.4) | 0 | 1 (10.0) | 1 (3.2) |
| Klebsiella pneumoniae | 6 (20.7) | 2 (15.4) | 8 (19.0) | 3 (14.3) | 1 (10.0) | 4 (12.9) |
| Pseudomonas aeruginosa | 19 (65.5) | 8 (61.5) | 27 (64.3) | 16 (76.2) | 8 (80.0) | 24 (77.4) |
| No. of patients with isolates with the following β-lactamasesa : | ||||||
| Class A | ||||||
| Older-spectrum β-lactamases | ||||||
| SHVb | 4 (14.3) | 2 (15.4) | 6 (14.6) | 2 (9.5) | 1 (10.0) | 3 (9.7) |
| TEM | 12 (42.9) | 3 (23.1) | 15 (36.6) | 7 (33.3) | 3 (30.0) | 10 (32.3) |
| ESBLs | ||||||
| CTX-M | 12 (42.9) | 6 (46.2) | 18 (43.9) | 7 (33.3) | 4 (40.0) | 11 (35.5) |
| SHVb | 2 (7.1) | 0 | 2 (4.9) | 1 (4.8) | 0 | 1 (3.2) |
| VEB | 0 | 0 | 0 | 0 | 0 | 0 |
| KPC serine carbapenemase | 5 (17.9) | 1 (7.7) | 6 (14.6) | 4 (19.0) | 1 (10.0) | 5 (16.1) |
| Class C | ||||||
| Chromosomal AmpC (PDC) | 19 (67.9) | 8 (61.5) | 27 (65.9) | 16 (76.2) | 8 (80.0) | 24 (77.4) |
| Plasmid-mediated AmpC | ||||||
| ACT | 0 | 0 | 0 | 0 | 0 | 0 |
| CMY | 1 (3.6) | 0 | 1 (2.4) | 1 (4.8) | 0 | 1 (3.2) |
| DHA | 1 (3.6) | 0 | 1 (2.4) | 1 (4.8) | 0 | 1 (3.2) |
| Class D, OXA-48 | 4 (14.3) | 2 (15.4) | 6 (14.6) | 0 | 1 (10.0) | 1 (3.2) |
Qualifying baseline pathogens could have had multiple β-lactamases detected.
Older-spectrum β-lactamases were SHV-1 and SHV-11; ESBLs were SHV-26 and SHV-28.
APACHE II, Acute Physiologic Assessment and Chronic Health Evaluation II; cIAI, complicated intra-abdominal infection; cUTI, complicated urinary tract infection; ESBL, extended-spectrum β-lactamase; HABP, hospital-acquired bacterial pneumonia; IMI, imipenem-cilastatin; mMITT, microbiological modified intent-to-treat; REL, relebactam; SmMITT, supplemental microbiological modified intent-to-treat; VABP, ventilator-associated bacterial pneumonia.
Among the qualifying baseline pathogens isolated from the 10 patients in the SmMITT population who were excluded from the mMITT population, the majority of differences in the MIC values obtained by the local laboratory versus those obtained by the central laboratory for imipenem alone, imipenem-REL, or colistin were limited to 1 to 2 dilutions for most qualifying baseline pathogens (Table 2). Of these 10 patients excluded from the mMITT population, central laboratory testing determined that 5 had baseline pathogens that were imipenem susceptible, 4 had baseline pathogens that were imipenem-REL nonsusceptible, and 1 had a pathogen that was colistin nonsusceptible.
TABLE 2.
Central versus local MICs of qualifying baseline pathogens for patients included in the SmMITT population but excluded from the mMITT populationb
| Patient no. | TG | Infection type | Qualifying pathogen name | Local interpretive criteria used | MIC (μg/ml [susceptibility])c
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem |
IMI-REL |
Colistin |
||||||||
| Local lab | Central lab | Local lab | Central lab | Local lab | Central lab | |||||
| 1 | 2 | HABP/VABP | E. aerogenes | CLSI | 2 (I) | 1 (S) | 0.5 (S) | 0.12 (S) | 0.25 (S) | <1 (S) |
| 2a | 1 | cIAI | K. pneumoniae | EUCAST | 8 (I) | 4 (I) | 2 (S) | 2 (S) | 2 (S) | 4 (R) |
| 3 | 2 | cIAI | E. cloacae | CLSI | 2 (I) | <0.5 (S) | 0.5 (S) | 0.12 (S) | 1 (S) | <1 (S) |
| 4 | 1 | cIAI | E. cloacae | CLSI | 2 (I) | <0.5 (S) | 1 (S) | 0.12 (S) | 1 (S) | <1 (S) |
| 5 | 1 | cIAI | K. pneumoniae | CLSI | 4 (R) | 4 (R) | 1 (S) | 2 (I) | 0.5 (S) | <1 (S) |
| 6 | 1 | cUTI | E. coli | CLSI | 2 (I) | 2 (I) | 1 (S) | 2 (I) | 0.12 (S) | <1 (S) |
| 7 | 2 | cUTI | K. pneumoniae | CLSI | 2 (I) | 2 (I) | 1 (S) | 2 (I) | 0.12 (S) | <1 (S) |
| 8 | 1 | cUTI | K. pneumoniae | CLSI | 16 (R) | 32 (R) | 1 (S) | 2 (I) | 0.25 (S) | <1 (S) |
| 9 | 1 | cUTI | P. aeruginosa | CLSI | 8 (R) | 2 (S) | 2 (S) | 0.5 (S) | 1 (S) | <1 (S) |
| 10 | 1 | cUTI | P. aeruginosa | CLSI | 16 (R) | 2 (S) | 1 (S) | 0.25 (S) | 0.12 (S) | <1 (S) |
The participant had a polymicrobial infection, including an infection caused by a qualifying P. aeruginosa isolate, but was unevaluable for the mMITT population due to the presence of colistin-nonsusceptible K. pneumoniae.
cIAI, complicated intra-abdominal infection; CLSI, Clinical and Laboratory Standards Institute; cUTI, complicated urinary tract infection; EUCAST, European Committee on Antimicrobial Susceptibility Testing; HABP, hospital-acquired bacterial pneumonia; I, intermediate; IMI-REL, imipenem-cilastatin plus relebactam; mMITT, microbiological modified intent-to-treat; R, resistant; S, susceptible; SmMITT, supplemental microbiological modified intent-to-treat; TG, treatment group; VABP, ventilator-associated bacterial pneumonia.
Data in boldface indicate differences in susceptibility test interpretation (susceptible versus intermediate versus resistant) between the local laboratory and central laboratory, based on MIC values.
The results for the SmMITT and mMITT populations were generally consistent, and most patients achieved a favorable overall response (Table 3). Favorable overall response rates in the IMI-REL treatment group were >70% for both the SmMITT and mMITT populations. Favorable clinical response rates at day 28 were 71.4% for IMI-REL and 40.0% for colistin plus IMI in the mMITT population, whereas they were 75.0% for IMI-REL and 53.8% for colistin plus IMI in the SmMITT population. Day 28 all-cause mortality rates were 9.5% for IMI-REL and 30.0% for colistin plus IMI in the mMITT population, whereas they were 10.7% for IMI-REL and 23.1% for colistin plus IMI in the SmMITT population. Among the 10 patients excluded from the mMITT population, a favorable overall response was reported for 6/7 (85.7%) in the IMI-REL-treated group and 3/3 (100.0%) in the colistin plus IMI-treated group; a favorable clinical response at day 28 was reported for 6/7 (85.7%) in the IMI-REL-treated group and 3/3 (100.0%) in the colistin plus IMI-treated group; all-cause mortality was 1/7 (14.3%) in the IMI-REL-treated group, and no patients in the colistin plus IMI-treated group died.
TABLE 3.
Treatment responses in patients in mMITT and SmMITT populationsc
| Response and patient group | n/m (%) |
% (90% CI) |
||
|---|---|---|---|---|
| IMI-REL | Colistin plus IMI | Unadjusted difference | Adjusted differencea | |
| Favorable overall response | ||||
| mMITT population | 15/21 (71.4) | 7/10 (70.0) | 1.4 | −7.3 (−27.5 to 21.4) |
| HABP/VABP | 7/8 (87.5) | 2/3 (66.7) | 20.8 | |
| cIAI | 0/2 | 0/2 | 0.0 | |
| cUTI | 8/11 (72.7) | 5/5 (100.0) | −27.3 (−52.8 to 12.8)b | |
| SmMITT population | 21/28 (75.0) | 10/13 (76.9) | −1.9 | −4.5 (−24.2 to 20.7) |
| HABP/VABP | 7/8 (87.5) | 3/4 (75.0) | 12.5 (−25.4 to 56.6)b | |
| cIAI | 2/5 (40.0) | 1/3 (33.3) | 6.7 | |
| cUTI | 12/15 (80.0) | 6/6 (100.0) | −20.0 (−41.4 to 14.2)b | |
| Favorable clinical response (day 28) | ||||
| mMITT population | 15/21 (71.4) | 4/10 (40.0) | 31.4 | 26.3 (1.3 to 51.5) |
| SmMITT population | 21/28 (75.0) | 7/13 (53.8) | 21.2 | 17.6 (−5.9 to 42.5) |
| All-cause mortality (through day 28) | ||||
| mMITT population | 2/21 (9.5) | 3/10 (30.0) | −20.5 | −17.3 (−46.4 to 6.7) |
| SmMITT population | 3/28 (10.7) | 3/13 (23.1) | −12.4 | −10.5 (−35.2 to 9.6) |
Adjusted differences and 90% confidence intervals are based on the values obtained by the Miettinen and Nurminen method (36) stratified by infection site.
The 90% confidence intervals were calculated by the Miettinen and Nurminen method (36).
CI, confidence interval; cIAI, complicated intra-abdominal infection; cUTI, complicated urinary tract infection; HABP, hospital-acquired bacterial pneumonia; IMI, imipenem-cilastatin; mMITT, microbiological modified intent-to-treat; n/m, number of patients with a favorable response or all-cause mortality/number of patients evaluable; REL, relebactam; SmMITT, supplemental microbiological modified intent-to-treat; VABP, ventilator-associated bacterial pneumonia.
DISCUSSION
The previously reported efficacy and safety results from the RESTORE-IMI 1 trial support IMI-REL as a suitable treatment option for serious Gram-negative bacterial infections, including those caused by carbapenem-nonsusceptible pathogens in high-risk patients (20). These results were obtained in the primary efficacy population (i.e., the mMITT population), determined by qualifying baseline pathogen susceptibility results from the central microbiology laboratory. Here, we describe the results of a secondary analysis conducted in the protocol-defined SmMITT population, in which eligibility was based on susceptibility results from the local microbiology laboratory. The results of this secondary analysis lend further support to the primary study findings from RESTORE-IMI 1, in that they are consistent, regardless of whether patients were included in the analysis based on local or central laboratory susceptibility testing. In both the secondary SmMITT population and the primary mMITT population, IMI-REL and colistin plus IMI yielded favorable overall response rates of >70%. In both analysis populations, there was an apparent trend toward more favorable clinical and mortality outcomes among patients treated with IMI-REL than among those treated with a colistin-based therapy.
In contrast to recent open-label clinical trials comparing the efficacy and safety of new antibacterial agents with the best available therapy (BAT) for the treatment of resistant infections (29–31), the RESTORE-IMI 1 trial was a double-blind study with a single active comparator. Limiting the study to a single comparator rather than BAT reduced treatment variability and ensured that all patients received early appropriate therapy, given that only patients with colistin-susceptible infections were enrolled. Generally, in antibacterial clinical trials, pathogen isolates are commonly cultured in local laboratories for initial pathogen identification and subsequently sent to a central laboratory for confirmatory pathogen identification and susceptibility testing. The use of standardized susceptibility panels in local laboratories for the RESTORE-IMI 1 trial further distinguishes this trial from other similar resistant-pathogen trials in 2 important ways: (i) by ensuring the consistency of the susceptibility results for qualifying baseline pathogens from all sites, thereby allowing the rigorous selection of patients eligible for inclusion into the study, and (ii) by enabling enrollment in the trial (with initiation of appropriate therapy for all patients) at a consistent time point for all patients (i.e., as soon as susceptibility results are known).
The use of microbiological criteria for the identification of study populations for enrollment in clinical trials evaluating antibacterial agents presents a substantial challenge (32). As with similar studies, the primary analysis (mMITT) population in RESTORE-IMI 1 was small due to the difficulty in identifying patients with the desired resistance profile who qualified for a clinical trial, as well as the impact of the global study design with variable clinical practices and clinical trial experience across institutions (29, 30). Also consistent with similar trials, the RESTORE-IMI 1 mMITT population included a limited number of enrolled patients with cIAI, primarily because there were fewer prescreening isolates from intra-abdominal specimens than from urinary and lower respiratory tract specimens (29, 30). This secondary analysis expanded the participant population analyzed and doubled the size of the cIAI cohort from 4 to 8 patients. Compared with the mMITT population, the SmMITT population may better represent patients seen in a real-world setting, where treatment decisions are based on pathogen identification and susceptibility data provided by the local microbiology laboratory.
In this analysis, susceptibility testing results obtained from the local laboratory were similar to those obtained from the central laboratory. Among the 10 patients who were excluded from the mMITT population, most qualifying baseline isolates had minor differences (1 to 2 dilutions) in MIC results between the 2 laboratory settings, and differences in susceptibility determinations occurred to a similar extent for all 3 tested drugs. The differences in susceptibility results between the 2 laboratory settings, with MICs generally being higher at the local laboratories, may be due to differences inherent to the 2 susceptibility panels used (i.e., Sensititre in local laboratories versus custom frozen panels in the central laboratory). Local laboratory testing identified a patient population consistent with the population in which IMI-REL is expected to be used: those with Gram-negative bacterial infections caused by carbapenem-resistant pathogens. The similarity of the results obtained from both laboratory settings suggests that clinicians can rely on local laboratory data to guide decision-making and the use of IMI-REL. In addition, this analysis offers insight into study design options for antibacterial trials conducted in this challenging patient population.
Overall, IMI-REL was effective for the treatment of adults with imipenem-nonsusceptible, serious Gram-negative bacterial infections, including cIAI, cUTI, and HABP/VABP, with the response rates being comparable to the response rate of a commonly used regimen (20). The results of this secondary analysis, in which treatment response was evaluated among patients with qualifying baseline pathogens identified based only on local microbiology laboratory culture and susceptibility results, provide support for the expected future clinical use of IMI-REL for the treatment of infections caused by multidrug-resistant Gram-negative bacteria, where therapeutic decisions will typically be made based on local laboratory data.
MATERIALS AND METHODS
Study design.
RESTORE-IMI 1 (protocol MK-7655A-013) was a phase 3, randomized, double-blind, active comparator-controlled, parallel-group, multicenter clinical trial that evaluated the efficacy and safety of IMI-REL compared with those of colistin plus IMI (ClinicalTrials.gov identifier NCT02452047). The protocol was approved by appropriate institutional review boards and regulatory agencies, and the trial was conducted in accordance with the principles of Good Clinical Practice and the Declaration of Helsinki. The full methodology was previously published (20).
Patients.
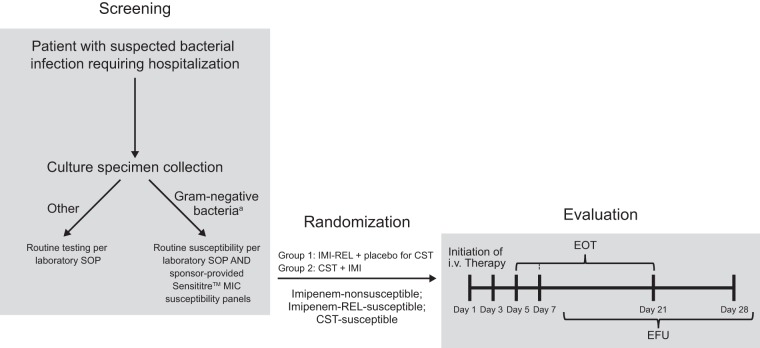
Inclusion and exclusion criteria have been previously described (20). Patients were adults (age, ≥18 years) who required hospitalization and treatment with intravenous (i.v.) antibiotics for serious infections (cIAI, cUTI, and HABP/VABP) caused by imipenem-nonsusceptible but imipenem-REL- and colistin-susceptible Gram-negative pathogens. As part of routine standard-of-care testing at each participating investigational site, all Gram-negative pathogens isolated from intra-abdominal, lower respiratory tract, and urinary tract specimens were evaluated for susceptibility to the 3 study drugs by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) guidelines (Fig. 2) (33). For this purpose, all local microbiology laboratories (i.e., those used by the individual investigational sites) were provided with Sensititre susceptibility panels (Thermo Fisher Scientific Inc., Waltham, MA, USA), a broth microdilution method, to standardize testing of the susceptibility of the isolates to imipenem, imipenem-REL, and colistin. This prescreening process identified patients with potentially eligible infections caused by imipenem-nonsusceptible, imipenem-REL-susceptible, and colistin-susceptible Gram-negative pathogens. The study investigators then decided whether to enter the identified patients into the formal screening process of this trial (i.e., obtaining informed consent and determining whether all other inclusion and exclusion criteria were met). In order to be eligible, a patient’s primary infection-site sample had to be collected within 1 week before study entry. Patients were eligible for enrollment into the study and randomization based on the local susceptibility test results for the Gram-negative isolates. In addition to local laboratory testing, all Gram-negative isolates from infection-site specimens from randomized patients were evaluated at a central microbiology reference laboratory (International Health Management Associates, Inc. [IHMA], Schaumburg, IL, USA). At the central laboratory, species identification was confirmed by matrix-assisted laser desorption ionization–time of flight spectroscopy (Bruker Daltonics, Billerica, MA, USA), and susceptibility testing was performed by broth microdilution according to CLSI guidelines using custom frozen panels prepared at IHMA (33). Repeat testing was performed for isolates for which the susceptibility results obtained at the central and local laboratories were not consistent. MIC values were interpreted using CLSI (34) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) 2017 (35) guidelines. Imipenem susceptibility breakpoints were also applied to imipenem-REL.
FIG 2.
Assessment schedule. a, from specimen types of interest: blood, urinary, intra-abdominal, or lower respiratory tract sources. CST, colistin; EFU, early follow-up; EOT, end of therapy; IMI, imipenem-cilastatin; i.v., intravenous; REL, relebactam; SOP, standard operating procedure.
Procedures.
Patients were randomized 2:1 (stratified by primary infection type) into 2 groups. The first group received i.v. IMI-REL (imipenem at 500 mg and cilastatin at 500 mg plus relebactam at 250 mg every 6 h) plus placebo to colistin; the second group received i.v. colistin (as colistimethate sodium at a loading dose to achieve 300-mg colistin base activity, followed by maintenance doses every 12 h up to 150-mg colistin base activity) plus IMI (imipenem at 500 mg and cilastatin at 500 mg every 6 h). Dosing of both study treatments was adjusted based on renal function. The treatment duration was ≥5 days for cIAI and cUTI and ≥7 days for HABP/VABP and did not exceed 21 days. Patients were screened for eligibility at ≤24 h before randomization (Fig. 2); study visits were performed on day 1 (randomization), on day 3 (on-therapy visit), and at the end of therapy (EOT; day 5/7 to day 21). Patients were also evaluated following completion of the i.v. study therapy at an early follow-up (EFU) visit (5 to 9 days post-EOT) and on day 28 (which could occur on the same day as the EFU visit).
For patients with HABP/VABP or cIAI, the collection of a baseline sample from the infection site was strongly preferred; however, collection at the time of study entry was not required for patients in whom infection-site specimen collection was not medically acceptable (e.g., a patient with cIAI in whom collection of an additional sample would require surgical intervention). Instead, a pure isolate of the suspected causative pathogen from a prior culture of a specimen collected within 1 week of enrollment was submitted to the central laboratory for evaluation. Additional unscheduled cultures of specimens from the infection site with pathogen susceptibility testing were performed at any time that there was clinical or laboratory evidence of infection persistence or progression (e.g., persistent fever, elevated white blood cell count, or significant changes in the patient’s clinical condition) and at the time of any surgical or drainage procedure in patients with HABP/VABP or cIAI. For patients with cUTI, infection-site specimens were required for visits on day 1, on day 3, at the EOT, and at the EFU visit.
Treatment response analysis.
Treatment response was defined using unique criteria for each infection type (Table 4). Key secondary endpoints included a favorable clinical response (sustained cure or cure) at day 28 after initiation of trial treatment and all-cause mortality (survival) at day 28 after trial treatment.
TABLE 4.
Primary endpoint (overall response) definition by infection typec
| Infection type | Time point | Outcome |
|---|---|---|
| HABP/VABP | Day 28 | All-cause mortality (survival) |
| cIAI | Day 28 | Favorable clinical response (sustained cure or curea ) |
| cUTI | EFU visit | Favorable microbiological response (sustained eradicationb ) and favorable clinical response (sustained cure or curea ) |
All pretherapy signs and symptoms of the index infection(s) had resolved (or returned to preinfection status), no additional intravenous antibiotic therapy was required, and, for patients with cIAI, no unplanned surgical procedures or percutaneous drainage procedures had been performed (cure was sustained if there was no evidence of a resurgence of the index infection).
A culture of urine taken at the EFU visit still showed eradication of the uropathogen found at study entry (i.e., a count of ≥105 CFU/ml was reduced to <104 CFU/ml).
cIAI, complicated intra-abdominal infection; cUTI, complicated urinary tract infection; EFU, early follow-up; HABP, hospital-acquired bacterial pneumonia; VABP, ventilator-associated bacterial pneumonia.
Treatment responses were evaluated in 2 protocol-prespecified analysis populations: the mMITT population, which was the primary efficacy population from the primary analysis, and the SmMITT population. Both the mMITT and SmMITT populations comprised patients who received ≥1 dose of study drug and had ≥1 qualifying baseline pathogen, a Gram-negative pathogen isolated from a culture of a specimen obtained from the primary infection site within 1 week of randomization and meeting protocol-specified criteria for susceptibility to imipenem, imipenem-REL, and colistin based on local laboratory susceptibility interpretive criteria (i.e., CLSI or EUCAST criteria). The eligibility of the baseline pathogens for the mMITT population was determined by MIC results obtained from the central laboratory. The SmMITT population included all patients in the mMITT population plus additional patients who had ≥1 qualifying baseline pathogen meeting susceptibility criteria according to local laboratory MIC results, regardless of the central laboratory MIC results.
This was an estimation trial; no formal statistical testing was performed for treatment response. Between-group 90% confidence intervals for the primary and key secondary endpoints were calculated using the stratified Miettinen and Nurminen method, an unconditional, asymptotic method (36). The between-group estimates were stratified by infection type, where appropriate.
Data availability.
The data sharing policy of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, including restrictions, is available at http://engagezone.msd.com/ds_documentation.php. Requests for access to the clinical study data can be submitted through the EngageZone site or via email to dataaccess@merck.com.
ACKNOWLEDGMENTS
This work was supported by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA (MSD). Medical writing and/or editorial assistance was provided by Todd Waldron and Alanna Kennedy of The Lockwood Group (Stamford, CT, USA). This assistance was funded by MSD.
All authors are responsible for the work described in this paper. All authors were involved in at least one of the following: conception, design of work or acquisition, analysis, interpretation of data, drafting the manuscript, and/or revising/reviewing the manuscript for important intellectual content. All authors provided final approval of the version to be published. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
M.L.B., A.A., I.K., H.-K.J., R.W.T., J.D., K.Y., J.R.B., and A.P. are current employees of MSD and may own stock and/or stock options in Merck & Co., Inc., Kenilworth, NJ, USA. K.S.K. is supported by the National Institute of Allergy and Infectious Diseases (Division of Microbiology and Infectious Diseases protocol 10-0065 and grant R01 1R01AI119446-01), has served as a consultant for MSD, and has received research funding from MSD. H.W.B. has been a (scientific advisory board) consultant to Merck & Co., Inc.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 16 January 2019.
- 2.Cai B, Echols R, Magee G, Arjona Ferreira JC, Morgan G, Ariyasu M, Sawada T, Nagata TD. 2017. Prevalence of carbapenem-resistant gram-negative infections in the United States predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect Dis 4:ofx176. doi: 10.1093/ofid/ofx176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin A, Fahrbach K, Zhao Q, Lodise T. 2018. Association between carbapenem resistance and mortality among adult, hospitalized patients with serious infections due to Enterobacteriaceae: results of a systematic literature review and meta-analysis. Open Forum Infect Dis 5:ofy150. doi: 10.1093/ofid/ofy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zilberberg MD, Nathanson BH, Sulham K, Fan W, Shorr AF. 2017. Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect Dis 17:279. doi: 10.1186/s12879-017-2383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasevic AT, Canton R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 2017. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 7.Tsuji BT, Pogue JM, Zavascki AP, Paul M, Daikos GL, Forrest A, Giacobbe DR, Viscoli C, Giamarellou H, Karaiskos I, Kaye D, Mouton JW, Tam VH, Thamlikitkul V, Wunderink RG, Li J, Nation RL, Kaye KS. 2019. International consensus guidelines for the optimal use of the polymyxins: endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 39:10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zavascki AP, Nation RL. 2017. Nephrotoxicity of polymyxins: is there any difference between colistimethate and polymyxin B? Antimicrob Agents Chemother 61:e02319-16. doi: 10.1128/AAC.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalfino L, Puntillo F, Ondok MJ, Mosca A, Monno R, Coppolecchia S, Spada ML, Bruno F, Brienza N. 2015. Colistin-associated acute kidney injury in severely ill patients: a step toward a better renal care? A prospective cohort study. Clin Infect Dis 61:1771–1777. doi: 10.1093/cid/civ717. [DOI] [PubMed] [Google Scholar]
- 10.Pogue JM, Lee J, Marchaim D, Yee V, Zhao JJ, Chopra T, Lephart P, Kaye KS. 2011. Incidence of and risk factors for colistin-associated nephrotoxicity in a large academic health system. Clin Infect Dis 53:879–884. doi: 10.1093/cid/cir611. [DOI] [PubMed] [Google Scholar]
- 11.Ariano RE, Zelenitsky SA, Kassum DA. 2008. Aminoglycoside-induced vestibular injury: maintaining a sense of balance. Ann Pharmacother 42:1282–1289. doi: 10.1345/aph.1L001. [DOI] [PubMed] [Google Scholar]
- 12.Paul M, Daikos GL, Durante-Mangoni E, Yahav D, Carmeli Y, Benattar YD, Skiada A, Andini R, Eliakim-Raz N, Nutman A, Zusman O, Antoniadou A, Pafundi PC, Adler A, Dickstein Y, Pavleas I, Zampino R, Daitch V, Bitterman R, Zayyad H, Koppel F, Levi I, Babich T, Friberg LE, Mouton JW, Theuretzbacher U, Leibovici L. 2018. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: an open-label, randomised controlled trial. Lancet Infect Dis 18:391–400. doi: 10.1016/S1473-3099(18)30099-9. [DOI] [PubMed] [Google Scholar]
- 13.Livermore DM, Meunier D, Hopkins KL, Doumith M, Hill R, Pike R, Staves P, Woodford N. 2018. Activity of ceftazidime/avibactam against problem Enterobacteriaceae and Pseudomonas aeruginosa in the UK, 2015–16. J Antimicrob Chemother 73:648–657. doi: 10.1093/jac/dkx438. [DOI] [PubMed] [Google Scholar]
- 14.Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young K, Painter RE, Raghoobar SL, Hairston NN, Racine F, Wisniewski D, Balibar CJ, Villafania A, Zhang R, Sahm DF, Blizzard T, Murgolo N, Hammond ML, Motyl MR. 2019. In vitro studies evaluating the activity of imipenem in combination with relebactam against Pseudomonas aeruginosa. BMC Microbiol 19:150. doi: 10.1186/s12866-019-1522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merck Sharp & Dohme Corp. 2019. Recarbrio™ (imipenem, cilastatin, and relebactam) for injection, for intravenous use: US prescribing information. Merck Sharp & Dohme Corp, Whitehouse Station, NJ. [Google Scholar]
- 17.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. 2015. Activity of imipenem with relebactam against gram-negative pathogens from New York City. Antimicrob Agents Chemother 59:5029–5031. doi: 10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore DM, Warner M, Mushtaq S. 2013. Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 68:2286–2290. doi: 10.1093/jac/dkt178. [DOI] [PubMed] [Google Scholar]
- 19.Carpenter J, Neidig N, Campbell A, Thornsberry T, Truex T, Fortney T, Zhang Y, Bush K. 2019. Activity of imipenem/relebactam against carbapenemase-producing Enterobacteriaceae with high colistin resistance. J Antimicrob Chemother 74:3260–3263. doi: 10.1093/jac/dkz354. [DOI] [PubMed] [Google Scholar]
- 20.Motsch J, Murta de Oliveira C, Stus V, Koksal I, Lyulko O, Boucher HW, Kaye KS, File TM, Brown ML, Khan I, Du J, Joeng HK, Tipping RW, Aggrey A, Young K, Kartsonis NA, Butterton JR, Paschke A. 10 August 2019. RESTORE-IMI 1: a multicenter, randomized, double-blind trial comparing efficacy and safety of imipenem/relebactam vs colistin plus imipenem in patients with imipenem-nonsusceptible bacterial infections. Clin Infect Dis doi: 10.1093/cid/ciz530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lob SH, Hackel MA, Kazmierczak KM, Young K, Motyl MR, Karlowsky JA, Sahm DF. 2017. In vitro activity of imipenem-relebactam against gram-negative ESKAPE pathogens isolated by clinical laboratories in the United States in 2015 (results from the SMART Global Surveillance Program). Antimicrob Agents Chemother 61:e02209-16. doi: 10.1128/AAC.02209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lob SH, Hoban DJ, Young K, Motyl MR, Sahm DF. 2018. Activity of imipenem/relebactam against gram-negative bacilli from global ICU and non-ICU wards: SMART 2015–2016. J Glob Antimicrob Resist 15:12–19. doi: 10.1016/j.jgar.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Karlowsky JA, Lob SH, Young K, Motyl MR, Sahm DF. 2018. Activity of imipenem/relebactam against Pseudomonas aeruginosa with antimicrobial-resistant phenotypes from seven global regions: SMART 2015–2016. J Glob Antimicrob Resist 15:140–147. doi: 10.1016/j.jgar.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 24.Lucasti C, Vasile L, Sandesc D, Venskutonis D, McLeroth P, Lala M, Rizk ML, Brown ML, Losada MC, Pedley A, Kartsonis NA, Paschke A. 2016. Phase 2, dose-ranging study of relebactam with imipenem-cilastatin in subjects with complicated intra-abdominal infection. Antimicrob Agents Chemother 60:6234–6243. doi: 10.1128/AAC.00633-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sims M, Mariyanovski V, McLeroth P, Akers W, Lee YC, Brown ML, Du J, Pedley A, Kartsonis NA, Paschke A. 2017. Prospective, randomized, double-blind, phase 2 dose-ranging study comparing efficacy and safety of imipenem/cilastatin plus relebactam with imipenem/cilastatin alone in patients with complicated urinary tract infections. J Antimicrob Chemother 72:2616–2626. doi: 10.1093/jac/dkx139. [DOI] [PubMed] [Google Scholar]
- 26.U.S. Food and Drug Administration. 2018. Guidance for industry: microbiology data for systemic antibacterial drugs—development, analysis, and presentation. U.S. Department of Health and Human Services, U.S. Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Silver Spring, MD. [Google Scholar]
- 27.European Medicines Agency. 2018. Guideline on the evaluation of medicinal products indicated for treatment of bacterial infections. https://www.ema.europa.eu/en/evaluation-medicinal-products-indicated-treatment-bacterial-infections. Accessed 27 January 2020.
- 28.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, Koeth L, Sei K, CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. 2018. CLSI Methods Development and Standardization Working Group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol 56:e01934-17. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmeli Y, Armstrong J, Laud PJ, Newell P, Stone G, Wardman A, Gasink LB. 2016. Ceftazidime-avibactam or best available therapy in patients with ceftazidime-resistant Enterobacteriaceae and Pseudomonas aeruginosa complicated urinary tract infections or complicated intra-abdominal infections (REPRISE): a randomised, pathogen-directed, phase 3 study. Lancet Infect Dis 16:661–673. doi: 10.1016/S1473-3099(16)30004-4. [DOI] [PubMed] [Google Scholar]
- 30.Wunderink RG, Giamarellos-Bourboulis EJ, Rahav G, Mathers AJ, Bassetti M, Vazquez J, Cornely OA, Solomkin J, Bhowmick T, Bishara J, Daikos GL, Felton T, Furst MJL, Kwak EJ, Menichetti F, Oren I, Alexander EL, Griffith D, Lomovskaya O, Loutit J, Zhang S, Dudley MN, Kaye KS. 2018. Effect and safety of meropenem-vaborbactam versus best-available therapy in patients with carbapenem-resistant Enterobacteriaceae infections: the TANGO II randomized clinical trial. Infect Dis Ther 7:439–455. doi: 10.1007/s40121-018-0214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuti JL, Kim A, Cloutier DJ, Nicolau DP. 2019. Evaluation of plazomicin, tigecycline, and meropenem pharmacodynamic exposure against carbapenem-resistant Enterobacteriaceae in patients with bloodstream infection or hospital-acquired/ventilator-associated pneumonia from the CARE study (ACHN-490–007). Infect Dis Ther 8:383–396. doi: 10.1007/s40121-019-0251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.U.S. Food and Drug Administration. 2017. Guidance for industry: antibacterial therapies for patients with an unmet medical need for the treatment of serious bacterial diseases. U.S. Department of Health and Human Services, U.S. Food and Drug Administration, Center for Drug Evaluation and Research (CDER) Silver Spring, MD. [Google Scholar]
- 33.Clinical and Laboratory Standards Institute (CLSI). 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed CLSI document M07-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Clinical and Laboratory Standards Institute (CLSI). 2017. Performance standards for antimicrobial susceptibility testing, 27th ed CLSI supplement M100 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.European Committee on Antimicrobial Susceptibility Testing. 2019. Breakpoint tables for interpretation of MICs and zone diameters, version 9.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf. Accessed 16 January 2019.
- 36.Miettinen O, Nurminen M. 1985. Comparative analysis of two rates. Stat Med 4:213–226. doi: 10.1002/sim.4780040211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sharing policy of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, including restrictions, is available at http://engagezone.msd.com/ds_documentation.php. Requests for access to the clinical study data can be submitted through the EngageZone site or via email to dataaccess@merck.com.